Abstract
Ligand binding assays (LBAs) have been the method of choice for protein analyte measurements for more than four decades. Over the years, LBA methods have improved in sensitivity and achieved larger dynamic ranges by using alternative detection systems and new technologies. As a consequence, the landscape and application of immunoassay platforms has changed dramatically. The introduction of bead-based methods, coupled with single molecule detection standardization and the ability to amplify assay signals, has improved the sensitivity of many immunoassays, in some cases by several logs of magnitude. Three promising immunoassay platforms are described in this article: Single Molecule Counting (SMC™) from Singulex Inc, Single Molecule Arrays (Simoa™) from Quanterix Corporation, and Immuno-PCR (Imperacer®) from Chimera Biotec GmbH. These platforms have the potential to significantly improve immunoassay sensitivity and thereby address the bioanalytical needs and challenges faced during biopharmaceutical drug development.
KEY WORDS: immunoassays, Immuno-PCR (Chimera Biotec GmbH), ligand binding assay (LBA), sensitivity, Single Molecule Array (Quanterix Corporation), Single Molecule Counting (Singulex Inc)
INTRODUCTION
Assay sensitivity is largely dependent on the use of reagents that have high affinity, selectivity, and specificity for their target. Incremental improvements in sensitivity may be made by optimizing assay conditions, such as buffers, reagent concentrations, and incubation conditions, but efficient bead-based binding kinetics combined with sensitive detection technologies may be required to achieve the required sensitivity. Enzyme-linked immunosorbent assays (ELISAs) have been the format of choice for protein analyte detection in a variety of biological matrices using ligand binding assays (LBA). Common ELISAs have a colorimetric readout based on absorbance measurements (1,2). Over the years, the dynamic range, robustness, and sensitivity of LBAs have been improved through the use of alternative detection systems, such as chemiluminescence and various fluorescent readouts (3). The emergence of new technologies has allowed for additional improvements in assay sensitivity and dynamic range beyond the capabilities of conventional LBAs. Many technologies have attempted to reach high sensitivities, but few have been able to achieve ultrasensitive detection at sub picogram or femtogram per milliliter levels (4–6). Herein, we review three emerging technologies, which we believe have the potential to significantly enhance the detection of analytes at low concentration. We define emerging technologies as technical improvements or novel applications of existing technologies that address assay sensitivity needs. Our evaluations are based on hands-on experience using these technologies as well as literature reviews. This article is not intended to be a comprehensive evaluation of every available platform currently addressing the sensitivity issue, but rather an overview of three emerging technologies, Single Molecule Counting (SMC™) on the Erenna® platform from Singulex, Inc., Single Molecule Arrays (Simoa™) from Quanterix Corporation, and Immuno-PCR (IPCR) on the Imperacer® platform from Chimera Biotec GmbH. The concepts of IPCR were initially described in 1992 (7). However, the commercially available Imperacer® IPCR platform from Chimera Biotec was introduced recently and provides optimized reagents and pre-synthesized DNA–detector conjugates (8,9) for best assay performance in a 96-well plate format.
We describe how ultra-sensitivity is achieved on each platform and provide information on their relative advantages and limitations. Table I summarizes the attributes of each platform that relates to sensitivity. Other attributes are discussed in the text and summarized in Table II. Lastly, we highlight demonstrated applications of each technology described in the literature.
Table I.
Comparison of Technology Attributes for the Three Platforms Related to Assay Sensitivity
| Sensitivity attributes | SMC™ (Singulex) | Imperacer® (Chimera) | Simoa™ (Quanterix) |
|---|---|---|---|
| Sensitivity level | fg–pg/ml | fg–pg/ml | fg–pg/ml |
| Functional read-out | Flow cytometry | Real-time qPCR | Enzyme/fluorescent substrate |
| Signal read-out | Digital and analog (at high concentrations) | Analog | Digital and analog (at high concentrations) |
| Bead vs. plate | Bead or plate | Plate | Bead |
| Sample volume requirementa | 100 μL | 30 μL | 100 μL |
| Signal amplification | No | Exponential | Enzymatic |
| Miniaturization | No | No | Yes |
| Typical minimum required dilution | Neat −1:10 | 1:2–1:10 | 1:2–1:4 |
| Detector label | Alexa Fluor label | DNA tag | β-Galactosidase enzyme |
| Automation | No | No | Fully automated |
aThis is the volume required at the appropriate dilution for one data point
qPCR quantitative polymerase chain reaction, DNA deoxyribonucleic acid
Table II.
Additional Assay Attributes
| Technology attributes | SMC™ (Singulex) | Imperacer® (Chimera) | Simoa™ (Quanterix) |
|---|---|---|---|
| Multiplexing capacity | No | 2-plex | 10-plex |
| Dynamic range | >4 logs | >4 logs | >4 logs |
| Throughputa | 4 × 96-well plates per day | 5 × 96-well plates per day | 5 × 96-well plates per day |
| Sample replication | Duplicates | Duplicates | Duplicates |
| Regulatory fit | Can be validated for GLP use | Can be validated for GLP use | Can be validated for GLP use |
| LIMS connectivity | Intermediate file generated for LIMS import | Intermediate file generated for LIMS import | Intermediate file generated for LIMS import |
| Technology vendor | Singulex | Chimera Biotec | Quanterix |
| Single vendor technology | Yes | Yes | Yes |
| Availability at CROs | Yes (limited) | Yes (limited) | Yes (limited) |
| Vendor business model | Options include: commercial kits, custom kits, or home-brews | Options include: commercial kits, custom kits, or home-brews | Options include: commercial kits, custom kits, or home-brews |
aFor single assay, not multiplex
ELISA enzyme-linked immunosorbent assay, GLP Good Laboratory Practice, LIMS Laboratory Information Management System, qPCR quantitative polymerase chain reaction, CROs clinical research organizations
Assay Technologies
Single Molecule Counting (SMC™)
The Erenna® immunoassay system from Singulex Inc. utilizes an LBA procedure followed by capillary flow analysis on an instrument that performs single molecule counting (SMC™) of fluorescent detector molecules (Fig. 1). Depending on the assay, picogram to femtogram per milliliter limits of detection for various protein analytes using this technology have been reported (10–13). Initially, a bead- or plate-based sandwich assay format is executed in either 96- or 384-well plates, using a biotinylated capture reagent and fluorescent labeled detector that are highly specific for the analyte of interest. The fluorescent detector is eluted off the beads or plate with a volume to concentrate the signal, and samples are then loaded onto the instrument in a 384-well plate. Sample analysis occurs using capillary fluidics through a flow cell where fluorescent detector molecules travel across a 5-μm chamber to be interrogated by a laser. The signal produced is translated into counts of single fluorescent molecules. The key advantages of this platform are enhanced sensitivity (sub-picogram per milliliter) and broad dynamic range.
Fig. 1.
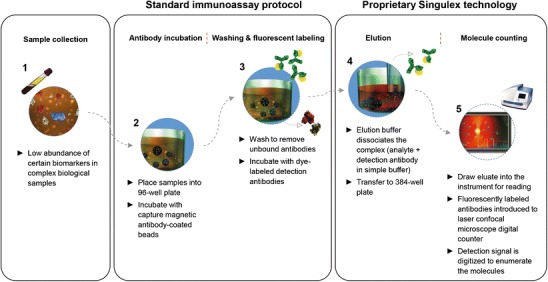
Singulex: Single Molecule Counting (SMC™) technology
The high sensitivity of this platform results from combining several elements, and each of these are discussed in the next section: (a) paramagnetic beads or plates enable efficient analyte capture and concentration by small volume elution prior to laser interrogation; (b) digital readout of single positive events compared with the analog readout typical of conventional LBAs; and (c) achieving low background by using low non-specific protein binding components including paramagnetic beads, plates, and multiple wash and transfer steps during the immunoassay process.
The relationship between volume and analyte concentration is important when measuring low abundance analytes, as described in detail by David Walt (14). An ultra-sensitive platform utilizing SMC™ needs to detect a statistically significant number of molecules in a short time frame, which may be difficult with samples at very low concentrations. The Erenna® immunoassay uses a pre-concentration step in which fluorescent labeled detector molecules are eluted off the beads or plate with a very small volume of elution buffer, thus concentrating the sample and ensuring adequate detection capabilities down to sub picogram per milliliter range.
The size of the laser interrogation space and sampling time through the detection space is precisely set so that only one fluorescent entity is present during the detection phase for lower concentrations (15). Fluorescence events are digitally counted by photon detectors within the instrument and are considered positive if the signal is 5–6 standard deviations above the threshold of background fluorescence. Compared with analog signal read-outs used in traditional LBAs where a single data point is used to represent all the molecules present in a sample, this digital technology allows for multiple individual data points to be summed over a specified time interval to create total signal. At higher analyte concentrations, the instrument switches to analog mode, using a unique algorithm to calculate the total number of photons, and hence, a very broad dynamic range is possible using this technology (∼4.5 logs) (15,16).
Several methods are used to mitigate non-specific background. Bead microparticles are constructed with materials designed to minimize non-specific binding of the fluorescent detector molecules, and assay incubations are carried out in polypropylene microtiter plates with very low protein binding properties. Erenna® assay protocols incorporate a plate transfer step after detector addition, and all beads are transferred to a new plate. This ensures avoidance of contamination from small amount of non-specific detection reagent bound to the plate eluting off with the specific detection reagent during the pre-concentration elution step (16). The Erenna® curve-fitting software combines three different instrument readouts (Detected Events, Event Photons and Total Photons) to generate one final concentration for each sample. This unique algorithm allows for ultrasensitive detection and improved dynamic range compared with ELISA.
In addition to improved sensitivity and broad dynamic range, the Erenna® brings other positive attributes to the bioanalytical laboratory. The platform offers flexibility by using bead- or plate-based assays, in both 96- and 384-well configurations. The plate-based version is more user-friendly, does not use magnets for bead extraction, and is generally less cumbersome. Sensitivity may be sacrificed if the plate-based format is chosen, although lower limits of quantification in the low picogram per milliliter range have been reported for plate-based assays (17,18). Singulex offers additional flexibility, with off-the-shelf kits as well as services for assay development if a custom assay is preferred. Reagent labeling kits are available for custom in-house labeling in addition to assay development kits, which provide assay diluents, wash buffer, elution, and neutralization buffers for “home brew” assay development.
The Erenna® platform can be used for regulated bioanalysis. The software has security and access control functions to ensure data access is compliant with United States Food and Drug Administration (FDA) 21 Code of Federal Regulations (CFR) part 11, and the data may be exported and analyzed in a standard bioanalytical database, such as the Laboratory Information Management System (LIMS). Other considerations for using the Erenna® platform in a Good Laboratory Practice (GLP) environment are described elsewhere (19).
In terms of limitations, the SMC™ technology throughput is relatively low due to sampling one well at a time, and low concentration samples can take up to 1 min each to read. The platform cannot be used for multiplexing, and carry-over can be an issue. The Erenna® instrument has no internal automation capabilities for the immunoassay steps, though off-line automation may be utilized for washing and transfer steps. Since particulate matter in complex matrices may obstruct the capillary fluidics, the vendor suggests a filtration or centrifugation step be utilized to clarify the samples. However, the effect of filtration on analyte recovery should be evaluated with spike and recovery studies during assay development (20). Given the complicated fluidics, abundant tubing and laser functions associated with this instrument, use of diligent practices, with respect to instrument maintenance, is very important.
Applications of SMC™
The use and application of SMC™ for protein biomarker measurements in various studies has been published, and several of these will be used to highlight the key attributes of the technology.
Todd et al. utilized the Erenna® to quantify ten different clinically relevant biomarkers, including cardiac troponin I and various cytokines, and achieved limits of detection from 10 to 100 fg/mL in 100 μL of sample with good accuracy and precision (16). Esparza et al. developed a highly sensitive (low picogram per milliliter), precise, and specific assay on the Erenna® that distinguished oligomerized amyloid-beta protein from the monomeric form in human brain tissue homogenates (17), and the ratio of oligomerized amyloid-beta levels to plaque density fully differentiated Alzheimer patients with and without dementia. The authors used small sample volumes in a 384-well plate format to increase speed and throughput. Shukla et al. (12) evaluated interleukin IL-22 as a potential inflammatory biomarker, utilizing a bead-based Erenna® assay with adequate sensitivity (0.2 pg/mL lower limit of quantification (LLOQ)) to obtain reportable IL-22 baseline values in all clinical samples tested. In addition, to increase throughput and ensure the robustness of the procedure, Tecan (Männedorf, Switzerland) liquid handling automation was incorporated for the wash and transfer steps. Tarasow et al. (18) faced challenges in measuring almost 100 proteins in a large number of patient samples to identify and stratify key biomarkers to predict early stage type 2 diabetes. With only 0.5 mL of sample available to measure almost 100 proteins, it was critical to conserve sample while retaining sensitivity and throughput. Because of the sheer number of assays to be developed and the different expected ranges of concentration, ease of assay development was a desired feature to make the workload manageable. Therefore, the authors chose to use a singleplex approach instead of multiplexing. The sensitivity capabilities and flexibility of the Erenna® platform in using 384-well plates enabled them to achieve their goal of measuring 89 different proteins from each sample and successfully identified critical biomarkers to predict early onset of the disease.
Single Molecule Arrays (Simoa™)-Based Digital ELISA
The Simoa™ digital ELISA from Quanterix Corporation is a promising platform, which can enable protein quantification at femtogram per milliliter levels (21). This technology uses a microscopic paramagnetic bead-based immunoassay coupled with a unique signal detection and data acquisition system. The digital ELISA is performed in two stages. In the first stage of antigen capture, the microscopic beads coated with capture reagent are mixed with samples followed by addition of biotin-labeled detection antibody and streptavidin–β-galactosidase. The beads are then loaded on a disc with an array of femtoliter-sized microwells. The diameter of each microwell (4.5 μm) can only accommodate zero to one bead with a diameter of 2.7 μm. The microwells are sealed against a silicone gasket in the presence of fluorogenic substrate for signal generation and amplification (Fig. 2). Simoa™ technology employs a unique signal detection system in the second stage of the assay. Relatively high concentration of fluorescent product in a 50-fL microwell (positive events) and total bead load by white light scattering can be easily detected and captured by a standard CCD camera. The Simoa™ platform achieves high sensitivity by improving several assay steps. With ∼80,000 capture antibody molecules per bead and ∼200,000 beads in a 100-μL reaction volume, the antibody–analyte ratio and equilibrium allows a high efficiency capture of analyte (>70%) in a given sample. With so many beads in solution, capture efficiency is further achieved because each target protein encounters a bead in less than 1 min. This is contrary to the slow binding in conventional LBAs. The optimized ratio between biotin-labeled detection antibody and streptavidin–β-galactosidase reduces assay background and maximizes specific signal acquisition. The fluorescent signal can be further amplified with enzymatic digestion of a fluorogenic substrate coupled with Simoa™ detection system. The Simoa™ detection system allows the detection of a single binding event, which can be acquired digitally at low concentration (improved sensitivity; digital ELISA) or in analog mode at high concentration (improved assay dynamic range). Fluorescence is used to detect enzyme activity, and the average enzyme per bead level is measured. As with the Erenna®, a very broad dynamic range can be achieved by combining the digital and analog readout capabilities (22,23).
Fig. 2.
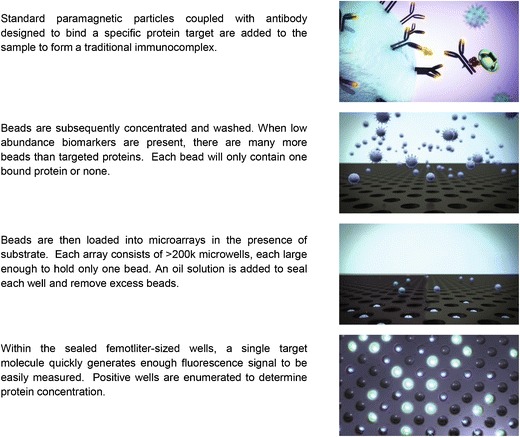
Quanterix: Single Molecule Arrays (Simoa™) technology
Because the final signal acquisition is done on a planar array with standard imaging, data acquisition is a much faster process compared with the digital readout with a fluidic system like the Erenna®.
Although the Erenna® and Simoa™ platforms share commonalities such as digital readout and broad dynamic range, the Simoa™ platform is a fully automated system. The instrument performs sample dilution (up to 1:10 dilution), mixing, washing, incubation, and data acquisition steps. The Simoa™ platform is capable of measuring up to ten different analytes simultaneously (multiplexing). This is achieved by using different dye linkers for each individual capture antibody (24). The array is then imaged using fluorescent detection at multiple wavelengths to identify subpopulations of different dyes and to determine the presence or absence of enzyme reporter labels.
Like Singulex, Inc, Quanterix Corporation has a number of manufactured kits to choose from. Custom development and “home brew” capabilities are also available options. With enhanced sensitivity, multiplexing, and automation capabilities, this platform seems ideal; however, it is a nascent technology, and the instruments are only recently becoming available for evaluation. Therefore, there are few examples of applications of this technology, particularly for multiplexing.
The Simoa™ platform is compatible with a LIMS system, and the vendor is currently developing software to meet the compliance requirements for regulated bioanalysis according to FDA 21 CFR part 11.
Applications of Simoa™ Digital ELISA
Most of the proof-of-concept studies have been performed by the vendor, Quanterix Corporation, by measuring clinical diagnostic biomarkers. However, these publications have demonstrated its potential in the field of clinical development. Rissin et al. (21) showed that a measurable sensitivity of two serum proteins, prostate-specific antigen (PSA) and tumor necrosis factor alpha, can be reached at 30 fg/mL (1 fM) and 10 fg/mL (∼600aM), respectively. The sensitivity improvement is at least ∼15,000- to 30,000-folds over the standard published ELISA methods used for these two proteins. Rissin et al. (21) tested 30 serum samples from patients who had undergone radical prostatectomy for the presence of PSA levels with the improved Simoa™ PSA method. While a leading clinical diagnostic assay could not detect any PSA present in these 30 serum samples with a method sensitivity of 100 pg/mL or 3 pM, the Simoa™-based PSA method was able to detected PSA levels in the same samples with improved assay sensitivity of 0.006 pg/mL or ∼200 aM. More recently, Shahim et al. (25) developed a sensitive method (0.002 pg/mL) for measuring the level of the axonal injury biomarker, total Tau, in human serum with Simoa™ technology. The improved method has a ∼1,000-fold improved sensitivity over the conventional immunoassay used for this protein. By using the improved method, Shahim’s group was able to monitor the Tau levels in plasma samples from professional ice hockey players prior to playing (N = 47) and post-concussion (N = 28) and demonstrated a statistically significant increase in Tau protein levels in the samples from players who suffered concussion. The vendor also published a successful feasibility study on multiplexed digital immunoassays for simultaneously detecting several human cytokines from human plasma samples (26). However, interference or “cross talk” among analytes would need to be evaluated as performed similarly for other immunoassay platforms.
Immuno-PCR (Imperacer®)
The immuno-polymerase chain reaction (IPCR) method combines the high specificity of antibody binding with the exponential signal amplification of a downstream quantitative-PCR (qPCR) read out (Fig. 3) capable of measuring low quantities of bound analytes (27). Due to the exponential signal amplification, the detectable levels of target molecules can be significantly improved from the –pico- to micromolar range in conventional immunoassays down to femtomolar analyte concentrations. IPCR case studies demonstrate sensitivities close to the theoretical detection limit of LBAs, down to a few thousand analyte molecules and wide dynamic range of up to five logs (9,28–31). Given a protein of 50–100 kD in size, this corresponds to LLOQ levels in the sub-picogram per milliliter levels in biological samples (8,32). Typical sample volumes are 15–30 μL for duplicates. The vendor provides flexible options including contract services for full assay development and bioanalytical support, as well as assay kits for home brew development. Standardized laboratory equipment is available with the option of 21CFR part 11-compliant software. Data analysis software is provided. Alternatively, data export and analysis in a data management system, e.g., LIMS, is possible. The Imperacer® workflow is a manual process at this time, and it is essential that IPCR assays are performed by a trained operator. Therefore, as with most analytical platforms, the vendor offers a training session with the purchase of Imperacer® platform. Although IPCR provides an ultrasensitive method for assay readout, highly sensitive detection may also pose challenges with regard to antibody specificity. The amplification of even lower-affinity binding events of the antibodies to alterative targets may result in assay background issues. Choosing high-quality antibodies and carefully optimizing the assay conditions and reagents is critical when using IPCR. Using highly diluted samples may reduce nonspecific matrix interactions, especially in cases where the background interactions decrease faster than specific interactions of interest upon sample dilution. In this regard, IPCR allows higher sample dilution ratios without loss in sensitivity compared with other LBA technologies due to exponential qPCR signal amplification. Imperacer® technology thus works well in applications when the analyte is known (e.g., well characterized biomarkers or a drug molecule). However, it may be more challenging to identify optimal assay conditions when the analyte is not fully characterized, such as the variety of anti-drug antibody (ADA) responses that are measured in immunogenicity assays. Imperacer® has a potential for assay multiplexing, typically limited to two analytes per reaction (33). The vendor recommends “polyplexing” as a sturdy alternative for multiplexing. Even small sample volumes can be split into multiple IPCR assays subsequent to dilution and used to separately measure individual analytes-of-interest at high sensitivity and no antibody cross-talk.
Fig. 3.
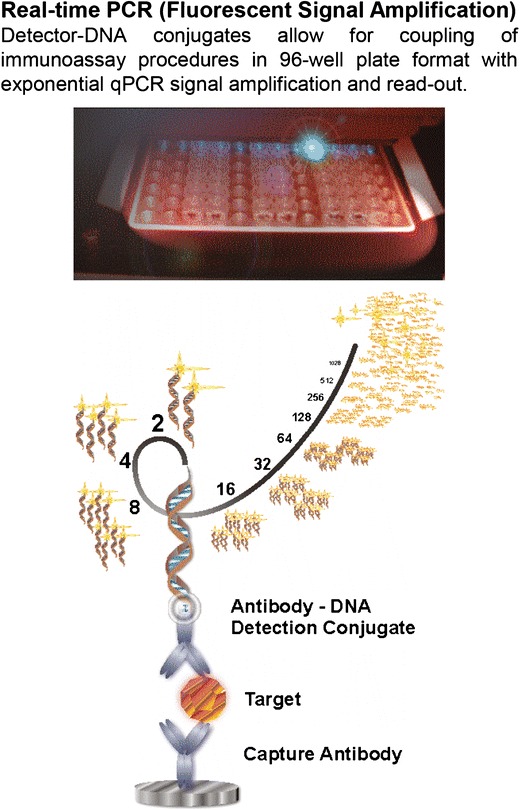
Chimera: Imperacer® ImmunoPCR (IPCR) technology
Applications of Imperacer® IPCR
Although the IPCR technology has been around for quite some time and appears to be a well-established immunoassay technology (28,31,33–35), technical improvements and novel IPCR-based applications addressing the assay sensitivity needs in bioanalysis are still emerging. Many new IPCR applications have been reported for biomarkers.
Case studies of the Imperacer® IPCR include: measuring biomarkers for neurodegenerative disorders in the periphery rather than brain tissue or cerebrospinal fluid (36); micro-sampling bioanalytics for metabolic biomarkers (9,37); and quantifying cytokines in the sub-picogram per milliliter concentration range (27,32). Sample testing for pharmacokinetic studies can typically be supported using less sensitive methods such as ELISA. However, measuring drug levels of certain drug candidates such as cytotoxic compounds (29,38) or potent toxins (39,40) that are administered at very low doses may require highly sensitive assays as well. Imperacer® sensitivity can also be employed to reduce matrix effects or avoid interference by endogenous compounds during drug quantification, e.g., to support replacement therapy trials (41). In relation to immunogenicity testing for biologics, it was demonstrated that Imperacer® assays can be optimized to achieve high drug tolerance in ADA detection (42). Similar to other LBA platforms, the pH-driven dissociation of ADA/drug-complex may also be employed to further improve drug tolerance in Imperacer® methods (43). However, acid dissociation may not be required when using the Imperacer® platform as high drug tolerance of 1:1,000 (ADA/drug) and even better was demonstrated with Imperacer® without any sample pre-treatment (44). Highly sensitive IPCR assays can also be used to detect desired immune responses in vaccination programs. For example, Imperacer® was successfully used to support bioanalytical sample testing in a phase-I clinical trial of the virosome vaccine program for protection from mucosal transfected diseases like HIV (45,46). Imperacer® enabled the quantification of vaccine-specific as well as total IgA and IgG responses in mucosal and serum samples with 100-fold less abundant antibody concentrations in mucosal tissue compared with serum, as well as with limited availability of the mucosal sample.
DISCUSSION
Developing protein therapeutics for complex disease indications poses unique bioanalytical challenges and highlights the need for additional tools. In many cases, serum is used as a surrogate matrix for evaluating drug levels and biomarkers at the site of action and, therefore, the circulating analyte levels can be very low. In addition, the concentrations of many important biological biomarkers are frequently below the detection capability of ELISA, which has been the method of choice for quantification of protein analytes and is still the gold standard. However, the ability to quantify protein therapeutics as well as biomarker levels in clinical samples is extremely important for selecting doses, evaluating efficacy, as well as gaining a better understanding of the diagnosis and prognosis of many diseases (47). Emerging technologies offering improved assay sensitivity by combining antibody-based molecular recognition with various amplification and detection techniques may aid in filling this gap.
Erenna®, Simoa™, and Imperacer® are three technology platforms in the immunoassay toolbox that promise improved assay sensitivity at sub-picogram to femtogram per milliliter levels, large dynamic range of ∼4 logs, with acceptable accuracy and precision. Each of these technologies utilizes a unique innovative approach to achieve high sensitivity and therefore has its benefits and constraints as summarized in Tables I and II. The decision to utilize one bioanalytical platform over another is multifaceted and will depend not only on the sensitivity requirements but also on platform accessibility, sample volume availability, ease of development, platform flexibility, and throughput needs. Another important consideration in selecting an appropriate technology is availability of off-the-shelf kits, custom kits by the vendor, as well as ability to develop home brew assays.
The ability to develop in-house or home brew assays are essential to users when considering a platform as it is much more convenient and economical and provides greater control over the assay, as well as utility of the technology. In many cases, the user is interested in evaluating a number of reagents and analytes, and it would be impractical to rely on a third party for reagent preparation and assay development. In addition, transferring reagents requires legal documents which can be time-consuming and could jeopardize project timelines.
The initial business model of Singulex required the vendor to carry out all reagent labeling and assay feasibility testing. One rationale for this decision was to ensure good quality reagents that would deliver the sensitivities that were claimed by the vendor. However, reagent labeling with biotin and Alexa fluorophore are well-established methods across industry and can be executed successfully by customers. Singulex has since changed its business practice by allowing customers to do their own reagent labeling and assay feasibility work. The latter has greatly enhanced the utility of Singulex platform. In the case of IPCR, DNA labeling of detection reagents can be performed using commercially available kits such as Antibody-Oligonucleotide All-in-One Conjugation Kit from Solulink™, though vendor-offered labeling services specializing in such labeling may be preferred to ensure optimal IPCR assay performance. The current business model of Imperacer® provider, Chimera Biotec, is to provide full bioanalytical clinical research organization (CRO) services, as well as to supply customers with labeled reagents and assay kits. Although antibody–DNA conjugation kits are not available through Chimera Biotec, home brew assay development is supported with the use of a generic anti-biotin–DNA or streptavidin–DNA conjugate and analyte specific kits. As for Simoa™, home brew assay development has not yet been instituted, although this platform is meant to be an open platform, and reagent labeling in this case (bead coupling and biotinylation) should be relatively straightforward.
In addition to sensitivity requirements, quantification of multiple low abundant analytes (such as biomarkers) often necessitates relatively large sample volumes, which may be hard to come by. Therefore, technologies requiring low sample volumes or technologies offering multiplexing capabilities would be desirable. Both Imperacer® and Simoa™ technologies are capable of using less sample volumes compared with ELISA. Imperacer® achieves its sensitivity through an amplification step enabling use of more diluted samples, which also tends to reduce matrix interference. Chimera Biotec has developed an extensive sample dilution buffer library to complement their IPCR technology. Erenna® technology generally requires larger volumes compared with Imperacer® and Simoa™ technologies to achieve the same sensitivity. While this technology does offer a plate-based format requiring lower volumes with higher throughput, it is at the expense of lower assay sensitivity.
Throughput is another important factor to consider. Imperacer® and Erenna® technologies tend to have lower throughput compared with the Simoa™ technology. The Simoa platform is completely automated, so there is less hands-on work compared with either Imperacer® or Erenna® technologies, has quicker signal detection and data acquisition system, and is capable of multiplexing (up to 10-plex), which also further increases throughput. Simoa™ is the only one of the three platforms that takes advantage of miniaturization to improve sensitivity and throughput. The miniaturized nature of the array lends itself to small diffusion distance, as opposed to the large volumes in LBA microwell plates, and this, in turn, promotes single molecule resolution and faster data acquisition.
Another very important consideration is the assay reagents. In principle, the higher the affinity of an antibody to its target, the higher is the assay sensitivity that may be achieved. However, high-quality reagents are not always available. In general, bead-based technologies such as Erenna® and Simoa™ seem to be more forgiving in situations where the assay reagents are not optimal. This is due to the three-dimensional surface of the beads, which allows for greater ligand coating and subsequent analyte capture compared with the traditional two-dimensional surface such as microtiter plates.
Simoa™ technology is the newest of the three technologies, and therefore, there is limited published information on its applications. There is also currently limited bioanalytical CRO presence for this technology. Nevertheless, based on the current information, this technology holds substantial promise for broad application in the future. However, a more thorough comparison of its capabilities is needed once it is available and widely used.
One word of caution with all ultrasensitive platforms is that special care should be taken to avoid contamination of tubes, plates, or reservoirs with particulates such as liquid aerosols or dust components. Best laboratory practices include using filtered pipette tips and plate covers, and keeping tubes/plates sealed or covered whenever possible during experiment in order to avoid high background and incorrect quantification. There is also a need to ensure that the signal detected is a “true” measurement of an analyte. It is therefore essential to demonstrate specificity of the signal through competition or immunodepletion step and illustrate the ability to inhibit a specific signal.
CONCLUSIONS
This evaluation is a snapshot of how these technologies perform today. Technologies evolve at a fast rate, and in many instances, the changes/improvements are driven by user input. It is therefore critical to not only evaluate emerging technologies as they come into the market but also to re-evaluate and re-assess the currently available technologies as they evolve and as new data become available.
The results from the “proof-of-concept” studies discussed here have demonstrated that the three technologies—SMC™, Simoa™, and Imperacer®, have the potential to significantly change the landscape of clinical macromolecule analytics. They provide the opportunity to enable the detection of diagnostic, prognostic, and disease-related biomarkers at earlier stage. This could provide a rare opportunity for early disease intervention and, in turn, the enhancement of patient lives.
However, an important consideration with new technology implementation during drug development is the business and regulatory risk of using platforms that are supported by a single vendor. The impact was very evident by the recent discontinuation of the widely used BioVeris electrochemiluminescent-based platform and its effect on ongoing clinical trials that were using this platform (48). In order to minimize the risk from the impact of potential technology discontinuation, it would be imperative to have multiple platform alternatives with similar benefits as with the three technologies discussed in this evaluation.
REFERENCES
- 1.Engvall E, Perlmann P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry. 1971;8(9):871–4. doi: 10.1016/0019-2791(71)90454-X. [DOI] [PubMed] [Google Scholar]
- 2.Lequin RM. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA) Clin Chem. 2005;51(12):2415–8. doi: 10.1373/clinchem.2005.051532. [DOI] [PubMed] [Google Scholar]
- 3.Novo P, Prazeres DM, Chu V, Conde JP. Microspot-based ELISA in microfluidics: chemiluminescence and colorimetry detection using integrated thin-film hydrogenated amorphous silicon photodiodes. Lab Chip. 2011;11(23):4063–71. doi: 10.1039/c1lc20362b. [DOI] [PubMed] [Google Scholar]
- 4.Kuo HT, Yeh JZ, Wu PH, Jiang CM, Wu MC. Application of immunomagnetic particles to enzyme-linked immunosorbent assay (ELISA) for improvement of detection sensitivity of HCG. J Immunoass Immunochem. 2012;33(4):377–87. doi: 10.1080/15321819.2012.655820. [DOI] [PubMed] [Google Scholar]
- 5.Li H, McGuire TC, Muller-Doblies UU, Crawford TB. A simpler, more sensitive competitive inhibition enzyme-linked immunosorbent assay for detection of antibody to malignant catarrhal fever viruses. J Vet Diagn Investig Off Publ Am Assoc Vet Lab Diagnosticians Inc. 2001;13(4):361–4. doi: 10.1177/104063870101300417. [DOI] [PubMed] [Google Scholar]
- 6.Tang S, Hewlett I. Nanoparticle-based immunoassays for sensitive and early detection of HIV-1 capsid (p24) antigen. J Infect Dis. 2010;201(Suppl 1):S59–64. doi: 10.1086/650386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sano T, Smith CL, Cantor CR. Immuno-PCR: very sensitive antigen detection by means of specific antibody-DNA conjugates. Science. 1992;258(5079):120–2. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- 8.Adler M, Spengler M. Novel strategies and tools for enhanced sensitivity in routine biomolecule analytics. Curr Pharm Anal. 2009;5(4):390–407. doi: 10.2174/157341209789649104. [DOI] [Google Scholar]
- 9.Adler M, Wacker R, Niemeyer CM. Sensitivity by combination: immuno-PCR and related technologies. Analyst. 2008;133(6):702–18. doi: 10.1039/b718587c. [DOI] [PubMed] [Google Scholar]
- 10.Dunn ME, Coluccio D, Hirkaler G, Mikaelian I, Nicklaus R, Lipshultz SE, et al. The complete pharmacokinetic profile of serum cardiac troponin I in the rat and the dog. Toxicol Sci. 2011;123(2):368–73. doi: 10.1093/toxsci/kfr190. [DOI] [PubMed] [Google Scholar]
- 11.Gauvreau GM, Boulet L-P, Cockcroft DW, FitzGerald JM, Carlsten C, Davis BE, et al. Effects of interleukin-13 blockade on allergen-induced airway responses in mild atopic asthma. Am J Respir Crit Care Med. 2011;183(8):1007–14. doi: 10.1164/rccm.201008-1210OC. [DOI] [PubMed] [Google Scholar]
- 12.Shukla R, Santoro J, Bender FC, Laterza OF. Quantitative determination of human interleukin 22 (IL-22) in serum using Singulex-Erenna® technology. J Immunol Methods. 2013;390(1–2):30–4. doi: 10.1016/j.jim.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 13.St. Ledger K, Agee SJ, Kasaian MT, Forlow SB, Durn BL, Minyard J, et al. Analytical validation of a highly sensitive microparticle-based immunoassay for the quantitation of IL-13 in human serum using the Erenna® immunoassay system. J Immunol Methods. 2009;350(1–2):161–70. doi: 10.1016/j.jim.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Walt DR. Optical methods for single molecule detection and analysis. Anal Chem. 2012;85(3):1258–63. doi: 10.1021/ac3027178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu AHB, Fukushima N, Puskas R, Todd J, Goix P. Development and preliminary clinical validation of a high sensitivity assay for cardiac troponin using a capillary flow (single molecule) fluorescence detector. Clin Chem. 2006;52(11):2157–9. doi: 10.1373/clinchem.2006.073163. [DOI] [PubMed] [Google Scholar]
- 16.Todd J, Freese B, Lu A, Held D, Morey J, Livingston R, et al. Ultrasensitive flow-based immunoassays using single-molecule counting. Clin Chem. 2007;53(11):1990–5. doi: 10.1373/clinchem.2007.091181. [DOI] [PubMed] [Google Scholar]
- 17.Esparza TJ, Zhao H, Cirrito JR, Cairns NJ, Bateman RJ, Holtzman DM, et al. Amyloid-beta oligomerization in Alzheimer dementia versus high-pathology controls. Ann Neurol. 2013;73(1):104–19. doi: 10.1002/ana.23748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarasow TM, Penny L, Patwardhan A, Hamren S, McKenna MP, Urdea MS. Microfluidic strategies applied to biomarker discovery and validation for multivariate diagnostics. Bioanalysis. 2011;3(19):2233–51. doi: 10.4155/bio.11.224. [DOI] [PubMed] [Google Scholar]
- 19.Dudal S, Baltrukonis D, Crisino R, Goyal MJ, Joyce A, Österlund K, et al. Assay formats: recommendation for best practices and harmonization from the Global Bioanalysis Consortium harmonization team. AAPS J. 2014;16(2):194-205. [DOI] [PMC free article] [PubMed]
- 20.Dyleski L, Joyce, A., et al. Method development and validation of an alternative immunoassay platform for PK studies in a regulated environment. Poster session presented at: AAPS National Biotechnology Conference, May 16-18, 2011; San Francisco, CA.
- 21.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotech. 2010;28(6):595–9. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rissin DM, Fournier DR, Piech T, Kan CW, Campbell TG, Song L, et al. Simultaneous detection of single molecules and singulated ensembles of molecules enables immunoassays with broad dynamic range. Anal Chem. 2011;83(6):2279–85. doi: 10.1021/ac103161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang L, Rissin DM, Fournier DR, Piech T, Patel PP, Wilson DH, et al. Single molecule enzyme-linked immunosorbent assays: theoretical considerations. J Immunol Methods. 2012;378(1–2):102–15. doi: 10.1016/j.jim.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rissin DM, Kan CW, Song L, Rivnak AJ, Fishburn MW, Shao Q, et al. Multiplexed single molecule immunoassays. Lab Chip. 2013;13(15):2902–11. doi: 10.1039/c3lc50416f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shahim P, Tegner Y, Wilson DH, Randall J, Skillback T, Pazooki D, et al. Blood biomarkers for brain injury in concussed professional ice hockey players. JAMA Neurol. 2014;71(6):684-92. [DOI] [PubMed]
- 26.Rissin DM, Kan CW, Song L, Rivnak AJ, Fishburn MW, Shao Q, et al. Multiplexed single molecule immunoassays. Lab Chip. 2013;13(15):2902–11. doi: 10.1039/c3lc50416f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niemeyer CM, Adler M, Wacker R. Detecting antigens by quantitative immuno-PCR. Nat Protoc. 2007;2(8):1918–30. doi: 10.1038/nprot.2007.267. [DOI] [PubMed] [Google Scholar]
- 28.Niemeyer CM, Adler M, Wacker R. Immuno-PCR: high sensitivity detection of proteins by nucleic acid amplification. Trends Biotechnol. 2005;23(4):208–16. doi: 10.1016/j.tibtech.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Adler M, Langer M, Witthohn K, Wilhelm-Ogunbiyi K, Schoffski P, Fumoleau P, et al. Adaptation and performance of an immuno-PCR assay for the quantification of Aviscumine in patient plasma samples. J Pharm Biomed Anal. 2005;39(5):972–82. doi: 10.1016/j.jpba.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Zhao Q, Li XF, Le XC. Ultrasensitive assays for proteins. Analyst. 2007;132(8):724–37. doi: 10.1039/b704256f. [DOI] [PubMed] [Google Scholar]
- 31.Schiavo S, El-Shafey A, Jordan N, Orazine C, Wang W, Zhou X, et al. Pushing the limits of detection with Immuno-PCR. PharmaGenomics. 2004:36–45.
- 32.Spengler M, Adler M. Development and validation of an ultra-sensitive Imperacer (Immuno-PCR) method for quantification of human granulocyte macrophage colony Stimulating factor (GM-CSF). American Association of Pharmaceutical Scientists, National Biotechnology Conference; San Diego, USA: American Association of Pharmaceutical Scientists; 2012.
- 33.Adler M. Immuno-PCR as a clinical laboratory tool. Adv Clin Chem. 2005;39:239–92. doi: 10.1016/S0065-2423(04)39009-8. [DOI] [PubMed] [Google Scholar]
- 34.Janssen KP, Knez K, Spasic D, Lammertyn J. Nucleic acids for ultra-sensitive protein detection. Sensors. 2013;13(1):1353–84. doi: 10.3390/s130101353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nong RY, Gu J, Darmanis S, Kamali-Moghaddam M, Landegren U. DNA-assisted protein detection technologies. Expert Rev Proteomics. 2012;9(1):21–32. doi: 10.1586/epr.11.78. [DOI] [PubMed] [Google Scholar]
- 36.Luk C, Compta Y, Magdalinou N, Marti MJ, Hondhamuni G, Zetterberg H, et al. Development and assessment of sensitive immuno-PCR assays for the quantification of cerebrospinal fluid three- and four-repeat tau isoforms in tauopathies. J Neurochem. 2012;123(3):396–405. doi: 10.1111/j.1471-4159.2012.07911.x. [DOI] [PubMed] [Google Scholar]
- 37.Diel P, Schiffer T, Geisler S, Hertrampf T, Mosler S, Schulz S, et al. Analysis of the effects of androgens and training on myostatin propeptide and follistatin concentrations in blood and skeletal muscle using highly sensitive immuno PCR. Mol Cell Endocrinol. 2010;330(1–2):1–9. doi: 10.1016/j.mce.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 38.Schoffski P, Riggert S, Fumoleau P, Campone M, Bolte O, Marreaud S, et al. Phase I trial of intravenous aviscumine (rViscumin) in patients with solid tumors: a study of the European Organization for Research and Treatment of Cancer New Drug Development Group. Ann Oncol. 2004;15(12):1816–24. doi: 10.1093/annonc/mdh469. [DOI] [PubMed] [Google Scholar]
- 39.Tarcha EJ, Chi V, Munoz-Elias EJ, Bailey D, Londono LM, Upadhyay SK, et al. Durable pharmacological responses from the peptide ShK-186, a specific Kv1.3 channel inhibitor that suppresses T cell mediators of autoimmune disease. J Pharmacol Exp Ther. 2012;342(3):642–53. doi: 10.1124/jpet.112.191890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Londono LM, Tarcha EJ, Adler M, Punnamoottil B, Iadonato SP. Development of an Imperacer (Immuno-PCR) method for clinical phase Ia human trial of ShK-186, a specific and potent peptide inhibitor of the Kv1.3 potassium channel. EBF 6th Open Meeting-Moving Forward Together -; Barcelona: The European Bioanalysis Forum (EBF); 2013.
- 41.Spengler M, Adler M, Jani D, Plavina T, Sommer J, Goyal J. Development of an Immuno-PCR method with extreme robustness against the presence of endogenous counterpart for clinical PK study bioanalytical support of replacement therapy Fc-fusion protein. American Association of Pharmaceutical Scientists, National Biotechnology Conference; San Diego, USA: American Association of Pharmaceutical Scientists; 2012.
- 42.Spengler M, Adler M, Jonas A, Niemeyer CM. Immuno-PCR assays for immunogenicity testing. Biochem Biophys Res Commun. 2009;387(2):278–82. doi: 10.1016/j.bbrc.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 43.Vermet L, Spengler M, Schroeder H, Cortez P. Technology comparison of two ligand-binding assay platforms for anti-drug–antibody sample testing support for a non-disclosed lead compound: MSD vs. Imperacer. American Association of Pharmaceutical Scientists, National Biotechnology Conference; San Diego, USA: American Association of Pharmaceutical Scientists; 2014.
- 44.Jani D, Sobolowski J, Keefe J, Adler M, Goyal J. From ELISA to a novel Immuno-PCR (i-PCR) based immunogenicity assay in quest to improve the drug tolerance. American Association of Pharmaceutical Scientists, National Biotechnology Conference; San Francisco, USA: American Association of Pharmaceutical Scientists; 2010.
- 45.Leroux-Roels G, Maes C, Clement F, van Engelenburg F, van den Dobbelsteen M, Adler M, et al. Randomized phase I: safety, immunogenicity and mucosal antiviral activity in young healthy women vaccinated with HIV-1 Gp41 P1 peptide on virosomes. PLoS One. 2013;8(2):e55438. doi: 10.1371/journal.pone.0055438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spengler M, Adler M, Chalifour A, Fleury S. Development and GLP-validation of Imperacer® (Immuno-PCR) methods for ultrasensitive quantification of mucosal antibodies in support of phase-I clinical trial for a novel vaccination strategy in preventing HIV infection. American Association of Pharmaceutical Scientists, National Biotechnology Conference; San Diego, USA: American Association of Pharmaceutical Scientists; 2012.
- 47.Cao Z-J, Peng Q-W, Qiu X, Liu C-Y, Lu J-Z. Highly sensitive chemiluminescence technology for protein detection using aptamer-based rolling circle amplification platform. J Pharm Anal. 2011;1(3):159–65. doi: 10.1016/j.jpha.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yohrling J. Ligand-binding assays: risk of using a platform supported by a single vendor. Bioanalysis. 2009;1(3):629–36. doi: 10.4155/bio.09.46. [DOI] [PubMed] [Google Scholar]


