Abstract
An HIV-infected patient presenting an unexpected viral escape under combined antiretroviral treatment is described. The virus isolated from plasma contained a large deletion in the HIV-1 integrase gene but no known resistance mutation. Nested polymerase chain reactions (PCRs) with patient virus integrase-specific primers and probes were developed and used to detect the mutant from plasma, blood, rectal biopsies, and sperm. The variant progressively emerged during a period of therapy-induced virosuppression, and persisted at a low but detectable level for at least 5 years. Surprisingly, proviral DNA from lymphocytes, rectal cells, and sperm cells was, and remained, mainly wild type. Cellular HIV RNA with the deletion was detected only once from the rectum. The origin and mechanisms underlying this so far not described production at a detectable level are largely hypothetical. This observation raised concern about the ability of defective viruses to spread.
Introduction
The accumulation of defective virus in peripheral blood mononuclear cells (PBMCs) has long been observed in human immunodeficiency virus (HIV) infection.1,2 These defective viruses reside in circulating PBMCs and in the viral reservoirs as integrated provirus and result from the high replication rate of HIV-1 and the low fidelity of reverse transcriptase (RT). Cells infected with defective HIV-1 have recently been estimated to outnumber by around 50-fold the reservoir of cells with replication-competent provirus.3 Generally, defective viruses are not detected as a circulating free viral population since they cannot be assembled or secreted and cannot perpetuate the infection.
However, defective provirus can be detected in PBMCs and other reservoir sites. Cells actively replicating the virus rapidly die due to the viral cytopathic effect and cytotoxic immune response. Cells that are latently infected or infected with defective proviruses are less susceptible to cytotoxicity and may survive during their natural life span.4 Among reservoir cells are resting memory as well as naive T cells and cells from the monocyte/macrophage lineage.5,6 The half-life of these cells is variable, with central memory cells having the greatest long-term persistence.4
The existence of a reservoir of infected cells, not detected by routine techniques, in treated patients with suppressed viral load for decades is now generally admitted. However, the source of this reservoir has not yet been discovered. It can be restricted to certain cells or sanctuary sites.
We report the case of a patient with an optimally suppressed viremia under combination antiretroviral therapy (cART) for several years in whom a variant emerged in the plasma, carrying a deleted integrase (IN) gene without RT or protease resistance mutations, that persisted for several years at a low level while the proviral HIV DNA in PBMCs was predominately wild type (WT). This case raises the problem of the productive source of this defective variant.
Materials and Methods
Patient characteristics
The patient was diagnosed with HIV-1 in February 2004 in the context of a zoster eruption. Serology for human T cell lymphotropic virus (HTLV) was negative and serology for hepatitis B virus was positive for a cured infection. The patient was anti-hepatitis C virus (HCV) negative during the follow-up period. Positive polymerase chain reactions (PCRs) for herpes simplex virus (HSV) and Epstein–Barr virus (EBV) were observed in 2012 while cytomegalovirus (CMV) DNA remained undetectable.
Viral load determination and sequencing were done from plasma samples collected during the patient's routine follow-up and stored at −80°C. PBMCs were isolated from June 2011, September 2011, and October 2012 blood samples. Total blood was stored in January 2012. Two rectal biopsies and sperm were collected in March 2012.
The patient has been subjected to positron emission tomography (PET) imaging and a thorough hematologic examination in January 2013.
Viral load measurement
From 2004 to 2006 viral loads were monitored with the bDNA 3.0 kit (Bayer Versant HIV 3.0, Bayer Diagnostics). The Nasba test was used (NucliSens HIV-1 EasyQ v1.1, BioMerieux) from the end of 2006 until August 2007, and was replaced afterward by the Abbott m2000 Real Time HIV assay (Abbott Diagnostics).
Cell immunophenotyping and sorting
Cellular immunophenotyping of CD4+ and CD8+ T cells subtypes, including Tregs, was performed on a Navios flow cytometer (Beckman Coulter, Miami, FL) from fresh blood sampled in October 2012. Treg cells were characterized based on a CD4+CD25highFoxP3+ phenotype. Activated T cells were identified using (1) increased HLA-DR expression for CD4+ T cells and (2) increased expressions of HLA-DR, or CD38, or both (HLA-DR+CD38+ cells) for CD8+ T cells. Stainings were performed using anti-CD4-Pacific Blue (PB), anti-CD8-AlexaFluor 750 (AF750), anti-CD45RA-fluoresceine isothiocyanate (FITC), anti-HLA-DR-krome orange (KO), anti-CD38-phycoerythrin-cyanine 5.5 (PC5.5), anti-CD25-phycoerythrin (PE), and anti-CD127-PC7 antibodies (Beckman Coulter). Antihuman FoxP3 antibodies (clone 259D) were purchased from BioLegend (San Diego, CA). CD4+ lymphocyte cells were isolated from blood sampled on October 2012. Cell sorting was performed on a FACS Aria II (Becton-Dickinson, San Jose, CA).
DNA and RNA extraction from plasma, PBMCs, and tissue cells
PBMCs were isolated by centrifugation on a Ficoll layer, washed, and the pellet was stored at −80°C following the standard procedure. Sperm was centrifuged, seminal plasma was collected, the cell pellet was washed and both were stored at −80°C. Two fragments of rectal biopsies were directly stored at −80°C until extraction was performed.
All nucleic acid extractions were done with the NucliSens Easy Mag instrument (BioMerieux). Cellular nucleic acids were quantified with a NanoDrop spectrophotometer (ThermoScientific).
Before extraction, seminal cells were treated with 11 μl of proteinase K (Roche Diagnostics, ref 03 115 844 001) for 10 min at 56°C, followed by inactivation at 95°C. Rectal biopsies were treated with 140 μl of a trypsin/EDTA solution (Sigma, ref T3924) at 56°C for 30 min, then 95°C for 10 min.
The cell extracts used to amplify viral RNA were first digested with 10 U of DNase (Roche Diagnostics, ref 04 536 282 001) for 30 min at 37°C, which was then inactivated for 5 min at 95°C.
Population sequencing
Protease, reverse transcriptase, integrase, and V3 sequences were obtained by population-based sequencing (CEQ2000 sequencer, Beckman) using primers and PCR and sequencing conditions described in the ANRS (Agence Nationale de Recherches sur le SIDA et les Hépatites Virales, Paris, France) consensus techniques (www.hivfrenchresistance.org).
Specific detection of the patient integrase gene
The deleted integrase gene was detected and its proportion estimated with specific techniques developed for this particular sequence, described in details in the Supplementary Methods section (Supplementary Data are available online at www.liebertpub.com/aid). The differential nested PCR with primers specific for deleted or WT regions could detect one copy of the deleted gene in 103 copies of the wild-type gene (Supplementary Fig. S1). Copy number was estimated by real time PCR with specific probes, using a standard curve obtained from mixtures of plasmids containing the patient viral WT and deleted IN sequence, in increasing proportion (0, 1%, 10%, 50%, 90%, 99%, and 100%), at 100 copies/μl.
Results
Patient history
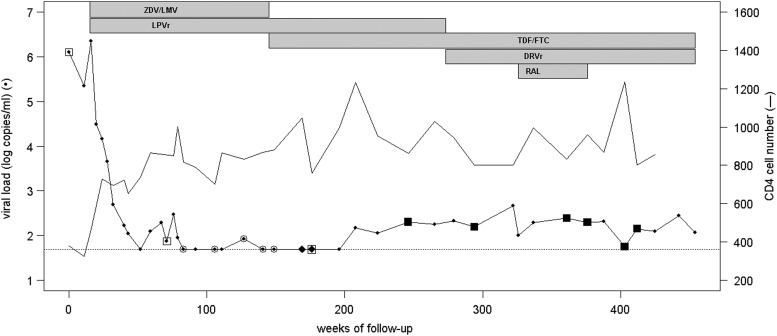
Antiretroviral treatment with zidovudine/lamivudine and lopinavir was initiated 15 weeks after diagnosis (time 0 of follow-up). HIV viral load decreased from more than 6 log to below 50 copies/ml and CD4 count increased from less than 400 to about 800/mm3 in 16 months, as shown in Fig. 1.
FIG. 1.
Schematic graph of the patient's follow-up data. Evolutions of viral load and CD4 cell count are represented before and during treatment periods with drugs shown at the top of the graph. Detection of the wild-type (WT) and deleted integrase by nested polymerase chain reaction (PCR) is indicated by different symbols: open square, WT detected by PCR for sequencing; open circle, negative nested PCR; black diamond, deleted variant detected by real time PCR with specific probes; black square, deleted variant detected by PCR for sequencing. ZDV, zidovudine; LMV, lamivudine; LPVr, ritonavir-boosted lopinavir; TDF, tenofovir; FTC, emtricitabine; DRVr, ritonavir-boosted darunavir; RAL, raltegravir.
Viral load remained below 50 copies/ml for more than 2 years (from week 83 to week 196), except for one observed blip at 84 copies/ml at week 127. After that HIV RNA increased slightly but persistently with values fluctuating from 56 to 466 copies/ml for more than 5 years.
Reverse transcriptase and protease genes had been sequenced for resistance genotyping before therapy and for the time on treatment, before and after the undetectable period. No known RT or protease resistance mutation was found at any time. The virus subtype was a recombinant CRF02.AG, with a predicted CCR5 tropism by V3 region genotyping (the geno2pheno algorithm). The treatment regimen was changed during follow-up (Fig. 1) by replacing zidovudine/lamivudine with tenofovir/emtricitabine at week 145, and then lopinavir with darunavir at week 273. As viremia persisted, the integrase inhibitor raltegravir was added to the tenofovir/emtricitabine/darunavir regimen (week 326) without any improvement, and then removed (week 376). Serum drug concentrations at the time of plasma HIV RNA detection were in the therapeutic ranges and were considered efficacious. Adherence to the treatment was good at all times.
Analysis of HIV-1 integrase by population sequencing
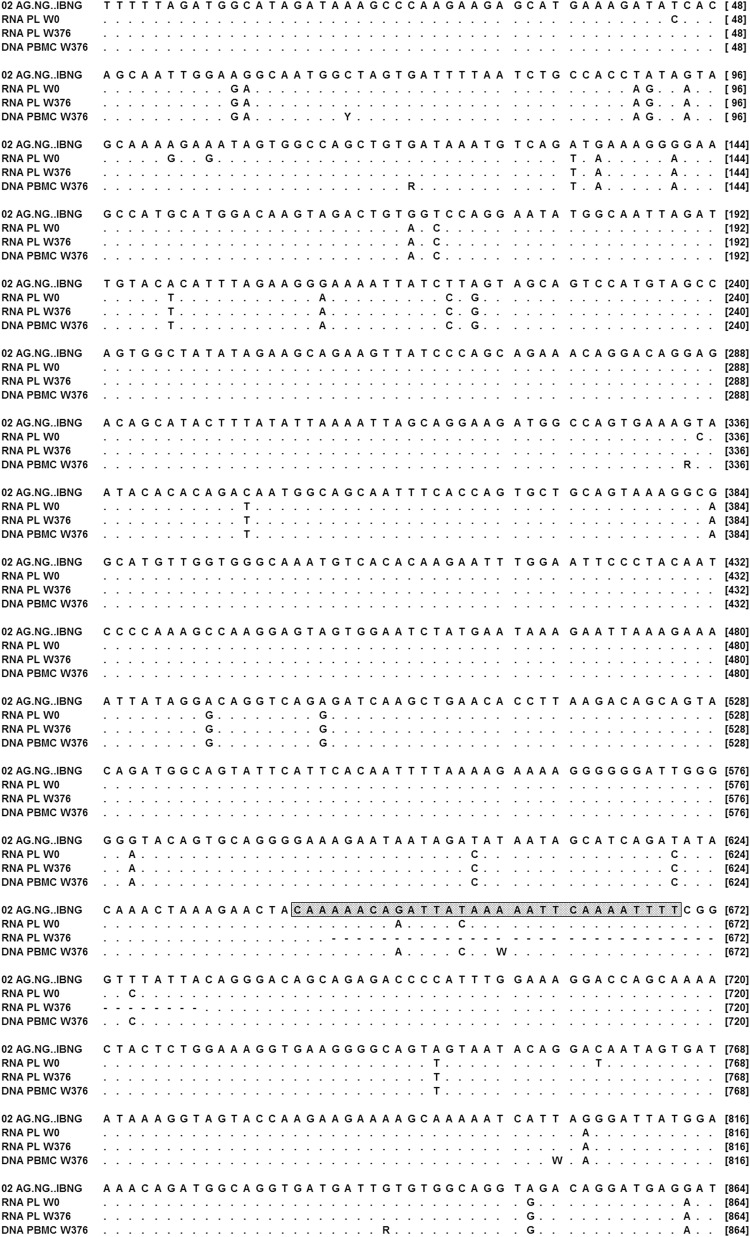
Since a low level of HIV-1 production persisted in the presence of raltegravir, the IN gene was sequenced from plasma virus. A 38-nucleotide deletion was observed in the IN C-terminal domain (CTD) encoding sequence (residues 215 to 227 of the WT IN). The deletion also induces a −1 frameshift leading to a stop codon at residue 245 on the WT IN (Fig. 2). Thus, the C-terminal end of IN is composed of a stretch of 18 residues that does not correspond to any known protein sequence. The resulting deleted protein is 233 residues long (instead of 288 for the WT IN).
FIG. 2.
Integrase sequence of the patient virus during follow-up. Consensus sequences obtained by population sequencing from plasma RNA at the time of diagnosis, before treatment (RNA PL W0), and at week 376 of follow-up, after the emergence of the deletion, as well as in the proviral DNA from peripheral blood mononuclear cells (PBMCs) at the same time (DNA PBMC W376) were aligned to the integrase sequence of the CRF02.AG reference virus (NG.IBNG). Symbols are points for identities to the reference sequence, and dashes represent deleted bases. A box is drawn around the central termination sequence.
Several previous plasma samples were sequenced to evaluate the period of emergence of this variant (Fig. 1). The first sample available at the time of diagnosis did not contain the deletion as well as those tested until week 176. At this time, the HIV RNA level was still below 50 copies/ml (using the NucliSens EasyQ HIV v1.1 kit, which was later found to underestimate this CRF02.AG virus). Attempts to amplify the integrase gene by nested PCR from plasma between week 83 and week 145 were unsuccessful. The deletion was first detected at week 246 when the HIV RNA level was above the Abbott Real Time HIV RNA kit threshold.
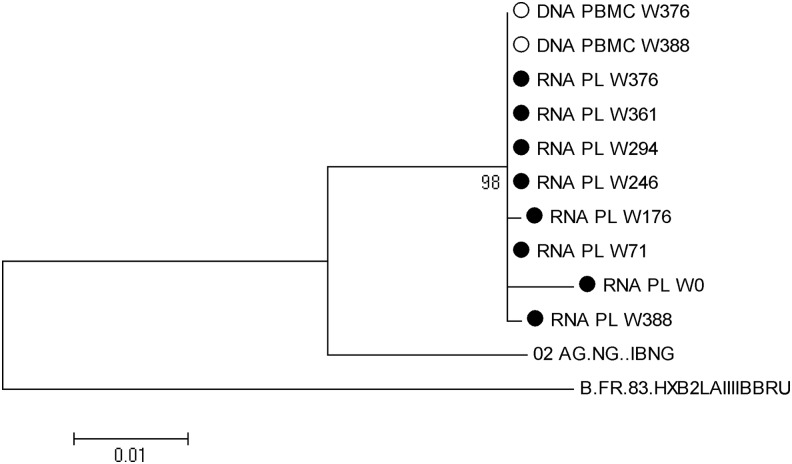
Surprisingly, at week 376, population sequencing from PBMCs showed a WT proviral integrase gene DNA, without evidence of the deletion (Fig. 2). A phylogenic analysis confirmed that all the integrase sequences clustered together (Fig. 3).
FIG. 3.
Phylogenic tree of the integrase sequences from the patient samples taken at different times during follow-up. Integrase genes from CRF02.AG and B reference viruses were included in the alignment. Maximum likelihood was used to construct the tree, with 500 bootstrap replications, using the MEGA version 5 software. Only branch frequencies above 70% are noted. RNA PL represents HIV RNA from plasma (closed symbol), and DNA PBMC proviral DNA from peripheral blood lymphocytes (open symbol). W indicates the week of follow-up at which the sample was collected.
Search for the source of deleted variant by specific PCR in different compartments
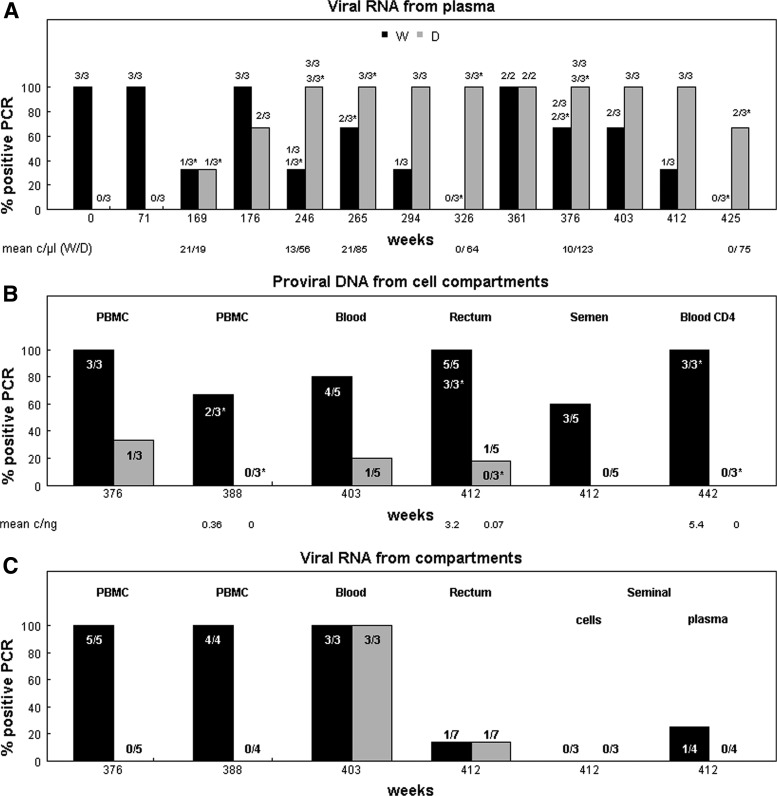
The apparent discrepancy between the viral and proviral circulating populations triggered a search to determine the origin of the variant virus. The presence of the deletion from plasma RNA, as well as RNA and DNA extracted from PBMCs and other compartments (rectal biopsy and sperm), was measured with two specific PCRs adapted to the patient's viral sequences. Qualitative results obtained with the two techniques, differential nested PCR with primers specific for the deleted or the WT regions and real time PCR with specific probes, are shown in Fig. 4. Quantitative results from the real time PCR are indicated below the corresponding samples in Fig. 4A and B. The results are expressed per μl of extract for plasma RNA and ng of nucleic acids in the extracts from cellular compartments. PCR from plasma was performed with an extract volume equivalent to 100 μl of plasma.
FIG. 4.
Evolution with time of the proportion of positive PCR for WT and deleted integrase in different virological compartments using specific primers and probes. Positive PCRs were recorded from each experiment using primers or probes specific for WT (W, black bars) and deleted (D, gray bars) integrase. Data are expressed in percent and the number of corresponding positive PCRs over the number of performed PCRs is indicated above or inside each bar. Asterisks correspond to real time PCR with specific probes, while other numbers are from primer-specific PCR. The collection time of samples is indicated in weeks from diagnostic, and samples types are indicated above the bar. Below the A and B graphs are noted, in copies/μl for plasma RNA, and copies/ng of nucleic acid for cellular proviral DNA, the quantitative results obtained by real time PCR for WT and deleted fragments, respectively. PBMC, peripheral blood mononuclear cells.
Replicates were tested to increase the chance of detecting low levels of variant, generally in triplicate but varying from two to five times.
The deleted variant likely emerged in the plasma around week 169, while HIV RNA was not detected by routine viral load assay (Fig. 1). Indeed, a positive result was obtained from RNA extracted at this time point with one of the three RT-PCR using the WT probe and another one using the deletion-specific probe in a similar amount. Later, the WT IN from plasma RNA, which was no longer detected by bulk sequencing, was still present, since some, but not all, RT-PCR using WT-specific primers were positive. However, the proportion of deleted plasma variant tended to increase with time, while WT did not (Fig. 4A).
The HIV DNA with WT integrase was amplified from each cell extract, with the lowest positivity rate occurring from seminal cells (Fig. 4B). The deleted provirus was infrequently detected in blood cells (2 over 14 PCRs from different time points) and rectal biopsy (1 over 8 PCRs). It was not amplified from PBMCs obtained at week 388, blood CD4 T cells at week 442, seminal cells, and one rectal biopsy specimen. The only proviral DNA-positive PCR using the deletion-specific probe was from the second rectal biopsy sample, while the WT was found in each DNA PCR from the two rectal biopsies.
WT viral RNA was detected in all RT-PCR from PBMCs and total blood, infrequently in the rectal cells and seminal fluid and not in the seminal cells (Fig. 4C). RT-PCRs specific for the deleted HIV RNA integrase gene were negative from PBMCs, seminal cells, and fluid. Detection of deleted IN from HIV RNA occurred once in the second rectal biopsy, and each time in whole blood, which was expected since it contains plasma virus.
Additional results
The enzymatic activities of the deleted IN could not be evaluated because of its known poor solubility.7,8 By using previously described techniques9 the WT IN was mainly detected in the pellet (insoluble fraction) and a small quantity was solubilized in the supernatant (soluble fraction). By contrast, the deleted IN protein was detected only in the bacteria pellet as an insoluble form and we were not able to purify it in sufficient quantity. No virus could be obtained by infecting phytohemagglutinin (PHA)-stimulated PBMCs from a seronegative subject with plasma from the HIV-1-infected patient in vitro.
Immunophenotyping of the different T cell populations did not detect any particular abnormality compared to 87 HIV-infected patients, virologically controlled for at least 1 year of therapy, when considering CD4+ T cells, activated CD4+ T cells, and Treg cells.10 Only an increase in the percentage of activated CD8+ T cell subsets (i.e., CD8+HLA-DR+, CD8+CD38+, and CD8+CD38HLA-DR+ T cells) was observed.
PET imaging did not reveal any evidence of malignancy.
Discussion
We report the case of a patient with a detectable viremia during antiretroviral treatment, without known resistance mutations in the RT, protease, and IN genes of HIV, but whose virus harbored a large deletion and a frameshift in the IN gene.
Although not confirmed by testing the IN enzymatic activity, it is likely that the deleted IN is not functional. Indeed, the CTD domain is required for IN enzymatic activities and oligomerization,11 whereas only five IN residues of the CTD are conserved in the IN mutant. It is unlikely that the nonstructured stretch of the 18-residue might replace the entire functions of the CTD domain as numerous residues known to bind to DNA or to participate in the oligomerization of the entire IN are lacking.8,12
This likely defective virus emerged while replication was profoundly suppressed by therapy. The emergence of the mutant was associated with a small but persistent increase in viremia while adherence to treatment remained good.
This variant appeared in the presence of the WT virus but was later predominantly produced and released in the circulation, considering a proportion of WT virus below 20–25%, the detection threshold of population sequencing. The overproduction of deleted virus was confirmed by quantitative RT-PCR.
However, we could not precisely determine the source of this defective virus. Most of the circulating mononuclear cells were infected with WT virus. The detection of the variant provirus in rectal tissue suggested the presence of lymphoid cells producing the deleted virus, but the detection was so rare that it is difficult to determine whether it is the main source. The presence of such infected lymphocytes might have been underestimated because biopsy sizes were small and because lymphoid zones are not uniformly distributed on the intestinal surface.13,14 Quasispecies can also vary along the gut as previously reported.15,16 Other potential sanctuaries could not be tested (central nervous system, spleen, other gut regions, progenitor cells, etc.).
Important questions arising from this case are how and why this defective variant emerged during efficient virosuppression (113 weeks of viral load <50 copies/ml) and how it could be preferentially and stably produced.
In the context of viral replication blocked by antiretrovirals, residual viremia can result from on-going replication in a remote reservoir or from viral production by latently infected cells. There is debate about which of these mechanisms is the source of viral persistence, and probably both may exist.4,17–20 In the patient described here an on-going replication in any compartment would be possible only by continual coinfection with the WT virus.
Synthesis of this defective variant by chronically infected cells, carrying the deleted provirus, is a likely hypothesis for its persistence.18 In this scenario the chronically infected cells should be long lived and duplicate the integrated proviral DNA during cell division.
The residual viremia on which research now focused is generally undetectable by routine HIV RNA assays and can be detected only by ultrasensitive tests detecting one copy/ml. This production of a predominant circulating free variant, absent from the circulating CD4 T cells, could have the same origin as the predominant plasma clone (PPC) previously described as one potential source of residual viremia by Siliciano's team.21,22 However, a difference is that this variant is defective and is produced above the detection threshold of viral load assays, a unique and so far unexplained event.
Josefson et al.23 described a subject with a defective provirus, deleted in the protease gene, expanding in the circulating cell population. They speculate that the PPC identified by Bailey et al.21 arises from proliferating cells with replication-competent HIV. We show here that production of defective virus can be maintained several years by clonal expansion. Such long-term activation-associated virus production highlights the potential difficulties of therapies aimed at purging the reservoir. We can speculate that production of this defective virus could persistently stimulate an immune response to HIV (as suggested by the persistent CD8 T cells activation and high CD4 cell number), contributing to a natural control of infection. However, the source of this virus has not been eliminated, and infected cells with intact integrase gene provirus remain detectable 5 years after the detection of the interfering variant.
Clonal expansion with production of a low level of virus in a specific compartment could be possible if the cells are continuously activated, for example, by a persistent antigen such as a herpes virus. Positive amplifications of type 2 herpes virus and Epstein–Barr virus were observed in this patient (not shown), but whether there is any relationship between a specific immune response and low-level viremia has not been determined. Another hypothesis is an integration of the defective proviral DNA near a cellular oncogene as supported by results presented at the 2014 CROI conference.24,25 However, the HIV LTR might not be strong enough to activate an oncogene. Another possibility could be its insertion close to a strong and constitutive cellular promoter allowing the persistent expression and release of the deleted virus.
Considering the continuous proliferation of a cell population raises fear of malignancy, but neither clinical nor imaging signs have been found in this patient despite intensive exploration.
Another potential mechanism could be derived from a recent report showing that HIV-1 can be produced in vitro from unintegrated 2-LTR circles DNA after infection of nonactivated and nonproliferating cells in a cytokine environment.26 Virus production was detected only when activation was induced several days postinfection, with the help of Vpr-transactivating and cell growth-inhibiting activities.
A similar case of an accumulation of defective variants with a truncated RT has been described.27 However, there are differences between this patient and ours. Resistance mutations were found in the patient with defective RT, and the deletions were detected in the proviral DNA before their emergence in the plasma. The deleted variants persisted thereafter in the two compartments at a proportion reaching 100% by clonal analysis. However, after several years of poor ART compliance, the proportion of defective free virus declined, but not that of the defective provirus.28 The differences between these two patients might be linked to the site at which the cells producing these variants resided. The selective pressure for the RT-defective variants was suspected to be a strong T cell response against the C-terminal part of the protein. We did not evaluate the T cell response to the integrase of our patient, but the deleted sequence is in a region potentially containing CTL epitopes.29
However, even if a specific immune response to this region on the WT virus developed, the deleted variant emerged while viral replication was optimally suppressed by antivirals in this adherent patient, which was not the case for the patient with an RT-defective virus.
The location of the deletion indicates that the event might have occurred late in the reverse transcription step or just after integration in the cellular DNA. Indeed, the deletion begins at the central termination sequence (CTS) where plus-strand synthesis, initiated with the polypurine tract (PPT) genomic RNA (gRNA), stopped, and downstream of the central PPT (cPPT) initiating 3′-half plus-strand synthesis (see Fig. 2). This is thus a region at which the plus strand is composed of two molecules with a triple-stranded “flap” at the junction and a gap repaired before or after integration in the nuclear DNA.30,31 It can be hypothesized that the repair did not proceed correctly and more than the exceeding region has been removed from the plus strand, with a necessary parallel excision from the minus strand. Another possibility is recombination at the level of the TACA homology at the deletion junction, with the insertion of an A, as previously shown with other retroviruses.32
Other cases of adherent-treated patients with a low but detectable viral load for a long period, and without a resistance mutation, were reported.33 Such instances have not previously been associated with a defective virus, but it would be advisable to further explore these cases. A defect can occur almost anywhere in the virus genome, but defects have been observed mainly in the central part of the genome,2 where the integrase gene is located. Finding the location and cell type producing such viruses with this distinguishable trait can help target a compartment other than blood in which the virus replicates.
Supplementary Material
Acknowledgments
The authors thank M-H. Teillon-Beranger, M-F. Rabel, J. Driussi, M. Carteron, Kathy Gallay, and Catherine Luengo for their technical assistance. We also thank the biological resources center “Virobiotec” for storing and handling patient's samples. This work was supported by Hospices Civils de Lyon (grant D50624).
The GenBank accession numbers of the WT and deleted IN sequences are KJ173816 and KJ173817.
These data have been presented at the 2nd ANRS Basic Research Symposium on HIV and Hepatitis Viruses and at the 2014 International Symposium on HIV and Emerging Infectious Diseases (ISHEID).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Coffin JM: HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science 1995;267:483–489 [DOI] [PubMed] [Google Scholar]
- 2.Sanchez G, Xu X, Chermann JC, and Hirsch I: Accumulation of defective viral genomes in peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected individuals. J Virol 1997;71:2233–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho YC, Shan L, Hosmane NN, et al. : Replication competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013;155:540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomont N, El-Far M, Ancuta P, et al. : HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009;15:893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahl V, Josefsson L, and Palmer S: HIV reservoirs, latency, and reactivation: prospects for eradication. Antiviral Res 2010;85:286–294 [DOI] [PubMed] [Google Scholar]
- 6.Pace MJ, Agosto L, Graf EH, and O'Doherty U: HIV reservoirs and latency models. Virology 2011;411:344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins TM, Engelman A, Ghirlando R, and Craigie R: A soluble active mutant of HIV-1 integrase: Involvement of both the core and carboxyl-terminal domains in multimerization. J Biol Chem 1996;271:7712–7718 [DOI] [PubMed] [Google Scholar]
- 8.Dolan J, Chen A, Weber IT, Harrison RW, and Leis J: Defining the DNA substrate binding sites on HIV-1 integrase. J Mol Biol 2009;385:568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cellier C, Moreau K, Gallay K, Ballandras A, Gouet P, and Ronfort C: In vitro functional analyses of the human immunodeficiency virus type 1 (HIV-1) integrase mutants give new insights into the intasome assembly. Virology 2013;439:97–104 [DOI] [PubMed] [Google Scholar]
- 10.Saison J, Ferry T, Demaret J, et al. : Association between discordant immunological response to highly active anti-retroviral therapy, regulatory T cell percentage, immune cell activation and very low-level viraemia in HIV-infected patients. Clin Exp Immunol 2014;176:401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelman A: Reverse transcription and integration. In: Retroviruses: Molecular Biology Genomics and Pathogenesis (Kurth R, Bannert N, eds.). Caister Academic Press, Norfolk, UK, 2010, pp. 129–159 [Google Scholar]
- 12.Krishnan L, Li X, Naraharisetty HL, Hare S, Cherepanov P, and Engelman A: Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc Natl Acad Sci USA 2010;107:15910–15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mowat AM: Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol 2003;3:331–341 [DOI] [PubMed] [Google Scholar]
- 14.Ling B, Mohan M, Lackner AA, et al. : The large intestine as a major reservoir for simian immunodeficiency virus in macaques with long-term, nonprogressing infection. J Infect Dis 2010;202:1846–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Marle G, Gill MJ, Kolodka D, McManus L, Grant T, and Church DL: Compartmentalization of the gut viral reservoir in HIV-1 infected patients. Retrovirology 2007;4:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yukl SA, Shergill AK, McQuaid K, et al. : Effect of raltegravir-containing intensification on HIV burden and T-cell activation in multiple gut sites of HIV-positive adults on suppressive antiretroviral therapy. AIDS 2010;24:2451–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buzón MJ, Massanella M, Libre , et al. : HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat Med 2010;16:460–465 [DOI] [PubMed] [Google Scholar]
- 18.Chun T-W, Murray D, Justement JS, et al. : Relationship between residual plasma viremia and the size of HIV proviral DNA reservoirs in infected individuals receiving effective antiretroviral therapy. J Infect Dis 2011;204:135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poles MA, Boscardin WJ, Elliott J, et al. : Lack of decay of HIV-1 in gut-associated lymphoid tissue reservoirs in maximally suppressed individuals. J Acquir Immune Defic Syndr 2006;43:65–68 [DOI] [PubMed] [Google Scholar]
- 20.Evering TH, Mehandru S, Racz P, et al. : Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog 2012;8:e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey JR, Sedaghat AR, Kieffer T, et al. : Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol 2006;80:6441–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennan TP, Woods JO, Sedaghat AR, Siliciano JD, Siliciano RF, and Wilke CO: Analysis of human immunodeficiency virus type 1 viremia and provirus in resting CD4+ T cells reveals a novel source of residual viremia in patients on antiretroviral therapy. J Virol 2009;83:8470–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Josefson L, von Stokenstrom S, Faria NR, et al. : The HIV reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci USA 2013;110:E4987–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maldarelli F, Wu X, Kearney M, et al. : The role of HIV integration sites in extensive clonal expansion of infected cells in patients. Top Antivir Med 2014;22(e-1):193 [Google Scholar]
- 25.Wagner TA, McLaughlin S, Garg K, et al. : Proliferation of cells with HIV integrated into regulatory genes is a mechanism of persistence. Top Antivir Med 2014;22(e-1):69 [Google Scholar]
- 26.Trinite B, Ohlson EC, Voznescensky I, et al. : An HIV-1 replication pathway utilizing reverse transcription products that fail to integrate. J Virol 2013;87:12701–12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paolucci S, Foli A, Gulminetti R, et al. : HIV-1 plasma variants encoding truncated reverse transcriptase (RT) in a patient with high RT-specific CD8+ memory T-cell response. Curr HIV Res 2009;7:302–310 [DOI] [PubMed] [Google Scholar]
- 28.Paolucci S, Gulminetti R, Maserati R, Dossena L, and Baldanti F: Accumulation of defective HIV-1 variants in a patient with slow disease progression. Curr HIV Res 2011;9:17–22 [DOI] [PubMed] [Google Scholar]
- 29.Brockman MA, Chopera DR, Olvera A, et al. : Uncommon pathways of immune escape attenuate HIV-1 integrase replication capacity. J Virol 2012;86:6913–6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freed EO: HIV-1 replication. Somat Cell Mol Genet 2001;26:13–33 [DOI] [PubMed] [Google Scholar]
- 31.Poeschla E: The importance of becoming double-stranded: Innate immunity and the kinetic model of HIV-1 central plus strand synthesis. Virology 2013;441:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Girod A, Drynda A, Cosset FL, Verdier G, and Ronfort C: Homologous and nonhomologous retroviral recombinations are both involved in the transfer by infectious particles of defective avian leukosis virus-derived transcomplementing genomes. J Virol 1996;70:5651–5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laprise C, de Pokomandy A, Baril J-G, Dufresne S, and Trottier H: Virologic failure following persistent low-level viremia in a cohort of HIV-positive patients: Results from 12 years of observation. Clin Infect Dis 2013;57:1489–1496 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.