Abstract
During endometriosis, a breakdown occurs in endometrial and peritoneal homeostasis caused by cytokine-induced cell proliferation and dysregulation of apoptosis. We studied tumor necrosis factor (TNF)-α, TNF receptor (TNFR) 1, and TNFR2 gene expression at both messenger RNA (mRNA) and protein levels in peritoneal fluid (PF) mononuclear cells (PFMCs), the percentages of these cells bearing the same markers, and soluble TNF-α (sTNF-α) values in PF of 80 women with endometriosis. We found that TNFR1 mRNA and protein levels, the percentages of TNFR1-bearing PFMCs, and sTNF-α values decreased from minimal to severe stages of the disease. Instead, TNF-α and TNFR2 mRNA and protein levels, the percentages of membrane TNF-α (mTNF-α)- and TNFR2-bearing PFMCs increased as the disease worsened. These data allow us to hypothesize that, in early stages, the high percentages of TNFR1-bearing PFMCs and the high levels of sTNF-α could address signal toward complex I pathway, favoring the inflammatory response. With the worsening of the disease, the low percentages of TNFR1-bearing PFMCs are probably due to decreased TNFR1 mRNA transcription and protein translation rate. In early stages (minimal and mild), the percentages of both TNFR2- and mTNF-α–bearing PFMCs are so low, due to decreased mRNA transcription and protein translation rate, that subsequent cellular events may depend minimally by this interaction. The high levels of sTNF-α may be rerouted to bind TNFR1. In contrast, in the moderate and severe stages, the high percentages of TNFR2-bearing PFMCs may be saturated by high percentages of mTNF-α–bearing PFMCs, triggering death process. So, in endometriosis, each component of the TNF-α/TNFRs system may trigger opposite cellular fate.
Keywords: endometriosis, PFMCs, TNF-α, TNFR1, TNFR2
Introduction
Endometriosis is an estrogen-dependent disease1 defined by the ectopic presence and growth of functional endometrial tissue, glands, and stroma outside the uterus,2,3 primarily on the peritoneal surface of endopelvic structures and the ovaries. Several studies evidenced that peritoneal microenvironment of women with endometriosis undergoes a number of local inflammatory-reparative phenomena, with the involvement of resident macrophages and the attraction and recruitment of peripheral mononuclear cells from the blood into the peritoneal cavity.4–8 These peritoneal fluid mononuclear cells (PFMCs), as well as endometriotic cells, secrete different patterns of cytokines,6–9 which may polarize immune cells toward effector or regulatory profiles and, in this way, alter the peritoneal homeostasis and cause dysregulation of apoptosis.6,10–13,14 The death ligand tumor necrosis factor (TNF)-α engages its receptors, named TNF receptor (TNFR) 1 and TNFR2, on the surface membrane of target cells and so activates extrinsic pathway of apoptosis. TNF-α, belonging to TNF superfamily, is synthesized as a 26-kDa transmembrane type II protein (membrane TNF-α [mTNF-α]) and its soluble form (sTNF-α, 17 kDa) rises by TNF-α-converting enzyme (TACE) action on mTNF-α and retains the biological activities. Both forms of TNF-α coexist as monomeric, dimeric, or trimeric proteins and the last form only of both cytokines is biologically active and exerts its effects interacting with the receptors of TNFR superfamily.15 This consists of 2 main groups of receptors: the first group or death receptors, including TNFR1, is characterized by the presence of the death domain (DD) in its intracellular region, whereas the second one, including TNFR2, does not have DD. Tumor necrosis factor receptor 1 is a 55-kDa protein and is expressed in almost all cell types. Tumor necrosis factor receptor 2 is a 75-kDa protein, expressed only in certain cell types, including T cells.15,16 The soluble forms of both TNFR1 and TNFR2 in peritoneal fluid (PF) of women with endometriosis are higher with respect to healthy controls and can modulate the effects of TNF-α by acting as antagonists.16,17 In addition, sTNF-α binds both receptors but only efficiently activates TNFR1, whereas mTNF-α can bind and activate both TNFR1 and TNFR2.15 The aim of the current study is to investigate the TNF-α and TNFR1/TNFR2 system to evaluate its behavior throughout the different stages of endometriosis. Therefore, we will assess the values of sTNF-α in the PF, the percentages of m TNF-α-, TNFR1-, and TNFR2-bearing PFMCs, and also the levels of the gene expression of TNF-α, TNFR1, and TNFR2 at both messenger RNA (mRNA) production and protein synthesis in the same cells of women with endometriosis at minimal, mild, moderate, and severe stages.
Materials and Methods
Study Population
Ninety-six women of reproductive age attending the infertility outpatient clinic of the University of Messina were eligible for this study. Patients (aged 20-45 years) were diagnosed with endometriosis (n = 80) following laparoscopy. Diagnosis was later confirmed by histopathological examination. In accordance with the revised American Society for Reproductive Medicine (rASRM) classification for endometriosis,18 the patients were divided into 4 groups: minimal (n = 26), mild (n = 28), moderate (n = 14), and severe (n = 12) stages of endometriosis. The controls (n = 16) consisted of women (aged 21-43 years) affected by non-immunological infertility, who underwent explorative laparoscopy. All diagnostic laparoscopic procedures were performed during the proliferative phase of the menstrual cycle. All women involved in this study had no other pelvic disorders, chronic circulatory, autoimmune, or neoplastic disease and had not been taking any anti-inflammatory or hormonal or immunomodulatory medications in the preceding 6 months. Investigation protocols were approved by the local Ethics Committee, and prior to participating in this study all the patients involved signed an informed consent form allowing us to use their biological fluids and cells. None of the authors have any proprietary, financial, professional, or other personal interest of any nature or kind in any product, service, and/or company.
Sample Collection
Peritoneal fluid sample preparation
Peritoneal fluid samples were collected under sterile conditions at the time of the laparoscopy, prior to any operative manipulation in order to minimize blood contamination. Peritoneal fluid samples were centrifuged at 200g for 5 minutes, the supernatants removed, aliquoted, and stored at −80°C until use.
Cell preparation and identification
The cell pellet was suspended in phosphate-buffered saline (PBS), layered onto Histopaque-1077 (Sigma, St Louis, Missouri), and centrifuged at 150g for 30 minutes, as described previously by Yoshino et al.19 The cells were collected at the interface, washed twice in PBS, suspended, counted, and identified as PFMCs by light microscopy observation (purity > 97%) and indirect immunofluorescence (IIF) as described subsequently. Anti-CD3, anti-B19, anti-CD56, and anti-CD14 monoclonal antibodies (mAbs; Biosource, Camarillo, California) characterized the cells as T lymphocytes (controls: 10%-20% and patients: 35%-40%), B lymphocytes (controls: 0.8%-1% and patients: 0.5%-1%), natural killer lymphocytes (controls: 2%-3% and patients: 5%-7%), and macrophages (controls: 75%-80% and patients: 45%-60%), respectively, in accordance with Oosterlynck et al.20 An aliquot of these cells was immediately used to evaluate mTNF-α, TNFR1, and TNFR2 expression by IIF. The remaining cells were stored at −80°C until assay for TNF-α, TNFR1, and TNFR2 mRNA and protein by real-time polymerase chain reaction (PCR) and Western blotting, respectively.
Determination of TNF-α, TNFR1, and TNFR2 mRNA Levels by Real-Time PCR
Total RNA from 1 × 106 isolated PFMCs was extracted by Trizol reagent (Invitrogen, Carlsbad). After precipitation and cold ethanol washing, the RNA was dried and dissolved in an appropriate volume of Tris-EDTA buffer (10 mmol/L Tris–HCl, pH 8.0, and 1 mmol/L EDTA). All RNA samples were treated with DNase I (Invitrogen) to remove any contaminating DNA. The RNA solution was quantified at 260 nm by spectrophotometer reading (Biomate 3; Thermo Electron Corporation, Waltham, Massachusetts), and purity was assessed by the ratio of absorbance at 260 and 280 nm. Only samples with a ratio of >1.8 were used for this study. Total RNA (2.5 μg) was reverse transcribed in complementary DNA (cDNA) using hexameric random primers and a High Capacity cDNA archive kit (Applied Biosystem, Bedford) in accordance with the manufacturer’s instructions. Specific primer pairs were designed for human TNF-α, TNFR1, and TNFR2 mRNA target sequences acquired from the GenBank database (Table 1). Amplicons were less than 150 bp in length and expected amplicon size was validated by agarose gel electrophoresis and ethidium bromide fluorochromatization. In order to quantify TNF-α, TNFR1, and TNFR2 mRNAs, 0.25 μg of total cDNA was used for the real-time PCR method with β-actin as housekeeping control in monoplex for each target. The analysis was performed in triplicate on a modified 7500 real-time PCR system (Applied Biosystem), using Sybr Green Dye (Fast Sybr Green Mastermix; Applied Biosystem) and primers as shown in Table 1. The levels of each target were calculated by the relative quantification method. After β-actin normalization, the results are expressed as the n-fold differences with respect to the mean levels for controls (relative expression).
Table 1.
Sequences of Gene-Specific Primer Sets Used in PCR.
| Genes | Entrez Gene No. | Primer sequences | |
|---|---|---|---|
| TNF-α | 7124 | Forward | 5′-ATGAGCACTGAAAGCATGATC-3′ |
| Reverse | 5′-AAAGTGCAGCAGGCAGAAGA-3′ | ||
| TNFR1 | 7132 | Forward | 5′-TATTGGACTGGTCCCTCACC-3′ |
| Reverse | 5′-GTCATTGTACAAGTAGGTTC-3′ | ||
| TNFR2 | 7133 | Forward | 5′-GAACCAGCCACAGGCACCA-3′ |
| Reverse | 5′-ACGATGCAGGTGACATTGAC-3′ | ||
| β-Actin | 60 | Forward | 5′-TCTACAATGAGCTGCGTGTG-3′ |
| Reverse | 5′-ATAGCACAGCCTGGATAGCA-3′ |
Abbreviations: PCR, polymerase chain reaction; TNF-α, tumor necrosis factor-α; TNFR, tumor necrosis factor receptor.
Determination of TNF-α, TNFR1, and TNFR2 Protein Levels by Western Blotting
The PFMCs were lysed in ice-cold radioimmunoprecipitation assay buffer (50 μL for 1 × 106 cells) with a cocktail of protease inhibitors. The cell extracts were centrifuged at 12 000g at 4°C for 20 minutes, and the resultant supernatants were used for Western blotting analysis. Protein concentrations were determined using a protein assay kit (DC Protein; Bio-Rad Lab, Hercules, California) with bovine serum albumin as standard. The protein samples (40 μg per lane) were mixed with an equal volume of Laemmli buffer (Bio-Rad Lab) and boiled for 10 minutes before loading. This mixture was resolved on a 9% sodium dodecyl sulfate-polyacrylamide gel electrophoresis at 150 V, under reducing and nonreducing conditions. The separated proteins were transferred onto polyvinylidene difluoride membrane (Amersham Life Science, Arlington Heights, Illinois) and kept at a constant voltage of 100 V for almost 4 hours at 4°C in a transfer buffer. Membranes were blocked at room temperature for 1 hour with 5% nonfat milk in TTBS (50 mmol/L Tris, pH 7.5, 0.9% NaCl, and 0.1% Tween-20). Blots were then incubated under gentle agitation at 4°C overnight in a 1:200 dilution of mouse antibody against TNF-α (Abcam, Cambridge, United Kingdom), a 1:1000 dilution of mouse antibody against TNFR1 (BD Transduction Laboratories, Franklin Lakes New Jersey), a 1:500 dilution of rabbit antibody against TNFR2 (Cell Signaling, Beverly, Massachusetts), and a 1:5000 dilution of mouse antibody against β-actin (Sigma-Aldrich, St. Louis, Missouri), all diluted in TTBS. Membranes were then washed 3 times for 10 minutes each and incubated in blocking buffer containing a 1:2000 dilution of appropriate horseradish peroxidase -conjugated secondary antibody (Sigma-Aldrich). After 30-minute wash time, the bound secondary antibody was detected using the West-Pico chemiluminescent detection kit following the manufacturer’s protocol (Pierce Biotechnology, Rockford, Illinois) and by autoradiography. The quantification of the detected compound was carried out on a DIGI DOC apparatus for image acquisition using Quantity One software (Bio-Rad). Once again, the results are expressed as the n-fold difference with respect to the mean levels for controls (relative expression) after β-actin normalization.
Determination of mTNF-α, TNFR1, and TNFR2 Positive Cell Percentages by IIF
The fresh PFMCs were counted and suspended (1 × 107/mL) in PBS containing 0.5% NaN3. The cell preparations were incubated with an appropriate dilution of anti-mTNF-α (Abcam), anti-TNFR1 (CD120a), or anti-TNFR2 (CD120b) primary mAbs (Biosource) for 30 minutes and then washed twice with washing buffer (PBS containing 2% fetal calf serum and 0.1% NaN3). The cells were then reacted with fluorescein-conjugated goat antimouse immunoglobulin G3 antibodies (Biotest, Dreich, Germany) as a secondary antibody for 30 minutes and washed twice. Finally, the cells were suspended in PBS containing 30% glycerol. All procedures were performed at 4°C. Fluorescence was read at 450 nm wavelength using fluorescence microscopy. Membrane TNF-α-, TNFR1-, and TNFR2-positive PFMCs are expressed as percentages (%) of all assayed cells.
Determination of sTNF-α by Enzyme-Linked Immunosorbent Assay
Determinations of sTNF-α were performed by standard enzyme-linked immunosorbent assay using commercially available kits (Diaclone, France) according to the manufacturer’s instructions. Briefly, triplicate aliquots of each sample, including standards of known cytokine content, control specimens, and experimental specimens, were placed in 96-microtiter plates. During the first incubation, the cytokine antigen and a biotinylated polyclonal-specific antibody were simultaneously incubated. After washing, the enzyme (streptavidin-peroxidase) was added to the wells. After incubation and washing to remove all the unbound enzyme, a substrate solution was added and finally color development was stopped by the addition of H2SO4. The colored product was directly proportional to the concentration of the cytokine present in the samples. The absorbance of each well was read on a spectrophotometer using 450 nm as the wavelength. Assay sensitivities of the kit were approximately <10 pg/mL. The intra-assay coefficient of variation (CV) was approximately <7.4% and the inter-assay CV was <10.3%. Results were obtained by reference curve and expressed as pg/mL.
Statistical Analysis
The assumption of normal distribution for continuous variables was tested by Kolmogorov-Smirnov test for goodness of fit. All inferential analyses were made using nonparametric statistical tests.21 Nonnormally distributed variables between the groups were compared using the Kruskal-Wallis test. The range of statistical significance was P < .05. In addition, we used post hoc multiple comparison tests either versus controls or versus each stage to analyze data. Moreover, we used post hoc multiple comparison tests either versus controls or versus each stage to analyze data. In addition, we used the nonparametric correlation (Spearman) to value association between TNF-α, TNFR1, and TNFR2 mRNA and protein levels in PFMCs. Correlation was considered significant at the .01 level (2-tailed).
Results
Tumor Necrosis Factor-α Content of PFMCs
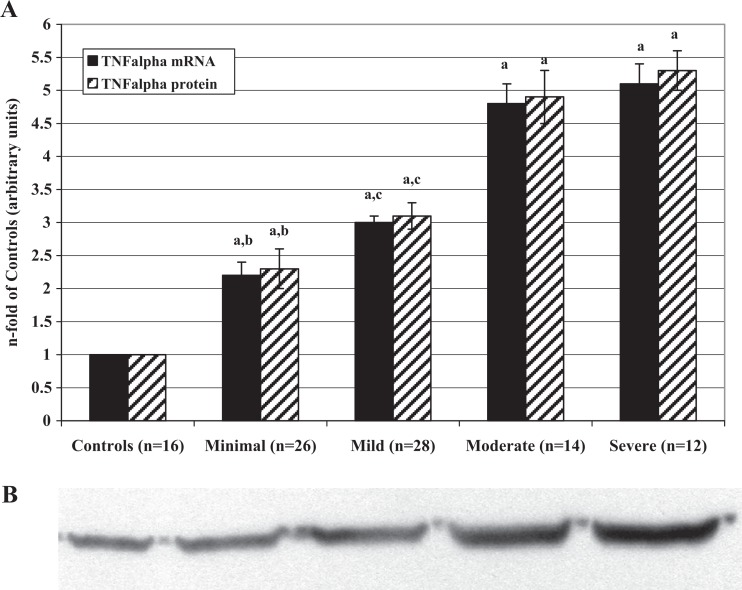
Tumor necrosis factor-α mRNA and protein levels (Figure 1) as well as the percentages of mTNF-α-positive PFMCs (Table 2) significantly increased from minimal to severe stage of the disease. All values of patient samples were significantly higher than those of control samples. Moreover, there is a significant correlation between mRNA and protein levels in controls (r = .815; P < .001), minimal (r = .820; P < .001), mild (r = .750; P < .005), moderate (r = .788; P < .002), and severe (r = .752; P < .005) stages.
Figure 1.
A, Levels of tumor necrosis factor-α mRNA and protein in PFMCs of women with and without endometriosis (controls). RNAs were extracted from isolated PFMCs and subjected to RT-PCR. The resulting cDNAs were then assayed by RT-PCR to determine expression of FasL as described in the text. Proteins were extracted from PFMC lysates and analyzed by Western blot using the antibodies against TNF-α. Values are the mean ± SD of n patients and are expressed as the n-fold increase in mRNA and protein levels in the stages of endometriosis with respect to the controls. Each sample was assessed in triplicate. The control group bar is set to 1. a P < .001 all the stages of endometriosis versus controls; b P < .001 minimal versus mild, moderate, and severe endometriosis; c P < .001 mild versus moderate and severe endometriosis. B, Representative Western blot image of the TNF-α proteins measured in PFMCs of women with and without endometriosis (controls). mRNA indicates messenger RNA; PFMCs, peritoneal fluid mononuclear cells; RT-PCR, real-time quantitative polymerase chain reaction; cDNA, complementary DNA; TNF-α, tumor necrosis factor α; SD, standard deviation.
Table 2.
Percentages of mTNF-α-, TNFR1-, and TNFR2-Positive PFMCs in Women With Endometriosis at Different Stages.
| Stage | n | mTNF-αa | TNFR1a | TNFR2a |
|---|---|---|---|---|
| Controls | 16 | 0.5 ± 0.02 | 1.6 ± 0.1 | 2.9 ± 0.2 |
| Minimal | 26 | 9.1 ± 0.8b,c,d | 9.3 ± 0.8b,c,d | 9.6 ± 0.9b,c,d |
| Mild | 28 | 11.7 ± 1.2b,e | 7.5 ± 0.7b,f | 12.9 ± 1.1b,f |
| Moderate | 14 | 15.1 ± 1.2b | 5.3 ± 0.4b | 14.8 ± 1.3b |
| Severe | 12 | 16.4 ± 1.7b | 4.9 ± 0.4b | 15.3 ± 1.4b |
Abbreviations: mTNF-α, membrane tumor necrosis factor-α; TNFR, tumor necrosis factor receptor; SD, standard deviation.
a Data are mean ± SD of triplicate analyses for each sample.
b P < .001 all the stages versus the controls.
c P < .05 at the minimal versus the mild stage.
d P < .001 at the minimal versus the moderate and severe stages.
e P < .001 at the mild versus the moderate and severe stages.
f P < .05 at the mild versus the moderate and severe stages.
Tumor Necrosis Factor Receptor 1 Content of PFMCs
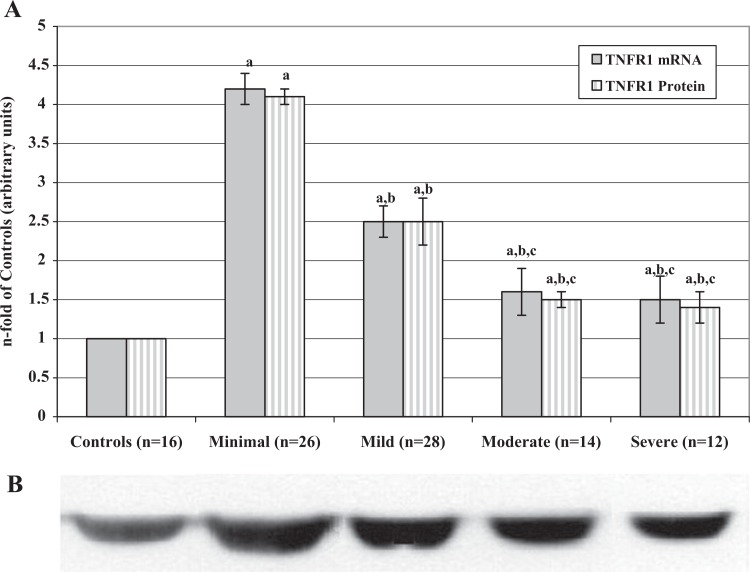
We observed that both TNFR1 mRNA and protein levels (Figure 2) were significantly higher in endometriosis groups than in controls and significantly decreased from minimal to severe stage of the disease. Also, the percentages of TNFR1-positive PFMCs (Table 2) in women with endometriosis were always significantly higher than those of controls and decreased with the worsening of the disease. Moreover, there is a significant correlation between mRNA and protein levels in controls (r = .830; P < .001), minimal (r = .818; P < .001), mild (r = .830; P < .001), moderate (r = .815; P < .005), and severe (r = .820; P < .005) stages.
Figure 2.
A, Tumor necrosis factor receptor 1 mRNA and protein levels in PFMCs of women with and without endometriosis. RNAs were extracted from isolated PFMCs and subjected to RT-PCR. The resulting cDNAs were then assayed by real-time quantitative PCR to determine expression of TNFR1 as described in the text. Proteins were extracted from PFMC lysates and analyzed by Western blot using the antibodies against TNFR1. Values are the mean ± SD of n patients and are expressed as the n-fold increase in mRNA and protein levels in the stages of endometriosis with respect to the controls. Each sample was assessed in triplicate. The control group bar is set to 1. a P < .001 all the stages of endometriosis versus controls; b P < .001 mild, moderate, and severe versus minimal endometriosis; c P < .001 moderate and severe versus mild endometriosis. B, Representative Western blot image of the TNFR1 proteins measured in PFMCs of women with and without endometriosis (controls). TNFR1 indicates tumor necrosis factor receptor 1; mRNA, messenger RNA; PFMCs, peritoneal fluid mononuclear cells; RT-PCR, real-time quantitative polymerase chain reaction; SD, standard deviation.
Tumor Necrosis Factor Receptor 2 Content of PFMCs
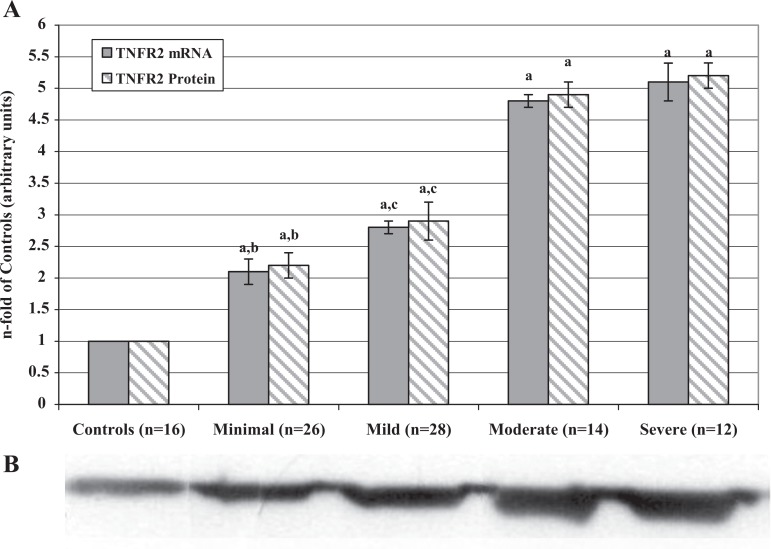
In contrast to TNFR1, TNFR2 mRNA and protein levels (Figure 3) as well as the percentages of TNFR2-positive PFMCs (Table 2) significantly increased from minimal to severe stage of the disease. All values of patient samples were significantly higher than those of the control samples. Moreover, there is a significant correlation between mRNA and protein levels in controls (r = .820; P < .001), minimal (r = .810; P < .001), mild (r = .815; P < .001), moderate (r = .807; P < .001), and severe (r = .812; P < .001) stages.
Figure 3.
A, Tumor necrosis factor receptor 2 mRNA and protein levels in PFMCs of women with and without endometriosis. RNAs were extracted from isolated PFMCs and subjected to RT-PCR. The resulting cDNAs were then assayed by real-time quantitative PCR to determine expression of TNFR2 as described in the text. Proteins were extracted from PFMC lysates and analyzed by Western blot using the antibodies against TNFR2. Values are the mean ± SD of n patients and are expressed as the n-fold increase in mRNA and protein levels in the stages of endometriosis with respect to the controls. Each sample was assessed in triplicate. The control group bar is set to 1. a P < .001 all the stages of endometriosis versus controls; b P < .001 minimal versus mild, moderate, and severe endometriosis; c P < .001 mild versus moderate and severe endometriosis. B, Representative Western blot image of the TNFR2 proteins measured in PFMCs of women with and without endometriosis (controls). TNFR2 indicates tumor necrosis factor receptor 2; mRNA, messenger RNA; PFMCs, peritoneal fluid mononuclear cells; RT-PCR, real-time quantitative polymerase chain reaction; SD, standard deviation.
Soluble TNF-α Content of PF
We found that sTNF-α concentrations in PF of women with endometriosis were always significantly higher than those of controls and significantly decreased from minimal to severe stage of the disease (Table 3).
Table 3.
Values of sTNF-α in Peritoneal Fluid of Women With Endometriosis at Different Stages.
| Stage | n | pg/mLa |
|---|---|---|
| Controls | 16 | 1.2 ± 0.1 |
| Minimal | 26 | 130.7 ± 8.9b,c |
| Mild | 28 | 73.2 ± 6.4b,d |
| Moderate | 14 | 19.7 ± 1.9b |
| Severe | 12 | 20.6 ± 1.8b |
Abbreviations: sTNF-α, soluble tumor necrosis factor α; SD, standard deviation.
a Data are mean ± SD of triplicate analyses for each sample.
b P < .001 all the stages versus the controls.
c P < .001at the minimal versus the mild, moderate and severe stages.
d P < .001at the mild versus the moderate and severe stages.
Discussion
The system of TNF-α/TNFRs forms a regulatory network that may control the immune response. The utilization of different signaling mechanisms by TNFR1 and TNFR2 is consistent with the ability of each receptor to trigger responses depending on the different forms of the ligand (soluble or mTNF-α), the cellular context, and the microenvironmental conditions. Our results show that TNFR1 mRNA and protein levels, percentages of TNFR1-bearing PFMCs, as well as sTNF-α concentration in PF were significantly higher in endometriosis groups than in controls. The most significant increase of this receptor and of this ligand occurs in minimal and mild stages, whereas these values decrease with the worsening of the disease (moderate and severe stages). Considering that TNFR1 is bound and efficiently activated by both sTNF-α and mTNF-α,15 we hypothesize that, in minimal and mild stages, the overexpression of TNFR1 on PFMC surface membranes and the high levels of sTNF-α in PF could address signal toward complex I pathway. This pathway controls the expression of antiapoptotic proteins that prevent the triggering of cell death processes,15 leading to the activation of the survival transcription factor nuclear factor kappa B, as proposed by Micheau and Tschopp.22 In this way, activation of complex I pathway could allow, at least in part, the survival of PFMCs, enhancing and favoring the inflammatory response that characterizes minimal and mild stages. The low percentages of mTNF-α–bearing PFMCs observed by us in the same stages may not allow PFMCs to play a role in this process. As suggested by Haider and Knofler,23 sTNF-α in the milieu could arise also from the endometriotic cells, since they as well as eutopic endometrial cells express mTNF-α which may be cleaved. Instead, with the worsening of the disease, the observed low percentages of TNFR1-bearing PFMCs are probably due to decreased TNFR1 low mRNA transcription and protein translation rate, according to our results, and not to its internalization or shedding. For these reasons, we hypothesize that, in the moderate and severe stages, TNFR1 may not play a key role in the PFMC fate.
TNFR2 behaves in opposite way with respect to TNFR1. In fact, TNFR2 mRNA and protein levels as well as percentages of TNFR2-bearing PFMCs increase from minimal to severe stages of endometriosis. TNFR2 may be bound either by soluble and membrane forms of TNF-α but, in contrast to TNFR1, it is efficiently activated by the last one only.15 In the minimal and mild stages, the percentages of both TNFR2- and mTNF-α−bearing PFMCs are so low that subsequent cellular events may depend only minimally by this interaction. We hypothesize that the decrease in TNFR2-bearing PFMCs may be due, also in this case, to low mRNA transcription and protein translation rate, observed by us, and not to increased shedding or internalization of the surface membrane receptor. In the same stages, the high levels of sTNF-α may be rerouted to bind TNFR1, due to the low presence of both mTNF-α- and TNFR2-bearing PFMCs. In the moderate and severe stages, in contrast, we found an overexpression of TNFR2 mRNA and protein, high percentages of TNFR2-bearing PFMCs, and of mTNF-α–bearing PFMCs. Considering that TNFR2 is efficiently activated by mTNF-α only,15 we hypothesize that TNFR2 on PFMCs may be saturated by mTNF-α, so triggering death process, in the context of limiting the immune response.24 Moreover, both surface membrane TNFR1 and TNFR2 are able to bind mTNF-α on the opposing cells and trigger signal transduction in the mTNF-α−bearing cells, the so-called “reverse signaling.”25 This, together with the low presence of TNFR1-bearing PFMCs, allows us to hypothesize that the binding between opposing TNFR2- and mTNF-α–bearing cells could induce apoptosis, paradoxically, in the latter. All together, our data allow us to point out the ability of each component of the system of TNF-α/TNFRs to trigger opposite cellular fate as proliferation or death, so underlying the coexistence of inflammatory and reparative phenomena in endometriosis. Additional studies on the role of TNF-α/TNFRs system are required for a better understanding of the pathogenesis of endometriosis.
Acknowledgments
The authors are grateful to Sandy Maria Cartella and Luigi Campanelli for providing language help and to Prof Giuseppe Trimarchi, PhD, for statistical analysis.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The work was supported by a grant by University of Messina, PRA 2004, Tit.2, Cat.21, CAP.01.
References
- 1. Bulun SE, Gurates B, Fang Z, et al. Mechanisms of excessive estrogen formation in endometriosis. J Reprod Immunol. 2002;55(1-2):21–33. [DOI] [PubMed] [Google Scholar]
- 2. Baldi A, Campioni N, Signorile PG. Endometriosis: pathogenesis, diagnosis, therapy and association with cancer. Oncol Rep. 2008;19(4):843–846. [PubMed] [Google Scholar]
- 3. Bulun SE. Endometriosis. N Engl J Med. 2009;360(3):268–279. [DOI] [PubMed] [Google Scholar]
- 4. Arici A, Oral E, Attar E, Tazuke SI, Olive DL. Monocyte chemotactic protein-1 concentration in peritoneal fluid of women with endometriosis and its modulation of expression in mesothelial cells. Fertil Steril. 1997;67(6):1065–1072. [DOI] [PubMed] [Google Scholar]
- 5. Braun DP, Ding J, Shen J, Rana N, Fernandez BB, Dmowski WP. Relationship between apoptosis and the number of macrophages in eutopic endometrium from women with and without endometriosis. Fertil Steril. 2002;78(4):830–835. [DOI] [PubMed] [Google Scholar]
- 6. Pizzo A, Salmeri FM, Ardita FV, Sofo V, Tripepi M, Marsico S. Behaviour of cytokine levels in serum and peritoneal fluid of women with endometriosis. Gynecol Obstet Invest. 2002;54(2):82–87. [DOI] [PubMed] [Google Scholar]
- 7. Cao X, Yang D, Song M, Murphy A, Parthasarathy S. The presence of endometrial cells in the peritoneal cavity enhances monocyte recruitment and induces inflammatory cytokines in mice: implications for endometriosis. Fertil Steril. 2004;82(suppl 3):999–1007. [DOI] [PubMed] [Google Scholar]
- 8. Tariverdian N, Siedentopf F, Rücke M, et al. Intraperitoneal immune cell status in infertile women with and without endometriosis. J Reprod Immunol. 2009;80(1-2):80–90. [DOI] [PubMed] [Google Scholar]
- 9. Kalu E, Sumar N, Giannopoulos T, et al. Cytokine profiles in serum and peritoneal fluid from infertile women with and without endometriosis. J Obstet Gynaecol Res. 2007;33(4):490–495. [DOI] [PubMed] [Google Scholar]
- 10. Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364(9447):1789–1799. [DOI] [PubMed] [Google Scholar]
- 11. Ulukus M, Arici A. Immunology of endometriosis. Minerva Ginecol. 2005;57(3):237–248. [PubMed] [Google Scholar]
- 12. Harada T, Taniguchi F, Izawa M, et al. Apoptosis and endometriosis. Front Biosci. 2007;12:3140–3151. [DOI] [PubMed] [Google Scholar]
- 13. Agic A, Djalali S, Diedrich K, et al. Apoptosis in endometriosis. Gynecol Obstet Inves. 2009;68(4):217–233. [DOI] [PubMed] [Google Scholar]
- 14. Sturlese E, Salmeri FM, Retto G, et al. Dysregulation of the Fas/FasL system in mononuclear cells recovered from peritoneal fluid of women with endometriosis. J Reprod Immunol 2011;92(1-2):74–81. [DOI] [PubMed] [Google Scholar]
- 15. Cabal-Hierro L, Lazo PS. Signal transduction by tumor necrosis factor receptors. Cell Signal. 2012;24(6):1297–1305. [DOI] [PubMed] [Google Scholar]
- 16. Richter C, Messerschmidt S, Holeiter G, et al. The tumor necrosis factor receptor stalk regions define responsiveness to soluble versus membrane-bound ligand. Mol Cell Biol. 2012;32(13):2515–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koga K, Osuga Y, Tsutsumi O, et al. Increased concentrations of soluble tumour necrosis factor receptor (sTNFR) I and II in peritoneal fluid from women with endometriosis. Mol Hum Reprod. 2000;6(10):929–933. [DOI] [PubMed] [Google Scholar]
- 18. American Society for Reproductive Medicine. Revised American society for reproductive medicine classification of endometriosis. Fertil Steril. 1996;67(5):817–821. [DOI] [PubMed] [Google Scholar]
- 19. Yoshino O, Osuga Y, Koga K, et al. Concentrations of interferon-γ-induced protein-10 (IP-10), an antiangiogenic substance, are decreased in peritoneal fluid of women with advanced endometriosis. Am J Reprod Immunol. 2003;50(1):60–65. [DOI] [PubMed] [Google Scholar]
- 20. Oosterlynck DJ, Meuleman C, Waer M, Vandeputte M, Koninckx PR. The natural killer activity of peritoneal fluid lymphocytes is decreased in women with endometriosis. Fertil Steril. 1992;58(2):290–295. [DOI] [PubMed] [Google Scholar]
- 21. Siegel S, Castellan NJ. Nonparametric Statistics for Behavioural Science. eds. London, UK: McGraw-Hill Education; 1988. [Google Scholar]
- 22. Micheau O, Tschopp J. Induction of TNF Receptor I-Mediated Apoptosis via Two Sequential Signaling Complexes. Cell. 2003;114(2):181–190. [DOI] [PubMed] [Google Scholar]
- 23. Haider S, Knofler M. Human tumour necrosis factor: physiological and pathological roles in placenta and endometrium. Placenta. 2009;30(2):111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tchikov V, Bertsch U, Fritsch J, Edelmann B, Schütze S. Subcellular compartmentalization of TNF receptor-1 and CD95 signaling pathways. Eur J Cell Biol. 2011;90(6-7):467–475. [DOI] [PubMed] [Google Scholar]
- 25. Rossol M, Meusch U, Pierer M, et al. Interaction between Transmembrane TNF and TNFR1/2 mediates the activation of monocytes by contact with T cells. J Immunol. 2007;179(6):4239–4248. [DOI] [PubMed] [Google Scholar]