SUMMARY
The intestinal epithelium harbors large populations of activated and memory lymphocytes, yet these cells do not cause tissue damage in the steady state. We investigated how intestinal T cell effector differentiation is regulated upon migration to the intestinal epithelium. Using gene loss- and gain-of-function strategies, as well as reporter approaches, we showed that cooperation between the transcription factors T-bet and Runx3 resulted in suppression of conventional CD4+ T helper functions and induction of an intraepithelial lymphocyte (IEL) program that included expression of IEL markers such as CD8αα homodimers. Interferon-γ sensing and T-bet expression by CD4+ T cells were both required for this program, which was distinct from conventional T helper differentiation but shared by other IEL populations, including TCRαβ+CD8αα+ IELs. We conclude that the gut environment provides cues for IEL maturation through the interplay between T-bet and Runx3, allowing tissue-specific adaptation of mature T lymphocytes.
INTRODUCTION
Interposed between intestinal epithelial cells, intraepithelial lymphocytes (IELs) constitute the most abundant T cell population in the body (Meresse et al., 2012). Developing T cells differentiate into IELs from precommitted thymic precursors (Gangadharan et al., 2006; Guy-Grand et al., 2013). Additionally, mature T cells can acquire IEL-like characteristics upon peripheral activation under appropriate conditions (Denning et al., 2007; Guy-Grand et al., 1991; Huang et al., 2011; Mucida et al., 2013; Reis et al., 2013). Thymic (TCRγδ+CD8αα+ and TCRαβ+ CD8αα+) and peripherally-converted (TCRαβ+ CD8αβ+CD8αα+ and CD4+−CD8αα+) IELs are commonly described as “natural” and “induced” IELs, respectively (Cheroutre et al., 2011). IELs are characterized by high expression of activation markers such as CD69; gut-homing integrins, including CD103 (αEβ7); NK-like receptors such as 2B4; cytotoxic (CTL)-related genes such as granzyme B and Runx3; and anti-inflammatory or inhibitory receptors like LAG-3; which define these cells as “activated yet resting” (Denning et al., 2007; Pobezinsky et al., 2012). Irrespective of their nature, tightly regulated control of IEL function is crucial for the maintenance of the epithelial cell barrier and gut physiological inflammation (Tang et al., 2009). Inappropriate activation of the CTL capacity of IELs can induce chronic inflammatory disorders such as celiac disease (Tang et al., 2009).
Another common characteristic of IELs is the surface expression of CD8αα homodimers, which can bind both to classical MHC-I and to epithelial cell-associated non-classical MHC-I molecules, such as mouse thymic leukemia antigen (TL), presumably working as TCR “corepressors” (Cheroutre and Lambolez, 2008; Cheroutre et al., 2011; Gangadharan et al., 2006; Guy-Grand et al., 1991; Guy-Grand et al., 2013). In addition, peripheral mature CD4+ T cells can acquire CD8αα expression upon migration to the intestine (Das et al., 2003; Mucida et al., 2013; Reis et al., 2013). This process is linked to acquisition of IEL characteristics, expression of the long-form (encoded by its distal promoter) of the transcription factor Runx3 and loss of expression of the key CD4+ T cell differentiation transcription factor ThPOK (also known as Zbtb7b and cKrox), which results in the loss of CD4+ T helper function (Mucida et al., 2013; Pobezinsky et al., 2012; Reis et al., 2013). However, despite the role of this pathway in different pathophysiological conditions (Mucida et al., 2013; Reis et al., 2013), the contributions of different signals and transcription factors to the induction of CD4+ IEL differentiation and suppression of T helper function are not yet understood.
In this study, we investigated the molecular mechanisms by which the intestinal environment mediates suppression of T helper (Th) programs and induces an IEL phenotype in peripheral CD4+ T cells. We discovered that upregulation of the transcription factor T-bet and interferon-γ (IFN-γ) or inter-leukin-27 (IL-27) signaling were required for IEL differentiation both in vitro and in vivo. The molecular requirements for the induction of this phenotype were distinct from those required for classical Th1 cell differentiation, typically associated with T-bet expression and IFN-γ production, and required synergistic effects of the transcription factors T-bet and Runx3. T-bet was found to bind Zbtb7b and Runx3 in differentiating IELs, assisting Runx3-dependent upregulation of genes associated with IELs, whereas T-bet-dependent suppression of T helper programs was largely Runx3 independent. We conclude that the gut environment provides cues for terminal IEL differentiation through the interplay between cytokine-regulated T-bet and Runx3, allowing tissue-specific adaptation and reprogramming of mature lymphocytes in the chronically stimulated mucosa.
RESULTS
T-bet Upregulation Is Linked to IEL Differentiation
Loss of ThPOK and acquisition of Runx3 by intestinal CD4+ T cells results in reduced T helper-associated gene expression, including that of Th2, Th17, and Treg cells. Moreover, CD4+ T cells undergoing this transition acquire cytotoxic and IEL gene-expression patterns, including high levels of CD8αα, CD103 (αEβ7), 2B4 (CD244), and granzyme B expression (Mucida et al., 2013; Reis et al., 2013). This pattern, hereafter referred to as “CD4-IEL differentiation,” resembles the pattern of effects on peripheral CD4+ and CD8+ T cells known to be mediated by T-bet, such as suppression of Th2, Th17, and Treg cell differentiation and enhancement of Th1 and CD8+ effector T cell differentiation (Cruz-Guilloty et al., 2009; Djuretic et al., 2007; Koch et al., 2009; Lazarevic et al., 2011; Zhu et al., 2012). Consistent with a potential link between T-bet and IEL differentiation, analysis of Tbx21 (encoding T-bet) mRNA expression in ex vivo sorted small intestine IELs from naive wild-type (WT) mice revealed that all IEL populations expressed increased amounts of Tbx21 when compared to peripheral T cells (Figure 1A). Corresponding to this increased expression in cells that have undergone IEL differentiation, mRNA analysis of sorted small intestine CD4+ IELs from naive Zbtb7b-GFP knockin reporter mice indicated that Tbx21 was upregulated in ThPOKlo cells (Figure 1B). Protein expression analysis further confirmed that expression of T-bet by CD4+ IELs closely mirrored the expression pattern of long-form Runx3 (Reis et al., 2013) and the expression pattern of other IEL-associated proteins, such as 2B4 and CD103, which were upregulated in CD4+CD8αα+ IELs (Figure 1C).
Figure 1. T-bet Upregulation Is Linked to IEL Differentiation.
(A) Expression of Tbx21 by sorted small intestine IEL populations, TCRαβ+CD4+, TCRαβ+CD8αα+ (CD4–, CD8β–), TCRαβ+CD8β+CD8αα+ (CD4–), TCRγδ+CD8αα+ (CD4–, CD8β–), and naive splenic CD4+ T cells from WT mice.
(B) Expression Tbx21 by sorted small intestine IEL populations, TCRβ+CD4+CD8β– ThPOKlo and ThPOKhi from naive Zbtb7b -GFP reporter mice.
(C) CD8α, T-bet, 2B4, and CD103 expression by TCRβ+CD4+CD8β– small intestine IELs of naive WT mice.
(D–H) In vitro CD4+CD8αα+ IEL induction. Sorted, naive Va2+CD4+ T cells isolated from WT reporter OTII mice (OTII Zbtb7b -GFP) (D–G) or OTII Runx3-YFP (G) were cocultured with DCs, OVA peptide, and indicated cytokines. (D) T-bet and IFN-γ, (E) Foxp3, and (F) CD8α and CD103 expression by cultured OTII cells. (G) Histograms of ThPOK (left) or Runx3 (right) expression by OTII cells from different culture conditions. (H) CD103, CD8α, and ThPOK expression by gated CD4+CD8β– OTII cells. (B) Error bars represent SEM of triplicates. All plots are representative of at least three independent experiments.
To evaluate whether T-bet upregulation correlated with induced CD4-IEL differentiation, we initially used an in vitro approach. We cultured naive, OVA-specific OTII CD4+ T cells with WT splenic dendritic cells (DCs) and OVA peptide in the presence of exogenous recombinant cytokines. This system allowed us to study the effects of a particular cytokine milieu on IEL differentiation in the context of a set T cell receptor (TCR) specificity. We compared conditions known to induce T-bet expression in CD4+ T cells (IL-12 plus anti-IL-4, Th1 cell conditions) with conditions associated with CD4+CD8αα+ differentiation (TGF-β and retinoic acid [RA]). We previously reported that the latter combination concomitantly induces a Foxp3+ (T-betlo) Treg cell population (Mucida et al., 2007) and a reduced CD4+CD8αα+ IEL-like population that exhibits increased Tbx21 mRNA expression compared to the Foxp3+ compartment (Reis et al., 2013). While Th1 cell conditions induced robust T-bet expression and IFN-γ production, this was not observed in TGF-β and RA-treated cells (Figure 1D). Contrary to TGF-β and RA alone, addition of direct T-bet inducers abundant in the intes tinal milieu, specifically IFN-γ and IL-27, also resulted in high T-bet and IFN-γ expression by OTII cells and, when added in combination with TGF-β and RA, led to intermediate to high T-bet and IFN-γ production (Figure 1D). Addition of IFN-γ or IL-27 also suppressed TGF-β and RA-mediated Foxp3 induction, consistent with the reported negative effect of T-bet on Treg cell induction (Figure 1E). In contrast, these T-bet-inducing cytokines boosted expression of the IEL-associated markers CD103 and CD8αα when added to the TGF-β and RA regimen (Figure 1F). Similar to ex vivo isolated IELs (Figure 1C), induction of CD8αα expression by these conditions was mostly confined to the CD103hi population (Figure 1F).
To better evaluate transcription-factor dynamics in different conditions and confirm this correlation with the CD4+ IEL signature, we used OTII cells from mice on a Zbtb7b-GFP or a Runx3-YFP background. We found that TGF-β and RA plus IFN-γ suppressed ThPOK expression and concurrently induced Runx3, whereas Th1 cell-polarizing conditions induced Runx3 upregulation without resulting in ThPOK loss (Figure 1G). In contrast, the combination of TGF-β and RA plus IFN-γ resulted in stronger suppression of ThPOK than TGF-β and RA alone (Figures 1G and 1H). Less significant results were obtained with IL-15, which has been also reported to induce T-bet (Intlekofer et al., 2005). When added in combination with TGF-β and RA, IL-15 slightly enhanced CD4+CD8αα+ induction, although we did not observe any additional ThPOK downmodulation (see Figure S1 available online). Similar to freshly isolated CD4+ IELs, CD8αα and CD103 expression was mostly found in the ThPOKlo population (Figure 1H). To confirm that qualitatively distinct differentiation programs were induced in these cells, we examined the gene-expression profile from in vitro-induced Th1 cells (ThPOKhi CD8αα−) and in vitro-induced CD4+ IELs (ThPOKlo CD8αα+). While sorted cells treated with IL-12 plus anti-IL-4 expressed hallmark Th1 cell genes, such as Il12rb1, Ccr5, Irf4, and Csf2r, TGF-β+RA+IFN-γ-treated cells expressed IEL-associated genes, such as Crtam and Runx3, but similar amounts of Tbx21 and Gzmb to Th1 cells (Figure S1). These observations demonstrate that an IEL-inducing cytokine milieu leads to concomitant upregulation of T-bet and Runx3, suggesting a potential role for this pathway in IEL maturation or differentiation.
T-bet Regulates IEL Development
We next examined the requirements for T-bet in the CD4-IEL differentiation process. First, we compared CD4+ T cells from WT OTII mice to those of OTII mice crossed with Tbx21–/– mice (OTII [Tbx21–/–]) for their IEL differentiation capacity. Because we have previously shown that Runx3 is required for this process in vivo (Reis et al., 2013), we compared these cells to CD4+ T cells isolated from conditional Runx3-deficient mice (OTII [DRunx3]) for CD4-IEL differentiation. We found that both OTII (ΔRunx3) and OTII (Tbx21–/–) cells failed to upregulate CD8αα and CD103 expression upon TGF-β+RA+IFN-γ treatment (Figures 2A and 2B). In similar experiments, we found that lack of either T-bet or Runx3 in polyclonal CD4+ T cells also impairs downmodulation of ThPOK upon culture in CD4-IEL-differentiating conditions (Figure 2C).
Figure 2. T-bet Regulates IEL Development.
Sorted naive Va2+CD4+ T cells isolated from WT (OTII), Tbx21–/– (OTII[DTbx21]) or Runx3 conditional knockout (OTII[DRunx3]) mice were cocultured with DCs, OVA peptide, and TGF-β+RA+IFN-γ (A and B). (A) CD8α (contour plot) and (B) CD103 (histogram) expression by gated CD4+CD8β–T cells.
(C) Sorted naive polyclonal CD4+ T cells isolated from WT, DTbx21 or Cd4(DRunx3) Zbtb7b -GFP reporter mice were cultured with plate-bound α-CD3 and soluble α-CD28 in the presence of TGF-β+RA+IFN-γ. Histogram of ThPOK expression by CD4+CD8β–T cells is shown.
(D–I) Ex vivo analysis of small intestinal IELs from 10- to 14-week-old (D–H) or 25- to 30-week-old (I) WT and Tbx21–/– mice. (D and I) TL-tetramer (CD8αα) and CD8β expression by gated TCRαβ+CD4– IEL. (E) Ratio TCRγδ+ to TCRαβ+ among CD45+ cells. (F) CD8αα quantification in natural IELs TCRγδ+ (left) and TCRαβ+CD8β–CD4– (right) T cells right. (G) CD8α and CD103 expression by TCRαβ+CD4+CD8β– IELs. (H) CD8αα quantification in induced IELs TCRαβ+CD4+ (left) and TCRαβ+CD8β+ (right).
(J and K) Ex vivo analysis of small intestine IELs from 10- to 14-week-old WT and Tbx21–/– Zbtb7b -GFP reporter mice. (J) ThPOK and CD103 expression by gated TCRαβ+CD4+CD8β– cells; (K) IL-17 and CD8α expression by gated TCRαβ+CD4+CD8β– cells, stimulated with PMA and ionomycin. Plots are representative of at least three independent experiments. Statistical significance analyzed by Student's t test of pooled experiments. Error bars represent SEM. *p < 0.05; **p < 0.01; ***p < 0.001.
To directly assess the role of T-bet in IEL maturation in vivo, we isolated intestinal cells from naive WT or Tbx21–/– mice. We found that in Tbx21–/– animals, all CD8αα-expressing IEL populations were reduced: TCRγδ+CD8αα+, TCRαβ+ CD8αα+, TCRαβ+ CD8β+CD8αα+, and TCRαβ+ CD4+ CD8αα+ (Figures 2D–2I). The ratio of TCRγδ+ to TCRαβ+ among CD45+ cells was also significantly reduced in mice lacking T-bet (Figure 2E). Another hallmark of IELs, CD103, was found to be reduced in the TCRαβ+ CD4+ IEL population, mostly due to a drastic reduction in the (CD103hi) CD4+CD8αα+ population in the Tbx21–/– mice (Figures 2G and 2H). This reduced frequency of CD8αα+ IELs in Tbx21–/– mice was maintained in older animals, indicating that T-bet requirement is likely not restricted to early waves of T cell egress from the thymus (Figure 2I).
The reduced frequency of CD4+CD8αα+ IELs in Tbx21–/– mice prompted us to investigate whether T-bet plays a role in the modulation of ThPOK expression by intestinal CD4+ T cells. To address this question, we interbred Tbx21–/– mice with Zbtb7b-GFP reporter mice. Analysis of naive Tbx21–/–Zbtb7b-GFP mice indicated that both CD8αα induction and ThPOK downmodulation were significantly impaired in the absence of T-bet (Figure 2J). Conversely, IL-17 production by CD4+ T cells from the IEL compartment was enhanced in mice without T-bet, which suggests that T-bet mediates suppression of the Th17 program and Th17-associated genes in differentiating CD4+ IELs (Figure 2K). These data indicate that T-bet is essential for both “natural” and “induced” IEL development and maturation.
Cell-Intrinsic Effects of T-bet on CD4-IEL Maturation
Tbx21–/– mice display immune-related defects associated with several immune cell populations, including NK cells, T helper cells, and CD8+ T cells (Szabo et al., 2002). To exclude cell-extrinsic effects of T-bet deficiency and evaluate CD4-IEL differentiation in an in vivo inflammatory setting, we used the T cell transfer model of colitis (Powrie et al., 1994). Sorted naive CD4+ T cells from Zbtb7b-GFP or Tbx21–/–Zbtb7b-GFP mice were adoptively transferred into Rag1–/– hosts. Similar to what had been previously described, Tbx21–/– donor naive T cells did not induce significant weight loss and colitis, likely due to impaired differentiation of Th1 or absence of another pathogenic CD4+ T cell population (Neurath et al., 2002; data not shown). In line with our ex vivo and in vitro data with Tbx21–/– mice, we found reduced loss of ThPOK and reduced CD8αα induction by donor Tbx21–/– CD4+ T cells (Figures 3A and 3B).
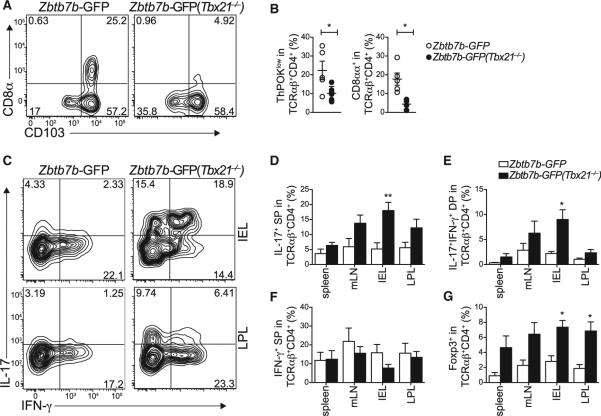
Figure 3. Cell-Intrinsic Effects of T-bet on CD4-IEL Maturation.
(A–G) Sorted naive CD4+GFPhi T cells from WT and Tbx21–/– Zbtb7b -GFP reporter mice were adoptively transferred to Rag1–/– recipient mice and analyzed 40 to 50 days later. (A) CD8α and CD103 expression, (B) ThPOK loss and CD8αα expression by gated TCRβ+CD4+CD8β– small intestine IELs of recipient mice. (C–F) Analysis of spleen, mesenteric lymph node (mLN), and large intestine IEL and LPL cells from recipient mice after PMA and ionomycin stimulation. (C) Plots for IL-17 and IFN-γ expression. (D–G) Frequency of (D) IL-17 single producer (SP), (E) IL-17-IFN-γ double producer (DP), (F) IFN-γ single producer, and (G) Foxp3 positive TCRβ+CD4+CD8β– cells from two pooled independent experiments. Error bars represent SEM. *p < 0.05; **p < 0.01. Plots are representative of three independent experiments.
In naive WT animals, IL-17 production is restricted to ThPOKhi or Runx3lo intestinal CD4+ T cells (Reis et al., 2013). Consistent with a role for T-bet in suppressing IL-17 production by CD4-IEL, we found that adoptive transfer of naive CD4+ T cells from WT or Tbx21–/– mice into Rag1–/– hosts resulted in increased frequency of IL-17-producing CD4+ T cells, particularly in the intestinal tissues of Tbx21–/– T cell hosts, whereas IFN-γ production was not significantly altered (Figures 3C–3F). This is in line with previous studies describing T-bet-independent development of IFN-γ-producing CD4+ T cells (Djuretic et al., 2007). Finally, similar to Th17 cell differentiation, Treg cell and CD4-IEL differentiation from mature naive CD4+ T cells also occurs in a mutually exclusive fashion (Mucida et al., 2013). Consistent with this pattern, Foxp3 induction was enhanced in several tissues from Tbx21-deficient CD4+ T cell hosts, suggesting a role for T-bet in the suppression of peripheral Treg differentiation (Figure 3G). Taken together, these data support a cell-intrinsic effect of T-bet on IEL programming.
Runx3 and T-bet Play Both Synergistic and Complementary Roles in Functional Differentiation of Intestinal T Cells
Transfer of primary CD4+ T cells from Cd4(DRunx3) mice into Rag1–/– hosts results in accelerated colitis and reduced loss of ThPOK or upregulation of CD8αα by these transferred CD4+ T cells (Reis et al., 2013). Furthermore, the congruence we observed in T-bet or Runx3-deficient CD4+ T cells in vitro suggests either interdependence or a hierarchy of these transcription factors in the IEL maturation process. To dissect the individual Runx3 and T-bet roles in IEL programming in vivo, we first overexpressed T-bet with Tbx21-encoding retrovirus (IRES-Thy1.1) in primary CD4+ T cells isolated from WT and Cd4(DRunx3) mice, both on Zbtb7b-GFP reporter backgrounds. CD4+ T cells were sorted based on Thy1.1 and GFP expression and adoptively transferred into Rag1–/– hosts. Supporting a role for T-bet in IEL differentiation, overexpression of Tbx21 resulted in enhanced CD8αα expression, CD103 induction, and ThPOK loss by WT Tbx21, when compared to WT (mock)-transferred cells (Figures 4A and 4B). However, in the absence of Runx3, these effects were abrogated, implying a nonredundant role for Runx3 in IEL programming (Figures 4A and 4B). Additionally, about 50% of ThPOKlo cells in the Cd4(DRunx3)(Tbx21) recipients did not express CD103, which indicates incomplete CD4-IEL programming even in a population that has already undergone ThPOK loss (Figure 4B).
Figure 4. Runx3 and T-bet Play Both Synergistic and Complementary Roles in Functional Differentiation of Intestinal T Cells.
(A–D) Sorted naive CD4+GFPhi T cells isolated from WT or Cd4DRunx3 Zbtb7b -GFP mice were adoptively transferred to Rag1–/– recipients, and analyzed 40 to 50 days later. Prior to transfer cells were transduced with Tbx21-expressing or empty control (mock) retrovirus. (A) CD8α, (B) CD103, and ThPOK expression by gated TCRβ+CD4+CD8β– small intestine IELs of recipient mice. (C) RORgt, T-bet, and (D) Foxp3 expression by TCRβ+CD4+CD8β– cells isolated from the mesenteric lymph node (mLN) and small intestine IEL compartment of recipient mice. Plots are representative of two independent experiments. (E) IL-17 and IFN-γ expression by sorted naive CD4+ T cells isolated from Tbx21–/– mice and infected with Tbx21-Thy1.1 (Tbx21), Runx3-GFP (Runx3), both (Tbx21+Runx3), or empty control (mock) retroviruses. Transduced cells were cultured in vitro for 4.5 days under Th17-polarizing differentiation conditions (TGF-β+IL-6+IL1β+IL-23). Plots are representative of two independent experiments. Error bars represent SEM.
In contrast to IEL differentiation, T-bet-mediated suppression of Th17 differentiation appeared to be Runx3-independent, because Tbx21 overexpression resulted in reduced levels of RORgt expression by CD4+ IELs even in transduced Cd4(ΔRunx3) cells (Figure 4C). Similarly, we found that T-bet inhibited Foxp3 expression whether or not Runx3 was present (Figure 4D). These results indicate that T-bet and Runx3 have complementary roles in the downregulation of ThPOK and the induction of the IEL differentiation program. This differentiation program is distinct from the T-bet-mediated suppression of Th17 and Treg differentiation, which was found to be independent of Runx3.
To further address whether the T-bet-Runx3 interplay is involved in the suppression of the CD4+ T helper program, we performed a rescue experiment in primary CD4+ T cells from Tbx21–/– mice and studied Th17 cell differentiation in vitro. Over-expression of T-bet by transduction with Tbx21-encoding retro-virus significantly suppressed IL-17 expression and allowed IFN-γ production by Tbx21–/– CD4+ T cells (Figure 4E). Consistent with previous reports (Djuretic et al., 2007; Wang et al., 2014), overexpression of Runx3 by infecting Tbx21–/– CD4+ T cells with retrovirus encoding the long-form of Runx3 led to an increased frequency of IFN-γ and IFN-γ-IL-17 double-producing CD4+ T cells but had very little effect on IL-17 single-producing cells (Figure 4E). Altogether, these data establish a role for T-bet in the suppression of Th17 and Treg cell induction in the context of IEL differentiation.
Hierarchical Roles for Runx3 and T-bet in IEL Differentiation
To investigate whether Runx3 could compensate for IEL-differentiation defects observed in T-bet-deficient cells, we performed rescue experiments with Tbx21–/– CD4+ T cells stimulated under TGF-β+RA+IFN-γ conditions. Because TGF-β has been shown to induce CD103 expression directly via Runx3 (Shi and Stavnezer, 1998), the absence of TGF-β-induced CD103 expression in Tbx21–/– cells (Figure 2B) indicated that T-bet plays a role in Runx3-dependent CD103 upregulation. Consistently, overexpression of Runx3 in Tbx21–/– CD4+ T cells efficiently rescued CD103 expression to the same level of WT(Runx3) cells (Figure 5A). Although Runx3 was essential for in vitro IEL differentiation, Runx1 overexpression was also able to rescue CD103 expression in Tbx21–/– CD4+ T cells (Figure 5A). To directly address the role of T-bet in Runx3 expression in vivo, we crossed Runx3-YFP knockin reporter mice with Tbx21–/– mice. We observed that IEL populations isolated from Tbx21–/– mice (red histograms) express lower amounts of Runx3 when compared to WT control (blue histograms) IELs (Figure 5B). These results were confirmed by quantitative PCR (qPCR) performed in sorted IEL populations, which showed drastically reduced levels of Runx3 mRNA expression by Tbx21–/– IELs when compared to WT controls (Figure 5C).
Figure 5. Hierarchical Roles for Runx3 and T-bet in IEL Differentiation.
(A) Histogram and mean fluorescence index (MFI) of CD103 expression by WT CD4+ T cells transduced with Runx1, Runx3, or an empty retrovirus (mock).
(B) Histogram of Runx3-YFP expression by small intestinal IEL populations from WT or Tbx21–/– (ΔTbx21) Runx3-YFP reporter mice.
(C) Runx3 expression by sorted small intestinal IEL populations from WT and Tbx21–/– mice.
(D and E) Anti-T-bet ChIP-qPCR for the −1 kb region of Up1, the −34 kb CNS region of Ifng, the −39 kb and the −17 kb Runx3 regulatory regions (D) or the Zbtb7b RBS 1 and 2 (E) in sorted peripheral CD4+ and CD8+ T cells from WT mice, Cd4(DRunx3) and Tbx21–/– mice. In (D), the regions −39 kb and −17 kb of Runx3 were only analyzed in cells from WT and Tbx21–/– mice. Error bars represent SD from pooled independent ChIP-qPCR experiments with biological replicates. (F and G) Anti-T-bet ChIP-qPCR (as in D and E) in sorted naive Vα2+CD4+ T cells isolated from WT OTII mice cocultured with DCs, OVA peptide, and IL-12 + α−IL-4-(Th1) or TGF-β+RA+IFN-γ (CD4CD8αα). Error bars represent SD from pooled independent ChIP-qPCR amplification analysis.
We next asked whether T-bet directly binds to the Runx3 locus, possibly regulating its expression. The −39 kb and the − 17 kb Runx3 regulatory regions were previously reported to contain a T-bet binding motif (Nakayamada et al., 2011). Therefore, we analyzed the binding of T-bet to these regions by chromatin immunoprecipitation (ChIP) in naive CD4+ and CD8+ T cells. We used T-bet binding to Up1 locus, as well as ChIP with Tbx21–/– cells as negative controls and T-bet binding to Ifng locus as positive control for the reactions. We found that T-bet binds to both −39 kb and −17 kb regions of Runx3 in CD4+ and CD8+ T cells, although T-bet binding was increased in CD8+ T cells when compared to CD4+ T cells particularly in the −17 kb region (Figure 5D). We also observed T-bet binding to Runx3 −39 kb and −17 kb regulatory regions of in vitro differentiated CD4+CD8αα IELs or Th1 cells (Figure 5F). These data provide a possible molecular mechanism behind the reduced CD103 and Runx3 expression observed in Tbx21–/– IELs, supporting a sequence of events where the binding of T-bet to Runx3 regulatory elements promotes Runx3 expression.
In addition to the modulation of Runx3 expression, our data also suggested T-bet-mediated effects on ThPOK downmodulation in developing CD4-IELs. We thus analyzed whether T-bet could also bind to the Zbtb7b regulatory binding sites 1 and 2 (RBS 1 and 2), known to silence Zbtb7b transcription upon Runx3 binding (Setoguchi et al., 2008). We found that in WT cells, T-bet binds to both RBS 1 and 2 of Zbtb7b in CD4+, but this is strongly enhanced in CD8+ T cells (Figure 5E). In line with reports showing protein cooperation between T-bet and Runx3 for CD8+ CTL differentiation (Cruz-Guilloty et al., 2009), the enhanced T-bet binding to Zbtb7b RBS in CD8+ T cells required Runx3 protein expression, because it was abrogated in CD8+ T cells isolated from Cd4(ΔRunx3) mice (Figure 5E). The above data suggest that reduced ThPOK and increased Runx3 expression are permissive for T-bet binding to Zbtb7b regulatory elements. Consistent with this hypothesis, we observed strong T-bet binding to RBS1 and 2 in CD4-IEL, but not in Th1 cells (Figure 5G). Altogether, the above data supports a Runx3-dependent, T-bet-mediated repression of Zbtb7b expression.
Retinoic Acid and T-bet-Inducing Cytokines Play Specific Roles in IEL Differentiation
Finally, we addressed the contribution and potentially differential effects of upstream T-bet-inducing signals involved in IEL differentiation. The T-bet inducing cytokine IL-15 and its receptor CD122 (IL-2Rβ or IL-15Rβ) have been shown to regulate crucial steps of terminal IEL development (Gangadharan et al., 2006; Pobezinsky et al., 2012; Suzuki et al., 1997). Because additional T-bet-inducing cytokines, IFN-γ and IL-27, were able to induce an IEL phenotype in vitro and because these cytokines are expressed in both healthy and inflamed intestine (Troy et al., 2009), we asked whether IFN-γ and IL-27 signaling are also required for IEL differentiation in vivo. Analysis of intestinal cells from naive Ifngr1–/– and Ifngr1+/– littermate and cage-mate control mice revealed a sharp decrease in the peripheral TCRαβ+CD4+CD8αα+ (TL-tetramer+, CD8β–) IELs in mice lacking IFN-γR (Figure 6A and B). Additionally, a significant decrease in the single-positive TCRαβ+CD4–CD8αα+ (TL-tetramer+, CD4–CD8β–) IEL population was found in mice lacking IFN-γR (Figures 6C and 6E). However, we did not observe significant differences in the frequency of TCRαβ+CD8β+CD8αα+ (CD4–) or TCRγδ+CD8αα+ (TL-tetramer+, CD4–CD8β–) IEL populations in Ifngr1–/– mice (Figures 6B–6E). Next, we analyzed intestinal cells from naive Il27ra–/– and Il27ra+/– littermate and cage-mate control mice. Contrary to Ifngr1–/– mice, animals lacking IL-27Rα (WSX-1) were found to have a decreased frequency of TCRαβ+CD8β+CD8αα+ and TCRγδ+CD8αα+ IELs, but had intact TCRαβ+CD4+CD8αα+ and single-positive CD8αα+ TCRαβ+ CD4− populations (Figures 6F–6J). Consistent with a T-bet-dependent process, we observed reduced T-bet expression in IEL populations isolated from either Ifngr1–/– or Il27ra–/– mice (data not shown). These observations suggest distinct and complementary roles for IFN-γ and IL-27 in the differentiation of several IEL populations.
Figure 6. Retinoic Acid and T-bet-Inducing Cytokines Play Specific Roles in IEL Differentiation.
(A–J) Ex vivo analysis of small intestinal IELs from 12- to 16-week-old Ifngr1–/– mice and Ifngr1+/– littermate and cage mate controls (A–E) or from 8- to 10-week-old Il27ra –/– mice and Il27ra+/– littermate and cage mate controls (F–J). (A and F) CD8α and CD103 expression by gated TCRαβ+CD4+CD8β– IELs. (B and G) TL-tetramer (CD8αα) quantification in induced IELs, TCRαβ+CD4+CD8β– (left), and TCRαβ+CD8β+ (right). (C and H) CD8αα and CD8β expression by gated TCRβ+CD4− IELs. (D and I) Ratio of TCRγδ+ to TCRαβ+ among CD45+ cells. (E and J) CD8αα quantification in natural IELs, TCRγδ+ (left), and TCRαβ+CD4–CD8β– (right).
(K–M) Sorted naive Va2+CD4+ T cells isolated from WT and (OTII) dominant-negative RA receptor (OTII [dnRara]) Zbtb7b -GFP reporter mice were cocultured with DCs, OVA peptide, and indicated cytokines. (K) Histograms of T-bet expression by OTII cells from WT (OTII) or (OTII[dnRara]).
(L) CD8β and CD8α expression by CD4+ T cells from OTII (left) or OTII (dnRara) (right) mice.
(M) Histogram of ThPOK (left) and CD103 (right) expression by gated CD4+CD8β– OTII cells. Plots are representative of three independent experiments. Statistical significance analyzed by Student's t test of pooled experiments. Error bars represent SEM. *p < 0.05; **p < 0.01; ***p < 0.001.
In addition to IFN-γ and IL-27, RA has also been shown to induce T-bet expression under conditions of TGF-β exposure in vitro (Takahashi et al., 2012). Furthermore, we recently showed that mice conditionally expressing a dominant-negative form of the RA receptor RARα in CD4+ T cells (Cd4[dnRaralsl/lsl mice]) (Rajaii et al., 2008) lack a CD4+CD8αα+ IEL population (Reis et al., 2013) suggesting an important role for RA signaling in the induction of the IEL phenotype in gut CD4+ T cells. To directly assess the contribution of RARa signaling to the induction of T-bet-dependent CD4-IEL maturation, we crossed OTII mice with Cd4(dnRaralsl/lsl) mice on a Zbtb7b-GFP background (OTII [dnRara]). We found reduced T-bet upregulation by OTII (dnRara) cells upon exposure to TGF-β and RA plus IFN-γ conditions when compared to WT OTII cells (Figure 6K). Consistent with this finding, TGF-β and RA plus IFN-γ enhanced CD8α and CD103 and suppressed ThPOK expression in WT OTII cells, but not in cells expressing dnRara (Figures 6L and 6M).
Overall, these results support a feedforward mechanism, where T-bet-inducing cytokines lead to T-bet-mediated upregulation of Runx3 via −39 kb and −17kb regulatory elements, after which T-bet and Runx3 have complementary roles in the down-regulation of ThPOK and the induction of the IEL differentiation program.
DISCUSSION
A single layer of epithelial cells separates the intestinal lumen from the lamina propria. IELs, which reside at this intersection between the external environment and the body, therefore constitute the lymphocyte population most exposed to environmental perturbations. This study provides insights into previously unknown molecular requirements for IEL maturation. We showed that a cytokine milieu resembling the gut environment induces an IEL program in T cells in a T-bet-dependent manner. This was confirmed by in vivo experiments in mice lacking T-bet, IL-27Rα, or IFN-γR, which displayed consequent defects in IEL development. Finally, we described synergistic and complementary effects of T-bet and Runx3 in this process.
The most pronounced effect of T-bet deficiency was the sharp decrease in the CD8αα-expressing IEL populations. Indeed, CD8αα expression represents a common feature of IELs (Cheroutre and Lambolez, 2008). The so-called “natural” or “thymic” IELs (TCRαβ+CD8αα+ and TCRγδ+CD8αα+) are present at birth and express several molecules associated with downregulated antigen reactivity and with anti-inflammatory activity (Cheroutre et al., 2011; Denning et al., 2007; Gangadharan et al., 2006). Induced IELs (TCRαβ+ CD4+8aa+ and CD8β+CD8αα+), similar to induced Treg cells, expand upon antigen encounter in the periphery (Huang et al., 2011; Mucida et al., 2013). The findings presented here link T-bet to these two temporally and geographically distinct IEL differentiation processes. The drastic decrease in the single-positive CD8αα+TCRαβ+ population in Tbx21–/– mice could indicate a role for T-bet during T cell agonist selection in the thymus (Leishman et al., 2002; Yamagata et al., 2004). It is possible that T-bet is required for developmental steps in pre-committed CD4+CD8β+CD8αα+ triple-positive (TP) thymocytes that differentiate toward a CD4–CD8– double-negative (TCRβ+) phenotype before leaving the thymus (Gangadharan et al., 2006). Alternatively, it is plausible that T-bet participates in later steps of IEL development when precommitted precursors arrive in the gut environment and upregulate Runx3, requiring IL-15 for their terminal differentiation (Gangadharan et al., 2006; Pobezinsky et al., 2012). Our data support the latter possibility, extending the range of T-bet inducers involved in this process to RA, IFN-γ, and IL-27. Additionally, Konkel et al. described an essential role for TGF-β in the development of single-positive CD8αα+ IELs (Konkel et al., 2011). Our data suggest that the cooperation between TGF-β, which suppresses T-bet but induces Runx3 (Shi and Stavnezer, 1998), and a T-bet-inducing environment, most likely containing RA, IFN-γ, and/or IL-27, in addition to IL-15, is involved in the differentiation of T cells into IELs. This IEL programing generates an “activated yet resting” state, which involves the expression of the TCR corepressor CD8αα and several molecules associated with both chronic activation and limited inflammatory capacity (Denning et al., 2007; Yamagata et al., 2004).
The significant decrease in the “induced” IEL populations also corroborates a role for T-bet in peripheral activation of mature T cells and their differentiation into IELs. We focused most of our studies on the intestinal process that redirects peripheral CD4+ T cells toward a CD8-like IEL phenotype (Mucida et al., 2013; Reis et al., 2013). We addressed how CD4+ T helper activity is suppressed during CD4-IEL differentiation and defined the molecular requirements for the induction of the IEL program. We found that a combination of environmental cues, particularly TGF-β, RA, T-bet-inducing cytokines, and transcriptional regulators, including T-bet and Runx3, can mimic signals from the gut microenvironment to induce CD4-IEL differentiation, a process distinct from T-bet-mediated Th1 differentiation or TGF-β-mediated Treg induction. Contrary to Treg-mediated control of inflammatory responses, the pathway addressed by this study relates to cell-intrinsic control of the inflammatory potential of T cells. A recent report by Fuchs and coworkers described a TGF-β-responsive, T-bet-dependent population of innate lymphoid cells present in the intraepithelial compartment of the gut that quickly responds to environmental stimuli by producing IFN-γ (Fuchs et al., 2013). This population also has features of IELs, and it is tempting to speculate that these cells could be involved in the tissue-specific, terminal differentiation of T cells or in their modulation within the epithelial layer. Several cell types could serve as additional sources of IFN-γ in the gut under physiological or pathological conditions, from IELs to lamina propria NK cells or T cells. Whether T-bet expression by intestinal T cells oscillates during events that trigger IFN-γ and additional T-bet-inducing cytokines, such as IL-15 and IL-27, and the consequences of such oscillations remain to be investigated. Nevertheless, our data indicate a differential requirement for IFN-γR- or IL-27Rα-mediated signaling by IEL subpopulations. Likewise, additional studies are needed to dissect the role of the indigenous microbiota in induction of the IEL phenotype. Although natural IELs are mostly stable under germ-free conditions, our recent report showing drastic reduction in the induced CD4+CD8αα+ IEL population in germ-free mice, but recovery upon microbial reconstitution, points to commensal bacteria as essential players (Mucida et al., 2013).
To undergo CD4-IEL differentiation, peripheral CD4+ T cells lose main features of CD4+ helper cells. We showed that T-bet suppresses both Th17 and Treg cell-associated genes during CD4-IEL differentiation, independent of Runx3 expression. Previous reports have shown analogous suppressive effects of T-bet during Th2, Th17, and Treg cell differentiation. These functions require T-bet-mediated silencing of respective “master regulator” transcription factors; i.e., GATA-3 in Th2 cells, RORgt in Th17 cells and Foxp3 in Treg cells (Djuretic et al., 2007; Koch et al., 2009; Lazarevic et al., 2011; Zhu et al., 2012). A recent study described that cooperation between T-bet and Runx transcription factors is required for IFN-γ production by pathogenic Th17 cells (Wang et al., 2014). Although we did observe increased frequency of IFN-γ-IL-17 double-producing cells in T-bet deficient animals, these cells did not induce intestinal inflammation in a murine model of transfer colitis. Further characterization of pathogenic CD4+ T cells in colitis and the role of T-bet in this process is therefore necessary.
In our studies, T-bet also supported an IEL program in CD4+ T cells by enhancing ThPOK loss and CD8αα expression. These effects could be direct, through T-bet binding to Zbtb7b regulatory elements, or indirect, by binding and modulation of Runx3 expression. Runx3 protein was required for T-bet binding to Zbtb7b regulatory elements, which implies significance for T-bet-Runx3 protein-protein interaction, similar to what was described for Th1 and CD8+ T cell effector differentiation (Cruz-Guilloty et al., 2009; Djuretic et al., 2007). T-bet is predicted to bind several other genes related to IELs, including Itgae (CD103) and Crtam (Nakayamada et al., 2011), although our data indicate that expression of Runx3 might be required for T-bet-mediated regulation of these genes, as well. This hypothesis is also supported by our in vivo analysis of Tbx21 overexpression in transferred WT versus ΔRunx3 CD4+ T cells and by the requirement for TGF-β in CD8αα+ IEL development (Konkel et al., 2011).
Master regulator transcription factors play defining roles in the initial differentiation process of all main T cell lineages, including CD4+ T cell helper and regulatory subsets. This study shows that cooperation between master regulators might also influence the late functional maturation of T cells, allowing for plasticity in the context of different environmental perturbations. In particular, we uncovered how diverse signals from the highly perturbed intestinal mucosa are integrated during intestinal lymphocyte functional differentiation, allowing cell-intrinsic control of inflammatory potential.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6, OTII TCR transgenic, Rag1–/–, Ifngr1–/–, Il27ra–/–, and Tbx21–/– mice were purchased from the Jackson Laboratories and maintained in our facilities. Cd4(ΔRunx3) mice were described previously (Reis et al., 2013). Dominant-negative Raralsl/lsl, Runx3-YFP, and Zbtb7b-GFP reporter mice were generously provided by R. Noelle (Rajaii et al., 2008), D. Littman, and I. Taniuchi, respectively. Several of these lines were interbred in our facilities to obtain the final strains described in the text. Mice were maintained at The Rockefeller University animal facilities under specific pathogen-free conditions, and sentinel mice from the Rag1–/– mouse colony were tested to be negative for Helicobacter spp. and C. rodentium. Mice were used at 7–12 weeks of age for all experiments except when otherwise indicated. Animal care and experimentation were consistent with the NIH guidelines and were approved by the Institutional Animal Care and Use Committee at The Rockefeller University.
In Vitro T Cell Culture
Naive (defined as CD4+CD25–CD62hiCD44lo) T cells were sorted with a FACS Aria cell sorter (Becton Dickinson) and cultured for 4.5 days in 96-well plates precoated with 2 mg/ml of anti-CD3ε (17A2) and 1 mg/ml of soluble anti-CD28 (37.51) with indicated cytokines (all from R&D). OTII cells were cocultured with magnetic bead-isolated (MACS, Miltenyi Biotec) CD11c+ splenic DCs in the presence of 500 nM OVA peptide and indicated cytokines. For details, see Supplemental Experimental Procedures.
Retroviral Transduction of CD4+ T Cells
Retroviral vectors for Runx1 and Runx3 were previously described (Muroi et al., 2008; Naoe et al., 2007). Retroviral vectors for T-bet were generously provided by L. Glimcher (Cornell) and were previously described (Lazarevic et al., 2011). Transduction of CD4+ T cells with retroviral vectors was performed as previously described (Muroi et al., 2008; Naoe et al., 2007). For details, see Supplemental Experimental Procedures.
Experimental Colitis Model
Colitis was induced after transfer of 5 × 105 sorted naive T cells into Rag1–/– mice, as previously described (Mucida et al., 2007). Recipient mice were monitored regularly for signs of disease, including weight loss, hunched appearance, piloerection of the coat, and diarrhea, and analyzed at various times after the initial transfer or when they reached 80% of their initial weight.
Chromatin Immunoprecipitation qPCR
ChIP was performed as previously described (Setoguchi et al., 2008; Wang et al., 2014). Briefly, chromatin was prepared from 5–10 × 106 ex vivo-isolated cells or from 3 × 106 in vitro differentiated Th1 and CD4+CD8αα+ cells. Cells were fixed with 1% (vol/vol) methanol-free formaldehyde and lysed. The chromatin was sonicated to obtain fragments around 500 bp–1 kb in length and incubated with Protein A beads coupled to anti-T-bet antibody (H-210) overnight at 4°C. Immunoprecipitated chromatin was treated with Proteinase K overnight at 56°C. DNA was extracted by phenol-chloroform extraction followed by ethanol precipitation. Extracted DNA was analyzed by real-time PCR with different primer sequences. For details, see Supplemental Experimental Procedures.
Statistics
Statistical analysis was performed in GraphPad Prism software. Data were analyzed by applying one-way ANOVA or unpaired Student's t test whenever necessary. A p value of less than 0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to K. Velinzon and Y. Shatalina for sorting cells and members of the Nussenzweig laboratory and The Rockefeller University employees for continuous assistance. We thank L. Glimcher (Cornell) and I. Taniuchi (Riken-RCAI) for vectors and H. Cheroutre (LIAI) for TL-tetramers. We especially thank A. Rogoz for outstanding technical support, Y. Gultenkin for help with in vitro assays, and V. Hou (FCCC) for help with mouse breeding and IL-27Rα ex vivo analysis. We thank G. Kim (LIAI) and members of our laboratory, particularly V. Pedicord and D. Esterhazy, for discussions, critical reading, and editing of the manuscript. D.M. is supported by an Ellison Medical Foundation New Scholar Award in Aging, an Irma T. Hirschl Award, a Crohn's & Colitis Foundation of America Senior Research Award, and a National Institutes of Health NIH R01 DK093674-02 grant. B.S.R. is supported by the Leona M. and Harry B. Helmsley Charitable Trust. D.P.H.v.K. is supported by the Dutch Digestive Foundation. S.I.G. is supported by a NIH R00DK088589, a FCCC-Temple University Nodal grant, and by the Pew Scholar in Biomedical Sciences Program.
Footnotes
AUTHOR CONTRIBUTIONS
D.M. conceived and supervised this study. B.S.R. and D.M. designed and performed experiments. S.I.G. helped with designing and performing Ifngr1 and Il27ra experiments. B.S.R. prepared figures and helped with manuscript preparation. D.P.H.v.K. assisted with performing experiments and helped with preparing figures and the manuscript. D.M. wrote the paper.
SUPPLEMENTAL INFORMATION
Supplemental Information includes one figure and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2014.06.017.
REFERENCES
- Cheroutre H, Lambolez F. Doubting the TCR coreceptor function of CD8alphaalpha. Immunity. 2008;28:149–159. doi: 10.1016/j.immuni.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 2011;11:445–456. doi: 10.1038/nri3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, Rao A. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 2009;206:51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G, Augustine MM, Das J, Bottomly K, Ray P, Ray A. An important regulatory role for CD4+CD8 alpha alpha T cells in the intestinal epithelial layer in the prevention of inflammatory bowel disease. Proc. Natl. Acad. Sci. USA. 2003;100:5324–5329. doi: 10.1073/pnas.0831037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning TL, Granger SW, Mucida D, Graddy R, Leclercq G, Zhang W, Honey K, Rasmussen JP, Cheroutre H, Rudensky AY, Kronenberg M. Mouse TCRalphabeta+CD8alphaalpha intraepithelial lymphocytes express genes that down-regulate their antigen reactivity and suppress immune responses. J. Immunol. 2007;178:4230–4239. doi: 10.4049/jimmunol.178.7.4230. [DOI] [PubMed] [Google Scholar]
- Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-g-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadharan D, Lambolez F, Attinger A, Wang-Zhu Y, Sullivan BA, Cheroutre H. Identification of pre- and postselection TCRalphabeta+ intraepithelial lymphocyte precursors in the thymus. Immunity. 2006;25:631–641. doi: 10.1016/j.immuni.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Guy-Grand D, Malassis-Seris M, Briottet C, Vassalli P. Cytotoxic differentiation of mouse gut thymodependent and independent intraepithelial T lymphocytes is induced locally. Correlation between functional assays, presence of perforin and granzyme transcripts, and cytoplasmic granules. J. Exp. Med. 1991;173:1549–1552. doi: 10.1084/jem.173.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy-Grand D, Vassalli P, Eberl G, Pereira P, Burlen-Defranoux O, Lemaitre F, Di Santo JP, Freitas AA, Cumano A, Bandeira A. Origin, trafficking, and intraepithelial fate of gut-tropic T cells. J. Exp. Med. 2013;210:1839–1854. doi: 10.1084/jem.20122588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Park Y, Wang-Zhu Y, Larange A, Arens R, Bernardo I, Olivares-Villagómez D, Herndler-Brandstetter D, Abraham N, Grubeck-Loebenstein B, et al. Mucosal memory CD8+ T cells are selected in the periphery by an MHC class I molecule. Nat. Immunol. 2011;12:1086–1095. doi: 10.1038/ni.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel JE, Maruyama T, Carpenter AC, Xiong Y, Zamarron BF, Hall BE, Kulkarni AB, Zhang P, Bosselut R, Chen W. Control of the development of CD8aa+ intestinal intraepithelial lymphocytes by TGF-b. Nat. Immunol. 2011;12:312–319. doi: 10.1038/ni.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgt. Nat. Immunol. 2011;12:96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leishman AJ, Gapin L, Capone M, Palmer E, MacDonald HR, Kronenberg M, Cheroutre H. Precursors of functional MHC class I- or class II-restricted CD8alphaalpha(+) T cells are positively selected in the thymus by agonist self-peptides. Immunity. 2002;16:355–364. doi: 10.1016/s1074-7613(02)00284-4. [DOI] [PubMed] [Google Scholar]
- Meresse B, Malamut G, Cerf-Bensussan N. Celiac disease: an immunological jigsaw. Immunity. 2012;36:907–919. doi: 10.1016/j.immuni.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- Mucida D, Husain MM, Muroi S, van Wijk F, Shinnakasu R, Naoe Y, Reis BS, Huang Y, Lambolez F, Docherty M, et al. Transcriptional reprogramming of mature CD4+ helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat. Immunol. 2013;14:281–289. doi: 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, Kawamoto H, Taniuchi I. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat. Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, Taniuchi I. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J. Exp. Med. 2007;204:1749–1755. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurath MF, Weigmann B, Finotto S, Glickman J, Nieuwenhuis E, Iijima H, Mizoguchi A, Mizoguchi E, Mudter J, Galle PR, et al. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn's disease. J. Exp. Med. 2002;195:1129–1143. doi: 10.1084/jem.20011956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobezinsky LA, Angelov GS, Tai X, Jeurling S, Van Laethem F, Feigenbaum L, Park JH, Singer A. Clonal deletion and the fate of autoreactive thymocytes that survive negative selection. Nat. Immunol. 2012;13:569–578. doi: 10.1038/ni.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powrie F, Leach MW, Mauze S, Menon S, Caddle LB, Coffman RL. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity. 1994;1:553–562. doi: 10.1016/1074-7613(94)90045-0. [DOI] [PubMed] [Google Scholar]
- Rajaii F, Bitzer ZT, Xu Q, Sockanathan S. Expression of the dominant negative retinoid receptor, RAR403, alters telencephalic progenitor proliferation, survival, and cell fate specification. Dev. Biol. 2008;316:371–382. doi: 10.1016/j.ydbio.2008.01.041. [DOI] [PubMed] [Google Scholar]
- Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, Mucida D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4+ T cell immunity. Nat. Immunol. 2013;14:271–280. doi: 10.1038/ni.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- Shi MJ, Stavnezer J. CBF alpha3 (AML2) is induced by TGF-beta1 to bind and activate the mouse germline Ig alpha promoter. J. Immunol. 1998;161:6751–6760. [PubMed] [Google Scholar]
- Suzuki H, Duncan GS, Takimoto H, Mak TW. Abnormal development of intestinal intraepithelial lymphocytes and peripheral natural killer cells in mice lacking the IL-2 receptor beta chain. J. Exp. Med. 1997;185:499–505. doi: 10.1084/jem.185.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kanno T, Nakayamada S, Hirahara K, Sciumè G, Muljo SA, Kuchen S, Casellas R, Wei L, Kanno Y, O'Shea JJ. TGF-b and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat. Immunol. 2012;13:587–595. doi: 10.1038/ni.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Chen Z, Ciszewski C, Setty M, Solus J, Tretiakova M, Ebert E, Han J, Lin A, Guandalini S, et al. Cytosolic PLA2 is required for CTL-mediated immunopathology of celiac disease via NKG2D and IL-15. J. Exp. Med. 2009;206:707–719. doi: 10.1084/jem.20071887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy AE, Zaph C, Du Y, Taylor BC, Guild KJ, Hunter CA, Saris CJ, Artis D. IL-27 regulates homeostasis of the intestinal CD4+ effector T cell pool and limits intestinal inflammation in a murine model of colitis. J. Immunol. 2009;183:2037–2044. doi: 10.4049/jimmunol.0802918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Godec J, Ben-Aissa K, Cui K, Zhao K, Pucsek AB, Lee YK, Weaver CT, Yagi R, Lazarevic V. The transcription factors T-bet and Runx are required for the ontogeny of pathogenic interferon-g-producing T helper 17 cells. Immunity. 2014;40:355–366. doi: 10.1016/j.immuni.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata T, Mathis D, Benoist C. Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha alpha lineage with characteristics of innate immune cells. Nat. Immunol. 2004;5:597–605. doi: 10.1038/ni1070. [DOI] [PubMed] [Google Scholar]
- Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, Hu G, Guo L, Yagi R, Yamane H, Punkosdy G, et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 2012;37:660–673. doi: 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.