Abstract
Receptor Tyrosine Kinase (RTK) signaling pathways induce multiple biological responses, often by regulating the expression of downstream genes. The HMG-box protein Capicua (Cic) is a transcriptional repressor that is downregulated in response to RTK signaling, thereby enabling RTK-dependent induction of Cic targets. In both Drosophila and mammals, Cic is expressed as two isoforms, long (Cic-L) and short (Cic-S), whose functional significance and mechanism of action are not well understood. Here we show that Drosophila Cic relies on the Groucho (Gro) corepressor during its function in the early embryo, but not during other stages of development. This Gro-dependent mechanism requires a short peptide motif, unique to Cic-S and designated N2, which is distinct from other previously defined Gro-interacting motifs and functions as an autonomous, transferable repressor element. Unexpectedly, our data indicate that the N2 motif is an evolutionary innovation that originated within dipteran insects, as the Cic-S isoform evolved from an ancestral Cic-L-type form. Accordingly, the Cic-L isoform lacking the N2 motif is completely inactive in early Drosophila embryos, indicating that the N2 motif endowed Cic-S with a novel Gro-dependent activity that is obligatory at this stage. We suggest that Cic-S and Gro coregulatory functions have facilitated the evolution of the complex transcriptional network regulated by Torso RTK signaling in modern fly embryos. Notably, our results also imply that mammalian Cic proteins are unlikely to act via Gro and that their Cic-S isoform must have evolved independently of fly Cic-S. Thus, Cic proteins employ distinct repressor mechanisms that are associated with discrete structural changes in the evolutionary history of this protein family.
Author Summary
Understanding the evolution of developmental regulatory mechanisms is a central challenge of biology. Here we uncover a newly evolved mechanism of transcriptional repression by Capicua (Cic), a conserved sensor of Receptor Tyrosine Kinase (RTK) signaling. In Drosophila, Cic patterns the central regions of the embryo by repressing genes induced by Torso RTK signaling at the poles. We show that Cic performs this function by recruiting the Groucho (Gro) corepressor and that this mechanism is an evolutionary innovation of dipteran insects. Indeed, we find that recruitment of Gro depends on a short motif of Cic (N2) specific to dipterans. Strikingly, moreover, the form of Cic that existed before the origin of dipterans is completely inactive in fly embryos, whereas the equivalent form carrying N2 displays significant function. This suggests that evolution of the N2 motif caused a fundamental change in Cic repressor activity, which we propose has enabled the complex roles of Cic, Gro and Torso signaling in fly embryonic patterning. In contrast, Cic functions independently of Gro in other Drosophila tissues and probably also in mammals, where Cic lacks the N2 sequence. Thus, our results illustrate the structural and evolutionary origins of essential functional variations within a highly conserved family of developmental regulators.
Introduction
Receptor Tyrosine Kinase (RTK) signaling pathways regulate tissue development and morphogenesis in all metazoans [1]. RTKs often signal through the conserved Ras-Raf-MAPK cascade, leading to phosphorylation of nuclear transcription factors which then elicit changes in target gene expression. The HMG-box protein Capicua (Cic) has recently emerged as a general nuclear sensor of RTK signaling pathways [2]. Originally discovered downstream of the Torso RTK in Drosophila embryogenesis, Cic has been subsequently shown to function downstream of other RTKs at multiple stages of fly development [3]–[11]. In all cases, Cic represses transcription of RTK-responsive genes in unstimulated cells, whereas activation of RTK signaling results in phosphorylation and downregulation of Cic and this causes derepression of its target genes [7], [10], [12], [13].
Cic is highly conserved from cnidarians to vertebrates and is implicated in several human pathologies such as spinocerebellar ataxia type 1 (SCA1) and oligodendroglioma (OD) [14]–[17]; reviewed in [2]. Indeed, Cic proteins from Drosophila and mammals share many functional and structural properties: they repress transcription by binding to related DNA sites in target genes, appear to be similarly downregulated by RTKs and are expressed as two main isoforms, short (Cic-S) and long (Cic-L), which differ in their N-terminal regions [7], [9], [10], [14], [15], [17]–[19]. However, despite these similarities, it is currently unclear whether all Cic family proteins employ a common mechanism of repression. Studies in mouse and human cells have shown that Cic associates with Ataxin1 (Atxn1), a co-repressor involved in SCA1 [14], [15], [17], [20], [21]. On the other hand, previous studies in Drosophila have suggested that Cic functions together with Groucho (Gro) [3], [10], a WD-repeat co-repressor that associates with multiple repressors, including Hairy/Hes, Nkx, Lef/Tcf and Runx family proteins (reviewed in [22], [23]). However, the functional links between Cic and Gro remain unclear, since no molecular interaction between these proteins has been validated in vivo [2], [24].
Here, we investigate the mechanism of Drosophila Cic repression and its relationship with Gro. We find that Cic functions via Gro in the early embryo but not at other developmental stages. The Gro-assisted mechanism depends on a previously unrecognized motif of Cic (N2), which is essential for recruitment of Gro in vivo. Remarkably, the N2 motif is highly conserved among Cic orthologues in flies and mosquitoes, but is absent in all other species, suggesting that it originated in ancestral dipterans. Furthermore, the N2 domain appears to be a structural innovation associated with the emergence of fly Cic-S isoforms from a pre-existing Cic-L-like isoform. This implies that mammalian Cic proteins, which lack the N2 motif, probably function independently of Gro, and that their Cic-S isoforms must have evolved independently of fly Cic-S. Thus, Cic proteins exhibit context-dependent repressor activities that are partly associated with key structural changes that have occurred during the evolution of this protein family.
Results
Context-dependent activities of Cic in Drosophila development
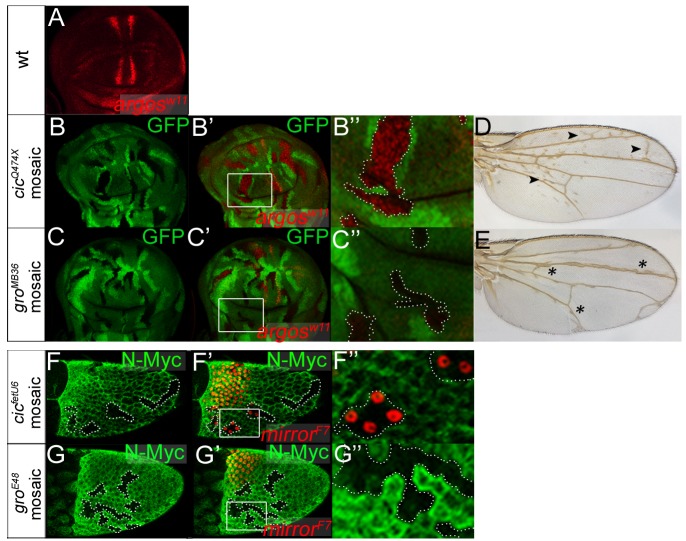
Cic and Gro are both essential for repression of two terminal gap genes, tailless (tll) and huckebein (hkb), in central regions of the blastoderm embryo; this repression is normally relieved by Torso RTK signaling at the embryonic termini, thereby enabling localized induction of tll and hkb by broadly distributed activators [3], [4], [25]. These shared requirements of Cic and Gro in the terminal system have led to the idea that both proteins act in a common repressor complex (see refs. [2], [24]). However, we have assayed the requirement of Gro for Cic repressor functions in other developmental contexts and found that Gro is dispensable for such functions (Fig. 1). Specifically, we examined two systems -the developing wing and the ovarian follicular epithelium- where Cic represses specific target genes such as argos and mirror, respectively, under the control of the EGFR pathway [4]–[6], [10]–[12]. In these experiments, we compared the effects caused by the loss of Cic or Gro function using mosaic analyses. Unexpectedly, we found that loss of Gro function does not impair Cic repression in any of those systems, indicating that Cic represses argos and mirror independently of Gro (Fig. 1).
Figure 1. Cic functions independently of Gro in the ovary and in the wing.
(A) Expression of argos in a third instar wing imaginal disc as revealed by LacZ (β-galactosidase) immunostaining using the argosW11–lacZ enhancer trap. Expression is detected in presumptive vein stripes where EGFR signaling is active, and is absent in intervein regions where Cic represses argos. (B-C″) Mosaic wing imaginal discs carrying cicQ474X (B-B″) and groMB36 (C-C″) mutant clones marked by absence of GFP (green, outlined in B″ and C″). B′ and C′ show merged images of GFP signals and argosW11–lacZ expression (red); B″ and C″ show close-ups of boxed areas in panels B′ and C′. Note that loss of Cic function leads to full derepression of argosW11–lacZ in the mutant clones, whereas the loss of Gro causes derepression of argosW11–lacZ only in close proximity to its normal stripes of expression. This localized effect of Gro probably reflects its role together with Enhancer-of-split/Hes repressors in refining argos expression [45]. (D and E) Mosaic adult wings carrying cicQ474X (D) and groMB36 (E) mutant clones induced in third instar larvae as above. Consistent with the effects on argosW11–lacZ expression, the phenotypes of cicQ474X and groMB36 mosaic wings are clearly different: cic mosaic wings show patches of ectopic vein material throughout the wing blade (arrowheads), whereas gro mosaic wings display localized thickening of veins (asterisks). This indicates that Cic repression in the developing wing does not rely on Gro. (F-G″) Stage-10 mosaic egg chambers carrying cicfetU6 (F-F″) and groE48 (G-G″) mutant clones marked by absence of N-Myc immunofluorescence (green, outlined in F″ and G″). F′ and G′ show merged images of N-Myc signals and mirror expression visualized using the mirrorF7–lacZ enhancer trap and anti-LacZ staining (red). F″ and G″ show close-ups of boxed areas in panels F′ and G′. mirror is a key regulator of dorsoventral axis formation that is activated by EGFR signaling in dorsal-anterior follicle cells, and repressed by Cic in ventral follicle cells. cic loss-of-function clones in ventral regions cause derepression of mirrorF7–lacZ, although only in the anterior half of the follicular epithelium [4], [6]. In contrast, gro mutant clones do not show mirrorF7–lacZ derepression, suggesting that Cic also acts independently of Gro in this context.
In light of these results, we have re-evaluated the functional links between Cic and Gro in the early embryo. First, we asked if Cic-mediated repression of a synthetic reporter gene relies on Gro activity in the early embryo. To this end, we used a transgenic construct containing a minimal hunchback (hb) enhancer linked to a pair of individual Cic binding sites (hbC; ref. [10]) (Fig. 2A). The intact hb enhancer drives broad expression in the anterior third of the embryo (Fig. 2B), whereas hbC is repressed by Cic and drives expression only in the anterior pole of the embryo, where Cic is downregulated by Torso RTK signaling (Fig. 2C, D). As shown in Fig. 2E, we find clear derepression of hbC activity in embryos lacking Gro function, implying that Cic represses hbC via Gro in this assay. These results support the idea that Cic indeed acts through Gro in early embryonic patterning.
Figure 2. Cic binding sites are sufficient for recruitment of Gro in vivo.
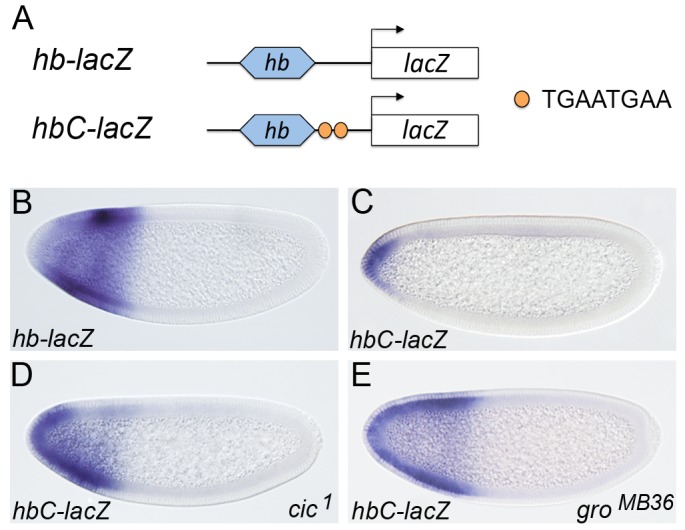
(A) Diagram of lacZ transgenes under the control of a minimal hb enhancer and canonical Cic binding sites (TGAATGAA). (B-E) mRNA expression patterns of hb-lacZ and hbC-lacZ in otherwise wild-type (B, C), cic1 (D) or groMB36 (E) mutant embryos; note the strong derepression of hbC-lacZ in both mutant backgrounds. In this and subsequent figures, anterior is to the left and dorsal is up.
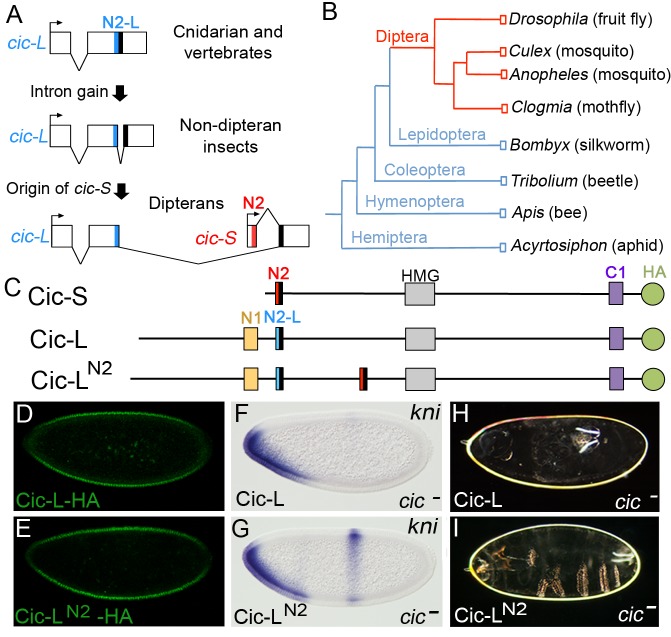
N2, a new motif of Cic that is essential for repression
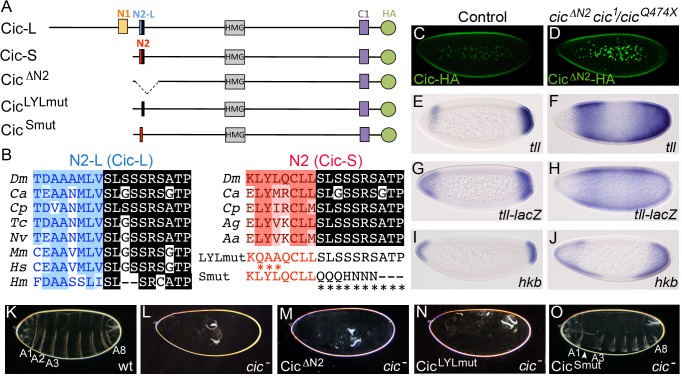
Cic does not contain either of the two previously defined Gro-binding motifs present in known Gro-dependent repressors, the WRPW- and eh1-like peptides [22], and we have not detected direct interactions between functionally important regions of Cic and Gro [10]. Therefore, we asked what sequences of Cic mediate its Gro-dependent repressor activity. Assuming that those sequences could be evolutionarily conserved, we noted a novel conserved motif present at the N-terminus of the Cic-S isoform (GenBank protein AAF55751), which we designate N2 (Fig. 3A, B). This motif is encoded in two adjacent exons: a 5′ exon specific of the cic-S transcript and a 3′ exon shared by both cic-S and cic-L transcripts (see also below). The sequence encoded by the cic-S-specific exon (LYLQCLL) is conserved in dipteran species (Fig. 3A, B, highlighted in red), whereas the peptide common to Cic-S and Cic-L isoforms (SLSSSRSATP) is conserved from hydra to humans (Fig. 3A, B, highlighted in black). To assess the functional significance of N2, we assayed the activity of a Cic-S derivative lacking this motif (CicΔN2). We find that CicΔN2 is expressed at normal levels in transgenic embryos but is unable to repress tll, a tll reporter or hkb (Fig. 3C-J). Accordingly, CicΔN2 does not provide any rescue of the cic embryonic mutant phenotype (Fig. 3K-M), indicating that N2 is critical for Cic function in the early embryo.
Figure 3. The N2 motif is essential for Cic embryonic function.
(A) Diagram of Drosophila Cic-L and Cic-S isoforms and three derivatives carrying mutations in the N2 motif. Cic-L and Cic-S are generated via use of alternative promoters and splicing sites, which produce different N-terminal domains. At the site of alternative splicing, Cic-L and Cic-S contain two different conserved motifs, N2-L and N2, which include different N-terminal sequences (shown in blue and red, respectively) and a common C-terminal peptide (highlighted in black). Other conserved domains (including the N1 and C1 domains of unknown function) are also indicated. The proteins are shown with an HA tag (green) to allow their visualization in transgenic embryos (see also below). (B) Alignment of Cic N2-L and N2 sequences from different species. Dm, Drosophila melanogaster; Ca, Clogmia albipunctata; Cp, Culex pipiens; Tc, Tribolium castaneum; Nv, Nasonia vitripennis; Mm, Mus musculus; Hs, Homo sapiens; Hm, Hydra magnipapillata; Ag, Anopheles gambiae; Aa, Aedes aegypti. Two different mutations of the N2 peptide (LYLmut and Smut) are also shown below the N2 alignment. (C and D) Expression of Cic and CicΔN2 proteins tagged with the HA epitope in embryos stained with anti-HA antibody; note that both proteins appear downregulated at the embryo poles. (E-M) mRNA expression patterns of tll, tll-lacZ and hkb in wt (E, G, I) and cic mutant (cic1/cicQ474X) embryos expressing CicΔN2 (F, H, J). Cuticle phenotypes of the same genetic backgrounds are shown in K and M, respectively; panel L shows a control cic1/cicQ474X mutant cuticle. (N and O) Cuticle phenotypes of cic1/cicQ474X mutant embryos expressing the CicLYLmut (N) and CicSmut (O) derivatives; only CicSmut rescues the cic phenotype, except for mild segmental defects (arrowhead). A1-A8, abdominal segments 1–8.
We also tested two mutations affecting each of the sub-elements of N2. Surprisingly, disruption of the Cic-S-specific element caused a complete loss of Cic-S function, whereas mutation of the second, highly conserved sequence had a minor effect on protein activity (Fig. 3N, O). Thus, only the dipteran-specific portion of N2 is essential for Cic embryonic function.
N2 is a Gro-dependent repressor element
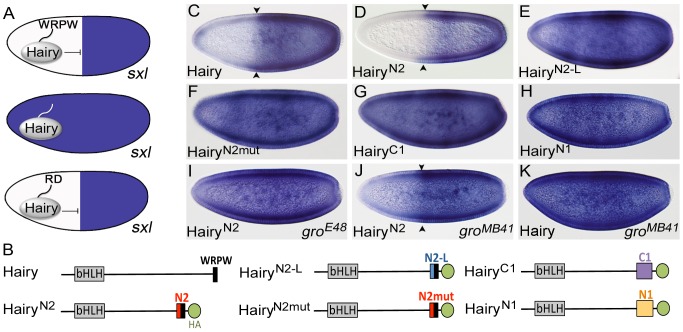
Based on the above results, we hypothesized that N2 could be involved in recruiting Gro to Cic target genes. In fact, the critical N2 sequence shares some similarity with the consensus eh1 motif (FxIxxIL) that binds directly to Gro, although it lacks the characteristic phenylalanine residue at position 1. We therefore tested if N2 functions as an autonomous, transferable Gro-dependent motif capable of imposing repressor activity on a heterologous DNA-binding domain. For this, we adopted the Sex-lethal (Sxl) repression assay, an in vivo strategy for analyzing the activity of known or potential repressor domains [26], [27]. In this assay, a domain under analysis is used to replace the Gro-binding WRPW motif of the Hairy repressor and tested for its ability to repress Sxl expression in the embryo (Fig. 4A). Using this approach, we found that a Hairy chimera carrying the N2 motif instead of the WRPW peptide (HairyN2) represses Sxl as efficiently as intact Hairy (Fig. 4B-D). In contrast, four control Hairy chimeras carrying a mutant version of N2 or other conserved motifs from Cic, did not (Fig. 4B, E-H; see also S1 Fig.). Moreover, repression by the HairyN2 chimera depends on Gro, as it is lost in groE48 mutant embryos that lack Gro activity (Fig. 4I). This indicates that N2 is a discrete, Gro-dependent repressor motif.
Figure 4. N2 is a Gro-dependent repressor motif.
(A) Schematic representation of the Sxl repression assay. In this assay, expression of Hairy under the control of the hb promoter in the anterior region of the embryo leads to repression of Sxl transcription (blue) in females (top). Repression depends on the WRPW motif of Hairy (middle), but replacement of this motif with autonomous repressor domains (RD) restores repression function (bottom). (B) Diagram of Hairy and Hairy fusion constructs tested in the Sxl assay; all fusions carry a C-terminal HA tag (see Materials and Methods). (C-K) Effects of Hairy constructs on Sxl expression in otherwise wild-type or gro mutant embryos; all images correspond to Sxl expression in female embryos. Arrowheads indicate borders of transcriptional repression. Note that HairyN2 does not cause complete repression of Sxl in groMB41 embryos (J); this may reflect a loss of function, in this genetic background, for endogenous Hairy-related factors such as Deadpan that normally contribute to Sxl repression [46].
We also analyzed the activity of HairyN2 in the presence of a Gro mutant protein, GroMB41, which can not bind to WRPW or eh1 motifs but retains normal function in the terminal system (potentially acting together with Cic) [28]. The GroMB41 mutant carries an amino acid substitution (R483H) affecting the central pore of the Gro β propeller domain, thereby preventing binding of WRPW or eh1 motifs across this pore [28]. We found that HairyN2 displays significant repressor activity in groMB41 embryos, whereas native Hairy is completely inactive in this background (Fig. 4J, K). Thus, GroMB41 is functional both in repressing terminal gap genes and in mediating repression by HairyN2, suggesting that it is recruited in each of these systems through similar interactions that involve the N2 motif.
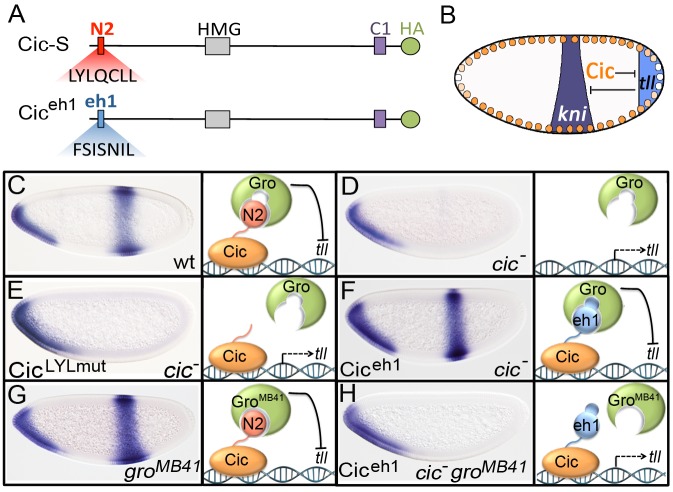
As an independent test of this idea, we analyzed a Cic derivative in which the N2 sequence was replaced by the eh1 motif (FSISNIL) from the Engrailed homeodomain protein (Ciceh1; Fig. 5A). If Gro is recruited to the terminal system through the N2 motif, replacing this motif by the eh1 element should render GroMB41 non-functional in that system. For these experiments, we monitored the expression of the central gap gene knirps (kni) as a sensitive readout of Cic and Ciceh1 repressor activities (Fig. 5B). kni is a target of the Tll repressor. When Cic is active, it restricts tll expression to the posterior pole of the embryo, thereby permitting expression of kni in the presumptive abdomen (Fig. 5B, C). In contrast, loss of Cic function causes derepression of tll and corresponding loss of the central kni stripe (Fig. 5D, E). We find that Ciceh1 is an active repressor capable of rescuing kni expression in cic mutant embryos (Fig. 5F), indicating that the eh1 peptide can compensate for the loss of endogenous N2 in its normal setting. We then compared kni expression in groMB41 embryos expressing either endogenous Cic or Ciceh1. As previously reported, kni expression is normal in the first case (ref. [28]; Fig. 5G), whereas there is clear loss of kni expression in the presence of Ciceh1 (Fig. 5H). Therefore, it is the presence of an intact N2 motif in Cic that enables GroMB41 to be functional in the terminal system, supporting our conclusion that N2 links Cic and Gro in the Drosophila embryo.
Figure 5. The N2 motif of Cic recruits Gro to the terminal patterning system.
(A) Diagram of Cic and Ciceh1 proteins; Ciceh1 carries the eh1 motif from Drosophila Engrailed instead of N2 and is tagged with an HA epitope at the C-terminus. (B) Schematic representation of cross-repressive interactions between Cic, tll and kni in the early blastoderm. (C-H) mRNA expression patterns of kni in wild-type (C), cic (D, E, F), groMB41 (G) and cic groMB41 (H) mutant backgrounds expressing the CicLYLmut (E) and Ciceh1 (F, H) products. A model diagram depicting the interactions of N2 and eh1 motifs with Gro proteins and the resulting repressor activities is shown next to each embryo; for simplicity, the interaction between N2 and Gro is modeled as being direct (see Discussion). The cic maternal mutant genotypes are cic1 for panels D, F and H, and cic1/cicQ474X for panel E.
Origin of N2 and Cic-S in dipterans
As indicated above, the key repressor element within the N2 motif is specific to the Cic-S isoform and is present only in dipterans. To get further insight into the evolution of this element, we examined the structure of the cic locus in different insect taxa, focusing on the region that spans the alternatively spliced exons of Drosophila cic-S and cic-L transcripts. We were able to perform these analyses given the high conservation of peptide sequences encoded by these alternative exon junctions (Fig. 3B). We found a similar cic genomic organization in Drosophila and four distant species of lower dipterans: Clogmia albipunctata, Culex pipiens, Anopheles gambiae and Aedes aegypti (Fig. 6A; S2 Fig.), implying that this organization was already present in an early common ancestor of dipterans. In contrast, a different structure, which lacks the first cic-S exon, is apparent across representative species of non-dipteran taxa, including Bombyx mori (Lepidoptera), Tribolium castaneum (Coleoptera), Apis mellifera (Hymenoptera), and Acyrthosiphon pisum (Hemiptera) (Fig. 6A, B). In this configuration, the two exons encoding the N2-L motif of Cic-L proteins are frequently separated by short (<150 pb) introns, which do not contain the first cic-S exon encoding the N2 repressor motif (LYLQCLL) nor its upstream promoter region (Fig. 6A). Thus, while we cannot rule out the possibility that other short isoforms of Cic exist in non-dipteran species (e.g. expressed from other alternative promoters within cic), a form equivalent to dipteran Cic-S (containing the N2 motif) is clearly absent in those species. Therefore, the simplest interpretation of these genomic organizations is that the Cic-S isoform and its N2 motif originated after the expansion of the above cic-L intron during the early radiation of dipterans; an alternative scenario, where the Cic-S isoform was already present in early insects, appears much less likely, since this would involve the independent loss of this isoform in each of the non-dipteran branches examined.
Figure 6. Recent origin of the N2 motif in dipterans.
(A) Schematic representation of proposed steps giving rise to the Cic-S isoform and the N2 motif. The diagrams show the regions spanning the alternatively spliced exons of cic-S and cic-L transcripts in selected species, which are represented graphically and are not drawn to scale. The size of introns splitting the N2-L coding sequences in non-dipteran species are as follows: Bombyx mori (758 bp), Tribolium castaneum (50 bp), Apis mellifera (86 bp) and Acyrthosiphon pisum (129 bp) (see also main text). The conserved protein motifs encoded at relevant exon junctions are shaded in red, blue or black as in Fig. 3. (B) Insect phylogeny illustrating the presence of the Cic-S and N2 motifs in dipterans (red). (C) Diagram of the Cic-S, Cic-L and Cic-LN2 proteins; Cic-LN2 carries the N2 motif inserted within a poorly conserved sequence of Cic. Cic-L and Cic-LN2 were expressed with an HA tag at the C-terminus. (D and E) Expression of HA-tagged Cic-L (D) and Cic-LN2 (E) proteins in embryos stained with anti-HA antibody. (F and G) mRNA expression patterns of kni in cic1/cicQ474X mutant embryos expressing Cic-L (F) and Cic-LN2 (G); only Cic-LN2 leads to significant rescue of the central kni stripe. (H and I) Cuticle phenotypes of the same genetic backgrounds shown in F and G, respectively.
These findings indicate that Cic-L represents the ancestral isoform of Cic in insects that gave rise to Cic-S in dipterans. To further test the significance of this evolutionary change, we compared the activities of the Drosophila Cic-L and Cic-S isoforms in early embryogenesis. The function of Cic-L has not been studied at the molecular level, and it is even unclear whether it functions as a repressor [2]. To assay Cic-L repressor activity in the early embryo, we generated a transgene expressing Cic-L under the control of the maternal cic-S promoter (Fig. 6C; Materials and Methods). This construct drives efficient accumulation of Cic-L in blastoderm nuclei (Fig. 6D), but does not rescue the embryonic cic phenotype (Fig. 6F, H), indicating that it cannot replace Cic-S in repressing the terminal gap genes. Since Cic-L lacks the N2 motif, we then tested a Cic-L derivative carrying the N2 sequence inserted N-terminal to the HMG-box (Fig. 6C). Strikingly, this protein (Cic-LN2) showed significant, although not complete, rescue of the embryonic cic mutant phenotype (Fig. 6E, G, I). This indicates that the Drosophila Cic-S and Cic-L isoforms have very different molecular activities, and that evolution of the N2 motif represented a key innovation for Cic repressor function in the early embryo.
Discussion
We have shown that Cic proteins exhibit both Gro-dependent and -independent activities, and that this functional diversity is associated with the origin of the Cic-S isoform and the N2 motif in dipterans, approximately 250 million years ago. By comparison, other functional attributes of Cic such as their sensitivity to RTK signaling and their binding to specific sites in DNA, are more broadly conserved and therefore probably more ancient. For example, the MAPK-interacting domain of Drosophila Cic (C2) is clearly recognizable outside the dipterans [7], and Cic is downregulated by RTK signaling in mammalian cells [15], [19]. Thus, while Cic proteins may have long served as sensors of RTK signaling, their mechanisms of repression appear to have evolved and adapted to fulfill new Cic functions in distinct transcriptional contexts. Below, we discuss the significance and implications of the newly evolved mechanism of Cic repression in fly embryogenesis.
Our results indicate that prior to the origin of dipterans, Cic was present in insects as a Cic-L-like isoform that lacked the N2 motif. Clearly, Drosophila Cic-L cannot function in the early embryo unless it carries the N2 motif from Cic-S (Fig. 6). This suggests that evolution of the N2 motif dramatically altered the mechanism of Cic repression by establishing a novel association with Gro. How, then, did the N2 motif appear? The comparison of different insect cic genes suggests that the N2 motif originated along with the Cic-S isoform, possibly through genomic rearrangements of intronic cic-L sequences that created a shorter cic-S transcript and subsequent evolution of a functional N2 motif via random drift. In this regard, it has been argued that short peptide sequences such as the WRPW and eh1 Gro-interacting motifs may be particularly easy to evolve by simple drift [29], [30].
The N2 motif is different from the WRPW and eh1 motifs, and we still do not know its precise mechanism of action. By analogy to the WRPW and eh-1 motifs, which bind the central pore of the Gro β propeller, it is possible that N2 also recognizes this region of Gro. If this is correct, the N2 motif should adopt a conformation across the pore that is insensitive to the MB41 mutation, just like another Gro mutation, MB31, prevents binding of WRPW but not eh-1 to the pore [28]. Another, non-exclusive possibility is that N2 binds the Gro β propeller with the help of auxiliary proteins. Consistent with this idea, the WRPY motif of Runx proteins binds very weakly to Gro and this interaction depends on other accessory proteins in vivo [28], [31], [32].
What could be the functional and evolutionary significance of the new Gro-dependent mechanism of Cic repression? It seems logical to assume that Cic employs qualitatively different mechanisms of repression in the embryo (via Gro) than when acting in other contexts (presumably with other corepressors; see below). We suggest that the combined activities of Cic-S and Gro may have facilitated the evolution of the complex transcriptional network regulated by Torso signaling in modern fly embryos. This network comprises multiple Cic target genes, including tll and hkb, whose boundaries of expression are regulated by Torso-dependent gradients of Cic repression at the embryo poles [9], [10], [33], [34]. In this system, Gro itself appears to exert a regulatory function beyond its obvious role as a component of the repression machinery. Indeed, Gro is directly phosphorylated and functionally downregulated in response to Torso signaling [35], [36], and even modest changes in Gro protein levels significantly affect the threshold concentrations at which Cic represses tll and hkb [37]. This suggests a model where Torso signaling controls the expression of Cic target genes via coordinate activity gradients of both Cic and Gro. These overlapping gradients might serve as a fail-safe mechanism to ensure the correct spatiotemporal response of target genes, buffering against random perturbations in either gradient. Furthermore, Gro is a highly versatile corepressor capable of functioning in different contexts of recruitment [22]–[24], [38], which may explain the ability of Cic to regulate multiple targets simultaneously. For example, tll and hkb are activated by different mechanisms that are either dependent (hkb) or independent (tll) of Lilliputian, a component of the super elongation complex (SEC) [39], [40], implying that Gro is capable of counteracting both activation mechanisms. Thus, the acquisition of Gro-mediated repression by Cic may have facilitated the precise, coordinated regulation of Cic target genes in response to Torso signaling.
In contrast, Gro is mostly dispensable for other Cic functions in the wing and the follicular epithelium (Fig. 1). The Cic-S isoform is sufficient for both of these functions [4], [5], [7], raising the possibility that Cic-S acts through other corepressors in those tissues. One potential candidate is the Drosophila ortholog of mammalian Atxn1 (dAtxn1; [41]). In mammals, Atxn1 and the related factor Ataxin1-Like (Atxn1L; also known as Brother of ATXN1, BOAT) potentiate Cic-S repressor activity in cultured cells [14], [42], and directly interact with a short motif of Cic that is conserved in Drosophila Cic-S [14], [21]. dAtxn1 has been mainly studied in models of SCA1 pathogenesis [41]. Thus, future studies should examine whether dAtxn1 also mediates Cic repressor functions in development.
Finally, our results suggest that mammalian Cic proteins probably function independently of Gro, unless they have evolved other specific Gro-interacting motifs different from N2. Similarly, the mammalian Cic-S isoform must have originated independently of the dipteran Cic-S isoform, resulting in coincidental presence of Cic-S isoforms in both taxa. It will be interesting to determine whether mammalian Cic-S and Cic-L proteins also exhibit differential functional properties in their ability to regulate gene expression.
Materials and Methods
Drosophila genetics and transgenic lines
The following alleles were used: cic1 [3], cicQ474X [8], groE48, groMB36 and groMB41 [28]. cic mutant embryos were obtained from cic1 or cic1/cicQ474X females, except in the experiments presented in Fig. 5D, F and H, which involved the generation of mosaic females whose germlines were homozygous for cic1 using the FRT/ovoD system [43]. All gro embryos were derived via the FRT/ovoD system. Transgenic lines were established by P-element-mediated transformation or using the ΦC31-based integration system [44]. The hb-h and hb-hN2 transgenes cause high levels (>98%) of female lethality and were maintained in males, either using an attached X chromosome [C(1)M3] (for X-chromosome insertions) or unbalanced (for autosomal insertions). In contrast, the hb-hN2-L, hb-hN2mut, hb-hC1 and hb-hN1 transgenes do not cause female lethality, even when present in two copies.
DNA constructs
Cic-expressing transgenes were based on the original cic rescue construct [3], which contains the cic-S transcription unit flanked by its natural 5′ and 3′ regulatory sequences, and were assembled in pCaSpeR4 or pattB vectors. The CicΔN2 construct lacks amino acids 3–77 of Cic-S. Ciceh1 contains the sequence VPLAFSISNIL instead of FQDFELGAKLYLQCLL. The Cic-L isoform used in this work is the product of cDNA LD17181 (GenBank accession number BT100233), a fully sequenced clone identified by the Berkeley Drosophila Genome Project (see S2 Fig.). The LD17181 product (LD17181p) was expressed from the ATG initiator codon present in the cic-S rescue construct, by replacing the sequence encoding amino acids 4–19 of Cic-S with the sequence encoding amino acids 3–487 of LD17181p; note that amino acid 20 of Cic-S corresponds to amino acid 488 of LD17181p. Cic-LN2 was constructed by inserting an N2-containing fragment (residues 4-35 of Cic-S) at amino acid position 852 of LD17181p. All Cic derivatives have a triple HA tag (YPYDVPDYA) inserted in the same position, corresponding to amino acid 1398 of Cic-S. Hairy fusion proteins contain amino acids 1-268 of Hairy fused to the following Cic sequences: amino acids 3-35 (HairyN2), and 1308-1396 (HairyC1) of Cic-S, and amino acids 376-437 (HairyN1) and 468-503 (HairyN2-L) of LD17181p. HairyN2mut, contains the sequence AYAQCLASQ instead of LYLQCLLSL.
Embryo analyses
Embryos were fixed in 4% formaldehyde-PBS-heptane using standard procedures. In situ hybridizations were performed using digoxigenin-UTP labeled antisense RNA probes, and anti-digoxygenin antibodies conjugated to alkaline phosphatase (Roche). Immunodetection of HA-tagged Cic proteins was performed using monoclonal antibody 12CA5 (Roche) at 1∶400 dilution and secondary Alexa488-conjugated antibodies (Molecular Probes). Cuticle preparations were mounted in 1∶1 Hoyer's medium/lactic acid and cleared overnight at 60°C.
Supporting Information
Expression of Hairy chimeras inactive in the Sxl assay. (A-D) Expression of HairyN2-L, HairyN2mut, HairyC1 and HairyN1 proteins under the control of the hb promoter (see Fig. 4). All proteins are readily detected by anti-HA immunostaining, indicating that their inability to repress Sxl is not due to inefficient accumulation in the embryo.
(TIF)
Structure of the Drosophila cic locus and two main transcripts, cic-S and cic-L, expressed from alternative promoters. White and grey boxes indicate transcribed untranslated regions and coding sequences, respectively. Sequences encoding the N1, N2, N2-L, HMG-box and C1 domains are highlighted in color. The structure of the cic-L transcript corresponds to the LD17181 cDNA (see Materials and Methods). The sequence of the first exon and its immediate upstream region is shown below to indicate the positions of the annotated transcription initiation site (TIS, bent arrow) and the 5′ end of LD17181 (arrowhead). The position of the TIS is based on RNA-seq profiles generated by the modENCODE project [47]. The translated peptide sequence is also shown in bold, with residues encoded by LD17181 highlighted in red; thus, the LD17181p product is 5 amino acid shorter than the corresponding predicted Cic-L protein (1871 vs. 1876 residues, respectively). Genomic sequences from Drosophila erecta (De) and Drosophila yakuba (Dy) are aligned below the melanogaster (Dm) sequence; note that both species contain in-frame stop codons (asterisks) immediately upstream of the N-terminal methionine, supporting the predicted initiation of translation.
(TIF)
Acknowledgments
We thank A. Olza for assistance with Drosophila injections, E. Jiménez-Guri and J. Jaeger for providing cic genomic sequences from C. albipunctata, S. Shvartsman, A. Veraksa, B. Jennings, A. Brehm, and D. Ish-Horowicz for discussions, M. Wainwright, B. Jennings and the Bloomington Drosophila Research Center for fly stocks, and S. Shvartsman for critical reading of the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by research grants from the Spanish Government (BFU2008-01875 and BFU2011-23611), Generalitat de Catalunya (2009SGR-1075) and Fundació La Marató de TV3 (20131730). GJ is an ICREA investigator. ZP is supported by grants from the National Institute of General Medical Sciences (NIH R01GM086537), the Israel Science Foundation (Center of Excellence; 1772/13) and the Król Charitable Foundation. ZP is an incumbent of The Lady Davis Chair in Experimental Medicine and Cancer Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lemmon MA, Schlessinger J (2010) Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jiménez G, Shvartsman SY, Paroush Z (2012) The Capicua repressor - a general sensor of RTK signaling in development and disease. J Cell Sci 125: 1383–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiménez G, Guichet A, Ephrussi A, Casanova J (2000) Relief of gene repression by Torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev 14: 224–231. [PMC free article] [PubMed] [Google Scholar]
- 4. Goff DJ, Nilson LA, Morisato D (2001) Establishment of dorsal-ventral polarity of the Drosophila egg requires capicua action in ovarian follicle cells. Development 128: 4553–4562. [DOI] [PubMed] [Google Scholar]
- 5. Roch F, Jiménez G, Casanova J (2002) EGFR signalling inhibits Capicua-dependent repression during specification of Drosophila wing veins. Development 129: 993–1002. [DOI] [PubMed] [Google Scholar]
- 6. Atkey MR, Lachance JF, Walczak M, Rebello T, Nilson LA (2006) Capicua regulates follicle cell fate in the Drosophila ovary through repression of mirror . Development 133: 2115–2123. [DOI] [PubMed] [Google Scholar]
- 7. Astigarraga S, Grossman R, Diaz-Delfin J, Caelles C, Paroush Z, et al. (2007) A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J 26: 668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tseng AS, Tapon N, Kanda H, Cigizoglu S, Edelmann L, et al. (2007) Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/Ras signaling pathway. Curr Biol 17: 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Löhr U, Chung HR, Beller M, Jackle H (2009) Antagonistic action of Bicoid and the repressor Capicua determines the spatial limits of Drosophila head gene expression domains. Proc Natl Acad Sci U S A 106: 21695–21700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ajuria L, Nieva C, Winkler C, Kuo D, Samper N, et al. (2011) Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila . Development 138: 915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Andreu MJ, González-Pérez E, Ajuria L, Samper N, Gonzalez-Crespo S, et al. (2012) Mirror represses pipe expression in follicle cells to initiate dorsoventral axis formation in Drosophila . Development 139: 1110–1114. [DOI] [PubMed] [Google Scholar]
- 12. Andreu MJ, Ajuria L, Samper N, González-Pérez E, Campuzano S, et al. (2012) EGFR-dependent downregulation of Capicua and the establishment of Drosophila dorsoventral polarity. Fly 6: 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grimm O, Sanchez Zini V, Kim Y, Casanova J, Shvartsman SY, et al. (2012) Torso RTK controls Capicua degradation by changing its subcellular localization. Development 139: 3962–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lam YC, Bowman AB, Jafar-Nejad P, Lim J, Richman R, et al. (2006) ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell 127: 1335–1347. [DOI] [PubMed] [Google Scholar]
- 15. Fryer JD, Yu P, Kang H, Mandel-Brehm C, Carter AN, et al. (2011) Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science 334: 690–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bettegowda C, Agrawal N, Jiao Y, Sausen M, Wood LD, et al. (2011) Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 333: 1453–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee Y, Fryer JD, Kang H, Crespo-Barreto J, Bowman AB, et al. (2011) ATXN1 protein family and CIC regulate extracellular matrix remodeling and lung alveolarization. Dev Cell 21: 746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kawamura-Saito M, Yamazaki Y, Kaneko K, Kawaguchi N, Kanda H, et al. (2006) Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35; q13) translocation. Hum Mol Genet 15: 2125–2137. [DOI] [PubMed] [Google Scholar]
- 19. Dissanayake K, Toth R, Blakey J, Olsson O, Campbell DG, et al. (2011) ERK/p90(RSK)/14-3-3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capicua. Biochem J 433: 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, et al. (2008) Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature 452: 713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim E, Lu H, Zoghbi HY, Song J (2013) Structural basis of protein complex formation and reconfiguration by polyglutamine disease protein Ataxin-1 and Capicua. Genes Dev 27: 590–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jennings BH, Ish-Horowicz D (2008) The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol 9: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Turki-Judeh W, Courey AJ (2012) Groucho: a corepressor with instructive roles in development. Curr Top Dev Biol 98: 65–96. [DOI] [PubMed] [Google Scholar]
- 24. Mannervik M (2014) Control of Drosophila embryo patterning by transcriptional co-regulators. Exp Cell Res 321: 47–57. [DOI] [PubMed] [Google Scholar]
- 25. Paroush Z, Wainwright SM, Ish-Horowicz D (1997) Torso signalling regulates terminal patterning in Drosophila by antagonising Groucho-mediated repression. Development 124: 3827–3834. [DOI] [PubMed] [Google Scholar]
- 26. Parkhurst SM, Bopp D, Ish-Horowicz D (1990) X:A ratio, the primary sex-determining signal in Drosophila, is transduced by helix-loop-helis proteins. Cell 63: 1179–1191. [DOI] [PubMed] [Google Scholar]
- 27. Jiménez G, Paroush Z, Ish-Horowicz D (1997) Groucho acts as corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev 11: 3072–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jennings BH, Pickles LM, Pearl LH, Ish-Horowicz D (2006) Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol Cell 22: 645–655. [DOI] [PubMed] [Google Scholar]
- 29. Hittinger CT, Carroll SB (2008) Evolution of an insect-specific Groucho-interaction motif in the Engrailed selector protein. Evol Dev 10: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carroll SB (2008) Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. [DOI] [PubMed] [Google Scholar]
- 31. Aronson BD, Fisher AL, Blechman K, Caudy M, Gergen JP (1997) Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol 17: 5581–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Canon J, Banerjee U (2003) In vivo analysis of a developmental circuit for direct transcriptional activation and repression in the same cell by a Runx protein. Genes Dev 17: 838–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen H, Xu Z, Mei C, Yu D, Small S (2012) A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell 149: 618–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Helman A, Lim B, Andreu MJ, Kim Y, Shestkin T, et al. (2012) RTK signaling modulates the Dorsal gradient. Development 139: 3032–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cinnamon E, Helman A, Ben-Haroush Schyr R, Orian A, Jiménez G, et al. (2008) Multiple RTK pathways downregulate Groucho-mediated repression in Drosophila embryogenesis. Development 135: 829–837. [DOI] [PubMed] [Google Scholar]
- 36. Helman A, Cinnamon E, Mezuman S, Hayouka Z, Von Ohlen T, et al. (2011) Phosphorylation of Groucho mediates RTK feedback inhibition and prolonged pathway target gene expression. Curr Biol 21: 1102–1110. [DOI] [PubMed] [Google Scholar]
- 37. Turki-Judeh W, Courey AJ (2012) The unconserved Groucho central region is essential for viability and modulates target gene specificity. PLoS One 7: e30610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cinnamon E, Paroush Z (2008) Context-dependent regulation of Groucho/TLE-mediated repression. Curr Opin Genet Dev 18: 435–440. [DOI] [PubMed] [Google Scholar]
- 39. Tang AH, Neufeld TP, Rubin GM, Muller HA (2001) Transcriptional regulation of cytoskeletal functions and segmentation by a novel maternal pair-rule gene, lilliputian . Development 128: 801–813. [DOI] [PubMed] [Google Scholar]
- 40. Luo Z, Lin C, Schilatifard A (2012) The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 13: 543–547. [DOI] [PubMed] [Google Scholar]
- 41. Tsuda H, Jafar-Nejad P, Patel AJ, Sun Y, Chen HK, et al. (2005) The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell 122: 633–644. [DOI] [PubMed] [Google Scholar]
- 42. Crespo-Barreto J, Fryer JD, Shaw CA, Orr HT, Zoghbi HY (2010) Partial loss of Ataxin-1 function contributes to transcriptional dysregulation in spinocerebellar ataxia type 1 pathogenesis. PLoS Genet 6: e1001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chou TB, Noll E, Perrimon N (1993) Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development 119: 1359–1369. [DOI] [PubMed] [Google Scholar]
- 44. Bischof J, Maeda RK, Hediger M, Karch F, Basler K (2007) An optimized transgenesis system for Drosophila using germ-line-specific ΦC31 integrases. Proc Natl Acad Sci U S A 104: 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Housden BE, Terriente-Felix A, Bray SJ (2014) Context-dependent enhancer selection confers alternate models of notch regulation on argos . Mol Cell Biol 34: 664–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Younger-Shepherd S, Vaessin H, Bier E, Jan LY, Jan YN (1992) deadpan, an essential pan-neural gene encoding an HLH protein, acts as a denominator in Drosophila sex determination. Cell 70: 911–922. [DOI] [PubMed] [Google Scholar]
- 47. Graveley BR, Brooks A, Carlson JW, Duff MO, Landolin JM, et al. (2011) The developmental transcriptome of Drosophila melanogaster . Nature 471: 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of Hairy chimeras inactive in the Sxl assay. (A-D) Expression of HairyN2-L, HairyN2mut, HairyC1 and HairyN1 proteins under the control of the hb promoter (see Fig. 4). All proteins are readily detected by anti-HA immunostaining, indicating that their inability to repress Sxl is not due to inefficient accumulation in the embryo.
(TIF)
Structure of the Drosophila cic locus and two main transcripts, cic-S and cic-L, expressed from alternative promoters. White and grey boxes indicate transcribed untranslated regions and coding sequences, respectively. Sequences encoding the N1, N2, N2-L, HMG-box and C1 domains are highlighted in color. The structure of the cic-L transcript corresponds to the LD17181 cDNA (see Materials and Methods). The sequence of the first exon and its immediate upstream region is shown below to indicate the positions of the annotated transcription initiation site (TIS, bent arrow) and the 5′ end of LD17181 (arrowhead). The position of the TIS is based on RNA-seq profiles generated by the modENCODE project [47]. The translated peptide sequence is also shown in bold, with residues encoded by LD17181 highlighted in red; thus, the LD17181p product is 5 amino acid shorter than the corresponding predicted Cic-L protein (1871 vs. 1876 residues, respectively). Genomic sequences from Drosophila erecta (De) and Drosophila yakuba (Dy) are aligned below the melanogaster (Dm) sequence; note that both species contain in-frame stop codons (asterisks) immediately upstream of the N-terminal methionine, supporting the predicted initiation of translation.
(TIF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.