Abstract
Aims/hypothesis
We aimed to identify microRNAs (miRNAs) associated with type 2 diabetes and risk of developing the disease in skeletal muscle biopsies from phenotypically well-characterised twins.
Methods
We measured muscle miRNA levels in monozygotic (MZ) twins discordant for type 2 diabetes using arrays. Further investigations of selected miRNAs included target prediction, pathway analysis, silencing in cells and association analyses in a separate cohort of 164 non-diabetic MZ and dizygotic twins. The effects of elevated glucose and insulin levels on miRNA expression were examined, and the effect of low birthweight (LBW) was studied in rats.
Results
We identified 20 miRNAs that were downregulated in MZ twins with diabetes compared with their non-diabetic co-twins. Differences for members of the miR-15 family (miR-15b and miR-16) were the most statistically significant, and these miRNAs were predicted to influence insulin signalling. Indeed, miR-15b and miR-16 levels were associated with levels of key insulin signalling proteins, miR-15b was associated with the insulin receptor in non-diabetic twins and knockdown of miR-15b/miR-16 in myocytes changed the levels of insulin signalling proteins. LBW in twins and undernutrition during pregnancy in rats were, in contrast to overt type 2 diabetes, associated with increased expression of miR-15b and/or miR-16. Elevated glucose and insulin suppressed miR-16 expression in vitro.
Conclusions
Type 2 diabetes is associated with non-genetic downregulation of several miRNAs in skeletal muscle including miR-15b and miR-16, potentially targeting insulin signalling. The paradoxical findings in twins with overt diabetes and twins at increased risk of the disease underscore the complexity of the regulation of muscle insulin signalling in glucose homeostasis.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-014-3434-2) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Keywords: Insulin signalling, Low birthweight, MicroRNA, miR-15b, miR-16, Muscle, Twins, Type 2 diabetes
Introduction
Type 2 diabetes is caused by multiple organ defects, with inadequate insulin secretion and insulin resistance being the most important. Skeletal muscle insulin resistance is of specific interest as muscle is the major site for glucose disposal [1]. The insulin signalling cascade is initiated by the binding of insulin to its receptor, which activates the key cellular processes of glucose metabolism, including glucose transport across the membrane and storage of glucose as muscle glycogen. Although the genetic influence on muscle insulin resistance cannot be disputed [2], non-genetic factors, including obesity, physical inactivity and low birthweight (LBW) [3–6], are important in controlling muscle insulin resistance [7–9].
Emerging evidence points towards microRNAs (miRNAs) as major players in the regulation of protein levels. These miRNAs are short single-stranded RNA molecules that regulate their target mRNAs mainly by translational repression and/or degradation [10]. We recently demonstrated that miR-483-3p, involved in adipose tissue development and lipid storage, is upregulated in adipose tissue from individuals born with LBW as well as in young adult rats exposed to a sub-optimal intrauterine environment [11]. Another recent study indicated that several miRNAs were differentially expressed in skeletal muscle from patients with type 2 diabetes [12], and Zhu et al recently demonstrated that overexpression of the miRNA let-7 results in insulin resistance and impaired glucose tolerance in rats, probably through repression of proteins in the insulin–phosphoinositide 3-kinase–mechanistic target of rapamycin (insulin–PI3K–mTOR) pathway [13]. Thus, miRNAs could therefore be important regulators of insulin signalling.
We have previously provided extensive proof-of-concept evidence for the powerful approach of using genetically identical monozygotic (MZ) twins discordant for type 2 diabetes to identify non-genetic metabolic and molecular defects in type 2 diabetes [4, 14]. When unravelling the complex underlying molecular mechanisms involved in the development of type 2 diabetes, it is important to distinguish between the primary mechanisms relevant to the development of type 2 diabetes and the secondary molecular alterations occurring as a consequence of the metabolic derangements including ‘glucotoxicity’ in hyperglycaemic patients with overt type 2 diabetes.
In the present study, we first aimed to identify altered miRNA levels of non-genetic origin in MZ twins discordant for type 2 diabetes. The top candidates identified were subsequently investigated in a larger group of phenotypically well-characterised twins without diabetes to uncover their role in skeletal muscle insulin action. Our comprehensive approach also included computational target prediction and in vitro target validation using antagomirs, while the effects of elevated glucose and insulin levels on miRNA expression level were studied in vitro. Finally, we studied miRNA expression in a rat model of LBW.
Methods
Participants
The MZ discordant twin population comprised 22 white MZ twins (11 pairs), aged 53–78 years, discordant for type 2 diabetes who were recruited through the Danish Twin Registry, University of Southern Denmark. Five pairs were from a previous cohort [15–17] where discordance for type 2 diabetes was recognised based on an OGTT. The additional six twin pairs were recruited based on their discordance for type 2 diabetes. The recruitment and clinical examinations of all 11 pairs has been described previously [18]. The basic clinical and anthropometric measures are presented in Table 1. Based on a 2 h OGTT, nine of the non-diabetic co-twins had impaired glucose tolerance (IGT) and two had normal glucose tolerance (NGT).
Table 1.
Clinical and metabolic characteristics of the study populations
| Characteristic | Discordant monozygotic twin study | Non-diabetic twin study | ||
|---|---|---|---|---|
| Diabetic twins | Non-diabetic co-twins | p value | ||
| n (female/male) | 11 (6/5) | 11 (6/5) | 164 (76/88) | |
| n (NGT/IGT/T2D) | 11 (0/0/11) | 11 (2/9/0) | — | |
| Zygosity (MZ/DZ) | — | — | 90/74 | |
| Age (years) | 68 ± 8 | 68 ± 8 | 42.9 ± 17.1 | |
| Birthweight (g) | — | — | 2,671 ± 468 | |
| BMI (kg/m2) | 32.5 ± 6.7 | 30.4 ± 6.5 | 0.04 | 25.3 ± 4 |
| Fasting glucose (mmol/l) | 10.6 ± 2.1 | 6.3 ± 0.5 | <0.001 | 5.4 ± 0.7 |
| Glu120 (mmol/l) | 19.5 ± 3.5 | 9.0 ± 1.7 | <0.0001 | 6.5 ± 1.7 |
| GIR (mg m−2 min−1)a | 153.8 ± 63.2 | 288.1 ± 175.4 | 0.05 | — |
| HbA1c (%)b | 7.7 ± 1.5 | 5.9 ± 0.5 | <0.003 | — |
| HbA1c (mmol/mol) | 61 ± 16.4 | 41 ± 5.5 | — | |
| Total fat (%) | — | — | 24.7 ± 9 | |
| R d (mg kgFFM −1 min−1) | — | — | 10.9 ± 3.4 | |
| (ml kg−2 min−1) | — | — | 33.9 ± 10.1 | |
Data are represented as mean ± SD
a n = 9 in each group; b n = 10 in each group
FFM, fat-free mass; Glu120, 120 min glucose values from an OGTT
The non-diabetic twin population was a subgroup of 164 MZ and same-sex dizygotic (DZ) twins (the number of individuals with available tissue material) from a previously described population [7] of 196 young and old twins (aged 25–32 and 58–66 years). Participant characteristics are displayed in Table 1.
All study participants gave informed consent and the study was carried out in accordance with the Declaration of Helsinki as revised in 2008.
Clinical examination
In the MZ discordant twin population, insulin sensitivity was measured by a 2 h euglycaemic–hyperinsulinaemic clamp (40 mU m−2 min−1) and expressed as the mean glucose infusion rate (GIR) during the final 30 min steady-state period of the clamp. In the non-diabetic twin population, 3-3H-glucose was infused for the calculation of the peripheral glucose disposal rate (Rd), and body composition was determined by dual-energy x-ray absorptiometry (DXA) scanning. A 2 h 75 g OGTT was performed in both populations and muscle biopsies (vastus lateralis) were excised under local anaesthesia (lidocaine), during basal fasting conditions using a Bergström needle with suction applied. The biopsies were frozen immediately in liquid nitrogen and stored at −80°C for later analysis. In the non-diabetic twin population, physical fitness was estimated as maximal oxygen consumption rate () calculated from the maximal load on ergometer bicycle [19]. More details can be found in previous publications of this cohort [7, 15–18].
LNA arrays
For each of the discordant twins (n = 22), 1 μg total RNA was isolated from a muscle biopsy (described in electronic supplementary material [ESM] methods) and each sample was run on a miRCURY locked nucleic acid (LNA) array (Exiqon, Vedbaek, Denmark) representing version 9.2 of the Sanger miRBase as described in the ESM Methods. Annotation was updated to miRBase version 14, and the data were normalised using normexp [20] and quantile normalisation [21] in R (version 2.9.1). The dataset has been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus [22], accession number GSE38835.
Familiar relationships and target analysis
The familiar relationships between miRNAs were based on miRBase version 17. TargetScan 4.2 [23–25] was used to predict mRNA targets. The lists of targets were imported into Ingenuity pathway analysis (IPA) version 8.0 (IPA, QIAGEN, Redwood City, www.qiagen.com/ingenuity) for canonical pathway analysis or the Database for Annotation, Visualization and Integrated Discovery (DAVID) 6.2. for a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis [26]. The IPA results were corrected for multiple testing using the Benjamini–Hochberg method.
Statistical methods
Differential expression levels in the discordant twins were assessed using a paired design matrix with linear models for microarray data (Limma) [27, 28] in R. Spot quality weights [29] were used and p values were adjusted with Benjamini–Hochberg’s false-discovery rate (FDR) [30]. Limma was used to analyse 170 miRNAs in a published online dataset [12] for differences between those with NGT, IGT and type 2 diabetes. Pearson intra-pair correlation coefficients were calculated using 2n, as previously recommended [31]. The SAS ‘proc mixed’ procedure (SAS Institute, Cary, NC, USA) was used to perform multiple regression analysis with adjustments for zygosity and twin-pair status. If not otherwise stated, additional adjustment for age, sex, , birthweight and total body fat percentage was performed. GraphPad Prism (GraphPad Software, La Jolla, CA, USA) was used to perform ANOVA.
Human myocytes
AntagomiR experiment
Proliferating myoblasts from five healthy donors (three men and two women) aged 59 ± 7 years were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) with 10 nmol/l antagomiRs (Exiqon) targeting miR-15b/miR-16 or a non-targeting antagomiR (Exiqon) for 48 h. Simultaneous knockdown of miR-15b and miR-16 was performed to hinder a possible rescue effect that could occur when inhibiting them separately. Cells were harvested after 5–10 min incubation with 0, 1, 10 or 100 nmol/l insulin (Actrapid, Novo Nordisk, Bagsværd, Denmark) in Trizol (Invitrogen) for RNA isolation or in lysis buffer (Cell Signaling Technology, Beverly, MA, USA) for protein isolation. See the ESM Methods for further details.
Short-term insulin experiment
See the ESM Methods for the description of this experiment.
Rat protocol
Adult females were fed ad libitum with either a control diet (20% protein) or an isoenergetic low-protein (LP) (8% protein) diet (Arie Blok, Woerden, the Netherlands) during gestation and lactation as described previously [32]. At 3 months of age, 11 control males and 11 males from LP litters, each of the 22 rats being from an independent litter, were killed and vastus lateralis muscle biopsies were excised and stored for later use. All animal procedures were approved by the local animal ethical review committee and were carried out under compliance with the UK Animal (Scientific Procedures) Act 1986. The investigation conforms to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication no. 85–23, revised 1996). See the ESM Methods for further details.
L6 cells
L6 myoblasts were obtained from the American Type Culture Collection (CRL-1458, ATCC) and cultured in DMEM (Sigma-Aldrich, Poole, UK) with 10% fetal bovine serum, penicillin (100 units/ml), streptomycin (100 mg/ml) and glutamine (0.2 mmol/l), as previously described [33]. Semi-confluent cultures were treated with four different combinations of insulin (Actrapid; Novo Nordisk Insulin, Bagsværd, Denmark) and glucose levels, 0 nmol/l insulin and 5.6 mmol/l glucose, 100 nmol/l insulin and 5.6 mmol/l glucose, 0 nmol/l insulin and 25 mmol/l glucose or 100 nmol/l insulin and 25 mmol/l glucose, every 24 h for a total of 96 h until harvesting.
Gene expression
RNA isolation
This is described in the RNA isolation section of the ESM Methods. ABI-PRISM 7900 Sequence Detection system ([ABI] Applied Biosystems, Foster City, CA, USA) was used for both miRNA and mRNA detection. Expression was quantified using TaqMan probes for miRNA and primers from ABI or the Roche Universal probe library for mRNA. Details are described in the ESM Methods.
Protein levels
Muscle cell cultures
Levels of phosphoinositide 3-kinase R1 regulatory subunit 1 α (PIK3R1), Akt and p-Akt were measured using western blotting (Invitrogen). Insulin receptor (INSR) levels were measured using Meso Scale Discovery analysis (Meso Scale Discovery, Rockville, MD, USA).
Muscle biopsies
The protein levels of INSR, IRS-1 and PIK3R1 were measured using western blotting as described in [2, 34]. Details of these methods are described in the ESM Methods.
Results
Differentially expressed miRNAs in MZ twins discordant for type 2 diabetes
To identify miRNAs related to type 2 diabetes in a non-genetic manner, we studied 571 human miRNAs in skeletal muscle from 11 pairs of MZ twins discordant for type 2 diabetes. The clinical and metabolic characteristics of these twins are summarised in Table 1. Twenty miRNAs were identified as significantly downregulated after correction for multiple comparisons (pFDR < 0.1) in twins with type 2 diabetes compared with their corresponding non-diabetic co-twins (Table 2). No miRNAs were significantly upregulated in the type 2 diabetes twins. No difference in the total miRNA expression levels were found between the diabetic twins and their non-diabetic co-twins when the medians of the log2 transformed raw expression values were compared using a paired t test (p = 0.25). Box and density plots are shown in ESM Figs 1 and 2, respectively. Intra-pair correlations between miRNA expression and HbA1c showed that ten of the 20 identified miRNAs, including miR-15b, were negatively correlated with HbA1c levels. Of these miR-15b, miR-451 and miR-363 remained significant after FDR correction for multiple comparisons (pFDR < 0.1) (Table 2). Similar intra-pair correlations showed that miR-15b alone was significantly negatively correlated with the 2 h post-OGTT glucose values. This result was not significant after FDR correction (pFDR = 0.2) (Table 2).
Table 2.
Differentially expressed miRNAs in skeletal muscle biopsies from genetically identical twins with type 2 diabetes when compared with their non-diabetic co-twins
| miRNA | Family | C | FC | p | p (FDR) | r Glu120 | p-r Glu120 | p- | |
|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-15b | miR-15 | a | −2.0 | 2.4 × 10−8 | 8.0 × 10−6 | −0.64 | 0.04 | −0.77 | 0.01 |
| hsa-miR-16 | miR-15 | a | −1.7 | 5.7 × 10−6 | 9.4 × 10−4 | −0.41 | 0.21 | −0.63 | 0.05 |
| hsa-miR-17 | miR-17 | b | −1.7 | 7.3 × 10−5 | 0.007 | −0.53 | 0.1 | −0.66 | 0.04 |
| hsa-miR-451 | −1.8 | 9.0 × 10−5 | 0.007 | −0.55 | 0.08 | −0.76 | 0.01 | ||
| hsa-miR-106b | miR-17 | c | −1.5 | 2.2 × 10−4 | 0.01 | −0.57 | 0.07 | −0.64 | 0.05 |
| hsa-miR-103 | miR-103 | −1.5 | 5.3 × 10−4 | 0.03 | −0.4 | 0.1 | −0.67 | 0.03 | |
| hsa-miR-425 | −1.6 | 6.2 × 10−4 | 0.03 | −0.52 | 0.22 | −0.52 | 0.12 | ||
| hsa-miR-106a | miR-17 | d | −1.7 | 8.9 × 10−4 | 0.04 | −0.52 | 0.1 | −0.67 | 0.03 |
| hsa-miR-10b | −1.5 | 1.0 × 10−3 | 0.04 | 0.1 | 0.77 | −0.1 | 0.77 | ||
| hsa-miR-25 | miR-25 | c | −1.7 | 1.3 × 10−3 | 0.04 | −0.4 | 0.11 | −0.64 | 0.05 |
| hsa-miR-93 | miR-17 | c | −1.5 | 1.5 × 10−3 | 0.04 | −0.51 | 0.23 | −0.55 | 0.1 |
| hsa-miR-363 | d | −1.7 | 1.6 × 10−3 | 0.04 | −0.43 | 0.19 | −0.76 | 0.01 | |
| hsa-miR-107 | miR-103 | −1.6 | 1.8 × 10−3 | 0.05 | −0.56 | 0.07 | −0.57 | 0.08 | |
| hsa-miR-191 | −1.4 | 1.9 × 10−3 | 0.05 | −0.56 | 0.07 | −0.56 | 0.09 | ||
| hsa-miR-30e* | −1.6 | 2.2 × 10−3 | 0.05 | −0.2 | 0.56 | −0.1 | 0.78 | ||
| hsa-let-7i | −1.4 | 3.2 × 10−3 | 0.07 | −0.25 | 0.46 | −0.37 | 0.3 | ||
| hsa-miR-20a | miR-17 | b | −1.7 | 3.4 × 10−3 | 0.07 | −0.41 | 0.2 | −0.52 | 0.12 |
| hsa-miR-223 | −1.6 | 4.1 × 10−3 | 0.08 | −0.55 | 0.08 | −0.65 | 0.04 | ||
| hsa-miR-92a | miR-25 | bd | −1.4 | 4.9 × 10−3 | 0.09 | −0.54 | 0.04 | −0.59 | 0.08 |
| hsa-miR-20b | miR-17 | d | −1.5 | 5.6 × 10−3 | 0.09 | −0.39 | 0.21 | −0.61 | 0.06 |
The miR family names from miRBase demonstrate the relatedness between the miRNAs
Letters in ‘C’ show which miRNAs are from the same transcriptional cluster
FC, fold change; p (FDR), p values adjusted with Benjamini–Hochberg’s false-discovery rate; r Glu120, intra-pair correlation coefficient between miRNA expression and 120 min OGTT glucose values; r HbA1c, intra-pair correlation coefficient between miRNA expression and HbA1c values; p-r HbA1c and p-r Glu120, the p values for the intra-pair correlation analyses
The diabetes-dysregulated miR-15 family is involved in insulin signalling
The 20 identified miRNAs belong to 11 different miRNA families (ESM Table 1); miRNAs within the same family are predicted to target the same mRNAs. As shown in Table 2, the top-ranking miRNAs according to statistical significance were miR-15b and miR-16. These both belong to the miR-15 family. The human miR-15 family consists of miR-15a, miR-15b, miR-16 and miR-195 (Fig. 1a), in addition miR-497 shares the seed sequence nucleotides 2–8. In humans, miR-15b and miR-16 originate from the same transcriptional cluster on chromosome 3 in the intron of SMC4, which encodes structural maintenance of chromosomes 4. In addition, miR-16 is transcribed from chromosome 13, where it is situated in the non-protein-coding region DLEU2 (deleted in lymphocytic leukaemia 2) clustered together with miR-15a. TargetScan predicts 968 conserved mRNA targets for the miR-15 family, and analysing these targets using IPA showed a total of 20 pathways that contained significantly higher numbers of the predicted miR-15 family targets than expected by chance (pFDR < 0.05) (Table 3). Among these, the insulin signalling pathway had the most significant over-representation of miR-15 family targets (pFDR = 0.004) (Fig. 1b and Table 3). This pathway analysis finding was confirmed using DAVID software, which was also used to perform additional pathway analyses on the predicted mRNA targets for each of the ten other miRNA families (ESM Table 1).
Fig. 1.
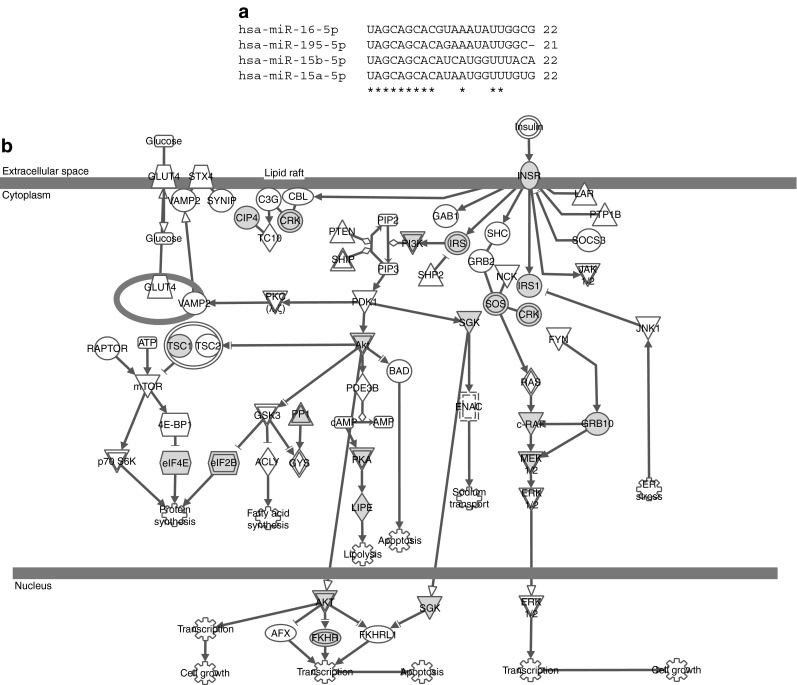
The miR-15 family and its targets in the INSR signalling pathway. (a) Alignment of the nucleotide sequences of the human miR-15 family members using CLUSTAL 2.0.12. Asterisks indicate conserved residues. (b) The INSR signalling pathway. Grey nodes indicate predicted targets of the miR-15 family (tuberous sclerosis 1 [TSC1], Raf-1 proto-oncogene, serine/threonine kinase [RAF1], serum/glucocorticoid regulated kinase 1 [SGK1], thyroid hormone receptor interactor 10 [TRIP10], v-crk avian sarcoma virus CT10 oncogene homolog-like [CRKL], PIK3R1, protein kinase, cAMP-dependent, regulatory, type II, α [PRKAR2A], lipase, hormone-sensitive [LIPE], protein phosphatase 1, regulatory [inhibitor] subunit 11 [PPP1R11], eukaryotic translation initiation factor 2B, subunit 2 β, 39 kDa [eIF2B2], eukaryotic translation initiation factor 4E [eIF4E], growth factor receptor-bound protein 10 [GRB10], forkhead box O1 [FOXO1], IRS1, son of sevenless homolog 1 [Drosophila] [SOS1], eukaryotic translation initiation factor 2B, subunit 5 ε, 82 kDa [EIF2B5], Akt3, IRS2, INSR and mitogen-activated protein kinase kinase 1 [MAP2K1]. Additional information on the nodes in the pathway can be found in ESM Table 2. ER, endoplasmic reticulum
Table 3.
Pathway analysis of the miR-15 family targets
| Ingenuity canonical pathways | p (FDR) |
|---|---|
| INSR signalling | 0.0039 |
| AMPK signalling | 0.0045 |
| PPARα/RXRα activation | 0.0045 |
| TGF-β signalling | 0.0068 |
| Wnt/β-catenin signalling | 0.0126 |
| IGF-1 signalling | 0.0126 |
| Molecular mechanisms of cancer | 0.0126 |
| mTOR signalling | 0.0126 |
| EIF2 signalling | 0.0126 |
| PI3K/Akt signalling | 0.0126 |
| BMP signalling pathway | 0.0155 |
| 14-3-3-mediated signalling | 0.0158 |
| PTEN signalling | 0.0245 |
| Protein ubiquitination pathway | 0.0339 |
| Cell cycle regulation by BTG family proteins | 0.0339 |
| Axonal guidance signalling | 0.0339 |
| Ceramide signalling | 0.0339 |
| Mitotic roles of polo-like kinase | 0.0355 |
| Myc-mediated apoptosis signalling | 0.0417 |
| Cell cycle: G1/S checkpoint regulation | 0.0417 |
IPA of the miR-15 family target genes predicted using TargetScan. Only pathways with p (FDR) <0.05 are shown
AMPK, protein kinase, AMP-activated, alpha 1 catalytic subunit; BMP, bone morphogenetic protein; BTG, B-cell translocation gene; EIF2, eukaryotic translation initiation factor 2; p (FDR), Benjamini–Hochberg’s multiple comparison adjusted p values; PPARα/RXRα, peroxisome proliferator-activated receptor α/retinoid X receptor α; PTEN, phosphatase and tensin homologue; WNT, wingless-type MMTV integration site family
miR-15b and miR-16 regulate protein levels of insulin signalling molecules
Three prominent players of the insulin signalling pathway (INSR, IRS1 and PIK3R1) were among the predicted miR-15b/-16 targets. To address the role of miR-15b and miR-16 in the control of insulin signalling proteins, we measured the protein and mRNA expression levels of these three predicted targets in muscle biopsies from the non-diabetic twin population.
We found miR-15b levels negatively associated with both INSR and IRS-1 but not with PIK3R1 protein levels when adjusted for confounding factors (Table 4). For miR-16, no significant associations with the levels of these proteins were observed. IRS1 mRNA levels was negatively associated with miR-15b, but not with miR-16. No significant associations between miR-15b or miR-16 and the mRNA levels of INSR and PIK3R1 were found (Table 4). Adjustments for fasting glucose levels and 2 h glucose values did not change any of the results.
Table 4.
Impact of miR-15b and miR-16 on specific molecular targets in the 164 non-diabetic twins
| Target | Effect | CI 95% | Change | p |
|---|---|---|---|---|
| INSR protein | ||||
| miR-15b | 0.91 | (0.86, 0.97) | 9%↓ | 0.004 |
| miR-16 | 0.93 | (0.86, 1.00) | 7%↓ | 0.06 |
| INSR mRNA | ||||
| miR-15b | 0.94 | (0.87, 1.02) | 0.1 | |
| miR-16 | 1.00 | (0.92, 1.09) | 1.0 | |
| IRS-1 protein | ||||
| miR-15b | 0.82 | (0.75, 0.88) | 18%↓ | <0.0001 |
| miR-16 | 0.95 | (0.87, 1.03) | 0.2 | |
| IRS-1 mRNA | ||||
| miR-15b | 0.92 | (0.86, 0.99) | 8%↓ | 0.03 |
| miR-16 | 0.95 | (0.88, 1.02) | 0.2 | |
| PIK3R1 protein | ||||
| miR-15b | 0.97 | (0.93, 1.02) | 0.2 | |
| miR-16 | 1.04 | (1.00, 1.08) | 4%↑ | 0.07 |
| PIK3R1 mRNA | ||||
| miR-15b | 0.92 | (0.85, 1.01) | 8%↓ | 0.07 |
| miR-16 | 0.98 | (0.90, 1.07) | 0.6 | |
Multivariate analysis of the effects of miR-15b or miR-16 on target mRNA and protein expression with adjustment for age, sex, total fat percentage, , birthweight, twin-pair and zygosity status
Effect, the effect on the response variable expressed as the change by 1 SD increase in either miR-15b or miR-16
To study the potential regulation of INSR and PIK3R1 protein and mRNA levels by miR-15b and miR-16, we transfected antisense inhibitors specific to these miRNAs into human primary myocytes, with insulin added 5–10 min prior to harvest. Using ANOVA models we found no general effect of insulin on protein levels and no interaction effects. We observed a general increase in the total INSR protein content following miR-15b/miR-16 inhibition (p = 0.01) and a general downregulation of PIK3R1 protein levels in response to miR-15b/16 inhibition (p = 0.0002) (ESM Fig. 3). The result on PIK3R1 was in line with the borderline positive association found between PIK3R1 protein levels and miR-16 in the non-diabetic twins. As miR-15b and miR-16 target multiple insulin signalling molecules (Fig. 1b), it is relevant to study the collective effect of their inhibition; to this end, we measured the levels of p-Akt/Akt after miR-15b/miR-16 inhibition (ESM Fig. 4). We did not observe any effect of miR-15b and miR-16 inhibition on p-Akt/Akt levels. However, this result was considered inconclusive due to large variations in the p-Akt measurements.
Effects of high glucose and insulin levels on miRNA expressions in cell culture
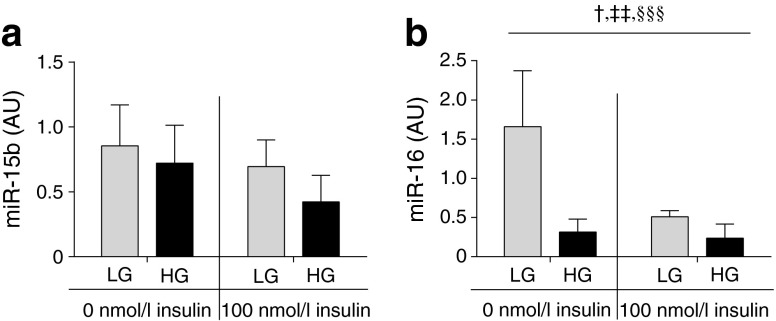
Elevated glucose and insulin levels defining a diabetogenic environment might regulate miR-15b and miR-16. Short-term incubation of cultured human myocytes with 10 nmol/l insulin for 5, 10, 30, 60 or 120 min did not affect miR-15b or miR-16 levels (data not shown). We also tested the effect of high glucose and high insulin levels in L6 cells (a rat myoblast cell line) using a higher insulin concentration (100 nmol/l) with or without high glucose (high 25 mmol/l, low 5.6 mmol/l) for a prolonged time period of 96 h. Although the effects of high glucose (p = 0.2) and high insulin levels (p = 0.2) on miR-15b expression did not reach statistical significance in these cells (Fig. 2a), we found that high insulin (p = 0.01) and high glucose (p = 0.003) levels reduced the miR-16 levels (Fig. 2b).
Fig. 2.
The effect of high insulin and high glucose on miR-15b and miR-16 expression levels in L6 cells. Levels of miR-15b (a) and miR-16 (b) were measured in L6 cells after 96 h of stimulation with either low glucose and no insulin, high glucose no insulin, low glucose with high insulin or high glucose with high insulin. Data are mean ± SD, n = 3. p values from two-way ANOVA: † p = 0.02 for interaction effect, ‡‡ p = 0.01 for insulin effect, §§§ p = 0.003 for glucose effect. AU, arbitrary expression unit; LG, low glucose (5.6 mmol/l); HG, high glucose (25 mmol/l)
Implications for intrauterine-dependent regulation of the miR-15 family
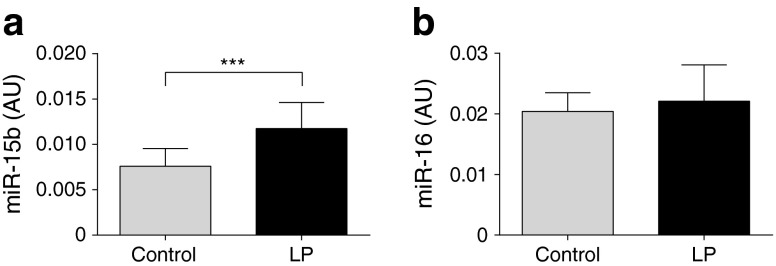
To further explore the potential reasons for dysregulation of the miR-15 family in type 2 diabetes, we measured the expression levels of miR-15b and miR-16 in the non-diabetic twins. In this cohort, we found no associations between miR-15b and miR-16 levels on one side and insulin action (Rd clamp) or the 2 h post-OGTT glucose values on the other (data not shown). Interestingly, multiple regression analysis showed a negative association between birthweight and both miR-15b and miR-16 expression levels (Table 5). Total body fat percentage was positively associated with miR-16 but not miR-15b expression. Age, sex or values were not associated with miR-15b or miR-16 expression levels (Table 5). To study further the effect of a poor intrauterine environment on the expression of miR-15b and miR-16, we studied muscle tissue from 3-month-old male rat offspring from dams fed either an LP or a control diet [35, 36]. The LP offspring had lower body weights both at day 3 after birth (p = 0.004) and at 3 months of age (p < 0.0001). Elevated miR-15b levels were found in the muscle of LP offspring (p < 0.001) (Fig. 3a), whereas the levels of miR-16 were similar in the two groups (Fig. 3b).
Table 5.
Factors associated with the expression of either miR-15b or miR-16 in skeletal muscle obtained from the non-diabetic twins
| Effect | 95% CI | Change | p | |
|---|---|---|---|---|
| miR-15b | ||||
| Age | 0.97 | (0.72, 1.28) | 0.8 | |
| Sex | 0.81 | (0.62, 1.05) | 0.1 | |
| 0.99 | (0.88, 1.11) | 0.9 | ||
| Birthweight | 0.87 | (0.79, 0.96) | 13%↓ | 0.005 |
| Total fat percentage | 1.11 | (0.93, 1.34) | 0.3 | |
| miR-16 | ||||
| Age | 1.28 | (0.99, 1.64) | 0.06 | |
| Sex | 1.06 | (0.83, 1.35) | 0.6 | |
| 1.08 | (0.97, 1.19) | 0.2 | ||
| Birthweight | 0.87 | (0.80, 0.95) | 13%↓ | 0.002 |
| Total fat percentage | 1.27 | (1.08, 1.48) | 27%↑ | 0.003 |
The effects of age, sex, , birthweight, and total body fat percentage on the expression of either miR-15b or miR-16, investigated by multiple regression analyses adjusted for twin-pair and zygosity status
Effect, the effect on the response variable expressed as the change by 1 SD increase in either miR-15b or miR-16
Fig. 3.
The effect of maternal protein restriction on skeletal muscle expression levels of miR-15b and miR-16 in rats. miR-15b (a) and miR-16 (b) expression levels were measured in vastus lateralis muscle biopsies from 3-month-old male offspring of dams fed with low protein (n = 11) and control dams (n = 11). Data are mean ± SD. ***p < 0.001 for the effect of LP from two-sided unpaired t tests. AU, arbitrary expression unit; LP, low protein
Discussion
In a unique cohort of 11 MZ twin pairs discordant for type 2 diabetes, we identified 20 miRNAs that were differentially expressed in the skeletal muscle of twins with type 2 diabetes compared with their non-diabetic identical twins. In silico analyses identified the two most significantly downregulated miRNAs in twins with diabetes, miR-15b and miR-16, as possible regulators of important metabolic functions including insulin signalling. The predicted insulin signalling targets of miR-15b and miR-16 included INSR, IRS-1 and PIK3R1, for which we demonstrated a likely biological role in vivo (for INSR and IRS-1) and in vitro (for INSR and p85). We found a significant positive association between LBW and both miR-15b and miR-16 in non-diabetic twins and elevated miR-15b levels in a rat model of LBW. Finally, we showed a downregulation of miR-16 by high glucose and insulin levels in cell cultures.
Interestingly, all of the 20 differentially expressed miRNAs in the MZ twins with overt type 2 diabetes were downregulated compared with their genetically identical twins, most of whom had IGT. In support of this we found eight of the 20 miRNAs, including miR-15b and miR-16, to be downregulated in singletons with overt type 2 diabetes when we compared their levels to those in singletons with IGT using an online dataset [12]. Differences between these studies may be explained by the selective identification of non-genetic differences in discordant MZ twins.
Diabetes is defined by the presence of hyperglycaemia, and we therefore tested the correlation between average glycaemic levels (HbA1c) and the expression levels of each of the 20 miRNAs. Although only three of these miRNAs, including miR-15b, showed a significant negative correlation with HbA1c level after correction for multiple testing, we found the same trend for all the 20 miRNAs, suggesting that hyperglycaemia may be involved in the coordinated downregulation of miRNAs in twins with overt diabetes. Whether this downregulation represents, for example, secondary potentially deleterious changes due to hyperglycaemia, or plays a role in the compensation for insulin resistance via hyperglycaemia-induced glucose uptake [37] remains to be investigated.
A previous study in pancreatic beta cells showed upregulated miR-15a levels after hyperglycaemic exposure for 1 h, with subsequent downregulation after 3 days of exposure [38]. We found that long-term exposure to elevated insulin and glucose levels was associated with a downregulation of miR-16 but not miR-15b in L6 cells, which suggests differential regulation. Acute insulin stimulation for up to 120 min in human myocytes using a concentration of 10 nmol/l did not show any effect, supporting previous in vivo clamp results in humans [39].
Based on the presence of complementary binding sites for the miR-15 family in the 3′UTR of mRNAs, we predicted insulin signalling proteins including INSR, IRS-1 and PIK3R1 to represent important targets for miR-15b and miR-16. Indeed, our experiments in human myocytes showed that the levels of INSR and PIK3R1 protein were altered following inhibition of miR-15b/miR-16. INSR was upregulated, indicating that miR-15b/miR-16 represses INSR translation, whereas PIK3R1 was downregulated, indicating enhanced translation. Both miRNA-mediated down- [10] and upregulation [40, 41] of translation have previously been described. Increased PIK3R1 levels were previously reported to be associated with insulin resistance [42, 43], thus elevated miR-15b and miR-16 levels could contribute to insulin resistance through downregulation of INSR and a parallel upregulation of PIK3R1. The biological relevance of our results was demonstrated in the non-diabetic twins, with a negative association between levels of miR-15b and INSR, a borderline significant positive association between miR-16 and PIK3R1 and a highly significant negative association between miR-15b and IRS-1 protein levels. 3′UTR luciferase studies could be instrumental in providing more insights into the extent to which miR-15b and miR-16 bind to the 3′UTR regions of their predicted targets, and to determine the extent to which these miRNAs—alone or in concert with other miRNAs—may influence the expression of one or more insulin signalling proteins. Given the complex and unknown interactions of miR-15b and miR-16 with many other miRNAs, including the 20 miRNA we found downregulated in type 2 diabetes, such experiments will need to be addressed in future papers.
There are multiple routes via which the insulin signal travels [44, 45], and the predicted targets of miR-15b and miR-16 suggest several that could be interesting. Here, we tested downstream signalling through p-Akt/Akt but reached no clear conclusion; thus, further studies will be needed to determine the specific route of action for these miRNAs as well as to determine their role in the other pathways containing their targets, several of which are known or suspected to influence glucose metabolism. We did not find any significant correlations between levels of miR-15b and miR-16 and in vivo peripheral insulin resistance in the non-diabetic twins. This may suggest that the absolute muscle expression level of these miRNAs plays only a minor role in the control of in vivo whole-body insulin action or that the potential effects need unmasking by adverse ‘diabetogenic’ lifestyle conditions.
In our large cohort of non-diabetic twins, we observed a negative association between birthweight and miR-15b and miR-16 levels, which was further supported by elevated miR-15b levels in an LBW rat model. The lack of significantly elevated miR-16 levels in the rat model could reflect interspecies differences in the regulation of the two clusters from which the miR-15 family are transcribed. The upregulation of miR-15b and miR-16 in twins with LBW, and the opposing downregulation in the MZ twins with diabetes, is to some extent paradoxical as LBW is associated with increased risk of developing type 2 diabetes. It suggests differential regulation as well as a potential impact of miR-15b and miR-16 at different stages in the life-long development of type 2 diabetes. This is supported by the finding of upregulated miR-15b and miR-16 levels in individuals with IGT when compared with both individuals with NGT and those with type 2 diabetes in an independent online dataset [12]. In addition, we previously reported a similar differential regulation of insulin signalling genes in patients with overt type 2 diabetes compared with their first-degree relatives which, similar to individuals with IGT and individuals born with LBW, have an increased risk of developing T2D [46]. Whether an adverse intrauterine environment directly alters miR-15b and miR-16 levels and the extent to which these miRNAs play a role in the development of insulin resistance in individuals with an increased risk of developing type 2 diabetes or whether they perhaps work to counteract the enhanced activity in the insulin signalling pathway that we have previously observed in insulin-resistant individuals [47] remains to be disclosed by future studies.
In conclusion, we identified 20 downregulated miRNAs including miR-15b and miR-16 in twins with overt type 2 diabetes compared with their genetically identical non-diabetic co-twins. In a larger cohort of non-diabetic twins, LBW was somewhat paradoxically associated with upregulation of miR-15b and miR-16 levels. Our results indicate that miR-15b and miR-16 may regulate key insulin signalling protein levels. Additional studies are needed to understand not only the role of the miR-15 family in the development of insulin resistance and type 2 diabetes, but also the distinct or interactive roles of the additional 18 downregulated miRNAs in type 2 diabetes.
Electronic supplementary material
(PDF 152 kb)
(PDF 145 kb)
(PDF 315 kb)
(PDF 153 kb)
(PDF 125 kb)
(PDF 190 kb)
(PDF 29.7 kb)
Acknowledgements
We thank all the study participants. In addition, the technical assistance of M. Modest, Steno Diabetes Center, Denmark, and the comments and suggestion to the discussion by E. A. Richter, Department of Exercise and Sport Sciences, University of Copenhagen, Denmark, are gratefully acknowledged.
Funding
JB-J was supported by a grant from the Danish PhD School of Molecular Metabolism. The study was supported by grants from the Danish Medical Research Council, the Danish Strategic Research Council. The Novo Nordisk Foundation, the Danish Ministry of Science, Technology and Innovation. DSF-T was supported by the Biotechnology and Biological Sciences Research Council project grant BB/F-15364/1. SEO is a British Heart Foundation Senior Fellow (FS/09/029/27902). K. Højlund was supported by grants from the Danish Medical Research Council and the Danish Council for Independent Research (Sapere Aude, DFF Starting Grant), and the Excellence Grant 2009 from the Novo Nordisk Foundation.
Duality of interest
JB-J has received a salary as an employee at Steno Diabetes Center. DVC has received salary as an employee at Novo Nordisk. JB-J, MF, PP, AV and LGG own Novo Nordisk shares. TL receives a salary as an employee at LEO Pharma. HB-N has received research funding from Novo Nordisk. PP receives a salary as an employee at Novo Nordisk. AV has received income from Novo Nordisk. The remaining authors have no conflicting interests.
Contribution statement
JB-J and AV are guarantors for this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
JB-J contributed to conception and design, analysed and interpreted data and wrote the manuscript. CS, DVC, EN, MF, DSF-T, K. Holmstrøm, BV, K. Højlund, PP and JW acquired and researched the data. TL, LGG, SEO, HB-N and BKP interpreted the data. AV contributed to the conception, design and interpretation. All authors have revised the manuscript critically for important intellectual content and given final approval of the version to be published.
Abbreviations
- DAVID
Database for Annotation Visualization and Integrated Discovery
- DZ
Dizygotic
- FDR
False-discovery rate
- GIR
Glucose infusion rate
- IGT
Impaired glucose tolerance
- INSR
Insulin receptor
- IPA
Ingenuity pathway analysis
- LBW
Low birthweight
- Limma
Linear models for microarray data
- LNA
Locked nucleic acid
- LP
Low protein
- miRNA
MicroRNA
- mTOR
Mechanistic target of rapamycin (serine/threonine kinase)
- MZ
Monozygotic
- NGT
Normal glucose tolerance
- PIK3R1
Phosphoinositide 3-kinase R1 regulatory subunit 1 α
- PI3K
Phosphoinositide 3-kinase
- Rd
Glucose disposal rate
Maximal oxygen consumption rate
Contributor Information
Jette Bork-Jensen, Email: jette.bork.jensen@sund.ku.dk.
Allan Vaag, Email: allan.vaag@regionh.dk.
References
- 1.Shulman GI, Rothman DL, Jue T, Stein P, DeFronzo RA, Shulman RG. Quantitation of muscle glycogen synthesis in normal subjects and subjects with non-insulin-dependent diabetes by 13C nuclear magnetic resonance spectroscopy. N Engl J Med. 1990;322:223–228. doi: 10.1056/NEJM199001253220403. [DOI] [PubMed] [Google Scholar]
- 2.Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet. 2009;41:1110–1115. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 3.Phillips DI, Barker DJ, Hales CN, Hirst S, Osmond C. Thinness at birth and insulin resistance in adult life. Diabetologia. 1994;37:150–154. doi: 10.1007/s001250050086. [DOI] [PubMed] [Google Scholar]
- 4.Poulsen P, Vaag AA, Kyvik KO, Moller JD, Beck-Nielsen H. Low birth weight is associated with NIDDM in discordant monozygotic and dizygotic twin pairs. Diabetologia. 1997;40:439–446. doi: 10.1007/s001250050698. [DOI] [PubMed] [Google Scholar]
- 5.Alibegovic AC, Sonne MP, Hojbjerre L, et al. Insulin resistance induced by physical inactivity is associated with multiple transcriptional changes in skeletal muscle in young men. Am J Physiol Endocrinol Metab. 2010;299:E752–E763. doi: 10.1152/ajpendo.00590.2009. [DOI] [PubMed] [Google Scholar]
- 6.Karakelides H, Irving BA, Short KR, O'Brien P, Nair KS. Age, obesity, and sex effects on insulin sensitivity and skeletal muscle mitochondrial function. Diabetes. 2010;59:89–97. doi: 10.2337/db09-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poulsen P, Levin K, Petersen I, Christensen K, Beck-Nielsen H, Vaag A. Heritability of insulin secretion, peripheral and hepatic insulin action, and intracellular glucose partitioning in young and old Danish twins. Diabetes. 2005;54:275–283. doi: 10.2337/diabetes.54.1.275. [DOI] [PubMed] [Google Scholar]
- 8.Mayer EJ, Newman B, Austin MA, et al. Genetic and environmental influences on insulin levels and the insulin resistance syndrome: an analysis of women twins. Am J Epidemiol. 1996;143:323–332. doi: 10.1093/oxfordjournals.aje.a008746. [DOI] [PubMed] [Google Scholar]
- 9.Florez JC. Newly identified loci highlight beta cell dysfunction as a key cause of type 2 diabetes: where are the insulin resistance genes? Diabetologia. 2008;51:1100–1110. doi: 10.1007/s00125-008-1025-9. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Ferland-McCollough D, Fernandez-Twinn DS, Cannell IG, et al. Programming of adipose tissue miR-483-3p and GDF-3 expression by maternal diet in type 2 diabetes. Cell Death Differ. 2012;19:1003–1012. doi: 10.1038/cdd.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher IJ, Scheele C, Keller P, et al. Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Med. 2010;2:9. doi: 10.1186/gm130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu H, Shyh-Chang N, Segre AV, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaag A, Henriksen JE, Madsbad S, Holm N, Beck-Nielsen H. Insulin secretion, insulin action, and hepatic glucose production in identical twins discordant for non-insulin-dependent diabetes mellitus. J Clin Invest. 1995;95:690–698. doi: 10.1172/JCI117715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poulsen P, Levin K, Beck-Nielsen H, Vaag A. Age-dependent impact of zygosity and birth weight on insulin secretion and insulin action in twins. Diabetologia. 2002;45:1649–1657. doi: 10.1007/s00125-002-0983-6. [DOI] [PubMed] [Google Scholar]
- 16.Poulsen P, Grunnet LG, Pilgaard K, et al. Increased risk of type 2 diabetes in elderly twins. Diabetes. 2009;58:1350–1355. doi: 10.2337/db08-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ling C, Poulsen P, Carlsson E, et al. Multiple environmental and genetic factors influence skeletal muscle PGC-1alpha and PGC-1beta gene expression in twins. J Clin Invest. 2004;114:1518–1526. doi: 10.1172/JCI21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribel-Madsen R, Fraga MF, Jacobsen S, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS One. 2012;7:e51302. doi: 10.1371/journal.pone.0051302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clausen JP, Oxhoj H, Hansen JF, Pedersen-Bjergaard O, Henningsen P, Uhrenholdt A. Exercise test and exercise ECG in the diagnosis and treatment of ischemic heart disease. Ugeskr Laeger. 1980;142:1743–1749. [PubMed] [Google Scholar]
- 20.Ritchie ME, Silver J, Oshlack A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 21.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 22.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed]
- 28.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 29.Smyth GK, Yang YH, Speed T. Statistical issues in cDNA microarray data analysis. Methods Mol Biol. 2003;224:111–136. doi: 10.1385/1-59259-364-X:111. [DOI] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 31.Bring J, Wernroth L. Inefficient analysis of twin data: is there an association between diabetes and birth weight? Diabetologia. 1999;42:898–899. doi: 10.1007/s001250051244. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez-Twinn DS, Ekizoglou S, Gusterson BA, Luan J, Ozanne SE. Compensatory mammary growth following protein restriction during pregnancy and lactation increases early-onset mammary tumor incidence in rats. Carcinogenesis. 2007;28:545–552. doi: 10.1093/carcin/bgl166. [DOI] [PubMed] [Google Scholar]
- 33.Loizou CL, Ozanne SE, Hales CN. The effect of insulin on delta5 desaturation in hepG2 human hepatoma cells and L6 rat muscle myoblasts. Prostaglandins Leukot Essent Fat Acids. 1999;61:89–95. doi: 10.1054/plef.1999.0076. [DOI] [PubMed] [Google Scholar]
- 34.Ribel-Madsen R, Poulsen P, Holmkvist J, et al. Impact of rs361072 in the phosphoinositide 3-kinase p110beta gene on whole-body glucose metabolism and subunit protein expression in skeletal muscle. Diabetes. 2010;59:1108–1112. doi: 10.2337/db09-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez-Twinn DS, Wayman A, Ekizoglou S, Martin MS, Hales CN, Ozanne SE. Maternal protein restriction leads to hyperinsulinemia and reduced insulin-signaling protein expression in 21-mo-old female rat offspring. Am J Physiol Regul Integr Comp Physiol. 2005;288:R368–R373. doi: 10.1152/ajpregu.00206.2004. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez-Twinn DS, Ekizoglou S, Wayman A, Petry CJ, Ozanne SE. Maternal low-protein diet programs cardiac beta-adrenergic response and signaling in 3-mo-old male offspring. Am J Physiol Regul Integr Comp Physiol. 2006;291:R429–R436. doi: 10.1152/ajpregu.00608.2005. [DOI] [PubMed] [Google Scholar]
- 37.Vaag A, Damsbo P, Hother-Nielsen O, Beck-Nielsen H. Hyperglycaemia compensates for the defects in insulin-mediated glucose metabolism and in the activation of glycogen synthase in the skeletal muscle of patients with type 2 (non-insulin-dependent) diabetes mellitus. Diabetologia. 1992;35:80–88. doi: 10.1007/BF00400856. [DOI] [PubMed] [Google Scholar]
- 38.Sun LL, Jiang BG, Li WT, Zou JJ, Shi YQ, Liu ZM. MicroRNA-15a positively regulates insulin synthesis by inhibiting uncoupling protein-2 expression. Diabetes Res Clin Pract. 2011;91:94–100. doi: 10.1016/j.diabres.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Granjon A, Gustin MP, Rieusset J, et al. The microRNA signature in response to insulin reveals its implication in the transcriptional action of insulin in human skeletal muscle and the role of a sterol regulatory element-binding protein-1c/myocyte enhancer factor 2C pathway. Diabetes. 2009;58:2555–2564. doi: 10.2337/db09-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srikantan S, Marasa BS, Becker KG, Gorospe M, Abdelmohsen K. Paradoxical microRNAs: individual gene repressors, global translation enhancers. Cell Cycle. 2011;10:751–759. doi: 10.4161/cc.10.5.14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 42.Terauchi Y, Tsuji Y, Satoh S, et al. Increased insulin sensitivity and hypoglycaemia in mice lacking the p85 alpha subunit of phosphoinositide 3-kinase. Nat Genet. 1999;21:230–235. doi: 10.1038/6023. [DOI] [PubMed] [Google Scholar]
- 43.Draznin B. Molecular mechanisms of insulin resistance: serine phosphorylation of insulin receptor substrate-1 and increased expression of p85alpha: the two sides of a coin. Diabetes. 2006;55:2392–2397. doi: 10.2337/db06-0391. [DOI] [PubMed] [Google Scholar]
- 44.Sylow L, Kleinert M, Pehmoller C, et al. Akt and Rac1 signaling are jointly required for insulin-stimulated glucose uptake in skeletal muscle and downregulated in insulin resistance. Cell Signal. 2014;26:323–331. doi: 10.1016/j.cellsig.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 45.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 46.Palsgaard J, Brons C, Friedrichsen M, et al. Gene expression in skeletal muscle biopsies from people with type 2 diabetes and relatives: differential regulation of insulin signaling pathways. PLoS One. 2009;4:e6575. doi: 10.1371/journal.pone.0006575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storgaard H, Song XM, Jensen CB, et al. Insulin signal transduction in skeletal muscle from glucose-intolerant relatives of type 2 diabetic patients [corrected] Diabetes. 2001;50:2770–2778. doi: 10.2337/diabetes.50.12.2770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 152 kb)
(PDF 145 kb)
(PDF 315 kb)
(PDF 153 kb)
(PDF 125 kb)
(PDF 190 kb)
(PDF 29.7 kb)