A large variety of organisms are capable of synthesizing hard matter in a process called biomineralization [1]. The transformation of a genetic blueprint into minerals such as, for example, calcium phosphate in bones and calcium carbonate in eggs or seashells provides a mechanical support for organismic growth and protection against predators, respectively. Iron oxides formed by fishes and birds provide them with magnetic properties used for magnetoreception and orientation [2], [3]. The biomineralization processes are remarkable for numerous reasons: organisms, contrary to engineers, have to form these biological materials with a limited subset of biologically available chemical elements and at physiological conditions. Still, these reduced means are not at the detriment of their function, which often surpasses man-made materials based on equivalent elements [4]. Therefore, understanding how biomineralizing organisms process chemical elements based on their genetic program is of primary interest. However, the biological mechanisms behind biomineralization have remained unclear, partly because of limited genetic knowledge: model organisms are limited to a few unicellular organisms [5], [6]. Therefore, the question has arisen of what genetic approach to use to get genetic information about the large majority of organisms that have remained intractable.
Magnetotactic Bacteria: Simply Microorganisms, but Not So Simple
The recent advances in sequencing techniques now offer the opportunity to bypass some of the restrictions associated with the unavailability of genetic systems to get novel insights into important microbial processes such as those associated with biomineralization. In their study, Rahn-Lee et al. [7] combine established genetic techniques (random mutagenesis) with modern sequencing platforms to understand magnetite biomineralization in the magnetotactic bacteria Desulfovibrio magneticus RS-1. Magnetotactic bacteria are microorganisms able to form intracellular magnetic nanoparticles made of the iron oxide magnetite (Fe3O4) or the iron sulfide greigite (Fe3S4) [8]. The nanoparticles together with their membrane envelope are called magnetosomes. They have strain-specific sizes and morphologies and are typically arranged in chains in order to form a magnetic dipole strong enough to passively orient the bacteria along the magnetic field lines of the Earth, a process called magnetotaxis [9].
The genomes of numerous strains have been sequenced [10]. However, genetic tools permitting the manipulation of the microorganisms are only available for two magnetospirilla strains: Magnetospirillum gryphiswaldense [11] and M. magneticum [12]. In these strains, the magnetosomes are formed and arranged thanks to a subset of genes called the magnetosome island [13], which, in particular, encompasses the mamAB, mamGFDC, mms6, and mamXY operons (Fig. 1A) [14], [15]. However, RS-1, the strain studied by Rahn-Lee et al. [7], forms elongated magnetosomes in contrast to magnetospirilla, which form cubooctaedral magnetosomes. Therefore, RS-1 would be a model organism to use to study the functional diversity of compartmentalization and biomineral formation in magnetotactic bacteria, providing tools were available to do so.
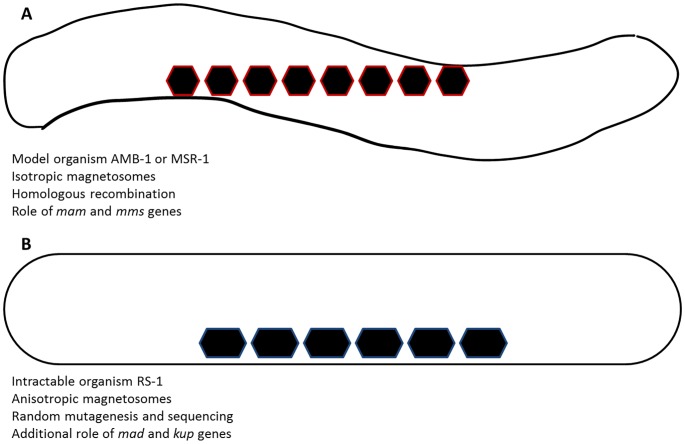
Figure 1. Sketch of magnetite biomineralization and associated differences.
In the model organisms (magnetospirilla, AMB-1 or MSR-1), isotropic magnetosomes are produced. Genetic studies have highlighted the roles of mam and mms genes in the process (A). In turn, intractable organisms such as RS-1, where elongated magnetosomes are produced, could so far not be genetically studied. By random mutagenesis and whole-genome sequencing, Rahn-Lee et al. (2015) showed the additional role of kup and mad genes in the process, possibly in their morphology control of the nanoparticles (B).
Getting Genetic Information in an Intractable Organism
The authors used random mutagenesis to generate nonmagnetic mutants and combined it with whole-genome re-sequencing to identify the mutated genes. In particular, Rahn-Lee et al. [7] first cultivated RS-1 in conditions where the microorganism no longer formed magnetosomes and then performed UV and chemical mutagenesis. Since screening a large number of colonies was impractical, they employed a two-step strategy that consisted of first selecting in liquid to increase the proportion of non-magnetic cells in the population and then only screening for single colonies of non-magnetic phenotypes [7]. These colonies of no- or low-magnetism were then analyzed by whole-genome sequencing to determine the causative genetic change. After the mutation for each strain was identified, the authors used PCR and Sanger sequencing to check for this change in the other strains isolated from the same outgrowth and analyzed those strains that were not clones by whole-genome sequencing to determine their mutation.
This approach led to the isolation of about 30 mutants, with mutations in genes shared amongst all magnetotactic bacteria, but also, more interestingly, with mad genes that are unique to the magnetotactic δ-proteo bacteria and even genes potentially unique to RS-1. The group of A. Komeili found that a potassium transporter (kup) is important for biomineralization of magnetite (Fig. 1B) [7], a surprising discovery since there is a priori no reason to expect the involvement of potassium in an iron oxide mineral. The authors, in addition, presented the first experimental proof of the involvement of mad genes in the control of the magnetosome morphology. This is an important confirmation since a bioinformatic study proposed earlier that these so-called mad genes could be responsible for the morphology control observed in some strains, since these genes are specifically found in magnetotactic δ-proteobacteria-forming elongated magnetosomes such as RS-1, and not in magnetospirilla [10].
In conclusion, the general methodology presented here will be of immediate relevance to other scientists working with fastidious and genetically intractable organisms, not limited to biomineralizing ones. In addition, the study delivers significant advancements for the understanding of biomineralization and its variety in prokaryotes by presenting the first genetic analysis of magnetotactic bacteria outside of the commonly studied α-proteobacteria. However, as random mutagenesis is stochastic and not directed, important genes might remain unprobed and, therefore, their role might possibly be overlooked by this method. Therefore, efforts in the development of genetic tools should not be abandoned. In addition, complementation of this approach by physical and chemical analytical techniques in the near future will enable the complete multidisciplinary understanding of biomineralization in different strains of magnetotactic bacteria.
Funding Statement
This work was supported by the European Research Council (Starting Grant MB2 n°256915 to DF). The funder had no role in the preparation of the article.
References
- 1. Lowenstam HA (1981) Minerals formed by organisms. Science 211: 1126–1131. [DOI] [PubMed] [Google Scholar]
- 2.Baeuerlein E (2007) The Biology of Biominerals Structure Formation; Baeuerlein E, editor. Weinheim: Wiley-VCH.
- 3.Baeuerlein E, Epple M (2007) Biomineralization in Medicine; Baeuerlein E, editor. Weinheim: Wiley-VCH.
- 4. Fratzl P (2007) Biomimetic materials research: what can we really learn from nature's structural materials? J R Soc Interface 4: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, et al. (2004) The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 306: 79–86. [DOI] [PubMed] [Google Scholar]
- 6. Matsunaga T, Okamura Y, Fukuda Y, Wahyudi AT, Murase Y, et al. (2005) Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp strain AMB-1. DNA Res 12: 157–166. [DOI] [PubMed] [Google Scholar]
- 7. Rahn-Lee L, Byrne M E, Zhang M, Le Sage D, Glenn D R, et al. (2015) A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation. PLoS Genet 11(1): e1004811 doi:10.1371/journal.pgen.1004811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Faivre D, Schüler D (2008) Magnetotactic Bacteria and Magnetosomes. Chem Rev 108: 4875–4898. [DOI] [PubMed] [Google Scholar]
- 9. Lefèvre C T, Bennet M, Landau L, Vach P, Pignol D, et al. (2014) Diversity of Magneto-Aerotactic Behaviors and Oxygen Sensing Mechanisms in Cultured Magnetotactic Bacteria. Biophysical Journal 107: 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lefevre C T, Trubitsyn D, Abreu F, Kolinko S, Jogler C, et al. (2013) Comparative genomic analysis of magnetotactic bacteria from the Deltaproteobacteria provides new insights into magnetite and greigite magnetosome genes required for magnetotaxis. Environ Microbiol 15: 2712–2735. [DOI] [PubMed] [Google Scholar]
- 11. Schultheiss D, Schüler D (2003) Development of a genetic system for Magnetospirillum gryphiswaldense. Archives Microbiol 179: 89–94. [DOI] [PubMed] [Google Scholar]
- 12. Matsunaga T, Nakamura C, Burgess JG, Sode K (1992) Gene transfer in magnetic bacteria: transposon mutagenesis and cloning of genomic DNA fragments required for magnetosome synthesis. J Bacteriol 174: 2748–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D (2005) A Hypervariable 130-Kilobase Genomic Region of Magnetospirillum gryphiswaldense Comprises a Magnetosome Island Which Undergoes Frequent Rearrangements during Stationary Growth. J Bacteriol 187: 7176–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lohße A, Borg S, Raschdorf O, Kolinko I, Tompa E, et al. (2014) Genetic Dissection of the mamAB and mms6 Operons Reveals a Gene Set Essential for Magnetosome Biogenesis in Magnetospirillum gryphiswaldense. J Bacteriol 196: 2658–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murat D, Quinlan A, Vali H, Komeili A (2010) Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc Natl Acad Sci USA 107: 5593–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]