One-dimensional nanostructures such as nanochannels (and nanotubes) are characterized by extremely small transverse size and resultant high degree of spatial confinement that endow them a unique set of properties. When patterned laterally, these nanostructures are widely used as critical transport devices for a variety of applications such as sensing, nanomanipulation, and information processing.[1–8] While numerous fabrication techniques have been developed, few can generate large and highly ordered arrays of both nanochannels and nanowires with no defects and low-cost. The most notable high-resolution lithographic techniques include electron beam lithography (EBL) and focused ion beam milling (FIB),[9–13] but they are associated with either low throughput or high-cost. Another lithographic technique, nanoimprint lithography (NIL), is of high throughput and relatively low-cost, but it requires the use of highly specialized equipment and molds prepared typically by EBL.[14– 17] Many inexpensive techniques have been developed, but they are inadequate in terms of high precision, low defect rate, or large area fabrication of both nanochannels/tubes and nanowires/strands.[7,18–25] Moreover, these nanostructures need to be connected to the micro/macroscale structures, such as reservoirs and channels, to form functional devices. This is not a trivial task and the lack of a low-cost solution to this problem significantly limits the applicability of many nanoconstructs.
Here we introduce a novel, simple and low-cost method capable of simultaneously forming sealed arrays of laterally ordered nanochannels with controllable sizes and rounded shape over arbitrarily large surface areas, but also interconnected micro/nanowell arrays. Associated micro- and macroscale inlets/outlets can also be formed in the same fabrication process or added later using low-cost, non-cleanroom methods.
This polymer based nanoscale transport system has great potential for many challenging biomedical applications.
The process for creating the aforementioned polymer nanoscale transport system consisting of arrays of nanochannels with interconnected micro/nanowells is illustrated in Figure 1. This nanostructure fabrication consists of two major procedures: generating DNA nanostrands[26–28] and translating these nanostrands to confined nanoscopic elements by imprinting. Briefly, a patterned PDMS stamp was placed on a small drop of DNA solution on a glass slide, followed by peeling off the stamp. As a result, DNA solution dewetted the stamp surface. Compared to the conventional molecular combing,[29] which is also based on a dewetting process but performed on a flat surface to generate stretched DNA molecules that are randomly distributed, the surface micro/nanofeatures here acted as wetting defects to hold the receding contact line locally. In the case of micro/nanopillars with a 2D cubic lattice (Figure 1), single droplets of the DNA solution formed on each pillars and the droplets were temporarily connected by a liquid bridge in the dewetting direction when it was in parallel to a lattice axis. Breakage of the liquid bridge and shrinkage of the droplets due to water evaporation and high surface tension of water led to stretching of DNA molecules once within the liquid bridges. As a result, suspended DNA nanostrands composed of stretched DNA molecules formed between adjacent pillars as shown in Figure 2a.
Figure 1.
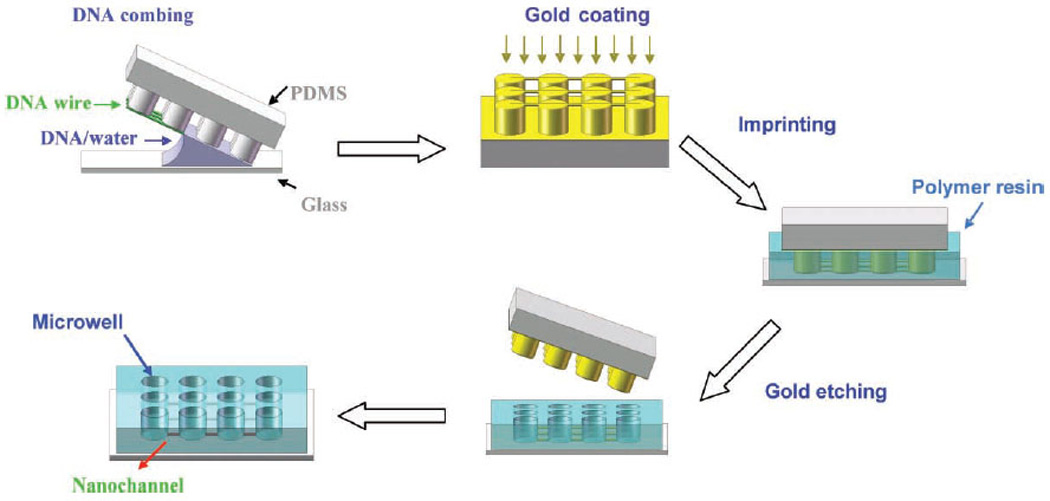
Fabrication schematic of arrays of nanochannels by DCI technique.
Figure 2.
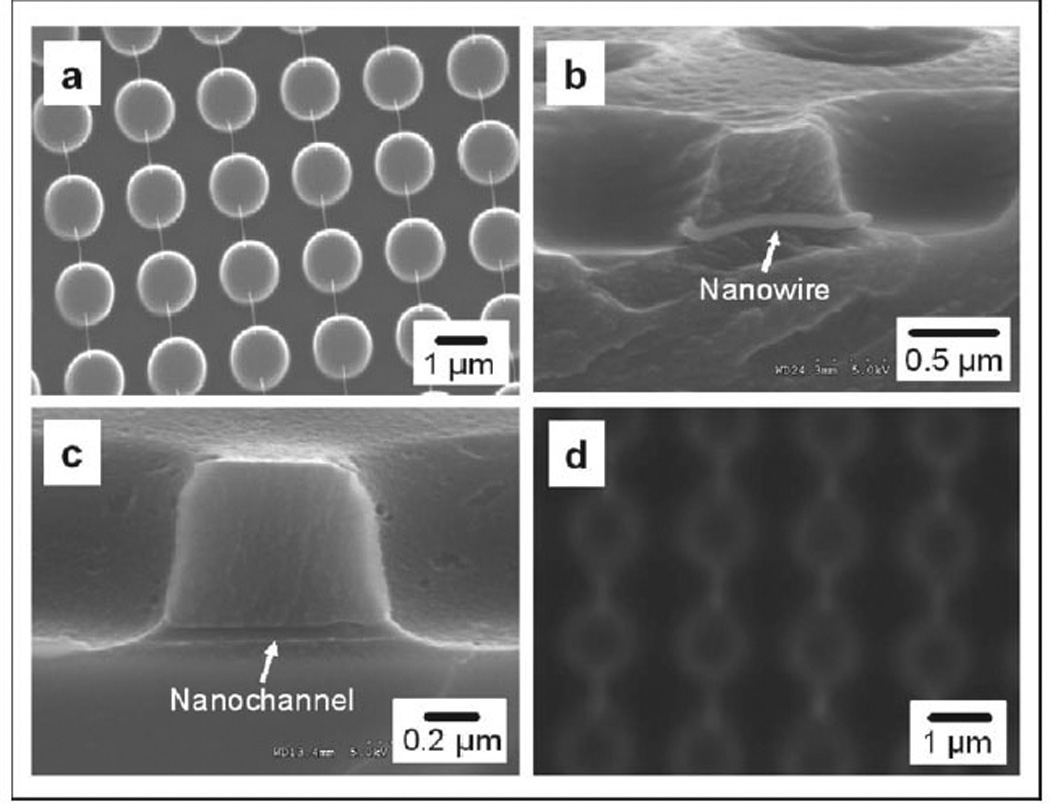
Scanning electron microscopy (SEM) images of (a) suspended gold-coated DNA nanostrands between 1 µm wide micropillars, (b) a gold-coated DNA nanostrand embedded in the cured polymer, (c) side view of a nanochannel connecting two microwells, and (d) fluorescence micrograph of nanochannels and microwells filled with Rhodamine.
For the formation of nanochannel arrays, the DNA nanostrands on pillars were sputter-coated with gold. Next, the gold-coated nanostrands on the PDMS pillars were placed on a solid substrate, in this case a glass slide. A low-viscosity prepolymer resin, e.g. ethyleneglycol dimethacrylate (EGDMA), was used to fill the space between the pillars by capillary flow and then was solidified by UV-induced cross-linking polymerization. The PDMS stamp was then removed, leaving the gold-coated nanostrands embedded in the matrix of the cured polymer. At the same time, microwells were formed from the molding of the micropillars. Embedding of a nanostrand in the cured polymer between two adjacent microwells is clearly shown in Figure 2b. The cured polymer tightly surrounds the nanostrand. As a result, the internal diameter of the nanochannels is equivalent to the diameter of the gold-coated nanostrands. To remove the gold-coated nanostrands and consequently form the nanochannels, the sample was soaked in a gold etchant. An example of one of the resulting nanochannels is shown in Figure 2c with an internal diameter around 60 nm. A fluorescent dye, Rhodamine, was used to confirm that the nanochannels were unobstructed as shown in Figure 2d. This simple DNA combing and imprinting (DCI) method can produce well-defined nanochannel and microwell arrays over an area larger than several square centimeters without any defects (Supporting Information, section S1). Lateral nanochannels alone do not constitute a functional device. Therefore, concurrent formation of the microwells offers significant benefits for device development. Since channel size is determined by the strand size, our method is able to generate sealed circular nanochannels with controllable internal diameter. Since naked DNA nanostrands below 5 nm in thickness have been prepared by this method[27] and polymers have been used to imprint sub-nanometer structures by the molecular imprinting technique,[30] DCI can thus generate nanochannel arrays with a range of channel diameters from <10 to >100 nm size. In order to generate the smaller sized nanochannels a highly photo-bleaching fluorescence dye, ethidium bromide was first conjugated with DNA molecules and the gold coating step was skipped. After DCI, the naked DNA nanostrands were cleaved by UV exposure[31] and removed by soaking in Piranha solution (7:3 v/v of conc. H2SO4 and 30% H2O2).
The EGDMA based nanochannel/microwell array is hydrophobic. Therefore, organic solvents and molecules can be loaded for nanoscale fluid transport and chemical reactions. For aqueous solutions the array can be soaked in Piranha to make it hydrophilic (Supporting Information, section S2), making the system convenient for sample loading in biomedical applications. Various nanofluidic applications such as molecular dynamics in confined nanoenvironment, bioseparation and gene delivery can be carried out using this system.
In the DCI process, we have used gold as the sacrificial material but many other materials can also be used. Moreover, a non-sacrificial layer can be coated around the sacrificial material to make nanochannels with properties different from the surrounding cured polymer. For example, if silica or aluminum is coated around gold nanostrands by sputter coating, the internal wall of the nanochannels will be made of silica or aluminum once the gold is etched away. Many other materials may also be used as the wall of nanochannels as long as they can be coated around the nanostrands from either liquid or gas sources. Since the surface-to-volume ratio is extremely high in nanochannels, such surface modification is crucial to achieve surface-force guided motion or assembly of molecules in the nanochannels and to avoid non-specific binding and clogging of the channels by undesirable molecular sticking on the channel wall.
Cross-networks of DNA nanochannels can also be fabricated by this approach. Figure 3a shows a cross-pattern of DNA nanostrands based on a 7 µm pillar array formed by double dewetting. First, DNA nanostrands were stretched on the PDMS pillars and coated with a thin layer of gold (~5 nm thick) to prevent the DNA from dissolving during the second dewetting step. Then, 1-octanethiol was used to create a hydrophobic surface for producing the second series of stretched DNA nanostrands perpendicular to the initial stretching. This network of nanostrands was converted to a cross-network of nanochannels with 7 µm interconnected wells by the same DCI approach and was confirmed by filling with Rhodamine dye in Figure 3b. Other patterns of nanostrands and consequent nanochannels can also be produced by controlling the surface patterns of the stamp as well as the combing direction. More examples are given in the Supporting Information, section S3. Figure 3c and 3d show a branch-like pattern of nanostrands and the corresponding nanochannels obtained using micropillars with a hexagonal lattice. Besides micropillars, we also applied our technique to nanopillars that are 150 nm wide and 220 nm in center-to-center distance. The nanostrands shown in Figure 3e have a width about 20 nm. Figure 3f shows nanowells and the ends of nanochannels converted from the nanopillars and nanostrands. Such high-density nanowell/nanochannel arrays are difficult to fabricate using other methods.
Figure 3.
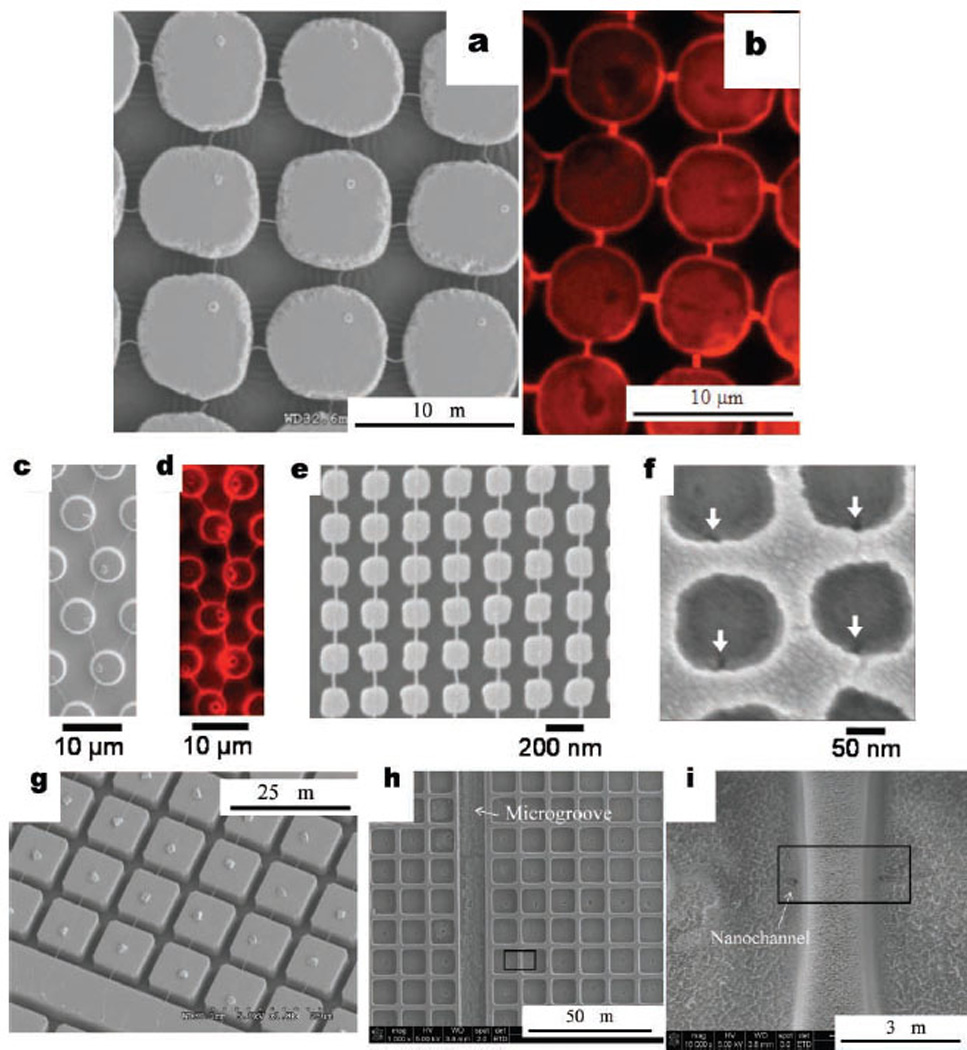
(a) SEM image of a cross-network of DNA nanostrands on 7 µm PDMS pillars coated with gold, (b) fluorescence photograph of a cross-network of EGDMA nanochannel/nanochannel interconnected with 7 µm wells imprinted from PDMS micropillars in (a), (c) SEM image of branch-like DNA nanostrands on micropillars with a hexagonal lattice. Particle on top of each micropillar is NaCl crystal. (d) Fluorescence micrograph of branch-like nanochannels. Cavities translated from NaCl crystals are at the bottom of each microwell. SEM image of (e) 20 nm-wide DNA nanostrands on 150 nm-wide PDMS nanopillars and (f) ends of nanochannels in nanowells (specified by arrows. Wells were slightly distorted by electron-beam exposure during SEM imaging). SEM images of (g) stretched DNA nanostrands between 10 µm square micropillars and a long microridge, (h) converted microwell array and a long microgroove by EGDMA imprinting, and (i) magnified 10 µm wells connected by a nanochannel (channel end specified by arrow).
In order to access the nanochannel/microwell arrays, it is essential to have inlet and outlet ports connected to microscale grooves leading into the arrays allowing the system to be externally filled with desired reagents or molecules. We have successfully employed several simple techniques for incorporating inlet/outlets and access microgrooves. After the nanoconstruct arrays are formed by DCI, we can add access microgrooves and inlet/outlets using micromilling (MM), reactive ion etching (RIE) or femtosecond laser ablation (FLB) to remove parts of the EGDMA (Supporting Information, section S4). We are also able to generate access microgrooves into the nanoconstruct arrays by incorporating the microgrooves in the micropillar design on the PDMS stamps that are used for dewetting. This is advantageous because it incorporates the access grooves in the dewetting process so that an additional fabrication step is not required. Figure 3g shows such a design where the stretched DNA nanostrands coated with gold are formed between square shaped micropillars and a long microridge. Figure 3h shows the converted microwell array and the long microgroove after EGDMA imprinting. The nanochannel between microwells can be seen in Figure 3i.
In summary, we have developed a novel and inexpensive method, DNA Combing and Imprinting (DCI), capable of simultaneously generating a large array of laterally ordered and sealed nanochannels, DNA nanostrands, and their combinations with interconnected micro/nanowell arrays and access microgrooves to link to external inlet/outlet. It allows for low-cost fabrication of comprehensive nanofluidics or nanoscale transport systems with no size limitation in principle. This easy-to-scale-up approach can be used to form useful devices for large scale molecular manipulation and various biomedical and nonbioapplications. Devices made using DCI are being tested in our laboratory for high-resolution protein and oligonucleotide separation and detection using functionalized DNA nanostrand arrays, gene and oligo delivery to individual cells using nanochannel-based local electroporation, multifunctional nanoparticle formation using nanochannel/nanostrand cross-network as a nanoreactor array, as well as molecular dynamics and ion transport in highly confined nanoenvironment.
Experimental Section
Fabrication of Micro-/Nanoscale Pillar
PDMS stamps with various micro-/nanopillar and micro-/macro-ridge designs were used in this study. One design consisted of circular micropillars that were wider at the base, which was 1 µm in diameter, than the top surface. The center-to-center distance of the pillars was 2 µm. The other type of stamps contained ~5 µm-wide circular pillars with a center-to-center distance of ~9.5 µm in a hexagonal lattice, as well as 7 µm and 10 µm circular pillars with a center-to-center distance of 8.2 µm and 30 µm respectively. Additionally, square 10 · 10 µm pillars with 2 µm spacing and a continuous micro-ridge were employed to produce microgrooves integrated to microwell/nanochannel arrays. Other patterns used for the dewetting process are given in SOM, section S3. Master molds for preparing the PDMS stamps were fabricated using standard photolithography techniques, and all micro-pillar stamps were prepared using standard soft lithography by casting PDMS (Sylgard® 184, Dow Corning) prepolymer and curing agent at 10:1 weight ratio on the master. Nano-sized pillars were also prepared using interference lithography. They were ~150 nm wide with the center-to-center distance around 220 nm.
Preparation of DNA Solutions
The DNA solutions were prepared by dissolving two types of commercially available DNA, Salmon testes (~2 kbp, Sigma-Aldrich) or Calf thymus (~75 kbp, USB Co) in TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH = 8) with appropriate concentrations. For micropillars with spacing between pillars larger than 1 µm, the longer DNA with 75 kbp was employed to produce stronger nanostrands. For the nanostrands shown in Figure 2a, the solution was 1 wt% DNA (2 kbp) in TE buffer. For generating nanostrands on nanopillars (Figure 3c), 0.1 wt% Salmon testes DNA (Sigma-Aldrich), with 5 wt% NaCl in TE buffer was used. For all other samples, 0.5 wt% DNA with 5 wt% NaCl in TE buffer was used.
Generating Nanostrand Array
To form DNA nanostrands across the pillars, a 30 µl drop of DNA solution was placed on a glass slide. The PDMS stamp was then gently placed face down on the DNA solution and immediately peeled off by pulling one end of the stamp up to produce directional stretching during the dewetting process. It is critical not to add external pressure on the stamp placed on the DNA solution so that the solution did not fill the space between the pillars. Otherwise, DNA solution would be trapped in the space rather than only on the pillars. The later case was essential to form the nanostrands. This method was relatively insensitive to the peeling speed, which presumably determined the dewetting speed. At a manually controlled rotational speed of ~90°/sec, centimeter-sized nanostrand arrays were repeatedly obtained. The stamp was then sputter coated with gold (Emitech K550X, Energy Beam Sciences Inc., CT, USA). The gold nanostrands were coated with 1-octane thiol by vapor deposition to facilitate demolding of the gold-coated stamp from the cured polymer.[32] Specifically, stamps with the gold-coated DNA nanostrands were placed in a 10 cm-diameter Petri dish next to 5 ml 1-octane thiol. A lid was placed on the dish and 2 hours were allowed for the vapor deposition of the 1-octane thiol onto the gold-coated stamps.
Surface Treatment
The cover slip glasses were subjected in a boiling Piranha (H2SO4:H2O2 7:3) etch/clean solution for 30 minutes and followed by rinsing with deionized (DI) water to make them ready for silane grafting. This Piranha solution should be handled with extreme caution and special care must be taken during the treatment. Then, pretreated glasses were soaked in 2 wt% 3-trimethoxysilylpropyl methacrylate in toluene for 12 hours at room temperature to graft methacrylate groups on the slide surface.
Fabrication of Nanochannels Arrays
A stamp with micropillars and gold-coated DNA nanostrands was placed face down on the treated glass slide. After ~2 ml of EGDMA resin (99 wt% EGDMA, 1 wt% Irgacure 651 initiator) was dropped on the slide near the stamp, the slide was placed in a vacuum desiccator. Once vacuum was established, the system was tilted to allow the EGDMA resin to flow towards the stamp. When the resin contacted the stamp, it immediately flowed into the space between the stamp and the slide by capillary force. Vacuum was used in this process to avoid trapped air bubbles. The slide was transferred from the vacuum chamber to a small nitrogen chamber. The resin was cured using UV light (wavelength: 365 nm, intensity: 4 mw/cm2) for 20 minutes under nitrogen. The adhesion between the cured polymer and the glass surface was enhanced by chemical bonds between surface-grafted methacrylate groups and EGDMA. The stamp was then peeled off from the slide leaving behind the array of EGDMA microwells connected by gold-coated DNA nanostrands. To remove gold-coated nanostrands in the nanochannels, the slide was soaked in gold etchant (GE8111, Transene Company Inc., Danvers, MA, USA) for 24 hours and then thoroughly rinsed with deionized water leaving behind embedded nanochannels connecting the microwells.
An approach similar to that described in the previous section was used to create cross-networks of nanochannels embedded in microwell arrays. To create a nanochannel/nanochannel network, first DNA was stretched on the micropillars in one direction by dewetting. The stretched DNA nanostrands were coated with gold and further treated with 1-octanethiol to produce sufficient hydrophobicity and strong strands for the second stretching step. The second dewetting step was carried out to stretch DNA perpendicular to the direction of the first stretching. These networks of stretched DNAs were coated with gold and treated with 1-octanethiol again. They were removed by gold etching after EGMDA imprinting. The final product is an EGDMA microwell array interconnected with a cross-network of nanochannels.
Fluorescence Imaging
To visualize the nanochannels and nanostrands using fluorescence microscopy, a nanochannel sample was immersed in 10−5 M Rhodamine in water for 5–12 hours allowing Rhodamine to diffuse into the nanochannels/nanostrands. The chip was then briefly washed with DI water to remove excess dye in the microwells but not in the nanochannels due to the limited diffusivity in the nanochannels. A high-speed confocal microscope system with a Yokogawa CSU-22 spinning disk and Hamamatsu EMCCD C9100 camera mounted on an Olympus IX81 inverted microscope with a 100× 1.4 numerical aperture oil immersion objective lens was used to capture fluorescent images of the nanochannels and micro/nanowells.
SEM Imaging
SEM images of all samples except the nanowell/nanochannel arrays were taken in a Hitachi S-4300 SEM. The high resolution SEM images of the nanowell/nanochannel arrays were taken by a FEI Nova nanoSEM 400 equipped with an additional through-the-lens detection system. Some nanowells were distorted by electron beam irradiation during the focusing procedure at high magnification. The distortion frequently caused cracks on the ridge, especially above the nanochannel where EGDMA is thinner. To image the nanowires or nanochannels in a cured EGDMA film, the film was bent to break roughly along the length direction of the nanowires/nanochannels. Since the fracture direction was not perfectly aligned with nanowires/nanochannels, most of the nanowires/nanochannels in the fracture surface were completely or partially covered by the cured EGDMA. Only a small portion of the nanowires/nanochannels were exposed as shown in Figure 2b and 2c.
Supplementary Material
Acknowledgements
The present work was supported by the National Science Foundation under NSF Grant No. EEC-0425626. We thank Carlos Pina and Professor L. Jay Guo at University of Michigan for providing PDMS nanopillar stamps, Daniel G. Perez for helping to produce master mold on silicone wafer, Wei-Ching Liao for drawing the fabrication schematic, HyunChul Jung and Professor Wu Lu for assisting RIE technique, and Yong C. Lim and Professor Dave Farson for producing microgrooves by femtosecond laser ablation at The Ohio State University.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Jingjiao Guan, Nanoscale Science and Engineering Center for Affordable, Nanoengineering of Polymeric Biomedical Devices, The Ohio State University, (USA).
Pouyan E. Boukany, Nanoscale Science and Engineering Center for Affordable, Nanoengineering of Polymeric Biomedical Devices, The Ohio State University, (USA)
Orin Hemminger, Department of Chemical and Biomolecular, Engineering The Ohio State University (USA).
Nan-Rong Chiou, Nanoscale Science and Engineering Center for Affordable, Nanoengineering of Polymeric Biomedical Devices, The Ohio State University, (USA).
Weibin Zha, Nanoscale Science and Engineering Center for Affordable, Nanoengineering of Polymeric Biomedical Devices, The Ohio State University, (USA).
Megan Cavanaugh, Nanoscale Science and Engineering Center for Affordable, Nanoengineering of Polymeric Biomedical Devices, The Ohio State University, (USA); Department of Chemical and Biomolecular, Engineering The Ohio State University (USA).
L. James Lee, Email: lee.31@osu.edu, Nanoscale Science and Engineering Center for Affordable, Nanoengineering of Polymeric Biomedical Devices, The Ohio State University, (USA); Department of Chemical and Biomolecular, Engineering The Ohio State University (USA).
References
- 1.Han J, Craighead HG. Science. 2000;288:1026. doi: 10.1126/science.288.5468.1026. [DOI] [PubMed] [Google Scholar]
- 2.Fu J, Schoch RB, Stevens AL, Tannenbaum SR, Han J. Nat. Nanotechnol. 2007;2:121. doi: 10.1038/nnano.2006.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang YM, Tegenfeldt JO, Reisner W, Riehn R, Guan XJ, Guo L, Golding I, Cox EC, Sturm J, Austin RH. Proc. Natl. Acad. Sci. USA. 2005;102:9796. doi: 10.1073/pnas.0502917102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riehn R, Lu M, Wang Y-M, Lim SF, Cox EC, Austin RH. Proc. Natl. Acad. Sci. USA. 2005;102:10012. doi: 10.1073/pnas.0503809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tegenfeldt JO, Prinz C, Cao H, Chou S, Reisner WW, Riehn R, Wang YM, Cox EC, Sturm JC, Silberzan P, Austin RH. Proc. Natl. Acad. Sci. USA. 2004;101:10979. doi: 10.1073/pnas.0403849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patolsky F, Lieber CM. Mater. Today. 2005;8:20. [Google Scholar]
- 7.Huang Y, Duan X, Wei Q, Lieber CM. Science. 2001;291:630. doi: 10.1126/science.291.5504.630. [DOI] [PubMed] [Google Scholar]
- 8.Zhong Z, Wang D, Cui Y, Bockrath MW, Lieber CM. Science. 2003;302:1377. doi: 10.1126/science.1090899. [DOI] [PubMed] [Google Scholar]
- 9.Bilenberg B, Jacobsen S, Schmidt MS, Skjolding LHD, Shi P, Bøggild P, Tegenfeldt JO, Kristensen A. Microelec. Engin. 2006;83:1609. [Google Scholar]
- 10.Harnett CK, Coates GW, Craighead HG. J. Vac. Sci. Technol. B. 2001;19:2842. [Google Scholar]
- 11.Campbell LC, Wilkinson MJ, Manz A, Camilleri P, Humphreys CJ. Lab Chip. 2004;4:225. doi: 10.1039/b312592k. [DOI] [PubMed] [Google Scholar]
- 12.Adeyeye AO, White RL. J. Appl. Phys. 2004;95:2025. [Google Scholar]
- 13.Stern E, Klemic JF, Routenberg DA, Wyrembak PN, Turner-Evans DB, Hamilton AD, LaVan DA, Fahmy TM, Reed MA. Nature. 2007;445:519. doi: 10.1038/nature05498. [DOI] [PubMed] [Google Scholar]
- 14.Cao H, Yu Z, Wang J, Tegenfeldt JO, Austin RH, Chen E, Wu W, Chou SY. Appl. Phys. Lett. 2002;81:174. [Google Scholar]
- 15.Li W, Tegenfeldt JO, Chen L, Austin RH, Chou SY, Kohl PA, Krotine J, Sturm JC. Nanotechnology. 2003;14:578. [Google Scholar]
- 16.Guo LJ, Cheng X, Chou C-F. Nano Lett. 2004;4:69. [Google Scholar]
- 17.Chou SY, Krauss PR, Renstrom PJ. Appl. Phys. Lett. 1995;67:3114. [Google Scholar]
- 18.Verbridge SS, Edel JB, Stavis SM, Moran-Mirabal JM, Allen SD, Coates G, Craighead HG. J. Appl. Phys. 2005;97:124317. [Google Scholar]
- 19.Zhang L, Gu F, Tong L, Yin X. Microfluid Nanofluid. 2008;5:727. [Google Scholar]
- 20.Harfenist SA, Cambron SD, Nelson EW, Berry SM, Isham AW, Crain MM, Walsh KM, Keynton RS, Cohn RW. Nano Lett. 2004;4:1931. [Google Scholar]
- 21.Fan R, Karnik R, Yue M, Li D, Majumdar A, Yang P. Nano Lett. 2005;5:1633. doi: 10.1021/nl0509677. [DOI] [PubMed] [Google Scholar]
- 22.Huh D, Mills KL, Zhu X, Burns MA, Thouless MD, Takayama S. Nat. Mater. 2007;6:424. doi: 10.1038/nmat1907. [DOI] [PubMed] [Google Scholar]
- 23.Kenis PJ, Ismagilov RF, Whitesides GM. Science. 1999;285:83. doi: 10.1126/science.285.5424.83. [DOI] [PubMed] [Google Scholar]
- 24.Heo K, Cho E, Yang J-E, Kim M-H, Lee M, Lee BY, Kwon SG, Lee M-S, Jo M-H, Choi H-J, Hyeon T, Hong S. Nano Lett. 2008;8:4523. doi: 10.1021/nl802570m. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Maspoch D, Zou S, Schatz GC, Smalley RE, Mirkin CA. Proc. Natl. Acad. Sci. USA. 2006;103:2026. doi: 10.1073/pnas.0511022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan J, Lee LJ. Proc. Natl. Acad. Sci. USA. 2005;102:18321. doi: 10.1073/pnas.0506902102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan J, Yu B, Lee LJ. Adv. Mater. 2007;19:1212. [Google Scholar]
- 28.Guan J, Ferrell N, Yu B, Hansford DJ, Lee LJ. Soft Matter. 2007;2:1369. doi: 10.1039/b709910j. [DOI] [PubMed] [Google Scholar]
- 29.Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, Bensimon A. Science. 1997;277:1518. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- 30.Wulff G. Angew. Chem. Int. Ed. 1995;34:1812. [Google Scholar]
- 31.Deniss IS, Morgan AR. Nucleic Acids Res. 1976;3:315. doi: 10.1093/nar/3.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLellan JM, Geissler M, Xia Y. Chem. Phys. Lett. 2005;408:80. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


