Abstract
The p53 tumor suppressor gene is one of the most commonly mutated genes in human cancers and the corresponding encoded protein induces apoptosis or cell-cycle arrest at the G1/S checkpoint in response to DNA damage. To date, previous studies have shown that antigens encoded by human tumor viruses such as SV40 large T antigen, adenovirus E1A and HPV E6 interact with p53 and disrupt its functional activity. In a similar fashion, we now show that EBNA3C, one of the EBV latent antigens essential for the B-cell immortalization in vitro, interacts directly with p53. Additionally, we mapped the interaction of EBNA3C with p53 to the C-terminal DNA-binding and the tetramerization domain of p53, and the region of EBNA3C responsible for binding to p53 was mapped to the N-terminal domain of EBNA3C (residues 130–190), previously shown to interact with a number of important cell-cycle components, specifically SCFSkp2, cyclin A, and cMyc. Furthermore, we demonstrate that EBNA3C substantially represses the transcriptional activity of p53 in luciferase based reporter assays, and rescues apoptosis induced by ectopic p53 expression in SAOS-2 (p53−/−) cells. Interestingly, we also show that the DNA-binding ability of p53 is diminished in the presence of EBNA3C. Thus, the interaction between the p53 and EBNA3C provides new insights into the mechanism(s) by which the EBNA3C oncoprotein can alter cellular gene expression in EBV associated human cancers.
Keywords: EBV, EBNA3C, p53, transactivation, apoptosis
Introduction
The p53 tumor suppressor gene is believed to be among the most frequently mutated genes in human cancer including lung, colorectal, and breast cancer, as well as many others. p53 responds to DNA damage or other various cellular stresses to reduce cell growth and promote programmed cell death (Gottlieb and Oren, 1996; Ko and Prives, 1996; Levine 1997; Vogelstein, Lane, and Levine, 2000). p53 accomplishes its antiproliferative properties through its action as a DNA-binding transcriptional activator, to induce expression of downstream target genes, which includes p21/waf1/cip1 (El-Deiry et al., 1993), GADD45 (Kastan et al., 1992), cyclin G (Okamoto and Beach, 1994), bax (Miyashita and Reed, 1995), IGF-BP3 (Buckbinder et al., 1995), and mdm2 (Wu and Levine, 1994). These target gene products are involved in cell-cycle arrest, apoptosis, and regulation of p53 function in cells exposed to genotoxic stresses.
The expression level of p53 is predominantly regulated through the ubiquitin-proteasome pathway by the specific E3 ubiquitin ligase, Mdm2 and is maintained at low levels during normal homeostasis (Tang et al., 2006). However, in response to cellular stresses such as DNA damage, hypoxia and oncogene activation, p53 is stabilized and activated through several mechanisms that block the Mdm2-p53 interaction (Sherr and Weber, 2000; Stommel and Wahl, 2004; Tang et al., 2006). Interestingly, mdm2 is one of the genes which is transactivated by p53 (Wu and Levine, 1994), thus not only is Mdm2 required for keeping p53 in check under non-stress conditions and releasing it when appropriate, it is also part of an auto-regulatory feedback loop (Prives, 1998).
Epstein-Barr virus (EBV) is a γ-herpesviruses that produces an asymptomatic infection in the majority of the global population (Rickinson and Kieff, 2002). However, EBV is also associated with several human malignancies of B-cell origin, including Burkitt’s lymphoma, nasopharyngeal carcinoma, Hodgkin’s disease, immunoblastic B lymphoma in AIDS patients, post-transplant associated lymphomas, and some gastric carcinomas (Ambinder, 1990; Rickinson and Kieff, 2002). Although, the precise mechanisms of these EBV mediated diseases are not yet clear, it has been suggested that interference with cell-cycle checkpoints and responses to DNA damage by EBV encoded oncoproteins may play vital roles in B-cell lymphomagenesis (O'Nions and Allday, 2004). In vitro, EBV can transform normal resting human B-cells to continuously proliferating lymphoblastoid cell lines (LCLs). The resulting LCLs express nine viral latency proteins, including six nuclear proteins (EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, and EBNALP) and three integral membrane proteins (LMP1, LMP2A, and LMP2B) (Rickinson and Kieff, 2002). Four viral antigens, EBNA2, LMP1, EBNA3A, and EBNA3C have been shown to be absolutely essential for EBV transformation of human B cells and establishment of latency in vitro (Cohen et al., 1989; Hammerschmidt and Sugden, 1989; Kaye, Izumi, and Kieff, 1993; Tomkinson, Robertson, and Kieff, 1993).
EBNA3C has been shown to play a complex regulatory role in the transcription of several viral and cellular genes. EBNA3C targets the cellular transcription factor, RBP-Jk to antagonize EBNA2 mediated transactivation (Johannsen et al., 1996; Robertson et al., 1995; Robertson, Lin, and Kieff, 1996). However, conversely, EBNA3C cooperates with EBNA2 in activation of the major viral LMP1 promoter via interaction with the cellular transcription factor, Spi-1/Spi-B (Zhao and Sample, 2000). EBNA3C is also involved in the regulation of chromatin remodeling by recruitng both histone acetylase and deacetylase activities (Knight et al., 2003; Radkov et al., 1999; Subramanian et al., 2002a). Furthermore, EBNA3C associates with Nm23-H1, a metastasis suppressor protein, and modulates the transcription of cellular genes involved in cell migration and invasion (Kaul et al., 2007; Subramanian, Cotter, and Robertson, 2001; Subramanian, Knight, and Robertson, 2002). In addition to its transcriptional functions, it has been reported that EBNA3C has cell-cycle regulatory functions, presumably mediated by direct protein–protein interactions (Knight et al., 2004; Knight and Robertson, 2004; Knight, Sharma, and Robertson, 2005a; Knight, Sharma, and Robertson, 2005b). EBNA3C expression stimulates cyclin A-dependent kinase activity (Knight et al., 2004; Knight and Robertson, 2004), and recruits the SCFSkp2 ubiquitin ligase complex and also regulates the stability of vital cell-cycle modulatory components, such as p27 (Knight, Sharma, and Robertson, 2005b) and Rb (Knight, Sharma, and Robertson, 2005a) in transiently or stably transfected cells. Paradoxically, a recent study showed that EBNA3C stabilizes another cellular oncoprotein, cMyc (Bajaj et al., 2008).
Overall, oncogenic viruses are able to disrupt cell-cycle checkpoints induced by genotoxic stress (O’Nions and Allday, 2004). Among the human DNA tumor virus oncoproteins, SV40 large T antigen, adenovirus E1A and HPV E6 have all been shown to form complex with p53 and disrupt p53-dependent transcriptional activity through distinct mechanisms, which can contribute to virus mediated oncogenesis (Lechner et al., 1992; Steegenga et al., 1996). Thus, it seems common for viruses to contribute to carcinogenesis by repressing the activities of p53. Several studies indicate that EBV does not specifically target p53 in LCLs (O'Nions and Allday, 2003; Wade and Allday, 2000), however, recent studies have demonstrated that EBV encoded antigens can interfere with cell-cycle checkpoints at both G1/S (Knight et al., 2004; Knight and Robertson, 2004) and G2/M (Choudhuri et al., 2007) to mediate cellular transformation and EBNA1 may contribute to this process by interacting with a cellular deubiquitination enzyme leading to reduction in p53 levels (Holowaty et al., 2003; Saridakis et al., 2005). Given the nature of p53 as a common target for several viral oncoproteins, it is reasonable to speculate that p53 may also be a target of EBNA3C.
In this report, we explored the effects of EBNA3C on the p53 tumor suppressor. In vitro binding assays as well as in vivo immunoprecipitation experiments in EBV infected LCLs showed that EBNA3C forms a stable complex with p53. The interaction occurs at two distinct regions of p53, which includes the central DNA binding domain and C-terminal regulatory domain. Additionally, the region of EBNA3C that binds to p53 was mapped at the amino-terminal region (residues 130–190), which includes the leucine zipper motif and was previously shown to include the binding site of numerous cellular components, such as RBP-Jκ, cyclin A, Rb, SCFSkp2 and cMyc (Bajaj et al., 2008; Knight et al., 2004; Knight, Sharma, and Robertson, 2005a; Knight, Sharma, and Robertson, 2005b; Robertson and Kieff, 1996). The addition of p53 molecule in this repertoire of cellular components further reveals the significance of this binding domain of EBNA3C in deregulating the cell-cycle. Similar to adenovirus E1A and HPV oncoprotein E6, here we demonstrate that EBNA3C can efficiently repress p53-mediated transcriptional activity as well as DNA-binding ability. Additionally, we confirm the manifestation of EBNA3C mediated repression on p53 functional activity by rescuing cells from p53-dependent apoptosis. Based on these results, we propose that EBNA3C modulates the functional activities of p53 in terms of its transactivation activity as well as induction of apoptosis, which may contribute to the oncogenic properties of EBNA3C in EBV infected cells.
Results
EBNA3C interacts directly with p53 in vitro
A number of tumor virus antigens have been shown to interact directly with p53 for disrupting its function. The role of EBV latent antigens in regulating p53 has so far been elusive in the context of latency III, however studies have sown that EBNA1 can protect infected cells from apoptosis by lowering the levels of p53 via interaction with a deubiquitination enzyme (Holowaty and Frappier, 2004; Holowaty et al., 2003; Saridakis et al., 2005). It would be interesting to investigate whether or not the essential EBV antigen, EBNA3C can similarly deregulate this cellular master regulator, p53 either by direct protein-protein interaction or via an indirect mechanism.
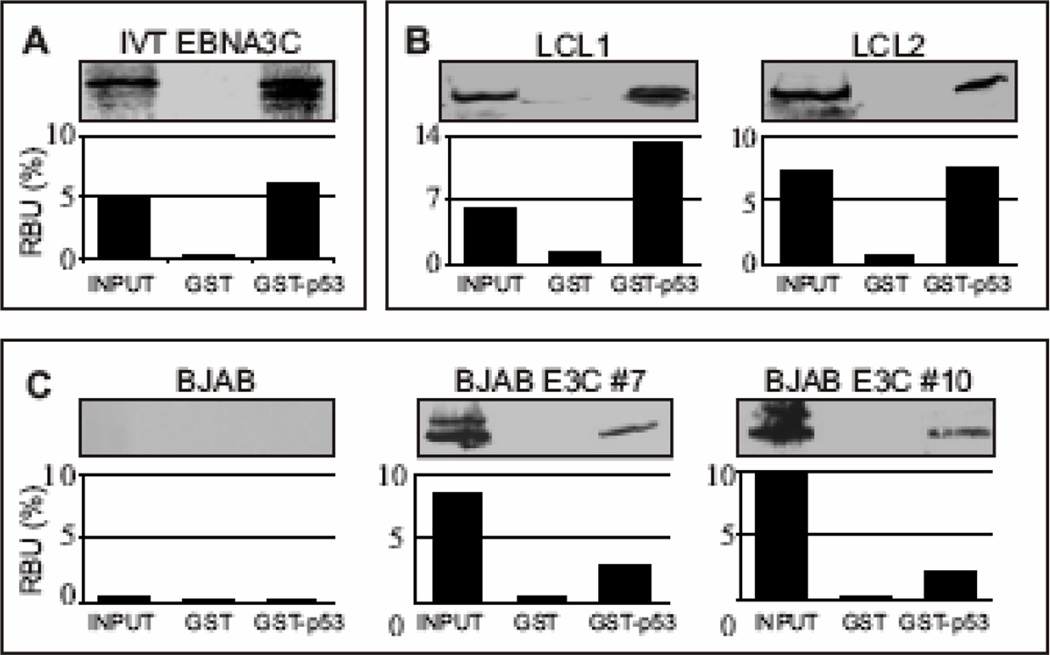
To determine whether EBNA3C is able to interact directly with p53, we performed in vitro binding assays. A GST-p53 expression construct was bacterially expressed and the fusion protein was incubated with in vitro translated 35S-labeled EBNA3C (Fig. 1A). GST-p53 beads precipitated a significant amount of radio-labeled EBNA3C, while negligible signal was observed with GST control beads (Fig. 1A; compare lane 3 with lane 2). To corroborate the result of the in vitro binding study in a more biologically relevant setting, we conducted parallel GST pull-down experiments, using cell lysates prepared from two transformed EBV positive lymphoblastoid cell-lines - LCL1 and LCL2, EBV negative Burkitt’s lymphoma cell line (BJAB) and BJAB cells stably expressing EBNA3C (two clones - BJAB E3C #7 and E3C #10). As shown in Figure 1B and 1C, EBNA3C strongly interacted with the GST-p53 fusion protein but not with the GST control in these pull-down assays. These results support the initial observation that EBNA3C interacts directly with p53 in vitro and can form a complex with p53 in lysates of human cells.
Figure 1.
EBNA3C forms a stable complex with p53 in vitro. (A) GST-p53 fusion protein was expressed in E. coli and purified with glutathione Sepharose beads. Full-length EBNA3C was labeled with 35S methionine by in vitro translation and incubated with either GST control or GST-p53 beads normalized by Coomassie staining. 5% of in vitro translation (IVT) input was used for comparison. Precipitated proteins were resolved by SDS-PAGE, and bands were visualized with a phosphorimager screen. Relative Density was quantified using Storm 850 imaging system. (B–C) Either GST control or GST-p53 beads were incubated with lysates prepared from either 50 million B) lymphoblastoid cell lines (LCL1 and LCL2) or C) BJAB cells and BJAB cells stably expressing EBNA3C (two clones – BJAB EBNA3C#7 and BJAB EBNA3C#10). Approximately 5% of the lysed cells were saved as input and precipitated protein complexes were resolved by 7% SDS-PAGE. EBNA3C was detected by western blot with the specific monoclonal antibody (A10) followed by an infrared tagged secondary antibody and scanned using Odyssey imager. All panels are representative gels from similar repeat experiments.
EBNA3C associates with p53 and forms a complex in human cells in vivo
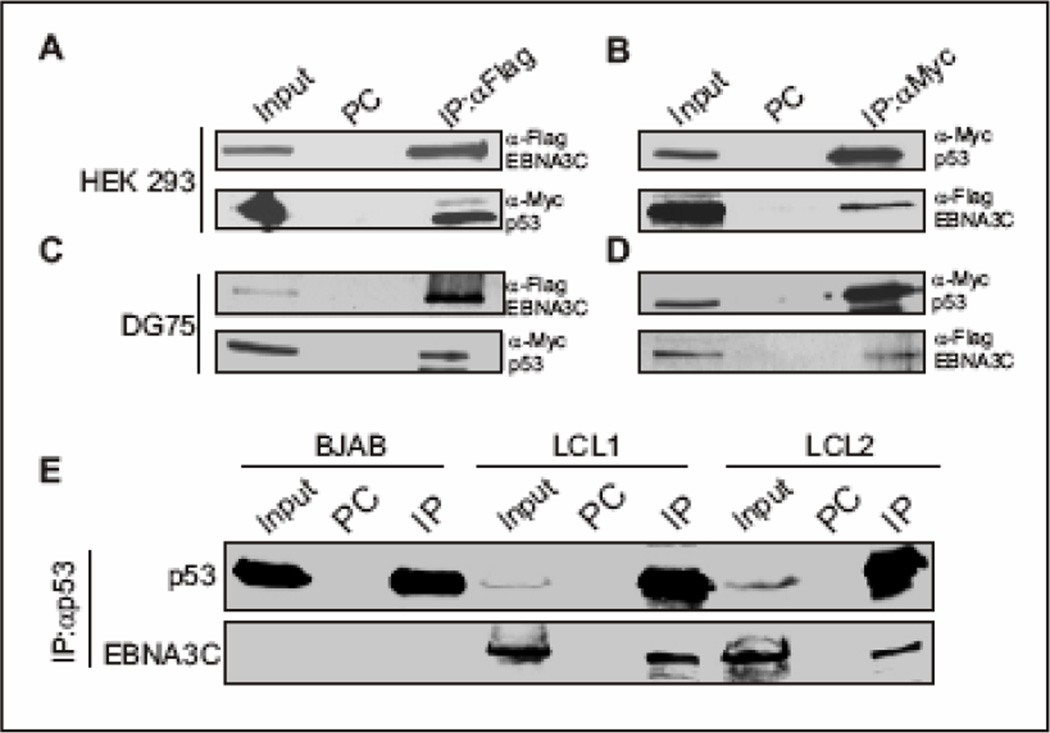
To corroborate the in vitro binding data next we performed a series of in vivo co-immunoprecipitation experiments. Both HEK 293 and DG75 cells were co-transfected with expression constructs for flag-tagged EBNA3C and p53 tagged with myc epitope. Immunoprecipitation analyses as shown in Figure 2A–D clearly demonstrated that EBNA3C strongly associated with p53 in vivo. A distinct band of myc-tagged p53 was detected in the anti-flag immunoprecipitated lane by immunoblot against myc-p53 (Fig. 2A and 2C bottom panels). In the reverse experiments, the flag-tagged EBNA3C was also clearly detected in the anti-myc immunoprecipitated sample (Fig. 2B and 2D, bottom panels). Importantly, EBNA3C or p53 binding was not detected in control samples using normal mouse serum (designated as PC in Fig. 2A–D) supporting the specificity of these complexes in cells. To verify further, if this binding was significant at endogenous levels, p53 was immunoprecipitated from EBV negative BJAB cells and two different EBV transformed lymphoblastoid cell lines (LCL1 and LCL2). Again, immunoprecipitation of p53 complexes resulted in a reproducible co-immunoprecipitation of EBNA3C, but not with the control serum, supporting the existence of this stable complex in EBV-transformed B lymphocytes (Fig. 2D). Hence, analysis of the data from the ectopic expression system as well as cell lines endogenously expressing p53 and EBNA3C demonstrated the strong association of p53 with EBNA3C in complex in EBV infected cells.
Figure 2.
EBNA3C interacts with p53 in vivo. (A–B) Either 10 million HEK 293 cells or (C–D) 20 million DG75 cells were co-transfected with flag-tagged EBNA3C and myc-tagged p53. Cells were harvested at 36h post-transfection and approximately 5% of the lysed cells were saved as input and remaining were immunoprecipitated (IP) with 1.5 µg of appropriate antibody. Lysates and IP complexes were resolved by SDS-PAGE and immuno blotted with the indicated antibodies. The same blots were stripped and reprobed with appropriate antibodies. PC: preclear; prior to set up IP, cell lysates were precleard with normal mouse serum with protein A/G beads. C) Either 50 million BJAB cells or LCLs (LCL1 and 2) were collected at exponential growth phase and lysed in RIPA buffer. Protein complexes were immunoprecipitated (IP) with 2.0 µg p53 specific antibody (DO-1, Santa Cruz). Samples were resolved by 7% SDS-PAGE and western blotting for the indicated proteins was done by stripping and reprobing the same membrane. PC: preclear. All panels are representative gels from similar repeat experiments.
The N-terminal domain of EBNA3C binds to the C-terminal domain of p53
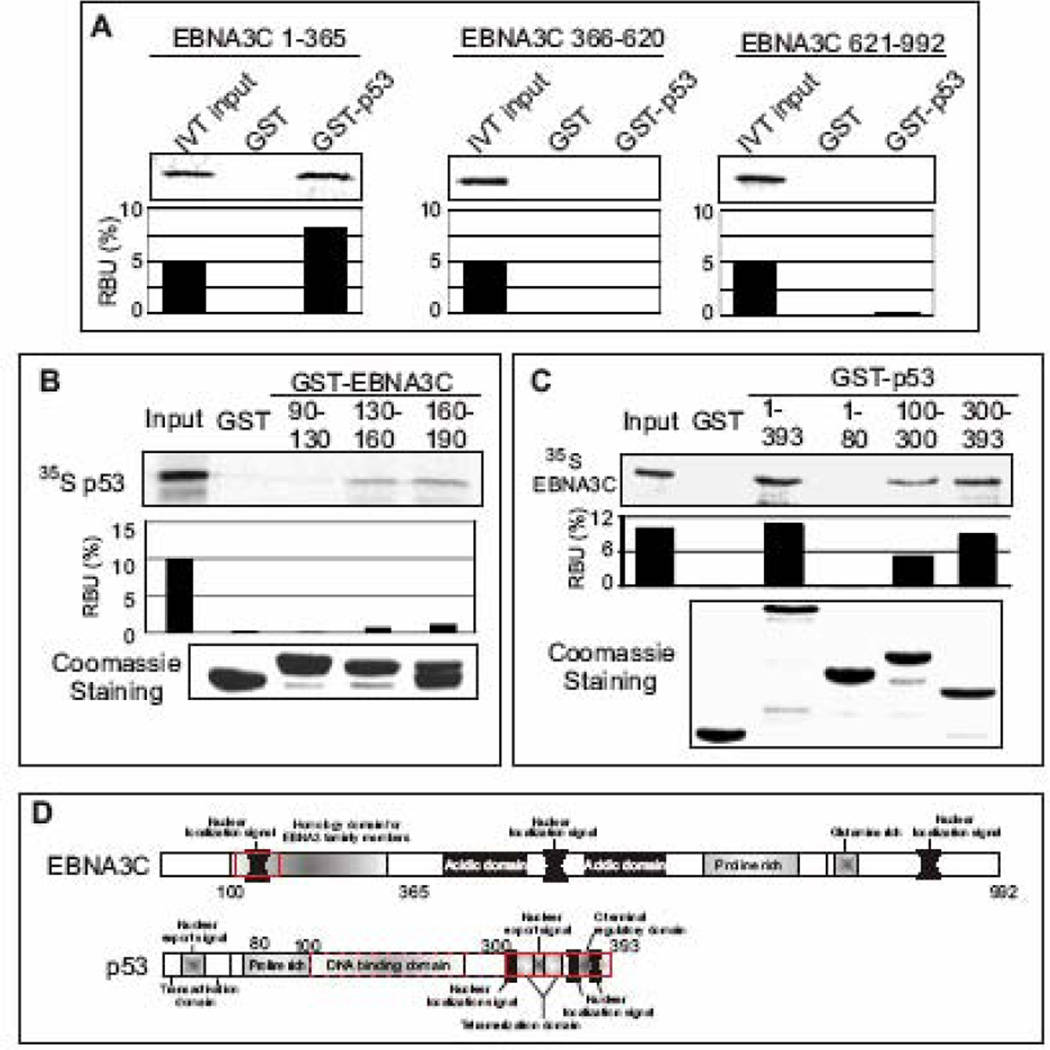
To determine the precise binding site of EBNA3C that interacts with p53, we utilized a series of EBNA3C truncated constructs (residues 1–365, 366–620, and 621–992) (Bajaj et al., 2008). These EBNA3C truncations were in vitro translated and subjected to GST-pulldown assays. As shown in Fig. 3A, GST-p53 strongly precipitated the N-terminal domain of EBNA3C (residues 1–365) (Fig. 3A, left panel), whereas neither the middle part (residues 366–620) nor the c-terminal domain (residues 621–992) was seen in GST-p53 pull-down samples (Fig. 3A, middle and right panels, respectively). All EBNA3C truncations failed to bind with the GST control, indicating that the observed interaction was specific for p53. To map the interacting domain within the N-terminal region of EBNA3C in more detail, bacterially expressed smaller truncations of GST-fused EBNA3C proteins were tested for their ability to bind with full-length p53. As shown Fig. 3B, in vitro translated 35S-radiolabeled p53 showed strong binding to GST-fused EBNA3C residues 130–160 and 160–190 (Fig. 3B, top panel, lanes 4 and 5, respectively), but no significant binding was observed either with the GST control or EBNA3C residues 90–130 (Fig. 3B, top panel, lanes 2 and 3, respectively). Coomassie staining of a parallel gel indicated the levels of various GST proteins used in this binding assay (Fig. 3B, bottom panel). These results subsequently indicated that the particular domain of EBNA3C which was previously shown to interact with several important cellular molecules, also forms a stable complex with p53, signifying the importance of this region in deregulation of the cell-cycle in EBV infected cells.
Figure 3.
N-terminal domain of EBNA3C binds to C-terminal domain of p53. (A–C) 35S-radiolabeled either full-length (C) or different EBNA3C truncated fragments (A) or full-length p53 (B) was in vitro translated using a T7 TNT translation kit. All 35S-radiolabeled in vitro translated proteins in binding buffer were precleared by rotating with GST-beads for 1h at 4°C. The binding reaction mixture was set up with either bacterially purified GST control or the indicated GST fusion proteins. Reactions were resolved by appropriate SDS-PAGE, exposed to phosphorimager plate, and scanned on a Storm 850 imaging system. The amount of protein bound in each GST-pulldown sample was quantified with ImageQuant software (Molecular Dynamics). Coomassie staining of SDS-PAGE resolved purified GST proteins is shown (B and C, bottom panels). All panels are representative gels from similar repeat experiments. (D) Schematics illustrate different structural domains of EBNA3C and p53. Red boxes indicate the respective binding domain(s) of two proteins.
EBNA3C binds p53 at the DNA binding and tetramerization domain
While little is known about the tertiary and quaternary structure of EBNA3C, p53 is well understood structurally in the context of structure-function relationships. The p53 protein can be divided roughly three distinct domains - the N-terminal domain containing the transactivation domain, the central core containing its sequence-specific DNA-binding domain, and the multifunctional C-terminal domain (Adler et al., 1997; Hupp et al., 1993). The acidic activation domain lies within residues 1–43, although neighboring sequences are also likely to contribute to the transcriptional activity of p53. The central core of p53 lies within residues 100–300, and the C-terminus of p53 lies within residues 300–393. To determine the interacting domain(s) of p53 with EBNA3C, different GST-fused truncated mutants of p53 were generated and tested for their ability to bind radio-labeled EBNA3C using in vitro binding experiments. The results of these binding studies clearly demonstrated that that full-length p53, the central domain (residues 100–300) and the C-terminal domain (residues 300–393) can tightly interact with EBNA3C (Fig. 3C, top panel, lanes 3, 5 and 6, respectively), whereas no binding was observed with either GST control or the N-terminal transactivation domain (Fig. 3C, top panel, lanes 2 and 4, respectively). Thus, the amino acid region 100–393 of p53 which encompasses both DNA binding and tetramerization domains, is responsible for interaction with EBNA3C, however, particularly, the tetramerization domain of p53 showed stronger (1.7 fold higher) interaction with EBNA3C. Coomassie staining of a parallel gel indicated the amounts of different GST-p53 proteins used in this binding assay (Fig. 3C, bottom panel).
EBNA3C colocalizes with p53 in the nucleus of EBV transformed LCLs
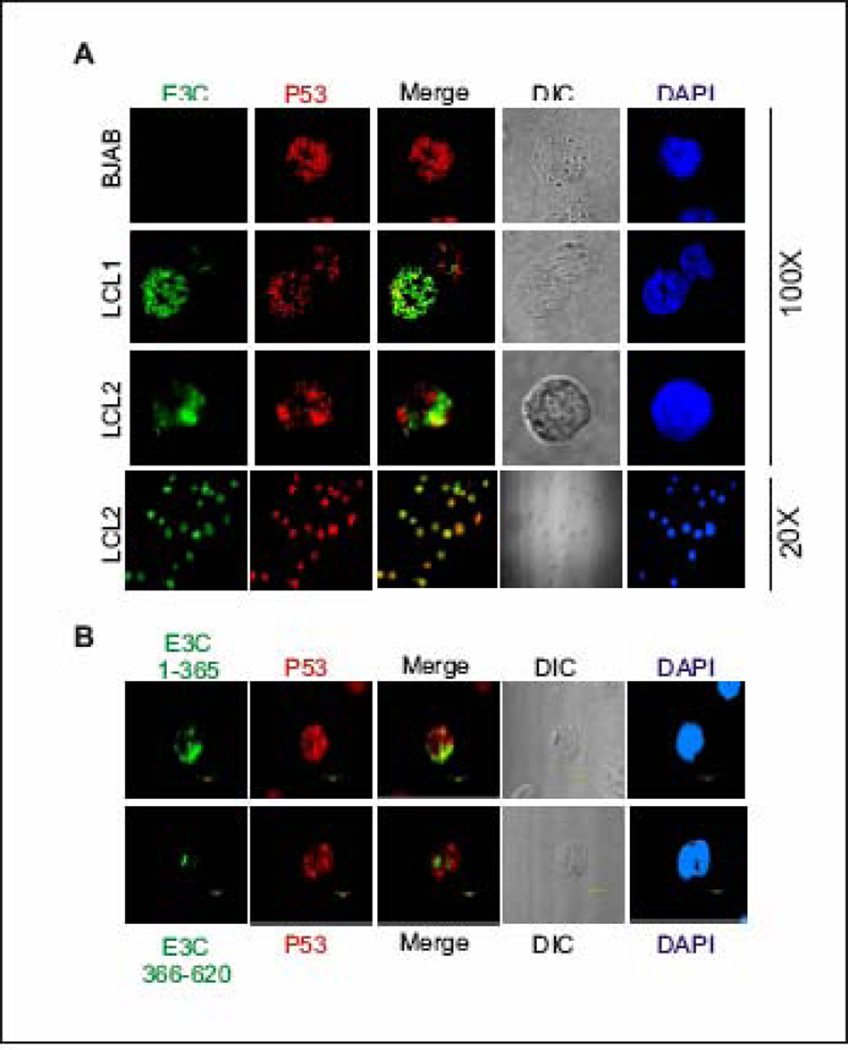
To further support the in vitro and in vivo binding data and also to visualize the interaction between p53 and EBNA3C under physiologically relevant conditions, colocalization experiments were performed using EBV infected LCL1 and LCL2 cells (Fig. 4, middle and bottom panels, respectively). BJAB cells represent EBV negative cells not expressing EBNA3C (Fig. 4, top panels). Immuno-fluorescence staining using specific monoclonal antibodies against EBNA3C and p53 showed that both proteins were tightly nuclear, demonstrating a stippled, punctate pattern staining with the exclusion of nucleoli (Fig. 4). In both EBV positive cell lines, p53 clearly colocalized with EBNA3C at a number of spots, as visualized by yellow fluorescence. These data corroborate the in vivo and in vitro association studies between p53 and EBNA3C showing that these molecules localized in part to similar compartments in the cell nucleus (Fig. 4, second, third and fourth panels, third column). For additional proof, the colocalization study was extended using the truncated domains of EBNA3C, the N-terminal domain (residues 1–365) and as a negative control, the middle part (residues 366–620), which did not interact with p53 in GST-binding assays or in in vivo co-immunoprecipitation assays. EBV negative Burkitt’s lymphoma cell line, BJAB was transiently transfected with EBNA3C constructs expressing GFP-tagged residues 1–365 and 366–620. Specific monoclonal antibody against p53 was used to detect endogenous p53. The results presented in Fig. 4B clearly demonstrated that p53 colocalized with N-terminal domain of EBNA3C (Fig. 4B, top panel), whereas in contrast, the central domain EBNA3C (residue 366–620) showed negligible colocalization with p53 (Fig. 4B, bottom panel).
Figure 4.
EBNA3C colocalizes with p53 in EBV positive cell lines. A) EBV negative burkitt lymphoma cell line, BJAB, and two EBV transformed cell lines - LCL1 and LCL2 were air-dried onto slides and fixed using a 1:1 mixture of acetone and methanol. Endogenously expressed p53 was detected using mouse monoclonal antibody (DO-1, 1:200 dilution) respectively, followed by anti-mouse Alexa Fluor 594 (red), and EBNA3C was detected using EBNA3C–reactive human serum (1:150 dilution) followed by anti-human Alexa Fluor 488 (green). EBV negative BJAB cells were used as EBNA3C null control cell lines. B) 10 million BJAB cells were transfected with GFP-tagged EBNA3C truncated mutants – residues 1–365 (top panels) and residues 366–620 (bottom panels). Endogenous p53 was detected using DO-1 antibody as B. The nuclei were counterstained using DAPI (blue). The images were sequentially captured using an Olympus confocal microscope. All panels are representative pictures from similar repeat experiments.
EBNA3C inhibits p53 mediated transactivation
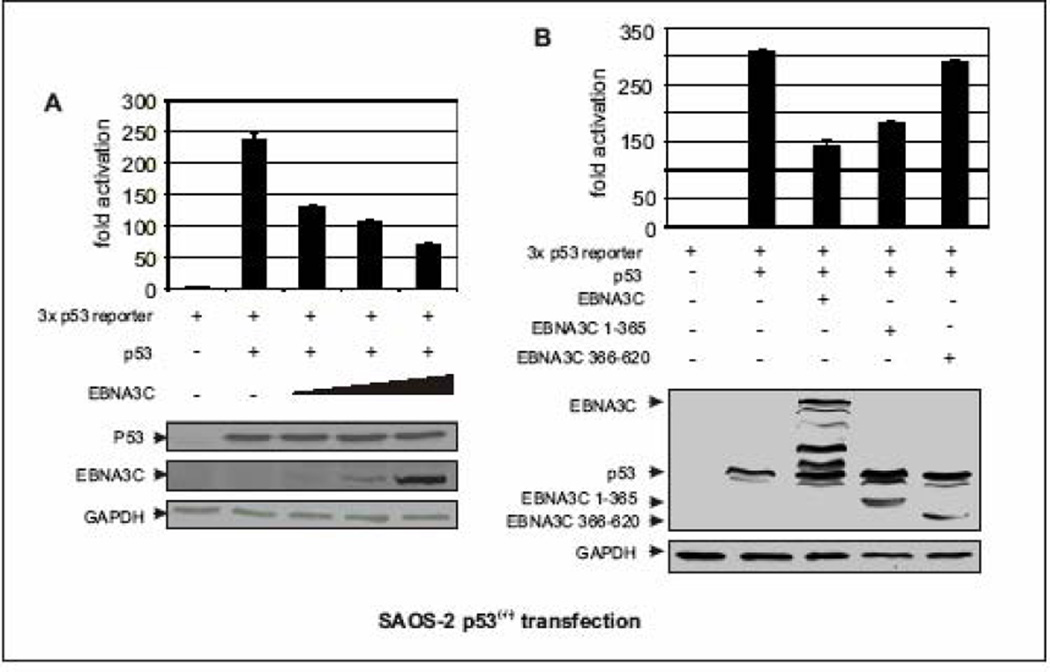
Since the transcriptional activity of p53 is important for p53-mediated regulation (Adimoolam and Ford, 2003), we investigated whether EBNA3C represses p53-dependent transcriptional activity. Reporter constructs containing thirteen consensus p53-binding sites promoter fused to the luciferase gene were transiently cotransfected into p53-null cells (SAOS-2) with the empty vector or plasmid expressing EBNA3C. In the presence of EBNA3C, p53 transactivation activity was significantly reduced in SAOS-2 cells (Fig. 5A). This reduction in p53 transactivation activity was not cell specific as in BJAB, a B-cell line, EBNA3C was also found to repress the transactivation function of p53 using the same reporter construct (data not shown). These results indicate that EBNA3C can suppress p53-dependent transcriptional activity. Moreover, increasing amounts of EBNA3C resulted in a dose-dependent inhibition of p53 transactivation (Fig. 5A, bar diagram). The expression levels of EBNA3C, p53, and GAPDH as a loading control, also were analyzed by Western blotting (Fig. 5A, bottom panels). EBNA3C expression did not interfere with the expression levels of p53 and GAPDH (Fig. 5A), and so indicates that EBNA3C mediated repression of p53 transactivation is not likely to occur by promoting the degradation of p53. To further define the domain or domains of EBNA3C important for this activity and also to determine if the binding domain of EBNA3C is essential for inhibition of p53-mediated transactivation, the aforementioned experiments were extended using different truncated domains of EBNA3C. Two EBNA3C truncated mutants were tested, the N-terminal binding region of EBNA3C (residues 1–365) showed almost similar ability to repress p53-dependent transcriptional activity as seen in case of wild-type EBNA3C (Fig. 5B, bar diagram, compare bars 3 and 4) whereas the non-binding middle region of EBNA3C (residues 366–620) had no significant effect (Fig. 5B, bar diagram, compare bars 4 and 5). Overall the data suggest that EBNA3C represses the p53 transactivation activity by forming a complex with p53 perhaps by interfering with its ability to access the target promoters.
Figure 5.
EBNA3C represses p53 mediated transcriptional activity. (A–B) Approximately 0.4 × 106 SAOS-2 (p53−/−) cells were cotransfected with 0.25 µg of the promoter construct containing p53 responsive element and 0.5 µg of myc-tagged p53 plus either vector control or A) increasing amount of flag-tagged EBNA3C expressing constructs (0, 0.25, 0.5, 1.0 µg respectively) B) flag-tagged different truncated mutants of EBNA3C using Lipofectamine 2000 (Invitrogen). At 24 h post-transfection, cells were harvested and lysed for luciferase assays. Total amount of proteins were normalized by Bradford assay. Increasing amounts of EBNA3C show proportional increment in luciferase activity. The representative plot is a mean of two independent experiments. Error bar represents standard deviation (SD). Bottoms panels indicate the fractions of the cell lysates were resolved by SDS-PAGE to demonstrate the expression levels of p53 and EBNA3C. GAPDH blot was done for loading control.
EBNA3C can inhibit p53 sequence-specific DNA-binding activity
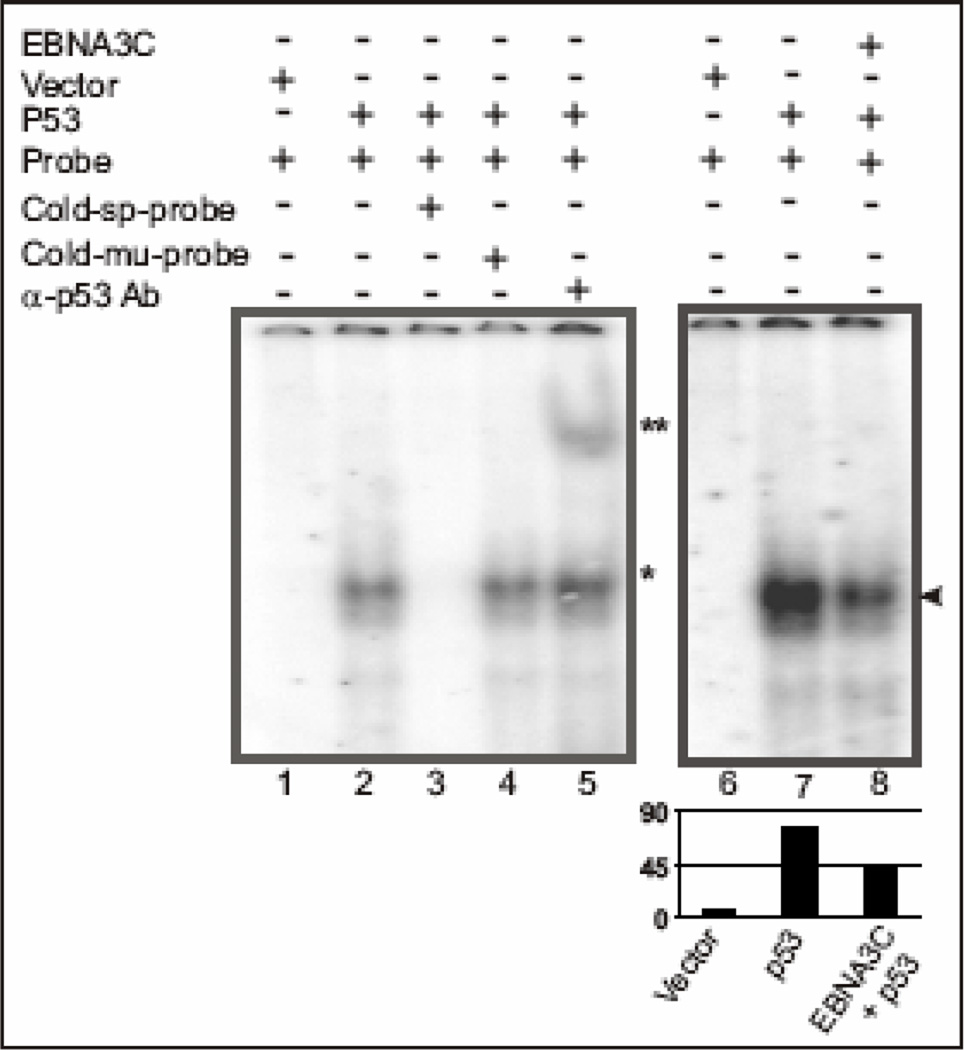
Repression of the p53 transactivation activity by EBNA3C plus the binding data showing that the central DNA binding domain of p53 is involved in p53-EBNA3C interaction, led us to further investigate whether EBNA3C exerts any potential inhibitory role on the DNA binding ability of p53. We performed electrophoretic mobility shift assay (EMSA) experiments to resolve whether any changes in DNA binding activities can be detected when p53 binds to its respective DNA-binding site, in presence of EBNA3C (Fig. 6). Nuclear extracts from SAOS-2 (p53−/−) cells transfected with either pA3M–p53, encoding myc-tagged p53 and pcDNA empty vector or pcDNA-EBNA3C, expressing full-length untagged EBNA3C, were incubated with 32P-labeled, consensus p53-binding sequences and analyzed using EMSA. Nuclear extracts containing p53 reduced the mobility of the radio-labeled probe, as expected (Fig. 6, comapare lane 2 with lane 1). The mobility was further super-shifted with an anti-p53 monoclonal antibody, suggesting that the specificity of the shift was due to the presence of p53 (Fig. 6, comapare lane 5 with lane 2). Additionally, the p53-specific shift was competed with the addition of a 200 X cold specific competitor (Fig. 6, compare lane 3 with lane 1), whereas, it was not affected by a similar amount of a mutant cold competitor (Fig. 6, compare lane 4 with lane 1). Importantly, in a separate experiment, addition of nuclear extracts containing both p53 and EBNA3C to the binding reaction, resulted in a significant decrease (~50%, Fig. 6 bar diagram) in the observed intensity of the p53-specific shift (Fig. 6, compare lane 8 with lane 7). These results strongly suggest that the EBNA3C-p53 complex may influence wild-type p53 activity by inhibiting the sequence-specific DNA binding activity of p53. Moreover, the complex between EBNA3C and p53 may alter the conformation of p53 in a way analogous to p53 missense mutants found in human cancers. Interestingly, the interaction between EBNA3C and another cellular transcription factor, cMyc - enhances its transcriptional activity probably via increasing its DNA-binding specificity (Bajaj et al., 2008). These contrasting results indicate that the functional consequence of EBNA3C on gene expression may depend on its interaction with specific cellular transcription factors.
Figure 6.
EBNA3C reduces DNA-binding ability of p53. A probe containing the p53 binding sequence (5’-AGGAAGAAGACTGGGCATGTCTGGGCA-3’) was labeled by Klenow fill-in reaction with [α-32P]dCTP and used for EMSA in presence and absence of EBNA3C. Lanes 1 and 6, probe incubated with nuclear extracts (NE) isolated from SAOS-2 (p53−/−) cells transfected with emprty vector control; lane 2 and 7, probe incubated with myc-tagged p53 expressing NE, showing band shift due to p53 binding (*); lane 3, probe with 200-fold molar excess of cold specific competitor, which abolished completely the p53 specific shift; lane 4, probe with 200fold molar excess of cold mutant probe competitor showing no effect on p53 specific shift; lane 5, probe incubated with p53 expressing NE and monoclonal antibody against p53 (DO-1), which super-shifted the p53 specific probe (**); lane 8, probe incubated with NE expressing both p53 and EBNA3C, reduced the p53 specific band intensity most likely due to formation of a stable inhibitory complex between p53 and EBNA3C in vivo (arrowhead).
p53-mediated apoptosis in SAOS-2 cells is suppressed by EBNA3C expression
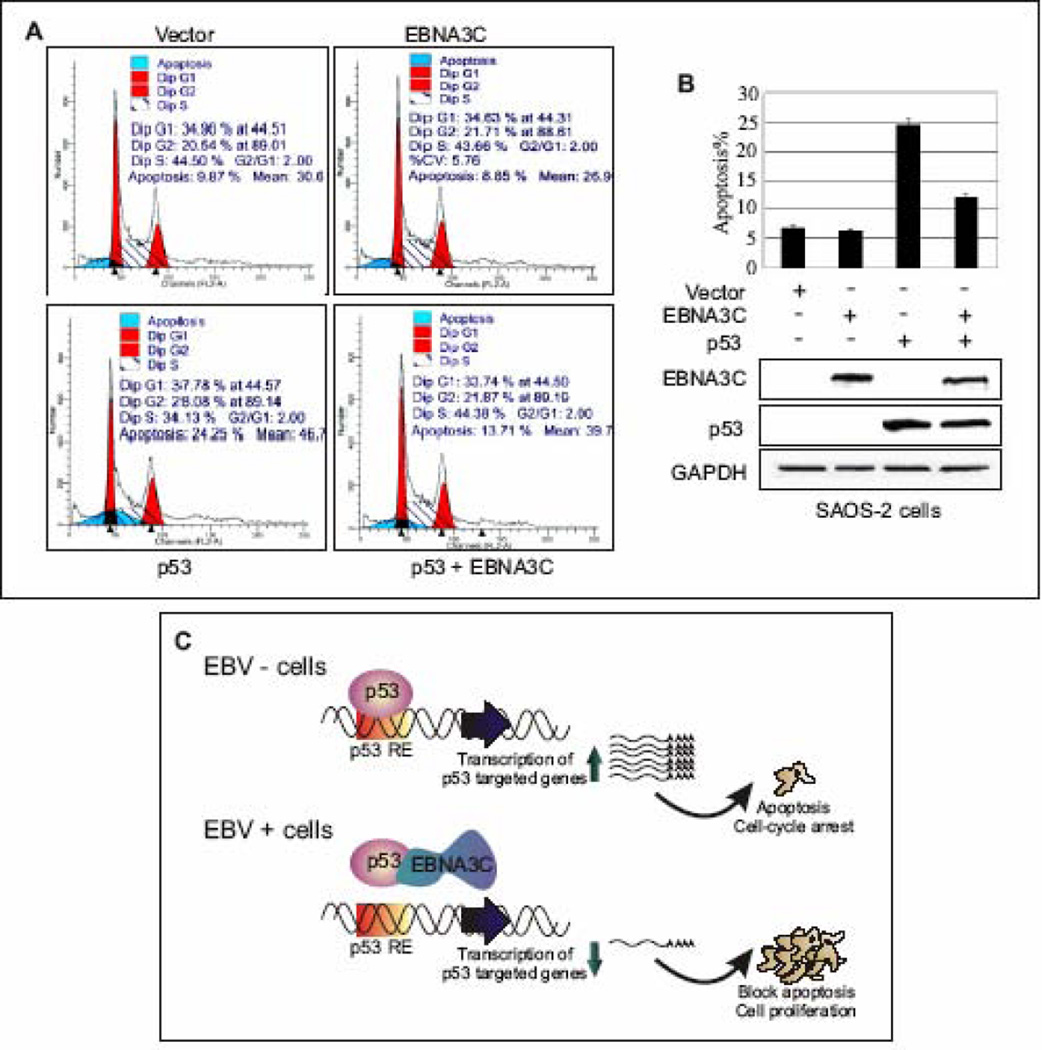
To determine the significance of EBNA3C in terms of its direct regulation of p53 function, we verified whether or not EBNA3C could affect p53 mediated apoptosis. It has been shown earlier that the over-expression of p53 can lead to apoptosis in p53-null SAOS-2 cells (Schuler et al., 2000). In order to determine if EBNA3C expression affects the ability of p53 to mediate apoptosis, the levels of apoptotic cells in the transiently transfected SAOS-2 cells with constructs expressing either wild-type p53 or EBNA3C alone, or p53 and EBNA3C together, were examined. Further, in order to monitor the transfection efficiency and viability of cells, a GFP expression construct was co-transfected and checked under fluorescent microscopy at 48 h post-transfection which revealed about 95% transfection efficiency (data not shown). Apoptosis was detected in approximately 25% of SAOS-2 cells expressing p53, which was significantly higher than the basal level of apoptotic cells (6–7%) expressing either vector control or EBNA3C alone (Fig. 7A–B, histogram and bar-representation). However, in presence of EBNA3C, p53 mediated apoptosis was drastically inhibited and was seen in only 10–12% of the cells, a decrease in approximately 50% (Fig. 7A–B, histogram and bar-representation). Thus, these data revealed that EBNA3C can provide a significant protection from p53-dependent apoptosis. The expression levels of EBNA3C, p53, and GAPDH as a loading control, were also analyzed by Western blotting (Fig. 7B, bottom panels).
Figure 7.
EBNA3C blocks p53 induced apoptosis in p53 null cell line SAOS-2. (A–B) SAOS-2 (p53−/−) cells were transiently transfected either with vector control, or constructs expressing untagged wild-type EBNA3C and p53 alone, or p53 and EBNA3C together. After 48 h of transfection, cells were harvested, fixed and levels of apoptotic cells in each samples were analyzed by flow cytometry (Becton-Dickinson). Total 20,000 events were analyzed for each sample. Data were analyzed using the ModFIT model program (Verity Software House). The representative plot is a mean of three independent experiments. Error bar represents SD. EBNA3C (top panel) and p53 (middle panel) protein expression levels were judged by western blot. Equal protein loading was analyzed GAPDH (bottom panel) blot. (C) A schematic representation of p53 mediated transcriptional regulation in EBV negative (-) and positive (+) cells. In response to genotoxic stress, p53 achieves its antiproliferative properties through its action as a DNA-binding transcriptional activator, to induce expression of numerous downstream target genes, involved in cell-cycle arrest and apoptosis. In EBV positive cells, EBNA3C potentially inhibits p53 mediated transcriptional activity via forming a stable complex with p53. p53 RE: p53 responsive element.
Discussion
The tumor suppressor p53 protects mammalian cells from malignant transformation by inducing either cell-cycle arrest or apoptosis in response to viral infection among other genotoxic stresses (Gottlieb and Oren, 1996; Ko and Prives, 1996; Levine 1997; Vogelstein, Lane, and Levine, 2000). Not surprisingly, either p53 itself or cellular factors involved in downstream activities are inactivated by various viral antigens either by releasing cells from cell-cycle checkpoints or by protecting cells from p53-dependent apoptosis (El-Deiry et al., 1993; Waldman, Kinzler, and Vogelstein, 1995; Yin et al., 1997). While cell-cycle arrest depends on the ability of p53 to induce the transcription of target genes such as the CDK inhibitor p21 (El-Deiry et al., 1993), apoptosis depends on induction of a distinct class of target genes, including bax, puma, perp, and many others (Vousden and Lu, 2002). Several virus encoded antigens including SV40 large T antigen, adenovirus E1A and E1B, hepatitis B virus (HBV) X protein, and HPV E6 and E7 interfere with p53 function via multiple mechanisms (Lechner et al., 1992; Martin and Berk, 1998; Mietz et al., 1992; Scheffner et al., 1990; Wang et al., 1994). Large T antigen and E1B was found to interact with p53 and enhance its stability, however surprisingly its functional activity was inhibited (Martin and Berk, 1998; Mietz et al., 1992). On the other hand, HPV E6 antigen induces the p53 degradation utilizing ubiquitin-proteasome mediated pathway (Mietz et al., 1992). In addition, the HBV X protein has been shown to interact with p53 and inhibits its functional activity in multiple ways (Wang et al., 1994).
Similar to smaller DNA viruses, herpesvirus family members also have been shown to manipulate p53. For instance, the CMV IE2 protein (Castillo, Yurochko, and Kowalik, 2000), the KSHV ORF K8 protein (Park et al., 2000), and the HHV6 ORF 1 protein (Doniger, Muralidhar, and Rosenthal, 1999) all inhibit the transactivation activity of p53. Also KSHV LANA and EBV encoded lytic antigen BZLF1 and latent antigen LMP1 have been shown to interfere with p53 functional activity (Friborg et al., 1999; Mauser et al., 2002; Fries, Miller, and Raab-Traub, 1996). Moreover, EBNA1 has also been shown to contribute to regulation of p53 level through interaction with a deubiquitination enzyme and so may contribute to regulation of its transcription and apoptotic activities (Holowaty et al., 2003; Saridakis et al., 2005). However, in comparison to the effects of the smaller DNA viruses, considerably less is known regarding the various mechanisms by which herpesviruses inhibit the transcriptional function of p53.
Recent studies have established that an essential EBV encoded latent antigen, EBNA3C (Rickinson and Kieff, 2002) strongly interferes the functional consequences of multiple components which are primarily involved in cell-cycle checkpoints at both G1/S (Knight et al., 2004; Knight and Robertson, 2004; Bajaj et al., 2008) and G2/M (Choudhuri et al., 2007). In contrast to the initial studies (Allday et al., 1995), we now show that EBNA3C has potential inhibitory effects on p53 functional activities. We speculate that the repressive effects of EBNA3C on p53 function provide a more effective platform which increases the efficiency of EBV mediated lymphomagenesis.
In vitro binding experiments suggested that EBNA3C and p53 physically interact, and this was supported by in vivo immunoprecipitation studies of cell lysates from EBV-positive cell lines (LCL1 and LCL2). Immuno-localization using specific antibodies against EBNA3C and p53 showed a speckled nuclear localization of these two molecules which were colocalized distinctly in EBV infected cells. Additional mapping of the interacting domain of EBNA3C showed that it associates with p53 via the same region, residues 130–190, which has been implicated previously for interaction with several important cellular components, including SCFSkp2, Rb, cMyc, cyclin A and RBPJκ (Bajaj et al., 2008; Knight et al., 2004; Knight, Sharma, and Robertson, 2005a; Knight, Sharma, and Robertson, 2005b; Robertson and Kieff, 1996). The inclusion of p53 in the group of cellular components modulated by this particular domain of EBNA3C further emphasizes the importance of this critical domain for bypassing the cell-cycle checkpoints in EBV-infected cells.
We further delineated the domains of p53 that are involved in binding to EBNA3C. In vitro binding experiment demonstrated that EBNA3C interacts with p53 at two different regions – the core DNA binding domain and the C-terminal oligomerization domain. Three major functional domains have been identified in p53: a N-terminal transactivation domain (residues 1–80) (Candau et al., 1997; Fields and Jang, 1990; Funk et al., 1992; Raycroft et al., 1990), a central sequence-specific DNA-binding domain (residues 94–293) (Bargonetti, et al., 1993; Halazonetis et al., 1993; Wang et al., 1993), and a C-terminal oligomerization domain (residues 325–355) (Clore et al., 1994; Jeffrey et al., 1995; Lee et al., 1994; Sakamoto et al., 1994; Sturzbecher et al., 1992). In addition to the oligomerization domain, the C-terminal domain contains two regions (residues 290–325 and residues 356–393) which negatively regulate its DNA-binding activity (Waterman et al., 1995; Hupp et al., 1992) through multiple posttranslational modifications, such as phosphorylation (Wang and Prives, 1995) and acetylation (Liu et al., 1999). In addition, evidence supports that the highly positively charged C-terminal regulatory domain may interact with the core DNA-binding domain and lock p53 in an inactive conformation (Muller-Tiemann et al., 1998). The critical oligomerization domain of p53 showed the higher affinity for EBNA3C, suggesting that EBNA3C may be involved in regulation of p53 mediated transcriptional activity and its DNA-binding ability.
Results from the binding experiments and the colocalization studies, prompted us to investigate further whether or not EBNA3C has any effect on p53 functional activity. The ability to regulate the luciferase expression from a reporter construct composing multiple units of p53 responsive element was tested in presence and absence of EBNA3C. Indeed the data showed that EBNA3C substantially blocks p53 mediated transactivation. Other viral proteins have also been shown to deactivate p53 mediated transactivation in a similar fashion (Park et al., 2000; Doniger, Muralidhar, and Rosenthal, 1999). The possibility of squelching was excluded as we have previously shown that EBNA3C can upregulate the transcription of the c-Myc target gene, telomerase reverse transcriptase (TERT) in promoter assays and that the effect was dose responsive (Bajaj et al., 2008).
Modifications to the C-terminal domain, including deletion of the last 30 amino acids (Hupp et al., 1992), protein binding (Hupp et al., 1992), phosphorylation of Ser-315, Ser-378, and Ser-392 (Hupp and Lane 1995; Takenaka et al., 1995; Wang and Prives, 1995), and most recently, acetylation by CREB-binding protein CBP/p300 (Gu and Roeder, 1997) were shown to enhance sequence-specific DNA-binding of wild-type p53, possibly by inhibiting p53’s C-terminal non-sequence-specific DNA-binding activity (Anderson et al., 1997). CBP/p300 along with p300/CBP-associated factor PCAF (Yang et al., 1996) interacts with p53 and enhances its transcriptional activity (Avantaggiati et al., 1997; Lill et al., 1997) through acetylation at multiple lysine residues at C-terminal site. However, on the other hand deacetylation of p53 by HDAC1-containing complex (Luo et al., 2000) led to transcriptional deactivation. Moreover, its major negative regulator, Mdm2 has also been shown to recruit HDAC1 activity towards p53, which subsequently increases its ubiquitination and degradation (Ito et al., 2002). Interestingly, EBNA3C has also been shown to interact with both p300 (Subramanian et al., 2002a) and HDAC1 (Cotter and Robertson, 2000). The interaction of EBNA3C with p300 and HDAC1 resulted in modulation of acetylation of cellular factors (Cotter and Robertson, 2000). These findings lead us to speculate that EBNA3C may regulate p53 acetylation for modulating its transcriptional activity and DNA binding ability via interaction with p300 and HDAC1. The precise mechanisms of how EBNA3C acts on specific lysine residues involved in acetylation or/and ubiquitination in EBV scenario is currently under investigation in our laboratory. It would also be interesting to investigate whether EBNA3C forms a ternary complex with p53 and Mdm2 or p300 or HDAC1 for critically modulating p53’s activity.
Abundant evidence indicates that p53’s ability to bind to DNA is tightly linked to its physiological functions in tumor suppression (Levine 1997). EMSAs using a DNA probe from the p21 promoter containing the p53 binding site in the presence of nuclear extracts containing p53 showed binding to the probe, which was super-shifted by the addition of an anti-p53 antibody. The addition of EBNA3C to the reaction largely reduced the p53-specific complex. Studies have shown that the C-terminal regulatory domain negatively regulates p53’s DNA-binding ability; perhaps by maintaining p53 in the low affinity DNA-binding state (Hupp et al., 1992; Waterman et al., 1995), raises the possibility that the inactivation of p53 activity by EBNA3C, is likely due to the formation of a stable complex between the p53 C-terminal negative regulatory domain and EBNA3C.
Our data also indicated that expression of EBNA3C was able to abrogate p53-mediated apoptosis in the p53 null cell-line, SAOS-2. Since EBNA3C interacts with complexes that include co-repressors and silencers of transcription, it is tempting to speculate that EBNA3C forms part of a multi-protein repression complex that inhibits transcription mediated by p53. Based on our findings in reporter assays, it is likely that EBNA3C interferes with p53-mediated apoptosis by potentially influencing its transcriptional activation. A recent study demonstrated that EBNA3C functionally cooperate to repress the expression of the proapoptotic Bcl-2-family member Bcl-2-interacting mediator of cell death (Bim) (Anderton et al., 2008). Alternatively EBNA3C may inhibit p53 dependent apoptosis by disrupting protein-protein interactions between p53 and other cellular factors involved in the apoptotic pathway or directly interacting with proteins involved. We are working on deciphering the mechanism(s) that leads to this inhibition.
This study is an initial report showing that EBNA3C directly interacts with p53 in vitro and associates in a complex in vivo. This interaction of EBNA3C with p53 warrants further investigation as p53 controls the progression of the cell-cycle by regulating the genetic integrity. The repression of p53 function by EBNA3C could result in increased genetic instability, which may directly contribute to EBV-mediated cellular transformation.
Materials and Methods
Plasmids, cell lines and antibodies
pA3F–EBNA3C constructs encoding either full-length or EBNA3C truncations residues 1–365, 366–620, and 621–992 with C-terminal flag tag have been described previously (Bajaj et al., 2008). The p53 reporter plasmid pGL-3 contains 13 x p53-binding sites upstream of the luciferase gene and was constructed by EcoRV insertion of p53-binding sequences into the pGL-3 luciferase reporter plasmid (Wang and El-Deiry, 2003). The p53 expression vector pC53-C1N3 carries a wild-type human p53 gene with a proline polymorphism at residue 73 and is controlled by the cytomegalovirus promoter (gift from Gary J. Nabel, National Institutes of Health, Bethesda, MD). pGEX-p53 expresses an N-terminal glutathione S-transferase (GST)-p53 fusion protein and was derived from pGEX-2T (Amersham Pharmacia, Inc., Piscataway, NJ) by insertion of human p53 cDNA (gift from Gary J. Nabel, National Institutes of Health, Bethesda, MD) at the BamHI and EcoRI sites. pA3M-p53 was generated by cloning PCR-amplified p53 cDNA using pGEX-p53 as a template into the previously described vector pA3M (Knight et al., 2003; Bajaj et al., 2008) at EcoRI and NotI sites. GST-p53 deletion constructs were constructed by insertion of PCR fragments into the pGEX-5x-1 backbone (gift from Shelley L. Berger, The Wistar Institute, Philadelphia, PA). Constructs expressing GFP-tagged either EBNA3C residues 1–365 or 366–620 mutants were prepared by cloning PCR-amplified fragments into pEGFP-C1 vector (BD Biosciences Clontech) at EcoRI and SalI restriction sites. All constructs and mutations were verified by DNA sequencing (University of Pennsylvania DNA sequencing facility).
Human embryonic kidney cell line, HEK 293 was obtained from Jon Aster (Brigham and Woman’s Hospital, Boston, MA) (Aiello et al., 1979). p53-null cell line SAOS-2 is derived from human osteosarcoma and was obtained from Jon Aster (Brigham and Women’s Hospital, Boston, MA, USA). EBV negative Burkitt’s lymphoma cell-lines BJAB and DG75 were provided by Elliot Kieff (Brigham and Woman's Hospital, Boston, MA). LCL1 and LCL2 are in vitro-transformed EBV-positive cell-lines. BJAB cells expressing EBNA3C and BJAB neo control were generated previously by transfecting with either pZipneo eukaryotic expression vector with EBNA3C cDNA or vector alone (Robertson et al., 1995). The adherent cell lines (HEK 293, U2OS and SAOS-2) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% bovine growth serum, 25 U/ml penicillin, 50 µg/ml streptomycin, and 2 mM L-glutamine. All B-cell lines were grown in RPMI 1640 medium (HyClone) supplemented as Dulbecco’s modified Eagle’s medium.
The monoclonal antibodies mouse anti-myc (9E10) and A10 reactive to EBNA3C were prepared from their respective hybridoma culture (Knight et al., 2003; Bajaj et al., 2008). Mouse monoclonal anti-p53 antibody DO-1 and mouse monoclonal anti-flag antibody were purchased from Santa Cruz Biotechnology, Inc and Sigma respectively.
Transfection
HEK 293, BJAB and DG75 cells were transfected by electroporation with a Bio-Rad Gene Pulser II electroporator. 12–15 × 106 cells were harvested at exponential growth phase, collected, washed in phosphate-buffered saline (PBS; Hyclone, Inc., UT) and resuspended in 400 µl of the appropriate medium containing DNA for transfection (Knight and Robertson, 2004). Resuspended cells were transferred to a 0.4-cm gap cuvette and electroporated at 975 µF and 210 V for HEK 293 cells or 975 µF and 220 V for B-cells. Transfected cells were transferred to a 100 mm Petri dish containing 10 ml of complete medium and incubated at 37°C in a humidified environment supplemented with 5% CO2. Unless otherwise indicated, transfected cells were harvested after 36h and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with 5% of the total normalized protein lysate.
Purification of GST fusion proteins
Purification of GST fusion proteins were performed essentially as described previously (Bajaj et al., 2008) with some modification. Briefly, Escherichia coli BL21 (DE3) cells were transformed with the plasmid constructs for each glutathione S-transferase (GST) fusion protein as described in the text. Single colonies were picked and grown overnight in 3 ml of Luria broth supplemented with 100 µg/ml ampicillin. 1 ml of the overnight culture was used to inoculate a 500-ml culture. The larger culture was incubated approximately 3 h until the OD600 reached approximately 0.6, at which point it was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 12 h at 30°C. The bacteria were pelleted, washed once with STE buffer (100 mM NaCl, 10 mM Tris, and 1 mM EDTA, pH 7.5), resuspended in 3 ml NETN buffer (0.5% NP-40, 100 mM NaCl, 20 mM Tris, 1 mM EDTA, pH 8.0) supplemented with protease inhibitors, and incubated on ice for 15 min. 150 µl of 1 M dithiothreitol (DTT) and 1.8 ml of a 10% solution of Sarkosyl in STE buffer was added, and the suspension was sonicated (for 3 min on ice) to solubilize the proteins. The lysate was centrifuged (12,000 × g, 10 min, 4°C) to separate the unsolubilized fraction. The clear supernatant was transferred to a fresh tube, to which 3 ml of 10% Triton X-100 in STE buffer and 200 µl of glutathione Sepharose beads were added. The tube was rotated overnight at 4°C, after which the purified protein bound to glutathione was collected by centrifugation (2 min, 600 × g, 4°C) and washed five times with NETN buffer supplemented with protease inhibitors. Level of purification was judged by SDS-PAGE and purified proteins were stored at 4°C.
Immunoprecipitation and western blotting
Transfected cells were harvested, washed in ice-cold PBS, and lysed in 0.5 ml ice-cold radioimmunoprecipitation assay (RIPA) buffer [0.5% NP-40, 10 mM Tris pH 7.5, 2 mM EDTA, 150 mM NaCl supplemented with protease inhibitors - 1 mM phenylmethylsulphonyl fluoride (PMSF), 1 µg/ml aprotinin, 1 µg/ml pepstatin and 1 µg/ml leupeptin]. Cellular debris was removed by centrifugation (21,000 × g, 10 min, 4°C), and the supernatant was transferred to a fresh tube. Approximately 5% of the lysate was saved as an input control. Lysates were then precleared with normal mouse serum and then rotated with 30 µl of a 1:1 mixture of protein A-and protein G- conjugated Sepharose beads for 1 h, at 4°C. Beads were spun out, and the supernatant was transferred to a fresh microcentrifuge tube and approximately 5% of the lysate was saved for input control. The protein of interest was captured by rotating overnight the remaining lysate with 1 µg of specific antibody at 4°C. Complexes were precipitated with 30 µl of 1:1 mixture of protein A- and protein G-Sepharose beads. The samples were washed three times with ice-cold RIPA buffer, fractionated by SDS-PAGE, and transferred to a 0.45 µm nitrocellulose membrane for Western blotting (WB). The membranes were probed with the appropriate antibodies followed by incubation with appropriate infrared-tagged secondary antibodies for 1 h at room temperature (RT) and viewed on an Odyssey imager (LiCor Inc., Lincoln, NE).
Immunofluorescence
BJAB, LCL1 and LCL2 cells were dried onto slides and fixed using a 1:1 mixture of acetone and methanol. After fixation cells were extensively washed in PBS and incubated in blocking buffer [PBS supplemented with 0.1% Triton-X 100, 0.2% fish skin gelatin (Sigma)] at room temperature for 30 min. Endogenous expression of p53 was detected using mouse monoclonal antibody DO-1 (1:200 dilution), and EBNA3C was detected using EBNA3C-reactive human serum (1:150 dilution). Primary antibodies were diluted in blocking buffer and incubated with fixed cells for 1 h at RT. Cover slips were washed three times (5 min each) with PBS and incubated with appropriate secondary antibody (1:2000) for 1 h at RT followed by three times washes (5 min each) with PBS. The last wash contained 4’, 6’-diamidino-2-phenylindole (DAPI; Promega Inc., Madison, WI) for nuclear staining. Goat anti-human antibody Alexa Fluor 488 and goat anti-mouse antibody Alexa Fluor 594 were purchased from Molecular Probes Inc. (Carlsbad, CA). Cover slips were then washed in PBS and mounted using Prolong anti-fade (Molecular Probes Inc, Carlsbad, CA). Fluorescence was viewed by confocal microscopy and analyzed with Fluoview 300 software from Olympus Inc. (Melville, NY).
Reporter assays
Reporter assay was essentially performed as described previously (Bajaj et al., 2008) with few modifications. Briefly, either 0.4 × 106 SAOS-2 or 1.0 × 106 BJAB cells were cotransfected with 0.25 µg of the promoter construct containing p53 responsive element and 0.5 µg pA3M-p53 and increasing amount of pA3F-EBNA3C constructs (0, 0.25, 0.5, 1.0 µg) expressing wild-type p53 and EBNA3C respectively using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The differences in the amounts of total DNA were equalized with vector control to keep the total amount of transfected DNA constant. At 24 h post-transfection, cells were collected and washed with ice-cold PBS, and lysed in 100 µl of reporter lysis buffer (Promega Inc., Madison, WI). A 40 µl aliquot of the lysate was transferred to a 96-well plate. Luciferase activity was measured using an LMaxII384 luminometer (Molecular Devices, Sunnyvale, CA) by automatically injecting 25 µl of luciferase substrate into each well and integrating the luminescence for 20 seconds postinjection. The results represent experiments performed in duplicate.
Electrophoretic mobility shift assay (EMSA)
EMSA was essentially performed as described previously (Lan et al., 2006). Equivalent amount of nuclear extract was used in each EMSA reaction. The probe containing the p53 responsive element (5’-GTCAGGAACATGTCCCAACATGTTGAGCTC-3’) of the p21 promoter (Fujioka et al., 2004) was labeled by Klenow fill-in reaction with α-32P dCTP and purified with NucTrap probe purification columns (Stratagene, Inc., La Jolla, CA). Reaction mixture consisted of 0.02 picomole of labeled probe and 5 µg of nuclear extract protein in a buffer consisting of 20 mM HEPES (pH 7.5), 0.01% NP-40, 40 mM KCl, 100 µg BSA, 2.5 mM MgCl2, 20 mM DTT, 5% glycerol. Poly(dI/dC) (Sigma-Aldrich, Inc., St. Louis, MO) was added as nonspecific competitor DNA, and the final volume (50 µl) of the reaction mixture was adjusted with appropriate amount of double-distilled H2O. Following incubation at 25°C for 25 min, the DNA-protein complexes were resolved by nondenaturing 4% polyacrylamide gel electrophoresis. The gel was run in 0.5 x TBE buffer (45 mM Tris-borate, 1 mM EDTA, pH 8.0) at a constant voltage of 120 V for 4 h at 4°C. Following electrophoresis, the gel was transferred to Whatman paper and dried for 1 h at 80°C. Dried gels were exposed to a phosphorimager screen for 12 h (Amersham Biosciences, Inc., Piscataway, NJ) and scanned by a PhosphorImager (Molecular Dynamics, Piscataway, NJ). For supershift experiment, monoclonal antibody against p53 was added (DO-1) after an initial 30-min incubation of probe with nuclear extract and incubated for 1 h at 4°C.
Apoptosis Assay
SAOS-2 cells were co-transfected with constructs expressing myc tagged p53 and flag tagged EBNA3C (as indicated in the text and figure) plus GFP expressing plasmid (pEGFP-C1; BD Biosciences Clontech) for checking transfection efficiency. At 48 h post-transfection, floating and attached cells were collected, washed in ice-cold PBS. Cells were then resuspended in PBS and fixed in 90% cold ethanol. Following incubation cells were stained with PBS containing 40 µg/ml of propidium iodide (PI), 200 µg/ml of RNase A (Sigma) and 0.05% Triton X-100 for 1 h at RT in dark. Stained cells were analysed on FACScan (Becton-Dickinson). Total 20,000 events were analyzed for each sample. Data was analyzed using the ModFIT model program (Verity software House).
ACKNOWLEDGMENTS
We thank Gary J. Nabel, Shelley L. Berger and Wafik S. El-Deiry for generously providing reagents. This work was supported by grants from the National Institutes of Health: NCI CA137894-01 to E.S.R. E.S.R. is a scholar of the Leukemia and Lymphoma Society of America. We also thank Richard Dzeng for helping in preparation of manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adimoolam S, Ford JM. p53 and regulation of DNA damage recognition during nucleotide excision repair. DNA Repair. 2003;2(9):947–954. doi: 10.1016/s1568-7864(03)00087-9. [DOI] [PubMed] [Google Scholar]
- Adler V, Pincus MR, Minamoto T, Fuchs SY, Bluth MJ, Brandt-Rauf PW, Friedman FK, Robinson RC, Chen JM, Wang XW, Harris CC, Ronai Z. Conformation-dependent phosphorylation of p53. Proc Natl Acad Sci U S A. 1997;94(5):1686–1691. doi: 10.1073/pnas.94.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello L, Guilfoyle R, Huebner K, Weinmann R. Adenovirus 5 DNA sequences present and RNA sequences transcribed in transformed human embryo kidney cells (HEK-Ad-5 or 293) Virology. 1979;94(2):460–469. doi: 10.1016/0042-6822(79)90476-8. [DOI] [PubMed] [Google Scholar]
- Ambinder RF. Human lymphotropic viruses associated with lymphoid malignancy: Epstein-Barr and HTLV-1. Hematol Oncol Clin North Am. 1990;4(4):821–833. [PubMed] [Google Scholar]
- Anderson ME, Woelker B, Reed M, Wang P, Tegtmeyer P. Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: implications for regulation. Mol Cell Biol. 1997;17(11):6255–6264. doi: 10.1128/mcb.17.11.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderton E, Yee J, Smith P, Crook T, White RE, Allday MJ. Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt’s lymphoma. Oncogene. 2008;27(4):421–433. doi: 10.1038/sj.onc.1210668. [DOI] [PubMed] [Google Scholar]
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89(7):1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- Bajaj BG, Murakami M, Cai Q, Verma SC, Lan K, Robertson ES. Epstein-Barr virus nuclear antigen 3C interacts with and enhances the stability of the c-Myc oncoprotein. J Virol. 2008;82(8):4082–4090. doi: 10.1128/JVI.02500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargonetti J, Manfredi JJ, Chen X, Marshak DR, Prives C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev. 1993;7(12B):2565–2574. doi: 10.1101/gad.7.12b.2565. [DOI] [PubMed] [Google Scholar]
- Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger BR, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377(6550):646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- Candau R, Scolnick DM, Darpino P, Ying CY, Halazonetis T, Berger SL. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene. 1997;15(7):807–816. doi: 10.1038/sj.onc.1201244. [DOI] [PubMed] [Google Scholar]
- Castillo JP, Yurochko AD, Kowalik TF. Role of human cytomegalovirus immediate-early proteins in cell growth control. J Virol. 2000;74(17):8028–8037. doi: 10.1128/jvi.74.17.8028-8037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri T, Verma SC, Lan K, Murakami M, Robertson ES. The ATM/ATR signaling effector Chk2 is targeted by Epstein-Barr virus nuclear antigen 3C to release the G2/M cell cycle block. J Virol. 2007;81(12):6718–6730. doi: 10.1128/JVI.00053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Omichinski JG, Sakaguchi K, Zambrano N, Sakamoto H, Appella E, Gronenborn AM. High-resolution structure of the oligomerization domain of p53 by multidimensional NMR. Science. 1994;265(5170):386–391. doi: 10.1126/science.8023159. [DOI] [PubMed] [Google Scholar]
- Cohen JI, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci U S A. 1989;86(23):9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter MA, 2nd, Robertson ES. Modulation of histone acetyltransferase activity through interaction of epstein-barr nuclear antigen 3C with prothymosin alpha. Mol Cell Biol. 2000;20(15):5722–5735. doi: 10.1128/mcb.20.15.5722-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger J, Muralidhar S, Rosenthal LJ. Human cytomegalovirus and human herpesvirus 6 genes that transform and transactivate. Clin Microbiol Rev. 1999;12(3):367–382. doi: 10.1128/cmr.12.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Fields S, Jang SK. Presence of a potent transcription activating sequence in the p53 protein. Science. 1990;249(4972):1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402(6764):889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- Fries KL, Miller WE, Raab-Traub N. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J Virol. 1996;70(12):8653–8659. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Schmidt C, Sclabas GM, Li Z, Pelicano H, Peng B, Yao A, Niu J, Zhang W, Evans DB, Abbruzzese JL, Huang P, Chiao PJ. Stabilization of p53 is a novel mechanism for proapoptotic function of NF-kappaB. J Biol Chem. 2004;279(26):27549–27559. doi: 10.1074/jbc.M313435200. [DOI] [PubMed] [Google Scholar]
- Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol. Cell. Biol. 1992;12(6):2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb T, Oren M. p53 in growth control and neoplasia. Biochim. Biophys. Acta. 1996;1287(2–3):77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90(4):595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Davis LJ, Kandil AN. Wild-type p53 adopts a 'mutant'-like conformation when bound to DNA. EMBO J. 1993;12(3):1021–1028. doi: 10.1002/j.1460-2075.1993.tb05743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340(6232):393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- Holowaty MN, Frappier L. HAUSP/USP7 as an Epstein-Barr virus target. Biochem Soc Trans. 2004;32(5):731–732. doi: 10.1042/BST0320731. [DOI] [PubMed] [Google Scholar]
- Holowaty MN, Zeghouf M, Wu H, Tellam J, Athanasopoulos V, Greenblatt J, Frappier L. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J Biol Chem. 2003;278(32):29987–29994. doi: 10.1074/jbc.M303977200. [DOI] [PubMed] [Google Scholar]
- Hupp TR, Lane DP. Two distinct signaling pathways activate the latent DNA binding function of p53 in a casein kinase II-independent manner. J Biol Chem. 1995;270(30):18165–18174. doi: 10.1074/jbc.270.30.18165. [DOI] [PubMed] [Google Scholar]
- Hupp TR, Meek DW, Midgley CA, Lane DP. Activation of the cryptic DNA binding function of mutant forms of p53. Nucleic Acids Res. 1993;21(14):3167–3174. doi: 10.1093/nar/21.14.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupp T, Meek D, Midgley C, Lane D. Regulation of the specific DNA binding function of p53. Cell. 1992;71(5):875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21(22):6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey PD, Gorina S, Pavletich NP. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science. 1995;267(5203):1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- Johannsen E, Miller CL, Grossman SR, Kieff E. EBNA-2 and EBNA-3C extensively and mutually exclusively associate with RBPJkappa in Epstein-Barr virus-transformed B lymphocytes. J Virol. 1996;70(6):4179–4183. doi: 10.1128/jvi.70.6.4179-4183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71(4):587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Kaul R, Murakami M, Choudhuri T, Robertson ES. Epstein-Barr virus latent nuclear antigens can induce metastasis in a nude mouse model. J Virol. 2007;81(19):10352–10361. doi: 10.1128/JVI.00886-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye KM, Izumi KM, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A. 1993;90(19):9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Robertson ES. Epstein-Barr virus nuclear antigen 3C regulates cyclin A/p27 complexes and enhances cyclin A-dependent kinase activity. J Virol. 2004;78(4):1981–1991. doi: 10.1128/JVI.78.4.1981-1991.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Lan K, Subramanian C, Robertson ES. Epstein-Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines. J Virol. 2003;77(7):4261–4272. doi: 10.1128/JVI.77.7.4261-4272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Sharma N, Robertson ES. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc Natl Acad Sci U S A. 2005a;102(51):18562–18566. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Sharma N, Robertson ES. SCFSkp2 complex targeted by Epstein-Barr virus essential nuclear antigen. Mol Cell Biol. 2005b;25(5):1749–1763. doi: 10.1128/MCB.25.5.1749-1763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Sharma N, Kalman DE, Robertson ES. A cyclin-binding motif within the amino-terminal homology domain of EBNA3C binds cyclin A and modulates cyclin A-dependent kinase activity in Epstein-Barr virus-infected cells. J Virol. 2004;78(23):12857–12867. doi: 10.1128/JVI.78.23.12857-12867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10(9):1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Lan K, Murakami M, Choudhuri T, Kuppers DA, Robertson ES. Intracellular-activated Notch1 can reactivate Kaposi's sarcoma-associated herpesvirus from latency. Virology. 2006;351(2):393–403. doi: 10.1016/j.virol.2006.03.047. [DOI] [PubMed] [Google Scholar]
- Lechner MS, Mack DH, Finicle AB, Crook T, Vousden KH, Laimins LA. Human papillomavirus E6 proteins bind p53 in vivo and abrogate p53-mediated repression of transcription. EMBO J. 1992;11(8):3045–3052. doi: 10.1002/j.1460-2075.1992.tb05375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Harvey TS, Yin Y, Yau P, Litchfield D, Arrowsmith CH. Solution structure of the tetrameric minimum transforming domain of p53. Nat. Struct. Biol. 1994;1(12):877–890. doi: 10.1038/nsb1294-877. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88(3):323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387(6635):823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19(2):1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408(6810):377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- Martin ME, Berk AJ. Adenovirus E1B 55K represses p53 activation in vitro. J Virol. 1998;72(4):3146–3154. doi: 10.1128/jvi.72.4.3146-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massimi P, Banks L. Repression of p53 transcriptional activity by the HPV E7 proteins. Virology. 1997;227(1):255–259. doi: 10.1006/viro.1996.8315. [DOI] [PubMed] [Google Scholar]
- Mauser A, Saito S, Appella E, Anderson CW, Seaman WT, Kenney S. The Epstein-Barr virus immediate-early protein BZLF1 regulates p53 function through multiple mechanisms. J Virol. 2002;76(24):12503–12512. doi: 10.1128/JVI.76.24.12503-12512.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietz JA, Unger T, Huibregtse JM, Howley PM. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. Embo J. 1992;11(13):5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80(2):293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- Muller-Tiemann B, Halazonetis T, Elting J. Identification of an additional negative regulatory region for p53 sequence-specific DNA binding. Proc Natl Acad Sci USA. 1998;95(11):6079–6084. doi: 10.1073/pnas.95.11.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Beach D. Cyclin G is a transcriptional target of the p53 tumor suppressor protein. Embo J. 1994;13(20):4816–4822. doi: 10.1002/j.1460-2075.1994.tb06807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Nions J, Allday MJ. Epstein-Barr virus can inhibit genotoxin-induced G1 arrest downstream of p53 by preventing the inactivation of CDK2. Oncogene. 2003;22(46):7181–7191. doi: 10.1038/sj.onc.1206838. [DOI] [PubMed] [Google Scholar]
- O'Nions J, Allday MJ. Deregulation of the cell cycle by the Epstein-Barr virus. Adv Cancer Res. 2004;92:119–186. doi: 10.1016/S0065-230X(04)92006-4. [DOI] [PubMed] [Google Scholar]
- Park J, Seo T, Hwang S, Lee D, Gwack Y, Choe J. The K-bZIP protein from Kaposi’s sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J Virol. 2000;74(24):11977–11982. doi: 10.1128/jvi.74.24.11977-11982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prives C. Signaling to p53: breaking the MDM2-p53 circuit. Cell. 1998;95(1):5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- Radkov SA, Touitou R, Brehm A, Rowe M, West M, Kouzarides T, Allday MJ. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol. 1999;73(7):5688–5697. doi: 10.1128/jvi.73.7.5688-5697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raycroft L, Wu HY, Lozano G. Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science. 1990;249(4972):1049–1051. doi: 10.1126/science.2144364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickinson A, Kieff E. Epstein-Barr virus. In: Knipe DM, Howley P, editors. "Fields virology 4th Ed". Vol. 2. Philadelphia, PA: Lippincott Williams & Wilkins; 2002. pp. 2575–2627. 2 vols. [Google Scholar]
- Robertson ES, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein J kappa. J Virol. 1995;69(5):3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson ES, Lin J, Kieff E. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJ(kappa) J Virol. 1996;70(5):3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H, Lewis MS, Kodama H, Apella E, Sakaguchi K. Specific sequences from the carboxyl terminus of human p53 gene product form anti-parallel tetramers in solution. Proc. Natl. Acad. Sci. USA. 1994;91(19):8974–8978. doi: 10.1073/pnas.91.19.8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, Nguyen T, Zhang RG, Liao J, Lee W, Edwards AM, Arrowsmith CH, Frappier L. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol Cell. 2005;18(1):25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63(6):1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Schuler M, Bossy-Wetzel E, Goldstein JC, Fitzgerald P, Green DR. p53 Induces Apoptosis by Caspase Activation through Mitochondrial Cytochrome c Release. J. Biol. Chem. 2000;275(10):7337–7342. doi: 10.1074/jbc.275.10.7337. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Weber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10(1):94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- Steegenga WT, van Laar T, Riteco N, Mandarino A, Shvarts A, van der Eb AJ, Jochemsen AG. Adenovirus E1A proteins inhibit activation of transcription by p53. Mol Cell Biol. 1996;16(5):2101–2109. doi: 10.1128/mcb.16.5.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. Embo J. 2004;23(7):1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzbecher HW, Brain R, Addison C, Rudge K, Remm M, Grimaldi M, Keenan E, Jenkins JR. A C-terminal α-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene. 1992;7(8):1513–1523. [PubMed] [Google Scholar]
- Subramanian C, Cotter MA, 2nd, Robertson ES. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat Med. 2001;7(3):350–355. doi: 10.1038/85499. [DOI] [PubMed] [Google Scholar]
- Subramanian C, Hasan S, Rowe M, Hottiger M, Orre R, Robertson Es. Epstein-Barr virus nuclear antigen 3C and prothymosin alpha interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J Virol. 2002a;76(10):4699–4708. doi: 10.1128/JVI.76.10.4699-4708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian C, Knight JS, Robertson ES. The Epstein Barr nuclear antigen EBNA3C regulates transcription, cell transformation and cell migration. Front Biosci. 2002;7:d704–d716. doi: 10.2741/subraman. [DOI] [PubMed] [Google Scholar]
- Takenaka I, Morin F, Seizinger BR, Kley N. Regulation of the sequence-specific DNA binding function of p53 by protein kinase C and protein phosphatases. J Biol Chem. 1995;270(10):5405–5411. doi: 10.1074/jbc.270.10.5405. [DOI] [PubMed] [Google Scholar]
- Tang J, Qu LK, Zhang J, Wang W, Michaelson JS, Degenhardt YY, El-Deiry WS, Yang X. Critical role for Daxx in regulating Mdm2. Nat Cell Biol. 2006;8(8):855–862. doi: 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67(4):2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat. Rev. Cancer. 2002;2(8):594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55(22):5187–5190. [PubMed] [Google Scholar]
- Wang W, El-Deiry WS. Bioluminescent molecular imaging of endogenous and exogenous p53-mediated transcription in vitro and in vivo using an HCT116 human colon carcinoma xenograft model. Cancer Biol Ther. 2003;2(2):196–202. doi: 10.4161/cbt.2.2.347. [DOI] [PubMed] [Google Scholar]
- Wang XW, Forrester K, Yeh H, Feitelson MA, Gu JR, Harris CC. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci U S A. 1994;91(6):2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376(6535):88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Reed M, Wang PJ, Stenger E, Mayr G, Anderson ME, Schwedes JF, Tegtmeyer P. p53 domains: identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993;7(12B):2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- Waterman J, Shenk J, Halazonetis T. The dihedral symmetry of the p53 tetramerization domain mandates a conformational switch upon DNA binding. EMBO J. 1995;14(3):512–519. doi: 10.1002/j.1460-2075.1995.tb07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Levine AJ. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci U S A. 1994;91(9):3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382(6589):319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- Yin C, Knudson CM, Korsmeyer SJ, Van Dyke T. Bax suppresses tumorigenesis and stimulates apoptosis in vivo. Nature. 1997;385(6617):637–640. doi: 10.1038/385637a0. [DOI] [PubMed] [Google Scholar]
- Zhao B, Sample CE. Epstein-barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an spi-1/Spi-B binding site. J Virol. 2000;74(11):5151–5160. doi: 10.1128/jvi.74.11.5151-5160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]