Summary
Propagation of episomally maintained KSHV genome into new daughter cells requires replication of its genome once every cell division. This study demonstrates a potential alternative mechanism of KSHV latent DNA replication independent of LANA expression. A cis-acting DNA element within the Long Unique Region is capable of initiating replication shown by DpnI analysis, Meselson-Stahl analysis, BrdU incorporation and single molecule analysis of replicated DNA. This DNA element supports replication of the plasmid which persists in its native form. Human ORC2 and MCM3 also associate with this region of the KSHV genome and depletion of cellular ORC2 by RNAi abolished replication of the plasmids. Thus recruitment of the host cellular replication machinery is important for latent replication. This autonomously replicating DNA element demonstrates that KSHV can initiate replication of its genome independent of any trans-acting viral factors and identifies a secondary replicator element for the initiation of KSHV DNA replication.
Introduction
Members of the herpesviridae family share the characteristic feature of persisting latently in their natural hosts and typically harbor viral genome as closed circular episomes (Roizman, 1996). Latent replication is thought to proceed from a defined origin, typically with the expression of a viral encoded protein for maintenance of the viral episomes (Roizman, 1996). Kaposi’s Sarcoma Associated Herpesvirus (KSHV/HHV8) primarily infects the human endothelial cells and B lymphocytes and shares similar genomic characteristics to Epstein Barr Virus (EBV) (Roizman, 1996). KSHV is tightly associated with the development of KS as well as two other lymphoproliferative diseases, primary effusion lymphoma (PEL) and Multicentric Castleman’s disease (MCD) (Verma et al., 2007).
KSHV is a member of the Rhadinovirus genera of the gammaherpesvirinae subfamily (Russo et al., 1996). The most closely related human herpesvirus is EBV a prototypic member of the other gammaherpesvirinae genera, Lymphocryptovirus (Kieff, 1996; Russo et al., 1996). Similar to EBV, KSHV infection within tumor cells is predominantly latent and expresses a limited number of viral genes which include LANA, viral cyclin and viral Fas-associated death domain (FAAD) interleukin-1B-converting enzyme (FLICE) inhibitory protein (v-FLIP) (Verma et al., 2007).
The KSHV latency-associated nuclear antigen (LANA) functions in both transcription (Verma and Robertson, 2003a) and plasmid maintenance (Ballestas et al., 1999; Cotter and Robertson, 1999). LANA is a large, 222-234 kDa nuclear protein expressed in virtually all KSHV latently infected cells (Kedes et al., 1997; Rainbow et al., 1997), protects against cell death by inhibition of p53-mediated transcription (Friborg et al., 1999; Verma and Robertson, 2003a) and regulates p53 and VHL through recruitment of the EC5S ubiquitin E3 ligase (Cai et al., 2006). Viral cyclin associates with cdk6 and phosphorylates human ORC1 promoting S phase entry for origin firing and replication (Laman et al., 2001a; Laman et al., 2001b).
Replication of latent EBV episomes requires the viral cis acting element, oriP, and a trans acting viral protein EBNA-1 bound to oriP DNA consisting of the family of repeats (FR) and dyad symmetry element (DS) and recruits the host cellular replication machinery (Rawlins et al., 1985; Reisman et al., 1985). The FR provides EBNA-1 binding sites important for anchoring the EBV episome to the nuclear matrix (Jankelevich et al., 1992). The DS element contains 4 EBNA-1 binding sites and replication appears to initiate at or immediately adjacent to the DS element although DS-independent EBV replication has been reported (Gahn and Schildkraut, 1989; Little and Schildkraut, 1995).
LANA has been suggested to play a role in partitioning of the viral genome during mitosis as a recombinant KSHV virus (BAC36ΔLANA) disrupted for LANA expression was inefficient in maintenance of the viral episomes (Ye et al., 2004). However, a recent report using short hairpin RNA (shRNA) to knockdown the oncogenic latent gene cluster including LANA, led to the reduction of KSHV genomic copies to a low but steady copy number (Godfrey et al., 2005).
Previous studies have shown that LANA is critical for replication of KSHV TR containing plasmids suggesting the presence of a LANA dependent origin within the TR. However, knock down of LANA was unable to eliminate KSHV episomes from the infected cells (Godfrey et al., 2005). Thus, additional ori sites within the KSHV genome may function independent of LANA. In this report we have demonstrated that a KSHV cis-acting DNA element within the KSHV long unique region (LUR) allowed for replication of the plasmid and suggests a functional oriP. Plasmids containing this region of the KSHV genome incorporate BrdU, precipitates with chromatin bound cellular replication machinery proteins ORC2 and MCM3, and replicates independent of any viral protein expressed in trans. Hence, this represents a potential unique mechanism for KSHV replication and suggests the possibility for development as a vector system for long term maintenance of episomal DNA without integration or gene transfer by ex-vivo strategies.
Results
A cis-acting element within the long unique region of KSHV genome replicates in transient replication assays
Previous studies demonstrated that Z6, the 33kbp left end of the KSHV genome, binds to LANA with high affinity therefore hypothesized to contain cis-acting elements important for DNA maintenance (Cotter and Robertson, 1999). The terminal repeat (TR), provides binding sites for LANA and is important for tethering of the viral episomes to the host chromosomes (Moore and Chang, 2003; Verma and Robertson, 2003a). TR supports replication of plasmids in eukaryotic cells (Grundhoff and Ganem, 2003; Hu et al., 2002; Verma et al., 2006). However, the replication potential of other regions of the genome was not extensively evaluated. Here we used a vector, pBSpuro which lacks a eukaryotic origin of replication and contain a puromycin acetyl transferase (Pac) gene, for the screening of replication potential of the Z6 fragments. Cloning of Z6 DNA fragments into pBSpuro allowed straightforward screening of the candidate viral cis-acting elements capable of replication. Only insert(s) which complemented the absence of an origin in the vector provided puromycin resistance to the transfected cells after replication and maintenance of the plasmids.
The Z6 fragments cloned into the pBSpuro vector at the respective enzyme sites (Figure 1A). Hirt DNA extracted from 293 cells with these plasmids at 96h post-transfection were digested with either BamHI or with BamHI and DpnI followed by electrophoresis and Southern detection of the replicated copies. Signal intensities of the linearized vectors were different because of the difference in recovery of each plasmid by Hirt extraction. Fragments of the Z6 region cloned into the pBSpuro vector showed various degrees of replication (Fig 1B). As expected, pBSpuroA3, containing three copies of TR, showed no detectable replication without LANA expression however replicated efficiently in LANA expressing cells (Grundhoff and Ganem, 2003; Hu et al., 2002). In contrast fragment G replicated in the absence of LANA and interestingly the replication efficiency was unchanged even in the presence of LANA (Fig 1B, and Supplementary data, Fig S1). However, it is important to note that in LANA expressing cells the replication efficiency of the TR containing plasmids was higher than the G fragment plasmids. Replication of the G fragment containing plasmid without LANA suggested the presence of a LANA independent origin referred to as an autonomous replication origin and was further analyzed.
Figure 1.
Evaluation of the replication potential of the Z6 fragments. (A) Schematic of the KSHV genome and the fragments of Z6 cloned into pBSpuro. (B) DpnI sensitivity assay of fragments A-H. Lanes B, 10% of the total Hirt’s DNA digested with BamHI and lanes N digested with NotI to linearize, Lane B+D or N+D, digested with BamHI and DpnI or NotI and DpnI. Arrow indicates the pBSpuro spiked plasmid to test the completion of digestion. Asterisks indicate the replicated copies of the plasmids. Relative quantities of DpnI bands are shown in a bar diagram after normalizing the inputs at 10%. (C) DpnI analysis of pBSpuroG and pBSpuro into human foreskin fibroblast (R2F), Rat-1 and DG-75. DpnI resistant band intensities were calculated based on input at 5% and the relative amounts of DpnI resistant bands are shown as bar diagram after normalizing against the background of individual lane.
Plasmids containing the KSHV G fragment replicated in primary cells
The G fragment containing vector (pBSpuroG) or empty vector (pBSpuro) were transfected into primary human foreskin fibroblasts (R2F), as well as Rat-1 and DG75 cell lines to evaluate the replication potential of the G fragment in primary cells and B cells. Hirt DNA extracted 96h post transfection followed by digestion with either EcoRI (E) or EcoRI and DpnI (E+D) revealed a DpnI resistant band suggesting replication of the plasmid (Fig. 1C). Lack of a DpnI resistant DNA band in empty vector (pBSpuro) transfected cells confirmed that the replication of pBSpuroG was due to the G fragment (Fig. 1C). DpnI resistant bands in all the lanes were quantified as the relative amount based on their respective inputs.
The pBSpuroG plasmid incorporates BrdU in synchrony with host cellular DNA during replication
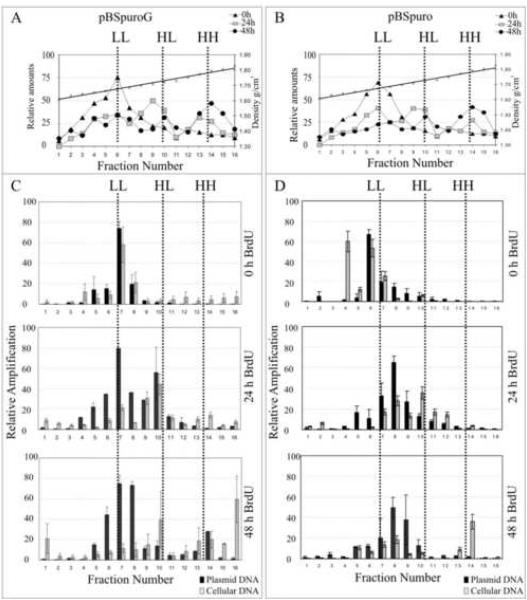
Short term replication assays demonstrated that the G fragment can support replication of the plasmid in various eukaryotic cell backgrounds. Therefore, we wished to determine whether replication mediated by the G fragment is synchronous to cellular DNA replication. We transfected 20 million 293 cells with pBSpuroG or pBSpuro vector and labeled the cells with 5-bromo-3-deoxyuridine (BrdU), to increase the buoyant density of the replicated DNA. During semi-conservative replication, BrdU is incorporated into newly synthesized DNA which forms a hybrid of light:heavy (H:L), after one or two rounds of replication and heavy:heavy (H:H), after two or more rounds of replication. HEK293 cells which typically divide in 24h were harvested at 0, 24 and 48h post BrdU pulse to allow sufficient time for no, one and two rounds of replication, respectively. Total DNA extracted from these cells were subjected to CsCl density gradient centrifugation and the distribution of total DNA, a measure of BrdU incorporation, on the gradient was analyzed.
Figure 2A and B showed a peak at H:L (hemi-substituted) position within 24h of pulsed period after a single round of cell division. DNA from the cells incubated with BrdU for 48h showed a peak at H:H along with H:L suggesting a second round of replication substituting both strands with H DNA. The distribution of density labeled DNA peaks were similar as seen in semi-conservative DNA replication (Aiyar et al., 1998; Schaarschmidt et al., 2004). Quantitation of pBSpuroG DNA copies (based on the relative amplification) in L:L, H:L and H:H fractions showed that plasmid DNA (Figure 2, black bars) peaked at L:L in a 0h BrdU pulsed cells similar to cellular DNA peaks detected based on the relative quantities of GAPDH in each fraction (Figure 2, gray bars). After 24h, pBSpuroG shifted to the H:L peak similar to the cellular DNA (Figure 2, gray bars) which was further shifted to the H:H peak after two rounds of replication (Fig 2C). Relative copies of the pBSpuroG and cellular DNA in these fractions demonstrated that the plasmid replicated in synchrony with cellular DNA, in a once-per-cell cycle manner. Empty vector, pBSpuro showed a slight shift of the plasmid copies (from fraction 6 to 8/9) below that expected for a single round of replication suggesting that empty vector was unable to replicate (Fig 2D). Cellular DNA in pBSpuro transfected cells showed similar peaks at L:L, H:L and H:H at 0h, 24h and 48h post BrdU pulse suggesting that the cells were actively growing. After 48h with two rounds of replication pBSpuroG showed definite incorporation of BrdU in H:H fraction, whereas little or no change was seen with the pBSpuro control. Therefore, this result strongly supports that G fragment can mediate replication.
Figure 2.
pBSpuroG plasmid replicated and incorporated the density label, BrdU in synchrony with the cellular DNA. 293 cells transfected with either pBSpuroG or pBSpuro were labeled with 30μg/ml BrdU for 0h, 24h or 48h. Total DNA extracted from these cells were sheared and separated on CsCl gradient centrifugation. The distribution of total DNA from pBSpuroG (A) and pBSpuro (B) transfected 293 cells in all 16 fractions of CsCl density gradient after 0h; triangles, 24h; squares and 48h; circles, were quantified using SYBR Green and plotted against density of each fraction. Relative copies of the pBSpuroG (C) and pBSpuro (D) in each fraction, quantified in a semi-quantitative PCR amplifying puro gene as a target sequence, are shown as black bars. Relative distribution of the cellular DNA quantified based on the amplification of GAPDH gene is shown as gray bars.
The pBSpuroG transfected in HEK293 cells persists for extended periods as episomal plasmids
HEK293 cells were transfected with pBSpuroG and empty vector. These plasmid yielded colonies after 3 weeks of selection with puromycin (2μg/ml). pBSpuroG yielded a significantly higher number (50-70 colonies) of colonies as compared to the vector (5-10 colonies). These HEK293 colonies were amplified in culture and subjected to in-situ gel lysis analysis to determine the episomal nature of the plasmid. Approximately 2 million cells from three independent colonies selected for 5 weeks were analyzed by in-situ gel lysis. Southern analysis detected episomal plasmid in the populations of pBSpuroG selected HEK293 cells similar to the bacterially replicated plasmid (Fig 3A). The majority of the plasmid copies detected were super-coiled suggesting that these plasmids were replicated and maintained independently (Fig 3A). Colonies selected with pBSpuro vector did not show the presence of plasmid bands but a band of genomic size was detected. This suggested integration of the puromycin plasmid DNA into the host genome providing puromycin resistance and colony outgrowth. As control purified pBSpuro plasmid DNA was shown in lane 6 (Fig. 3A).
Figure 3.
pBSpuroG plasmid persists as an episomal DNA detected by in-situ cell lysis. (A) In-situ cell lysis analysis of the long term selected clones. Left panel shows EtBr stained gel which was transferred onto the gene screen membrane, hybridized using 32P labeled puro probe (right panel) showed the presence of different forms of the plasmid. Lanes 1, 2 and 3 are the pBSPuroG plasmid selected clones, lane 4 is purified pBSpuroG (3ng), lane 5, clones with pBSpuro plasmid and lane 6 is the purified pBSpuro (3ng). Chr DNA; is the chromosomal DNA. Arrows indicates the plasmids pBSpuroG and pBSpuro in respective lanes. Triangles, indicates the hybridization signal of chromosomal DNA in pBSpuro plasmid selected clone. (B) Localization of pBSpuroG in long term selected colonies by Fluorescence in-situ hybridization. Chromosomes spreads of pBSpuroG selected cells were hybridized with biotin labeled probe and detected with streptavidin conjugated Alexa flour 594 (red dots). Chromosome spreads of HEK293 cells were used as a control. Average number of hybridizing dots per chromosome calculated based on 10 optical fields is presented. (C) Long term selected pBSpuroG plasmids replicated in its native form. DpnI sensitivity assay of Hirt DNA extracted from four long term selected clones (clone 1-4). Lane E (EcoRI) and E+D (EcoRI and DpnI) pBS was added in each reaction to test for the completion of digestion. Number of colonies yielded after transformation of the DpnI digested Hirt DNA extracted from pBSpuroG and pBSpuro clones. (D) Restriction patterns of the isolated plasmid from the transformants with BamHI and PstI. Lane 1 is pBSpuro, lane 2 is pBSpuroG, lanes 3-10 are plasmid DNA isolated from the colonies yielded with DpnI digested long term selected Hirt DNA.
To determine the approximate plasmid copies per cell, chromosome spreads from four independent populations of HEK293 containing pBSpuroG were prepared. In-situ hybridization using biotin labeled probe followed by detection with Streptavidin alexaflour 594 showed 6-12 copies of the plasmid per cell as an average of multiple counts per colony of HEK293 (Fig. 3B). The relative copy number was lower than for latent KSHV infected cells (Cotter and Robertson, 1999). This suggested that the plasmid containing G fragment was maintained as an episomal DNA element. Non transfected HEK293 cells used as a control did not show any hybridization (Fig 3B).
Long term maintenance is due to the replication of pBSpuroG with a functional cis acting replication origin
Hirt DNA isolated from pBSpuroG colonies after selection for five weeks were subjected to DpnI sensitivity analysis. This analysis showed DpnI resistant copies of the pBSpuroG plasmid whereas pBS, the spiked control DNA was completely digested (Fig. 3C). This suggests that the pBSpuroG plasmid persisted following replication during successive rounds of cell division. G fragment was cloned into pBluescript to test the replication mediated by the G fragment in the absence of the Pac gene to avoid selection of puromycin. The DpnI analysis of the isolated DNA without selection also showed similar replication efficiencies (Supplementary data, Fig S2). These data suggests that the G fragment can support replication and is likely to contribute to replication of the KSHV genome.
The pBSpuroG plasmid replicates in its native form under long-term selection
To determine whether episomally maintained pBSpuroG in long-term selected cells replicated in its native form, the DNA extracted from four long term selected colonies were subjected to DpnI digestion. Transformation of the digested plasmid DNA into E. coli yielded a significant number of ampicillin resistant colonies (Fig 3D). Restriction pattern of the plasmids isolated from these colonies matched the parental pBSpuroG demonstrating that the replicated plasmid was maintained in its native form (Fig. 3D). Furthermore, digested Hirt DNA from control pBSpuro selected HEK293 cells did not transform E. coli to produce ampicillin resistant colonies.
To determine if the recovered plasmids were defective or altered during our analysis, we sequenced 4 plasmids obtained from individual colonies. The analysis showed that recovered plasmids had identical nucleotide sequence as the parental input plasmid and confirmed the integrity of the plasmids (Supplementary data, Fig S3).
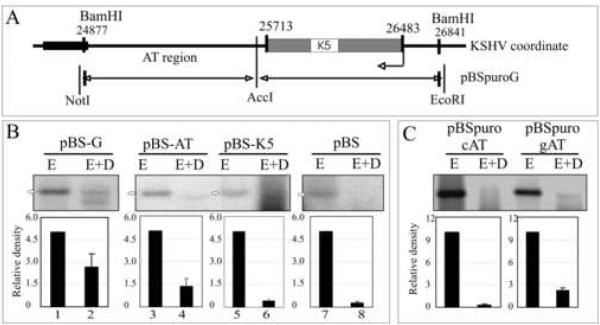
An AT rich region of the G fragment supports replication
Sequence analysis of the G fragment revealed the presence of a non-coding AT rich region adjacent to the coding sequence of K5 (Fig. 4A). The replication potential of the AT and K5 regions were analyzed and showed a DpnI resistant band in the AT region containing plasmid (Fig.4B, lane 4). A plasmid containing the K5 region did not show a DpnI resistant band (Fig.4B, lane 6). The pBS-G parental plasmid, as expected, showed almost similar efficiency of replication suggesting that the AT region is likely to be the predominant region responsible for replication. The pBS vector was used as a control for transfection and replication in this assay. The relative densities of the bands were plotted after substracting the respective background from each.
Figure 4.
The AT Region of the G fragment can support replication. (A) Schematic of the G fragment DNA sequence which lies between 24877-25713nt on KSHV coordinates (Russo et al., 1996). (B) Hirt DNA extracted from 293 cells transfected with G, AT or K5 fragments were digested with EcoRI (5%, to linearize) or EcoRI +DpnI and hybridized with 32P labeled ampicilin gene probe. The DpnI resistant bands were quantified. Intensities of input EcoRI lanes were set to 5 and relative intensities of DpnI band in each lane was calculated based on the respective inputs bands after normalizing against the background of individual lane. (C) AT region of the G fragment containing plasmid (pBSpuro gAT) and another similar length (760bp) control AT region of KSHV genome containing plasmids (pBSpuro cAT) were transfected into 293 cells for DpnI sensitivity assay. 10% of the total Hirt DNA was digested with EcoRI and rest 90% with E+D.
To confirm that the AT region of the G fragment can support replication, we cloned a similar size fragment of the Z6 fragment with similar AT content (~52%) into pBSpuro (pBSpuro cAT). Scanning analysis representing GC content of the control region is shown as supplemental data (Supplementary data, Fig S4). Comparison of the replication efficiencies of the control AT (pBSpuro cAT) and the AT region of the G fragment containing plasmids (pBSpuro gAT) revealed almost negligible signal in pBSpuro cAT indicating lack of replicated DNA as compared to the AT region of the G fragment (Fig 4B). This suggests that the DNA sequence present in the AT region of the G fragment not the AT content is important for DNA replication.
ORC2 and MCM3 associate with complexes containing the cis-acting AT rich element within the KSHV genome
To determine whether the human replication machinery accumulates in-vivo at the chromatin of KSHV AT rich region, we performed Chromatin Immunoprecipitation (ChIP) analysis on G1/S and G2/M cells fractionated using centrifugal elutriation (Fig 5A). KSHV positive PELs (2X109 cells) were subjected for centrifugal elutriation and approximately 2X108 cells collected in G1/S and G2/M cell cycle phases were subjected to chromatin immunoprecipitation using human α-ORC2, α-MCM3 antibody and a matched control. A fraction of immunoprecipitated chromatin was subjected to western blot analysis to detect the specific proteins (Fig. 5B). Quantitation of AT rich region DNA in a real-time PCR assay in the chromatins of G1/S cells immunoprecipitated with α-ORC2 and α-MCM3 antibodies showed significant copies of the AT rich region (Fig 5D). The three KSHV positive cell lines (BC3, BCBL-1 and JSC-1) showed similar pattern of ORC2 and MCM3 binding to the chromatin of the AT rich region. Interestingly, chromatin immunoprecipitated with α-ORC2 antibodies brought down AT rich DNA from G2/M phase cells suggesting that ORC2 remained associated with the DNA throughout the cell cycle. In contrast, MCM3 antibodies showed consistent reduction in the amount of immunoprecipitated AT rich region in the G2/M phase cells suggesting phase specific association of MCM. Higher number of AT rich DNA copies with MCM3 antibodies in G1/S phase cells suggests that MCM associate with the replication origin during the synthesis (S) phase, similar to cellular DNA replication (Bell and Dutta, 2002).
Figure 5.
The AT region of the G fragment immunoprecipitated with components of the cellular replication machinery in ChIP assay. (A) Representative FACS profile of the elutriated PEL cells in G1/S and G2/M phases. (B) Representative western blots showing immunoprecipitation of chromatin using α-ORC2 and α-MCM3 antibodies (lane 3). Lane 2, control IgG did not immunoprecipitate chromatins non-specifically. Lane 1; Input (10% total chromatin). Lane 4, proteins in post-ChIP. (C) Coordinates of the KSHV genome indicating E, and G regions. (D) Quantitation of the relative copies of AT region of the G fragment and control E fragment in α-ORC2 and α-MCM3 antibodies immunoprecipitated chromatin from PELs (BC-3, BCBL-1 and JSC-1). Immunoprecipitated AT and E copies were calculated based on the copies in input lane (10%). (E) The AT region of G but not the control AT region was immunoprecipitated with anti-ORC2 and anti-MCM3 antibodies in ChIP assay. Amplification of puro gene (adjacent to the G, AT or K5 region) in the DNA recovered from immunoprecipitated chromatins of 293 cells transfected with pBSpuroG, pBSpuroAT and pBSpuroK5 and pBSpuro using α-ORC2 and α-MCM3 antibodies. Relative amounts of amplicon in immunoprecipitated chromatins were quantified based on the intensity of amplification of the target region. (F) Cellular ORC2 and MCM3 do not binds to control AT region of KSHV genome. Western blot shows immunoprecipitation of chromatins with α-ORC2 and α-MCM3 antibodies from pBSpuro gAT and pBSpuro cAT. DNA from chromatins of pBSpuro cAT did not show any amplification (lanes 7 and 8) as compared to pBSpuro gAT (lanes 3 and 4) suggesting specific recruitment of cellular proteins at gAT region of the KSHV genome.
An adjacent region in the G fragment, K5 ORF did not show any amplification (Supplementary data, Fig S5) of the target. Thus the AT region is most likely the site for association of the replication proteins (Fig 5D). Another region of the Z6 cosmid, E fragment which did not replicate in our transient replication assays, was used as control for ChIP and failed to show amplification of the target region by ChIP with α-ORC2 and α-MCM3 antibodies. (Fig 5C and D, E region). Thus ORCs and MCMs binding were specific in terms of their association to the chromatin on the DNA of the G region fragment.
To further confirm the binding of host replication machinery at the AT region of G fragment we transfected pBSpuroG, pBSpuroAT, pBSpuro K5, and pBSpuro plasmids separately into 293 cells. 48h post transfection chromatins prepared from the above transfected cells were immunoprecipitated using α-ORC2 and α-MCM3 antibodies. Western blot analysis detected the immunoprecipitation of the respective proteins (Supplementary data, Fig. S6). PCR amplification of the specific regions G, AT and K5 on the plasmids immunoprecipitated by ChIP, revealed amplification of the AT region but not the K5 region of the G fragment (Fig 5E).
The AT rich region within the G fragment is specific for recruiting ORC2 and MCM3
To confirm the binding of cellular replication machinery proteins to the AT region of the G fragment, we performed chromatin immunoprecipitation from 293 cells transfected with either pBSpuro gAT or a control AT rich region from the KSHV LUR referred to as pBSpuro cAT. DNA extracted by ChIP using α-ORC2 and α-MCM3 antibodies revealed amplification of target DNA from cells with plasmids containing the G fragment AT rich region but not the control AT region plasmid (Fig 5F). Binding of ORC2 and MCM3 to the chromatin structure of the AT region of the G fragment was further reproduced in the B cell background (DG75) (Supplementary data, Fig S7).
The pBSpuroG plasmid actively incorporates BrdU due to replication in eukaryotic cells
BrdU was used to pulse HEK293 cells transfected with plasmids, pBSpuroG, pBSpuroTR with LANA expression (positive control) and pBSpuro (negative control). 24h post transfection cells were pulsed with BrdU (50μM) followed by extraction of the plasmid DNA using modified Hirt’s procedure. Figure 6A shows the schematic of the BrdU labeling and immunoprecipitation using α-BrdU antibody. A fraction of Hirt DNA (10%) was used as input (Fig 6A, lane 1). Relative copies of plasmid DNA quantified by semi quantitative PCR in the immunoprecipitated samples showed that α-BrdU antibodies immunoprecipitated the G fragment (Fig. 6A, pBspuroG, lane 3) but not the control IgG (Fig. 6A, pBSpuroG, lane 2). TR containing plasmid which supports replication in the presence of LANA (Grundhoff and Ganem, 2003; Hu et al., 2002) was immunoprecipitated by the α-BrdU antibody (Fig 6A, pBSpuroTR+LANA, lane 3). BrdU incorporation in the plasmids containing G and TR suggested that both contained an origin of replication. However, the G fragment plasmid incorporated BrdU at a lower efficiency than the TR with LANA (Fig. 6A). The pBSpuro vector transfected cells pulsed with BrdU failed to show detectable levels of vector DNA confirming that pBSpuro was unable to replicate (Fig. 6A, pBSpuro).
Figure 6.
The pBSpuroG actively incorporates BrdU and failed to replicate in ORC2 depleted cells. (A) A schematic of BrdU incorporation and immunoprecipitation of BrdU labeled DNA. α-BrdU antibody immunoprecipitated BrdU incorporated newly synthesized DNA from TR with LANA expression as well as G fragment containing plasmid detected by the amplification of immunoprecipiated DNA in lanes 3. Lane 1, Input, Lane 2 immunoprecipitation of BrdU labeled DNA with control IgG. Vector control (pBSpuro) did not show detectable level of amplification. (B) G fragment containing plasmid replicates in the absence of LANA. DpnI sensitivity assay of Hirt DNA extracted from 293 cells transfected with pBSpuroTR and pBSpuroG either with (+) or without (-) LANA expression. Quantitation of DpnI resistant copies based on input, 10% in EcoRI lanes. (C) Western blots showing efficient depletion of ORC2 expression in ORC2 specific siRNA and scrambled siRNA (SCR) transfected 293 cells. (D) Evaluation of pBSpuroG mediated replication by BrdU incorporation in 293 cells depleted with ORC2. α-BrdU antibody failed to immunoprecipitate G fragment containing plasmid from ORC2 siRNA treated cells (lane 3, ORC2 siRNA) whereas scrambled siRNA (SCR siRNA) treated cells showed immunoprecipitation of BrdU labeled DNA (lane 3). Lane 4 is control DNA for amplification and lane 5 had water as a negative control. (E) TR Sequence alignment to show deletion of 512-556bp for RE deletion. LBS1, LANA binding sequence. (F) DpnI sensitivity assay of the puro-GFP plasmid containing wt TR or ΔRE TR in LANA expressing cells. 96h post-transfection low molecular weight DNA was extracted and digested with either EcoRI (10%) or with EcoRI+DpnI. TR containing plasmids showed replication however ΔRE TR did not. (G) Schematic of the plasmids used in replication and complementation assay. (H) Real-time quantitation of the DpnI resistant plasmid copies. DpnI digested DNA was quantified using a primer pair flanking a DpnI restriction site. 10% of the total Hirt DNA was digested with EcoRI and used to calculate the relative amounts of DpnI resistant copies in respective lanes.
Since copies of BrdU labeled G fragment was lower compared to the TR plasmid with LANA, we wanted to determine the replication efficiencies of these two plasmids with and without LANA expression. DpnI sensitivity assay performed on Hirt DNA extracted 96h post-transfection revealed replication of pBSpuroG in the absence of LANA although at lower levels compared to pBSpuroTR plus LANA (Fig 6B). Interestingly, replication mediated by the G fragment was unaffected by LANA expression compared to TR mediated replication.
Depletion of ORC2 abolished replication of the G fragment containing plasmids
ORC2 specific siRNA was used to deplete endogenous ORC2 in 293 cells. Levels of ORC2 after 96h post transfection showed a significant reduction in ORC2 protein levels in specific ORC2 siRNA treated cells but not in a scrambled siRNA treated cells (Fig 6C, compare lanes 1 and 2). pBSpuroG was transfected into either specific or scrambled (SCR) siRNA treated cells and allowed for replication along with BrdU to label the replicating DNA. Hirt DNA extracted 96h post-transfection from these cells were digested with DpnI and subjected to α-BrdU immunoprecipitation. ORC2 depleted cells had no detectable levels of immunoprecipitated DNA suggesting absence of replication activity of the plasmid in ORC2 lacking cells. In contrast, scrambled siRNA transfected cells showed amplification suggesting involvement of cellular ORCs as a component of the replication machinery (Fig 6D).
Loss of replication efficiency was complemented by the G fragment in a replication deficient TR containing plasmid
To evaluate the biological significance of replication mediated by the G fragment, we used a plasmid based strategy. We used a replication deficient TR (ΔRE 512-556bp) (Fig 6E) (Hu and Renne, 2005) containing plasmid to determine whether the replication origin of the G fragment is able to complement the replication function of the ΔRE TR containing plasmid. We cloned ΔRE TR into a GFP pBSpuro (puro-GFP) plasmid followed by insertion of the G fragment to produce puro-GFP+ ΔRE TR+G (Fig 6F). These plasmids were transfected into HEK293 cells and assayed for replication after 96h. Extracted DNA was digested with EcoRI or EcoRI+DpnI followed by detection of DpnI resistant copies in a quantitative PCR assay using primers that amplified only DpnI resistant copies. The relative copies of the DpnI resistant plasmids were calculated by normalizing the input to 10 (10% of the total Hirt DNA). Puro-GFP+G yielded DpnI resistant copies similar to previous panels (Fig 5B). ΔRE TR containing plasmid was unable to replicate. Interestingly, the plasmid with ΔRE TR containing the G fragment (puro-GFP ΔRE TR+G) yielded DpnI resistant copies suggesting that the G fragment was able to complement the replication ability of the plasmid (Fig 6H).
HEK293 cells transfected with the above plasmids were subjected to puromycin selection for three weeks to evaluate long term replication by the G fragment in ΔRE TR plasmids. This was assayed by counting the puromycin resistant colonies produced after subsequent replication of the plasmid. Plasmids puro-GFP-G and puro-GFPΔRE TR+G yielded almost similar number of colonies (Supplemental data Fig S8) but not the puro-GFP-ΔRE TR plasmid.
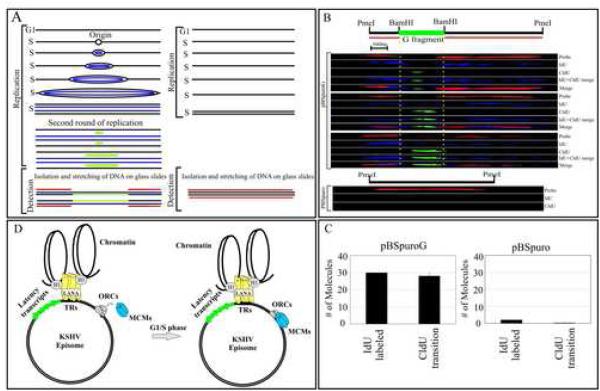
Single molecule analysis of replicated DNA showed replication initiation within the G fragment of the KSHV genome
To visualize the individually replicated DNA molecules we used a recently described technique, Single Molecule Analysis of Replicated DNA (SMARD) (Norio and Schildkraut, 2001). In this assay we used 5’-Iodo-2’-deoxyuridine (IdU) and 5’-chloro-2’-deoxyuridine (CldU) to label pBSpuroG and pBSpuro transiently transfected 293 cells. 72h post-transfection cells were labeled with IdU to completely label the entire plasmid length followed by removing IdU and pulsing with CldU for 2hours. A schematic illustration of double labeling is shown in figure 7A. Stretched DNA molecules were detected by hybridization with biotin labeled probes and further detected using Streptavidin conjugated Alexa Flour 594 (red). Incorporated IdU (first label) was detected using α-IdU antibody followed by Alexa Flour 350 (blue) (Fig. 7B). Transition from the first label (IdU) to the second label (CldU) was detected using rat α-CldU antibody and Alexa Flour 488 (green) antibody. Blue signal detecting the incorporation of IdU was seen throughout the entire length of the pBSpuroG and no detectable signal for IdU in pBSpuro vector (Fig.7B). Identification of transition sites due to incorporation of CldU at the replication initiation site was predominantly detected in the G region which may serve as a replication initiation site (Fig. 7B). Analysis of over one hundred DNA molecules in each of 4 independent experiments showed IdU incorporation in about 10% of the molecules as would be expected (Verma et al., 2006). However, the majority of molecules undergoing replication (IdU incorporation) showed transition from IdU to CldU indicating replication initiation within the G fragment. Three DNA molecules representing incorporation of IdU and CldU are shown (Fig. 7B). Pulsing with the second label was allowed only for a brief period (2h) so that a stretch of DNA can be predominantly labeled to identify the replication initiation sites. This replication initiation site was also confirmed on pBSpuroG molecules using different regions of the plasmid as probe (Supplementary data, Fig S9, A and B). Taken together the SMARD data supports our hypothesis that the G region of the KSHV DNA can also serve as a replication initiation site (Fig. 7C). The pBSpuro vector transfected and labeled in a similar manner did not show any detectable incorporation of the halogenated nucleotides.
Figure 7.
Single Molecule Analysis of the Replicated DNA. (A) Schematic showing incorporation of nucleotides in DNA containing origin of replication (ori) site. (B) Images of three PmeI linearized pBSpuroG DNA molecules. The hybridization signal detected pBSpuro backbone shown in red. G fragment cloned at BamHI sites of pBSpuro is flanked by regions of pBSpuro shows no hybridization signal. The incorporation of IdU was detected by α-IdU antibody followed by Alexa Flour 350 (blue) showed labeling of the entire length of the molecule after one round of replication. CldU detected by anti-CldU showed the transition site which is most likely the initiation site for the DNA replication. Varying length of CldU labeling was detected in these three molecules. pBSpuro lacking G region did not show incorporation of any halogenated nucleotides due to the lack of replication initiation sites. Hybridization signals detecting pBSpuro vector are shown in red. (C) Graphic representation of the number of molecules showing incorporation of IdU and transition to CldU in pBSpuroG and empty vector, pBSpuro. (D) Proposed model of KSHV persistence with a LANA independent origin of replication. LANA tethers the viral genome to the host chromosome and persists as highly ordered chromatin structure (Stedman et al., 2004; Verma et al., 2006). Binding of ORCs and MCMs at the G region of the KSHV most probably allows binding of other proteins of replication machinery to make pre-RC for latent replication of the KSHV genome.
Discussion
This study focuses on the identification of potential latent origin of replication at the left 33kbp region of the KSHV genome (Russo et al., 1996). Previous replication assays have identified a region in the 33 Kbp cosmid which included the terminal repeats that supported replication with concordant expression of LANA (Ballestas et al., 1999). Interestingly, the G fragment of this cosmid (the G fragment) did not show a dependence on LANA for replication. Thus it is considered to contain an autonomously replicating cis-acting DNA element. Replication mediated by the G fragment was consistent in transformed as well as primary cell lines suggesting the presence of a potential replication origin. Plasmids containing the G fragment incorporated BrdU and replicated in a cell cycle dependent manner. This region also supported long-term replication of plasmids as determined by selection of transfected HEK293 cells. Colonies yielded after selection showed the presence of replicated, un-rearranged episomal plasmids suggesting the presence of a cis-acting element capable of recruiting replication initiation proteins.
The precise function of LANA in replication of the KSHV genome is still unresolved. Recent studies suggest that LANA recruits the host cellular replication machinery at the TR and tethers the KSHV episomes to the host chromosomes (Barbera et al., 2006; Verma et al., 2006). Additionally, depletion of LANA did not completely abolish the episomes from PELs suggesting the presence of LANA independent replication origins (Godfrey et al., 2005). Similarly, HVS lacking identified replication origins are capable of establishing latent infection (Kung and Medveczky, 1996). This suggests that these herpesviruses do not have an absolute requirement for a particular origin, although they may contain a dominant element. There is likely to be flexibility where a number of additional sites can initiate replication. Additionally, Bovine Papillomavirus 1 persisted in a murine mammary tumor virus derived line independent of E1 suggesting the involvement of cellular proteins (Kim and Lambert, 2002). Incorporation of halogenated nucleotides and detection of single episomes of EBV have suggested the presence of non-conserved multiple replication initiation sites throughout the genome (Norio and Schildkraut, 2004). Identification of autonomous replication regions in the KSHV genome suggests that latent viral DNA replication can start at the identified sites which can progress to complete the replication of the entire genome. One primary function of LANA appears to be tethering of the viral genome to the host chromosome thus ensuring efficient segregation. KSHV mutants have yet to be identified which may yield interesting information regarding the viability of the virus in various cell types as well as under fluctuating immune status of the patients. Since LANA is a highly immunogenic protein the host immune system is likely to recognize infection by detection of LANA and thus set up defenses to inhibit viral propagation. However, it has been established that once KSHV infects the host B or endothelial cells it can persist indefinitely and reactivate if the immunity is compromised (Moore and Chang, 2003). To maintain viral DNA at a desired copy numbers in immune competent patients these autonomously replicating elements is likely to play a functional role in replication of the KSHV genome.
This study, for the first time demonstrates that the KSHV genome contains an autonomous replicating cis-acting sequence which can support replication of plasmids without a eukaryotic replication origin in the background of various cell types as determined by multiple assays. The replication of the G fragment containing plasmid requires the cellular replication machinery, as ORC2 depleted cells did not support replication performed in short term assays. Incorporation of halogenated nucleotides (IdU and CldU) suggests that DNA replication can initiate at the G region. This identified region (G) can also support the long-term replication of plasmid DNA in culture. This replication zone lies within a 750bp region between the 20/30bp GC repeats and the K5 ORF and is rich in AT with some specificity for this particular AT rich element. Further mutational analysis however, is needed to delineate the function of specific cis-elements within this region in the replication process. These results have important implications for understanding the functional variation associated with eukaryotic origins of replication and may also facilitate the construction of a first generation KSHV-based gene therapy vector for ex-vivo amplification and transfer into affected patients without the caveat of integration into the host genome.
Experimental Procedures
Cloning of Z6 fragments into pBSpuro
pBSpuro was constructed by subcloning the puromycin resistance expression cassette from pBABEpuro to the Sal I and Cla I sites of the pBluescript (Stratagene) multiple cloning site. Cloning procedure of sub-fragments of Z6 cosmid and short term replication assay is described in supplemental material.
Separation of BrdU labeled DNA in CsCl density equilibrium
Plasmids, pBSpuroG and pBSpuro were transfected into 2X107 293 cells and labeled with BrdU as described before with slight modification (Aiyar et al., 1998; Schaarschmidt et al., 2004). Briefly, 24h post-transfection cells were pulsed with BrdU (30μg/ml) and incubated for 24 and 48h to allow one or two rounds of replication, respectively. Total DNA from these cells were sonicated and resolved on CsCl gradient as described in supplementary material.
Long-term selection and in-situ gel lysis
10μg of pBSpuroG was transfected into 5 million of 293 cells and plated in 100mm plate. Transfected cells were selected using puromycin (1.0μg/ml) for three weeks which resulted in the outgrowth of colonies. These colonies were further sub-cloned and analyzed by in-situ lysis gels to determine the episomally maintained DNA (Gardella et al., 1984).
Preparation of chromosome spread and in-situ detection of plasmids
Colonies selected with pBSpuroG and 293 cells were mitotically arrested by colchicine treatment and the chromosome spreads were prepared as described previously (Verma and Robertson, 2003b). Chromosomes were aged at RT and pBSpuroG was detected using biotin labeled pBSpuro DNA as a probe. Probes were hybridized overnight at 37°C followed by detection with tyramide-rhodamine signal amplification system (Perkin-Elmer). Chromosomes were stained with DAPI; red dots which are the site of probe hybridization represent individual copies of plasmids.
Analysis of the plasmid isolated from long-term selected colonies
Low molecular weight DNA was extracted from pBSpuroG long term (5 weeks) selected clones and digested with either EcoRI or EcoRI and DpnI having been spiked with pBS plasmid as a control for digestion. The digested product was resolved on 0.8% agarose gel and transferred to gene screen for detection of replicated copies using pBSpuro backbone as probe. 10% of the digested product was transformed into E. coli and selected for the colonies on ampicilin plates. The plasmids from the transformants were isolated and the restriction patterns were analyzed using BamHI and PstI restriction endonucleases.
Analysis of pBSpuroG sequence and short term replication assay
Nucleotide sequence of G fragment (24877-26841) (Neipel et al., 1997; Russo et al., 1996) were analyzed. This region contains a unit of 20-30 bp GC repeats downstream to this region there is an AT rich region. Fragment G also encodes for the K5 ORF (25713-26483) which is BHV4-IE1 homolog of EBV (Russo et al., 1996). AT region and K5 ORF region were cloned separately in same vector backbone by digesting the G fragment with AccI.
Centrifugal elutriation and chromatin Immunoprecipitation Assay (ChIP)
Centrifugal elutriation was used to separate the cells in different cell cycle phases as described previously (Ritzi et al., 2003). For ChIP, experiments 2X108 cells in G1/S or G2/M were cross-linked with 1% formaldehyde for 10min. Chromatin immunoprecipotation (ChIP) assay was performed as described by Shang et al with slight modification (Shang et al., 2000) and a detailed method of chromatin immunoprecipitation and primers used for amplification of the respective region are described in supplemental material.
RNA interference
The siRNA for ORC2 was designed using BLOCK-iTTM RNAi express, siRNA designing tool (Invitrogen, Inc. Carlsbad, CA). siRNA ORC2 (5’- AUCCUGAGAUUACGAUAAA-3’) and control luciferase (5’- CUUACGCUGAGUACUUCGA-3’) (Prasanth et al., 2004) was transfected at 100nM final concentration with oligofectamine according to manufacturer’s instruction (Roche Inc.). 24h post transfection cells were trypsinized and re-plated followed by transfection with 100nM siRNA and pBSpuroG using Lipofectamine2000 as per manufacturer’s recommendation (Invitrogen).
BrdU labeling and immunoprecipitation of replicated DNA
293 cells (107) transiently transfected with pBSpuroG or pBSpuro was selected with puromycin for 72 hours followed by addition of 50μM BrdU in exponentially growing cells. Four hour later cells were harvested and washed twice in cold PBS. Plasmid DNA extracted from these cells was subjected for immunoprecipitation using anti-BrdU antibody described in supplemental material.
IdU and CldU labeling of replicating DNA and stretching of labeled DNA on glass slide
293 cells transfected with pBSpuroG or pBSpuro were used for detection of replicated DNA using SMARD. Exponentially growing cells were pulsed with 25μM of IdU (Sigma-Aldrich) for 20h followed by collecting the cells by centrifugation and incubating them with the medium containing 25μM CldU for 2h (Norio and Schildkraut, 2004). Low molecular weight DNA was extracted by modified Hirt’s procedure followed by digestion with PmeI to linearize the plasmids and purified on 0.6% agarose.
Gel purified dually labeled DNA was resuspended in printing buffer (3X SSC) and stretched on poly L-lysine coated slides (Erie Scientific). DNA molecules were immobilized on the slides by baking at 85°C for 1h. DNA was visualized by hybridizing with biotin labeled probe (NEB). Biotin was detected using Streptavidin conjugated with Alexa Flour 594 (red) (Molecular Probes). Incorporated IdU was detected using Mouse α-IdU (Becton-Dickinson Inc) as primary antibody (monoclonal), and Alexa Fluor 350 (blue) conjugated goat α-mouse (Molecular Probes) as secondary antibody. CldU was detected using rat anti-CldU (Accurate Chemicals, Westbury, NY) and goat anti-rat conjugated with Alexa Flour 488 (green) secondary antibody. DNA molecules were visualized using Olympus BX60 fluorescence microscope and photographs captured using Pixel Fly digital camera (Cooke Inc. Auburn Hills, MI).
Supplementary Material
Acknowledgements
This work was supported by Public Health service grant from NCI CA072510, CA091792 NIDCR DE014136, DE017338 and AI AI067037 (ESR). E.S.R. is a scholar of Leukemia and Lymphoma Society of America. S.C.V. is a fellow of the Lady TATA memorial Trust for research in leukemia. We would like to thank Carl Schildkraut and P. Norio for helpful hints with the SMARD technology Bill Sugden, Donald Ganem and Elliott Kieff for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. Embo J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Cai QL, Knight JS, Verma SC, Zald P, Robertson ES. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2006;2:e116. doi: 10.1371/journal.ppat.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter MA, Robertson ES. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. (2nd) 1999;264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- Friborg J, Jr., Kong W, Hottiger MO, Nabel GJ. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- Gahn TA, Schildkraut CL. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- Gardella T, Medveczky P, Sairenji T, Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J Virol. 1984;50:248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey A, Anderson J, Papanastasiou A, Takeuchi Y, Boshoff C. Inhibiting primary effusion lymphoma by lentiviral vectors encoding short hairpin RNA. Blood. 2005;105:2510–2518. doi: 10.1182/blood-2004-08-3052. [DOI] [PubMed] [Google Scholar]
- Grundhoff A, Ganem D. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J Virol. 2003;77:2779–2783. doi: 10.1128/JVI.77.4.2779-2783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Garber AC, Renne R. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J Virol. 2002;76:11677–11687. doi: 10.1128/JVI.76.22.11677-11687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Renne R. Characterization of the minimal replicator of Kaposi's sarcoma-associated herpesvirus latent origin. J Virol. 2005;79:2637–2642. doi: 10.1128/JVI.79.4.2637-2642.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankelevich S, Kolman JL, Bodnar JW, Miller G. A nuclear matrix attachment region organizes the Epstein-Barr viral plasmid in Raji cells into a single DNA domain. Embo J. 1992;11:1165–1176. doi: 10.1002/j.1460-2075.1992.tb05157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes DH, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J Clin Invest. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E. Field's Virology. Third Lippincott-Raven Publishers; Philadelphia: 1996. Epstein-Barr Virus and Its Replication; pp. 2343–2396. [Google Scholar]
- Kim K, Lambert PF. E1 protein of bovine papillomavirus 1 is not required for the maintenance of viral plasmid DNA replication. Virology. 2002;293:10–14. doi: 10.1006/viro.2001.1305. [DOI] [PubMed] [Google Scholar]
- Kung SH, Medveczky PG. Identification of a herpesvirus Saimiri cis-acting DNA fragment that permits stable replication of episomes in transformed T cells. J Virol. 1996;70:1738–1744. doi: 10.1128/jvi.70.3.1738-1744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laman H, Coverley D, Krude T, Laskey R, Jones N. Viral cyclin-cyclin-dependent kinase 6 complexes initiate nuclear DNA replication. Mol Cell Biol. 2001a;21:624–635. doi: 10.1128/MCB.21.2.624-635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laman H, Peters G, Jones N. Cyclin-mediated export of human Orc1. Exp Cell Res. 2001b;271:230–237. doi: 10.1006/excr.2001.5360. [DOI] [PubMed] [Google Scholar]
- Little RD, Schildkraut CL. Initiation of latent DNA replication in the Epstein-Barr virus genome can occur at sites other than the genetically defined origin. Mol Cell Biol. 1995;15:2893–2903. doi: 10.1128/mcb.15.5.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PS, Chang Y. Kaposi's sarcoma-associated herpesvirus immunoevasion and tumorigenesis: two sides of the same coin? Annu Rev Microbiol. 2003;57:609–639. doi: 10.1146/annurev.micro.57.030502.090824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neipel F, Albrecht JC, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P, Schildkraut CL. Visualization of DNA replication on individual Epstein-Barr virus episomes. Science. 2001;294:2361–2364. doi: 10.1126/science.1064603. [DOI] [PubMed] [Google Scholar]
- Norio P, Schildkraut CL. Plasticity of DNA replication initiation in Epstein-Barr virus episomes. PLoS Biol. 2004;2:e152. doi: 10.1371/journal.pbio.0020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. Embo J. 2004;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbow L, Platt GM, Simpson GR, Sarid R, Gao SJ, Stoiber H, Herrington CS, Moore PS, Schulz TF. The 222-to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins DR, Milman G, Hayward SD, Hayward GS. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzi M, Tillack K, Gerhardt J, Ott E, Humme S, Kremmer E, Hammerschmidt W, Schepers A. Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus. J Cell Sci. 2003;116:3971–3984. doi: 10.1242/jcs.00708. [DOI] [PubMed] [Google Scholar]
- Roizman B. Field's Virology. Third Lippincott-Raven Publishers; Philadelphia: 1996. Herpesviridae; pp. 2221–2230. [Google Scholar]
- Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci U S A. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaarschmidt D, Baltin J, Stehle IM, Lipps HJ, Knippers R. An episomal mammalian replicon: sequence-independent binding of the origin recognition complex. Embo J. 2004;23:191–201. doi: 10.1038/sj.emboj.7600029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Stedman W, Deng Z, Lu F, Lieberman PM. ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J Virol. 2004;78:12566–12575. doi: 10.1128/JVI.78.22.12566-12575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma SC, Choudhuri T, Kaul R, Robertson ES. Latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus interacts with origin recognition complexes at the LANA binding sequence within the terminal repeats. J Virol. 2006;80:2243–2256. doi: 10.1128/JVI.80.5.2243-2256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma SC, Lan K, Robertson E. Structure and function of latency-associated nuclear antigen. Curr Top Microbiol Immunol. 2007;312:101–136. doi: 10.1007/978-3-540-34344-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma SC, Robertson ES. Molecular biology and pathogenesis of Kaposi sarcoma-associated herpesvirus. FEMS Microbiol Lett. 2003a;222:155–163. doi: 10.1016/S0378-1097(03)00261-1. [DOI] [PubMed] [Google Scholar]
- Verma SC, Robertson ES. ORF73 of herpesvirus Saimiri strain C488 yethers the viral genome to metaphase chromosomes and binds to cis-acting DNA sequences in the terminal repeats. J Virol. 2003b;77:12494–12506. doi: 10.1128/JVI.77.23.12494-12506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye FC, Zhou FC, Yoo SM, Xie JP, Browning PJ, Gao SJ. Disruption of Kaposi's sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J Virol. 2004;78:11121–11129. doi: 10.1128/JVI.78.20.11121-11129.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.