Abstract
Umbilical cord blood transplant continues to increase in prevalence as a treatment option for various hematopoietic and immune disorders. Because of the limited number of cells available in a single cord blood unit, investigators have explored methods of increasing cell dose prior to transplant, including overexpression of the HOXB4 transcription factor. We have previously reported the development of leukemia in several nonhuman primate subjects transplanted with HOXB4-expanded bone marrow cells at approximately 2 years post-transplant. Here, we provide long-term data for a nonhuman primate receiving a HOXB4-expanded cord blood graft. Longitudinal follow-up included gene marking analysis, complete blood counts, morphological/pathological assessment, phenotypic analysis of subsets, and retroviral integration site analysis. In each of these independent assays, we saw no indication of clonal dominance, and all signs pointed toward normal, healthy hematopoiesis. Furthermore, in-depth clonal analysis of an animal that developed leukemia after transplantation of HOXB4-modiifed bone marrow cells showed that dominant clones could be detected as early as 6 months post-transplant using the genomic analysis technique detailed here. Parallel analysis of the cord blood transplant macaque showed no such sites. These findings demonstrate the ability to study the use of gene-modified and expanded cord blood cells in a NHP model.
Keywords: Cord Blood, Gene Therapy, Non-Human Primate, HOXB4, Leukemia
INTRODUCTION
Umbilical cord blood (CB) transplant is an increasingly common technique used in the treatment of malignant and non-malignant disease. Since 2009, the number of CB transplants performed worldwide has surpassed the number of bone marrow transplants performed, and there are over 20,000 patients who have received a CB graft. The rising number of CB transplants is likely due to the numerous advantages of using CB as a source of stem cells compared to the more traditional sources (bone marrow or mobilized peripheral blood obtained by leukapheresis). These advantages include ready availability, a less-stringent donor/host matching requirement, and reduced risk of transmitting infections.
However, the field continues to be limited by the low number of cells available in a single cord blood unit. This subsequently leads to a sub-optimal dose of stem cells per kilogram of body weight, which is directly correlated with transplantation success [1]. Consequently, double CB transplant (dCBT) is being explored as a means of overcoming the cell dose obstacle. However, in this setting it is typical to see one unit emerge as the source of long-term hematopoiesis [2]. In addition, patients continue to suffer from delayed engraftment and its repercussions, including increased infections. An ideal therapeutic approach involves identifying a safe, efficient way to expand repopulating cells within a single unit prior to infusion.
To this end, clinical trials involving transplant of cells expanded with cytokines have not yielded improved engraftment [3–5]. One method that has shown promise is overexpression of the transcription factor HOXB4. Our group has recently published the results of our studies establishing a nonhuman primate cord blood transplant model [6]. In this report, we used our model to compare the engraftment kinetics of HOXB4 green fluorescent protein (GFP)-transduced, expanded cells and control yellow fluorescent protein (YFP)-transduced, non-expanded cells in a competitive repopulation setting (Figure 1). We found that cells transduced with a gammaretroviral vector expressing HOXB4 and expanded ex vivo yielded superior engraftment of all hematopoietic lineages compared to untransduced controls that were not expanded.
Figure 1. Experimental designs for HOXB4-mediated stem cell transplant.
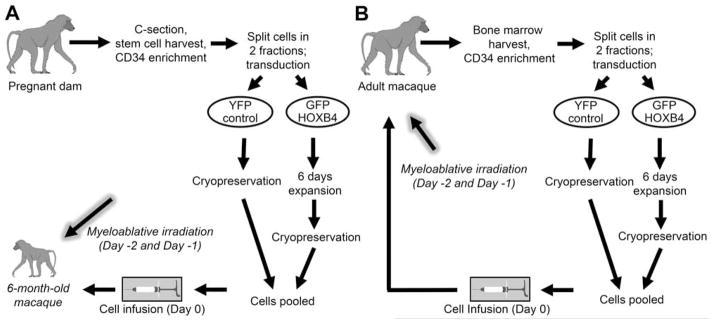
In the cord blood transplant setting (A), umbilical cord blood cells are harvested at time of birth and later infused into the infant macaque. In the bone marrow transplant setting (B), cells are harvested from the bone marrow of the adult macaque and later infused into the same animal. In both settings, the macaque receives autologous cells.
Though gammaretroviral gene therapy offers promise for repairing or regenerating the hematopoietic and immune systems, it does present risks. Several SCID-X1 patients developed T cell leukemia approximately 2 years after transplant with IL2Rγ-modified cells [7,8]. This discovery sparked concern and increased interest in understanding the role of integration site profile and the transgene itself in the development of leukemia.
Similarly, our laboratory has reported a high incidence of leukemia in large animals receiving grafts containing cells transduced with a HOXB4 expressing gammaretroviral vector and expanded ex vivo [9]. A total of six pig-tailed macaques were transplanted; four had successful hematopoietic reconstitution. Of these, two were followed-up for at least 2 years, and both developed myelodysplasia [10]. In a similar, related study, two of two dogs transplanted with HOXB4-expanded bone marrow cells developed leukemia [11]. In these cases, the leukemic cells exhibited dysregulated expression of oncogenes, overexpression of HOXB4, and a block in differentiation toward the myeloid lineage. By performing HOXB4 knockdown studies, we confirmed that normal differentiation was restored, which indicates a role for the HOXB4 transgene in the development of leukemia. In animals receiving gene-modified grafts that did not contain HOXB4, no leukemia was observed; in addition, leukemic symptoms were not observed in the control (non-HOXB4) portion of the graft [11]. Taken together, these findings provide more evidence that the transgene itself is partly responsible for leukemic outgrowth. Hence, one can conclude that the combination of genetic manipulation, along with a transgene with an intrinsic growth advantage, likely led to leukemia in these animals.
In the human and animal studies described above, leukemia was observed approximately 2 years post-transplant. This condition developed in two of two HOXB4 bone marrow transplant monkeys who lived at least 2 years following transplant. Interestingly, of three monkeys who received HOXB4-expanded cord blood cells, one is now more than 3 years post-transplant, and still remains healthy with no signs of leukemia. Engraftment of gene-modified cells remains stable, hematopoietic differentiation is similar to that of a non-transplanted control macaque, bone marrow analysis shows no signs of pathology, and integration site analysis confirms the absence of dominant clones. Here, we explore these findings and consider the possibility that the stem cell source (cord blood compared to bone marrow) may have contributed to improved outcome.
MATERIALS AND METHODS
Experimental design
We implemented our previously developed nonhuman primate competitive repopulation model [6] (Figure 1A) to study HOXB4-mediated expansion of CB cells in a clinically relevant setting. In accordance with this model, we perform a cesarean section approximately 1 week before the due date and collect umbilical CB cells. The resulting CD34-enriched cells are split into two fractions. Half are transduced with a YFP control gammaretroviral vector and immediately cryopreserved; the other half are transduced with a HOXB4GFP vector, expanded for 6 days and then cryopreserved. By differentially marking the two populations with independent reporter genes (YFP or GFP), we can easily detect the presence of transplanted cells as part of our routine follow-up. The two fractions of cells are cryopreserved for at least 6 months until the infant reaches an appropriate weight. On the day of transplantation, cells are thawed, pooled, and transplanted into the myeloablated recipient by intravenous infusion. This protocol is an adaptation of the protocol used for macaques receiving HOXB4-expanded bone marrow cells. The experimental design for macaque bone marrow transplant recipients is illustrated in Figure 1B and has been described previously [9]. In the current study, a total of three macaques were transplanted with HOXB4-expanded cord blood cells. Though all three achieved hematopoietic recovery, two were euthanized within the first 3 months due to viral pneumonia and TTP-like syndrome [6]. Thus, the study described here is an account of the third macaque, who continues to survive at greater than 3 years post-transplant.
Stem cell processing
The protocols for enrichment of CD34+ cells from umbilical cord blood and bone marrow are identical and have been described previously [6]. Briefly, red cells were lysed in ammonium chloride red cell lysis buffer. Nucleated cells were incubated for 20 minutes with the 12.8 IgM anti-CD34 antibody, washed, and incubated for another 20 minutes with MACS IgM microbeads (Miltenyi Biotec, Auburn, CA, USA). The cell solution was run through a magnetic column, allowing for enrichment of the CD34+ fraction to a purity of 80% to 99% CD34+ by flow cytometry.
HOXB4 transduction and expansion
CD34-enriched cells were prestimulated for 48 hours prior to being plated on fibronectin-coated, non-tissue culture-treated plates for transduction. Transduction consisted of two, 4-hour exposures to virus-containing media (one exposure per day for 2 consecutive days). Cells were transduced at a multiplicity of infection of 0.3 (previously determined to cause minimal toxicity). The generation of Phoenix GALV-pseudotyped MSCV-HOXB4-ires-GFP or MSCV-ires-YFP viral vectors has been described previously [9,12,13]. Virus titers were assayed on HT1080 cells, and titers were obtained in the range of 1 × 105 to 2 × 105 IU/mL. Vector supernatant was filtered through a 0.45-μm filter and frozen at −80°C until used for transduction. Before, during, and after transduction, cells were cultured in Stemspan serum-free media (Stem Cell Technologies, Vancouver BC, Canada) supplemented with 1% penicillin/streptomycin, and thrombopoietin, Flt3 ligand, and stem cell factor, all at 100 ng/mL.
Animal housing and care
All pig-tailed macaques (Macaca nemestrina) used in this study were housed at the University of Washington National Primate Research Center under conditions approved by the American Association for Accreditation of Laboratory Animal Care. All experimental procedures performed were reviewed and approved by the Institutional Animal Care and Use Committee. Macaques were conditioned with fractionated, myeloablative total body irradiation of 1020 cGy (for K00339) and 1100 cGy [14] (for T09214) from a 6 MV x-ray beam of a single-source linear accelerator located at the Fred Hutchinson Cancer Research Center South Lake Union Facility; irradiation was administered as a fractionated dose over the two days prior to cell infusion. During irradiation, animals were housed in a specially modified cage that provided unrestricted access for the irradiation while simultaneously minimizing excess movement. The dose was administered at a rate of 7 cGy/min delivered as a midline tissue dose. Granulocyte colony-stimulating factor (G-CSF) was administered daily from the day of cell infusion until the animals began to engraft; engraftment was considered to have occurred when the absolute neutrophil count (ANC) was equal to or greater than 500/μL for 3 consecutive days. Supportive care including antibiotics, electrolytes, fluids, and transfusions were given as necessary, and blood counts were analyzed daily to monitor hematopoietic recovery.
Flow cytometry
Gene marking analysis and subset stain analysis were performed using flow cytometry. Cells were prepared for FACS analysis following the instructions of the antibody manufacturer (Becton Dickinson, San Jose, CA, USA). Flow cytometric data were collected using a Canto I (Becton Dickinson, San Jose, CA, USA) and analyzed using FlowJo software (Tree Star Inc, Ashland, Oregon, USA). At least 10,000 events were collected for each sample. Parameters for analysis included expression of the reporter genes GFP and YFP, as well as expression of the surface markers CD3, CD4, CD8, CD13, CD14, CD20, CD34, and CD41. Non-transduced cells were used as a control for the gating of GFP+ and YFP+ cells, and isotype control antibodies were used as a control for gating of positive populations among antibody-labeled cells.
Pathology/Morphology
Bone marrow biopsies and peripheral blood smears were obtained from T09214 and K00339 and prepared for morphological examination. Bone marrow samples from T09214 were sectioned and stained with hematoxylin and eosin stain; these were imaged on a Leica microscope (Leica Microsystems, Wetzlar, Germany) at 64X and 320X using FireCam software (Leica Microsystems, Wetzlar, Germany). Peripheral blood smears from T09214 and K00339 were stained with Wright-Giemsa stain prior to viewing. All samples were analyzed by a pathologist and assessed on the basis of morphology, presence or absence of all 3 major cell lines, and presence or absence of blasts.
Southern blot and LAM-PCR
Original integration site analysis for K00339 performed in 2008 used a Southern blot and LAM-PCR approach. Briefly, DNA was extracted using the Puregene DNA Purification Kit (Quiagen, Inc., Valencia, CA, USA), followed by overnight digestion with BglII, EcoRI, or SacI. Samples were run on a 0.8% gel and transferred to a membrane. Next, samples were hybridized in Quick-Hyb solution (Amersham Biosciences Corp., Piscataway, NJ, USA) with 32P-labeled ires-GFP. Signals were detected using the Typhoon Phosphor Imaging System (Amersham Biosciences Corp., Piscataway, NJ, USA) LAM-PCR has been described previously [15] and involved the cloning of PCR products into the TOPO vector for sequencing. The collected sequences were aligned with the rhesus macaque genome assembly dated January 2006 from the University of Santa Cruz Genome Browser.
Acoustic mediated-Modified Genomic Sequencing (MGS)-PCR for amplification of retroviral integrations sites
Our lab has continued to improve upon our retroviral integration site processing technique since the procedures described above. Our improved technique was used for the re-processing of the samples from K00339 and for the processing of samples from T09214. Briefly, samples were DNA extracted using Qiagen Gentra Blood and Bodily Fluid Minikit (Qiagen, Inc., Valencia, CA, USA). Between 300ng to 3μg of gDNA samples were acoustically sheared to a mean fragment size of 1500bp using the M220 focused-ultrasonicator (Covaris Inc., Woburn, MA, USA). The sheared fragments were then isolated, polished to blunt-ended dsDNA, and ligated to modified linkers for PCR amplification. These modified linkers are ligated following the manufacturer’s directions (454/Roche-GS 20 DNA library preparation kit). Small fragments are removed using AMPure Beads (Beckman Coulter, Inc., Pasadena, CA, USA) and then sequential nested PCR is performed. The first PCR primer pair is biotin-tagged and is specific to the provirus Long Terminal Repeat (LTR). The biotin-tagged sequences are then captured and washed. The second nested PCR is performed to further amplify the vector-genome junction sequences and attached LTR specific barcodes. The product is then run on a 2% agarose E-gel (Invitrogen) and DNA fragments ranging from 300 and 800bp are then excised and purified. The final processed fragments are sent for multiplexed semiconductor sequencing by the Ion Torrent Personal Genome Machine (EdgeBio, Gaithersburg, MD, USA). Here, they were quality control checked and sequenced on a 318 chip. Samples were deconvoluted by barcodes in FASTA format on a secure server for downstream processing.
Processing of integration site data
DNA sequences isolated from MGS-PCR–based ampliSied vector LTR-chromosome junctions were processed as described previously [15–17]. Using a stand-alone version of BLAT [18], the LTR proximal genomic sequences were aligned to the rhesus genome (rheMac2). Rhesus genome alignments were converted to the human genome position (assembly GRCh37/hg19), and PERL programs were used to compare localized integration sites to various chromosomal features by using tables available from the University of California at Santa Cruz database as described previously [18].
RESULTS
No increase in HOXB4GFP marking long-term following cord blood transplant
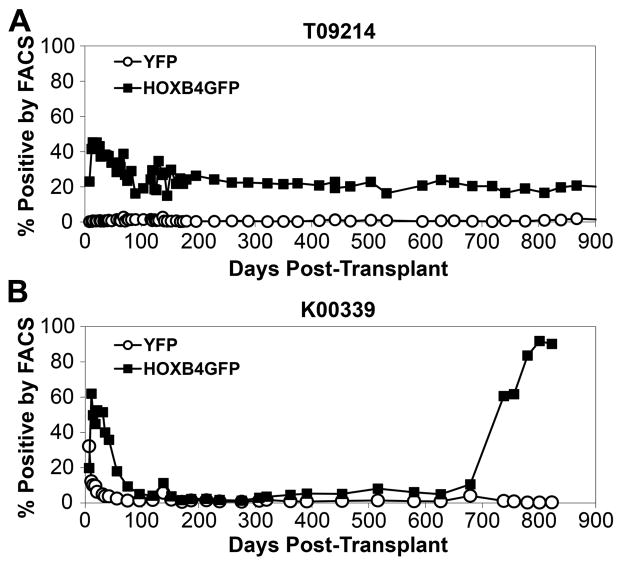
HOXB4GFP+ marking in cord blood transplant recipient T09214 reached a maximum within the first 30 days post-transplant at around 48%, followed by a gradual decrease over the following 6 months to approximately 20% percent. From this point on, HOXB4GFP+ marking has remained stable at around 20% with no drastic changes (Figure 2A). This is in sharp contrast to the marking pattern in bone marrow transplant recipient K00339. In this animal, HOXB4GFP+ marking peaked around 62% within the first 30 days, dropped to around 5%, and then began a steady increase to reach approximately 90% by 2.2 years post-transplant (Figure 2B). This rise in HOXB4GFP+ marking first became evident around 2 years post-transplant. Control YFP+ marking in both animals is comparable, with levels hovering around 0.5% to 1%.
Figure 2. Gene marking levels in granulocytes in a cord blood transplant monkey (A) and a bone marrow transplant monkey who later developed leukemia (B).
Marking in the cord blood transplant monkey remains stable, while the bone marrow transplant monkey demonstrated a rapid increase in GFPHOXB4 marking around 2 years post-transplant.
No evidence of abnormalities in blood panels of cord blood transplant recipient
Compete blood count (CBC) analysis was performed daily for the first 30 days post-transplant, weekly until 3 months post-transplant, and monthly thereafter. For cord blood transplant recipient T09214, CBC analysis revealed no abnormalities among any of the parameters tested including white blood cell (WBC) count and hematocrit. After engraftment was achieved, these values were consistently within the “normal” range for a macaque, which vary from 8.1 to 15.4 thousand WBC per μL and 33% to 44% for hematocrit. This is in contrast to CBC results from bone marrow transplant recipient K00339, who developed an increased white blood cell count and decreased hematocrit at 25 months post-transplant.
No signs of abnormal pathology in cord blood transplant recipient
A bone marrow biopsy was taken from cord blood transplant recipient T09214 at 3 years post-transplant (Figure 3A). Examination by a pathologist confirmed the presence of pockets of normal-appearing marrow and the existence of all three major cell lines (erythroblastic, granulocytic, and megakaryocytic). There was no evidence of an increased number of immature cells or blasts. Furthermore, the sample was normocellular and showed no obvious infiltrates. Peripheral blood smears from T09214 revealed a differential count of 42% neutrophils (normal range 34% to 71%), 44% lymphocytes (normal range 19% to 53%) and 14% monocytes (normal range 5% to 13%). Adequate numbers of platelets were detected, neutrophils appeared mature, and red cells seemed to be maturing normally. Platelets appeared to be present in normal amounts in the peripheral blood and there was no indication of atypical or dysplastic cells of any kind. In contrast, a biopsy and peripheral blood smear from bone marrow transplant recipient K00339 yielded a diagnosis of acute myeloid leukemia (Figure 3B).
Figure 3. Representative marrow morphology from a monkey receiving a HOXB4-expanded cord blood graft (A) and a monkey receiving a HOXB4-expanded bone marrow graft who later developed leukemia (B).
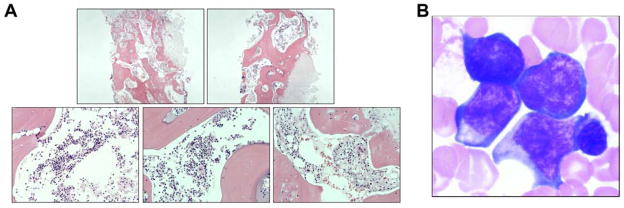
Slides were stained with Wright-Giemsa stain and analyzed by a pathologist. A high frequency of blast cells was observed in the marrow of the bone marrow transplant recipient. Image reprinted with permission from [11].
Phenotypic composition of long-term cord blood transplant recipient is normal
Subset stain analysis for T09214 is depicted in Figure 4A. The percentages of CD3+ T cells, CD20+ B cells, CD13+ granulocytes, CD14+ monocytes, and CD34+ hematopoietic stem cells were compared to those of a normal, non-transplanted control macaque. The composition of each phenotype was similar between T09214 and control. However, in our previous follow-up of bone marrow transplant recipient K00339 (Figure 4B), we observed a high frequency of CD34+ cells, nearly all of which were GFP+ indicating that they arose from a HOXB4-transduced stem/progenitor cell.
Figure 4. Subset analysis of bone marrow aspirates from a cord blood transplant monkey at nearly 3 years post-transplant (A) and bone marrow transplant monkey who later developed leukemia (B).
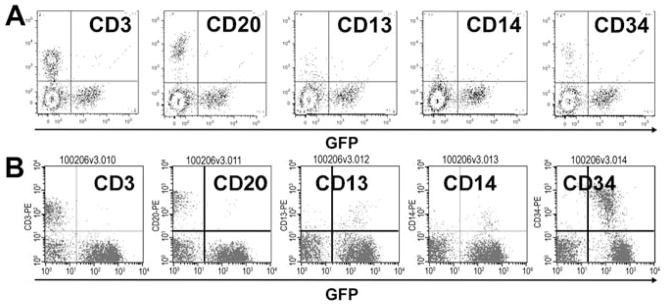
Nucleated marrow cells were analyzed for expression of CD3, CD20, cD13, CD14, and CD34 based on flow cytometric analysis. Image reprinted with permission from [11].
Leukemogenic clones can be detected by six months post-transplant
Previous analysis of retroviral integration sites by Southern Blot and LAM-PCR in bone marrow transplant recipient K00339 identified the presence of leukemic (or potentially oncogenic) clones PRDM16, SSBP2, STYK1, NSMCE1 and SOCS1 [11]. Please refer to the work of Zhang et al. [11] for a full, detailed description of how the leukemogenicity of each of these clones was assessed in detail. Using the improved integration site analysis technique described above, we went back and re-analyzed samples from K00339 at 3, 6, 12, and 24 months post-transplant (Figures 5B and 6). We were able to show that two of these leukemic clones, PRDM16 and SSBP2, were detectable as early as 6 months post-transplant. The other dominant clones detected in this animal (STYK1, NSMCE1, and SOCS1) could be picked up by 12 months post-transplant and increased their overall contribution drastically by 24 months post-transplant. None of the dominant clones identified in K00339, who developed leukemia, are present in T09214 at any time post-transplant (Figure 5A). Furthermore, analysis of clonal composition for macaque K00339 shows the presence of multiple clones near or above 20%, which is a threshold identified by the FDA as “potentially dominant” (Figure 5B). In T09214, however, no site is present at 20% and no site is continuously increasing over time (Figure 5A).
Figure 5. Longitudinal integration site analysis from a cord blood transplant monkey (A) and a bone marrow transplant monkey who later developed leukemia (B).
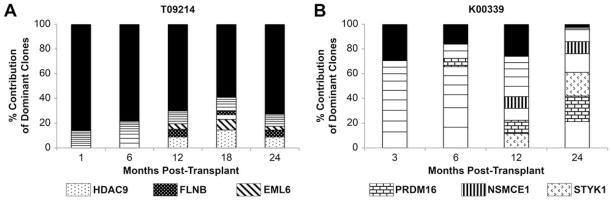
The relative frequency of the 9 most commonly identified integration sites is shown, along with the contribution of all others (black bars). Portions of columns with matching patterns illustrate site that were present at multiple time points.
Figure 6. Leukemic clones can be detected by 6 months post-transplant in macaques receiving bone marrow CD34+ cells.
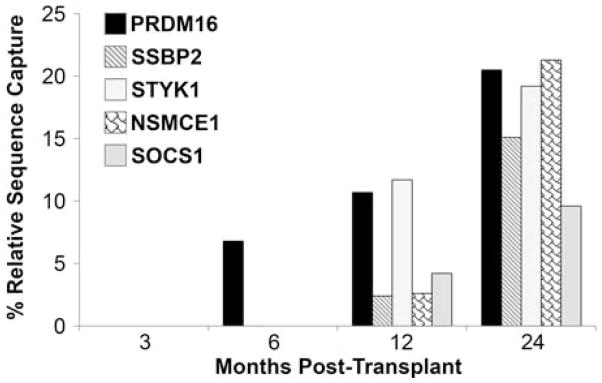
Integration sites contributing to leukemia outgrowth in a macaque receiving a bone marrow transplant were analyzed at regular time points in the period leading up to observation of clinical symptoms. Integration sites in PRDM16 and SSBP2 could be detected by 6 months post-transplant, while integrations within STYK1, NSMCE1, and 53 kb away from SOCS1 could be detected by 12 months post-transplant.
DISCUSSION
We have previously shown in large animal models that transplantation of HOXB4-expanded cells can lead to leukemia. In these circumstances, the graft was comprised of bone marrow-derived stem cells. Leukemia developed at around 2 years post-transplant in dogs and monkeys and was characterized by a spontaneous increase in HOXB4GFP marked cells, dysregulated expression of oncogenes, a block in myeloid differentiation, and an increase in circulating blasts. The observation of leukemia-associated insertional mutagenesis was determined by Southern blot and LAM-PCR.
Here, we have re-analyzed integration sites in macaque K00339, which later developed leukemia, using our modified genome sequencing approach and confirmed that dominant clones are present early on following transplant and can be detected as early as six months post-transplant.
Moving forward from our bone marrow transplantation studies, we have embarked on cord blood transplantation studies. After developing a large animal model in which to study cord blood expansion and transplantation [6], we tested our model using HOXB4 expansion based upon our previous experience with this modality. At the time we initiated the HOXB4 cord blood transplant studies, we had not yet observed leukemia in the HOXB4 bone marrow transplant monkeys. Of 3 monkeys transplanted with HOXB4-expanded cord blood cells, one is currently over three years post-transplant without evidence of clonal dominance, leukemia, or aberrant hematopoiesis in this animal. In addition, in-depth longitudinal retroviral integration site analysis on this animal confirms the absence of any integrations within the dysregulated genes (expected to be associated with leukemia) in K00339.
The other two macaques that received grafts of HOXB4-expanded cord blood cells survived for 1.5 months (K09175) and 3 months (L09025) post-transplant without evidence of leukemia. These animals succumbed to transplant-related complications including viral pneumonia and thrombotic thrombocytopenic purpura, neither of which is uncommon in the cord blood transplant setting. Because these animals did not survive long-term, clonal analysis could not be assessed longitudinally, but was assessed at early time points post-transplant. With both of these animals, there was no evidence of clonal dominance at time of analysis. As with long-term follow-up animal T09214, none of the potentially risky sites identified in K00339 were found in these animals. Based on our findings that leukemogenic clones can be detected early after transplant, it is likely that had these sites been present, we would have identified them during our analysis. This cannot be stated with certainly, as it took 6 months for these specific sites to be identified in K00339, but it does support our assertion that had K09175 and L09025 not succumbed to transplant related complications, they too may have been leukemia-free even several years following transplant. Though the numbers of animals studied in this investigation is small, we believe the findings are of value and provide a rationale to pursue these observations in more depth.
The interesting observation that multiple macaques receiving HOXB4-expanded bone marrow grafts developed leukemia, while none of the macaques receiving HOXB4-expanded cord blood grafts demonstrated any symptoms of myelodysplasia, is potentially valuable. The protocols for pre-transplant conditioning, cell processing, transduction, transplantation, and post-transplant follow-up for these animals were identical. The only difference between the bone marrow cohort and the cord blood cohort was the source of stem cells used.
There is a large body of literature exploring the differences between bone marrow CD34+ cells and cord blood CD34+ cells. For example, CD34+ cells from cord blood have a higher cloning efficiency, increased proliferation rate, respond more vigorously to cytokine stimulation, and generate approximately 7-fold more progeny than bone marrow derived CD34+ [19]. It has also been reported that the relative proportion of myeloid progenitors is higher in cord blood than in bone marrow, which has implications for engraftment kinetics; it has been suggested that the mean dose of myeloid progenitors required for timely engraftment from bone marrow is 2.2-fold higher than from cord blood [20]. Additionally, cell cycle kinetics vary between bone marrow and cord blood, as it has been found that at day 0, significantly fewer CD34+ cells from cord blood are in S + G2M phases of the cell cycle compared to bone marrow cells. Exposure to stem cell factor (SCF) and interleukin 3 (IL3) significantly increase cycling in cord blood CD34+ cells but have no effect on bone marrow CD34+ cells [21]. Furthermore, in studies comparing cytokine expression levels between cord blood and bone marrow, it was found that G-CSF was not detected in cord blood [22]. Other literature has cited differences in the immunogenicity and naivety of cord blood cells [23]. More recent analysis of the genomic and proteomic variation in the mitotic state of CD34+ cells derived from bone marrow, peripheral blood, and cord blood has also indicated the unique nature of cord blood cells. Analysis of 484 target genes and 646 proteins [24] showed that engraftment potential of bone marrow or peripheral blood cells was only evident in the G0 phase of the cell cycle, whereas in cord blood, cell cycle status was not a limiting factor in engraftment potential. Research by Jin et al (2013) confirms that mesenchymal stromal cells derived from cord blood have significantly higher clonality (and lower expression of cell senescence markers) than those derived from bone marrow [25]. Furthermore, researchers in the Terry Fox Laboratory recently published a study [26] in which they exposed cord blood CD34+ cells to a library of bar-coded viral vectors and used massively parallel sequencing to analyze the clonal distribution of cells from mice receiving transplants of these cells. They were able to detect a high number of clones, the majority of which were detected greater than 13 months post-transplant but with increasing stability. This study illustrated the ability of cord blood CD34+ cells to display varied clonal output patterns post-transplant. Taken together our data support previous findings that cord blood CD34+ cells have unique differences from bone marrow CD34+ cells. These include variations in proliferative rate, varying cell cycle kinetics, differing response to cell signaling molecules, and variations in the effect of cell cycle status on engraftment potential. But we hypothesize that perhaps the most significant factor among these differences is an inherent increase in clonality of cord blood cells compared to bone marrow cells. While the number of animals studied here is too small to draw any clear conclusions regarding the safety of gene modified cord blood CD34+ cells, the lack of clonal expansion or leukemia in the cord blood transplant animals is intriguing and warrant additional studies. Our data clearly establishes the potential of the nonhuman primate model to study cord blood CD34+ cells for gene therapy and cell expansion studies.
Acknowledgments
We would like to thank the staff at the University of Washington National Primate Center (Seattle, WA, USA) and thank Erica Wilson and Kelvin Sze for providing technical assistance. We also thank Grace Choi, Helen Crawford, Bonnie Larson, and Emily Menard for help with preparing the manuscript. This work was supported in part by the National Institutes of Health (Bethesda, MD) grants HL084345, HL098489, HL053750, and DK056465. H.-P.K. is a Markey Molecular Medicine Investigator and the recipient of the Jose Carreras/E.D. Thomas Endowed Chair for Cancer Research.
Footnotes
SUPPORT AND FINANCIAL DISCLOSURE DECLARATION:
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE LIST
- 1.Gluckman E, Rocha V, Arcese W, et al. Factors associated with outcomes of unrelated cord blood transplant: guidelines for donor choice. Exp Hematol. 2004;32:397–407. doi: 10.1016/j.exphem.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Ballen KK, Spitzer TR, Yeap BY, et al. Double unrelated reduced-intensity umbilical cord blood transplantation in adults. Biol Blood Marrow Transplant. 2007;13:82–89. doi: 10.1016/j.bbmt.2006.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pecora AL, Stiff P, Jennis A, et al. Prompt and durable engraftment in two older adult patients with high risk chronic myelogenous leukemia (CML) using ex vivo expanded and unmanipulated unrelated umbilical cord blood. Bone Marrow Transplant. 2000;25:797–799. doi: 10.1038/sj.bmt.1702222. [DOI] [PubMed] [Google Scholar]
- 4.Shpall EJ, Quinones R, Giller R, et al. Transplantation of ex vivo expanded cord blood. Biol Blood Marrow Transplant. 2002;8:368–376. doi: 10.1053/bbmt.2002.v8.pm12171483. [DOI] [PubMed] [Google Scholar]
- 5.Jaroscak J, Goltry K, Smith A, et al. Augmentation of umbilical cord blood (UCB) transplantation with ex vivo-expanded UCB cells: results of a phase 1 trial using the AastromReplicell System. Blood. 2003;101:5061–5067. doi: 10.1182/blood-2001-12-0290. [DOI] [PubMed] [Google Scholar]
- 6.Watts KL, Nelson V, Wood BL, et al. Hematopoietic stem cell expansion facilitates multilineage engraftment in a nonhuman primate cord blood transpalntation model. Exp Hematol. 2012;40:187–196. doi: 10.1016/j.exphem.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hacein-Bey-Abina S, von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1 [erratum appears in Science. 2003 Oct 24;302(5645):568] Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 8.Cavazzana-Calvo M, Fischer A. Gene therapy for severe combined immunodeficiency: are we there yet? (Review) J Clin Invest. 2007;117:1456–1465. doi: 10.1172/JCI30953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X-B, Beard BC, Beebe K, Storer B, Humphries RK, Kiem H-P. Differential effects of HOXB4 on nonhuman primate short- and long-term repopulating cells. PLoS Medicine. 2006;3:e173. doi: 10.1371/journal.pmed.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murnane R, Zhang XB, Hukkanen RR, Vogel K, Kelley S, Kiem H-P. Myelodysplasia in 2 pig-tailed macaques (macaca nemestrina) associated with retroviral vector-mediated insertional mutagenesis and overexpression of HOXB4. Veterinary Pathology. 2011;48:999–1001. doi: 10.1177/0300985810382673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X-B, Beard BC, Trobridge GD, et al. High incidence of leukemia in large animals after stem cell gene therapy with a HOXB4-expressing retroviral vector. J Clin Invest. 2008;118:1502–1510. doi: 10.1172/JCI34371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antonchuk J, Sauvageau G, Humphries RK. HOXB4 overexpression mediates very rapid stem cell regeneration and competitive hematopoietic repopulation. Exp Hematol. 2001;29:1125–1134. doi: 10.1016/s0301-472x(01)00681-6. [DOI] [PubMed] [Google Scholar]
- 13.Pineault N, Abramovich C, Ohta H, Humphries RK. Differential and common leukemogenic potentials of multiple NUP98-Hox fusion proteins alone or with Meis1. Mol Cell Biol. 2004;24:1907–1917. doi: 10.1128/MCB.24.5.1907-1917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watts KL, Beard BC, Wood BL, Kiem HP. Myeloablative irradiation in non-human primates. Journal of Medical Primatology. 2009;38:425–432. doi: 10.1111/j.1600-0684.2009.00368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beard BC, Dickerson D, Beebe K, et al. Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Mol Ther. 2007;15:1356–1365. doi: 10.1038/sj.mt.6300159. [DOI] [PubMed] [Google Scholar]
- 16.Beard BC, Keyser KA, Trobridge GD, et al. Unique integration profiles in a canine model of long-term repopulating cells transduced with gammaretrovirus, lentivirus, and foamy virus. Hum Gene Ther. 2007;18:423–434. doi: 10.1089/hum.2007.011. [DOI] [PubMed] [Google Scholar]
- 17.Trobridge GD, Miller DG, Jacobs MA, et al. Foamy virus vector integration sites in normal human cells. PNAS. 2006;103:1498–1503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao QL, Shah AJ, Thiemann FT, Smogorzewska EM, Crooks GM. A functional comparison of CD34 + CD38− cells in cord blood and bone marrow. Blood. 1995;86:3745–3753. [PubMed] [Google Scholar]
- 20.Fritsch G, Stimpfl M, Kurz M, et al. The composition of CD34 subpopulations differs between bone marrow, blood and cord blood. Bone Marrow Transplant. 1996;17:169–178. [PubMed] [Google Scholar]
- 21.De Bruyn C, Delforge A, Lagneaux L, Bron D. Characterization of CD34+ subsets derived from bone marrow, umbilical cord blood and mobilized peripheral blood after stem cell factor and interleukin 3 stimulation. Bone Marrow Transplant. 2000;25:377–383. doi: 10.1038/sj.bmt.1702145. [DOI] [PubMed] [Google Scholar]
- 22.Michejda M. Which stem cells should be used for transplantation? (Review) Fetal Diagnosis & Therapy. 2004;19:2–8. doi: 10.1159/000074252. [DOI] [PubMed] [Google Scholar]
- 23.Szabolcs P, Park KD, Reese M, Marti L, Broadwater G, Kurtzberg J. Coexistent naive phenotype and higher cycling rate of cord blood T cells as compared to adult peripheral blood. Exp Hematol. 2003;31:708–714. doi: 10.1016/s0301-472x(03)00160-7. [DOI] [PubMed] [Google Scholar]
- 24.Chitteti BR, Liu Y, Srour EF. Genomic and proteomic analysis of the impact of mitotic quiescence on the engraftment of human CD34+ cells. PLoS ONE. 2011;6:e17498. doi: 10.1371/journal.pone.0017498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin HJ, Bae YK, Kim M, et al. Comparative analysis of human mesenchymal stem cells from bone marrow, adipose tissue, and umbilical cord blood as sources of cell therapy. International Journal of Molecular Sciences. 2013;14:17986–18001. doi: 10.3390/ijms140917986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung AM, Nguyen LV, Carles A, et al. Analysis of the clonal growth and differentiation dynamics of primitive barcoded human cord blood cells in NSG mice. Blood. 2013;122:3129–3137. doi: 10.1182/blood-2013-06-508432. [DOI] [PMC free article] [PubMed] [Google Scholar]