Abstract
A meta-analysis was performed to evaluate the associations of uncoupling protein 2 (UCP2) gene polymorphisms (Ala55Val, 45-bp insertion/deletion, and -866G/A) with overweight and obesity. A total of 42 studies were included in our analysis. Pooled effect estimates and 95% confidential intervals of each polymorphism were calculated under different inherited models. Fixed or random effect model was selected based on the between-study heterogeneity evaluated with I2. Source of heterogeneity was explored by subgroup analysis and meta-regression analysis. Potential publication bias was assessed using funnel plot and Peters test. After excluding studies that deviated from the Hardy–Weinberg equilibrium, T allele of Ala55Val polymorphism was associated with an increased risk of overweight and obesity under recessive model in the overall (OR = 1.24, 95%CI = 1.06–1.45) and Asian (OR = 1.28, 95%CI = 1.06–1.55) populations; and A allele of -866G/A polymorphism had a protective effect on overweight and obesity, especially for European populations (dominant model: OR = 0.88, 95%CI = 0.81–0.96, co-dominant 1 model: OR = 0.89, 95%CI = 0.81–0.98, co-dominant 2 model: OR = 0.85, 95%CI = 0.74–0.94, additive model: OR = 0.88, 95%CI = 0.80–0.95, and allelic model: OR = 0.91, 95%CI = 0.86–0.97). No evidence was observed in the association of 45-bp insertion/deletion polymorphism with overweight and obesity susceptibility. We failed to fully explore the between-study heterogeneity regarding the association of Ala55Val polymorphism with overweight and obesity. Further studies are required to provide more convincing evidence.
Abbreviations: UCP, uncoupling protein; OR, odds ratio; CI, confidence interval; REM, random effect model; FEM, fixed effect model; HWE, Hardy–Weinberg equilibrium
Keywords: Obesity, Adiposity, Uncoupling protein, Polymorphism, Meta-analysis
Highlights
-
•
We evaluated the association of UCP2 gene polymorphisms with overweight and obesity.
-
•
Ala55Val polymorphism was associated with an increased risk in Asian populations.
-
•
No association was observed for 45-bp insertion/deletion polymorphism.
-
•
-866G/A polymorphism was associated with a decreased risk in European populations.
Introduction
Uncoupling proteins (UCP), a member of mitochondrial membrane transporters, were considered to be associated with energy homeostasis (Dalgaard and Pedersen, 2001). The UCP2, which is widely expressed in human tissues and serves as an uncoupler of oxidative phosphorylation, is involved in the regulation of lipid metabolism and ATP production (Diano and Horvath, 2012, Zhang et al., 2001). It has been reported that impaired expression of UCP2 mRNA in adipose tissue may be responsible for the pathophysiology of obesity (Oberkofler et al., 1998). Ala55Val (rs660339) in Exon 4, 45-bp insertion/deletion in Exon 8, and -866G/A (rs659366) in the promoter region, have been considered as the most interesting polymorphisms in UCP2 gene. The -866G/A and 45-bp insertion/deletion polymorphisms were found to be functional on mRNA expression (Esterbauer et al., 2001, Krempler et al., 2002). A variant allele of the Ala55Val polymorphism was reported to be associated with lower 24-h energy expenditure (Astrup et al., 1999). Associations of the three polymorphisms with the risk of overweight and obesity have been widely evaluated, but the conclusions remain controversial. In the present study, we performed a meta-analysis to assess the effect of UCP2 Ala55Val, 45-bp insertion/deletion, and -866G/A polymorphisms on the risk of overweight and obesity.
Methods
Search strategy
Relevant articles published in English and Chinese were searched from five databases: (1) PubMed (1990–2013); (2) Web of Science (ISI) (1990–2013); (3) China National Knowledge Infrastructure (CNKI) (1990–2013); (4) Database of Chinese Scientific and Technical Periodicals (VIP) (1990–2013); and (5) China Biology Medical literature database (CBM) (1990–2013). The search strategy involved the following keywords: ‘uncoupling protein 2’, ‘UCP2’, ‘polymorphism’, ‘obesity’, ‘adiposity’, ‘obese’, and ‘overweight’. References of the retrieved articles were checked for additional studies. The literature search was updated on February 22, 2013.
Eligibility criteria
A study that met the following criteria was included in the meta-analysis: (1) case–control, cohort study, or cross-sectional studies which evaluated the association of UCP2 gene polymorphisms with overweight and obesity; (2) provided sufficient data of genotypes in case (exposed) and control (unexposed) groups; (3) subjects were recruited from the same ethnicity during the same period; (4) if data were repetitively reported, the most complete one was included; (5) the genotype distribution of the controls obey Hardy–Weinberg equilibrium (HWE). The articles were independently assessed by two authors. Disagreements were resolved by consensus with a third reviewer.
Data extraction
Information of the studies was extracted by two independent authors, including first author, publication year, location, ethnicity of subjects, sample size, frequencies of genotype and allele, percentage of male subjects and mean age of subjects in case (exposed) and control (unexposed) groups.
Statistical analysis
HWE for UCP2 Ala55Val, 45-bp insertion/deletion, and -866G/A genotype distributions in controls was tested using Pearson χ2 test. Pooled odds ratios (ORs) with 95% confidence intervals (CIs) were calculated to assess the associations of the UCP2 Ala55Val, 45-bp insertion/deletion, and -866G/A polymorphisms with risk of overweight and obesity under dominant (CT+TT vs. CC for Ala55Val, ID+II vs. DD for 45-bp insertion/deletion, and GA+AA vs. GG for -866G/A), recessive (TT vs. CT+CC for Ala55Val, II vs. ID+DD for 45-bp insertion/deletion, and AA vs. GA+GG for -866G/A), co-dominant 1 (CT vs. CC for Ala55Val, ID vs. DD for 45-bp insertion/deletion, and GA vs. GG for -866G/A), co-dominant 2 (TT vs. CC for Ala55Val, II vs. DD for 45-bp insertion/deletion, and AA vs. GG for -866G/A), additive (2TT+CT vs. CC for Ala55Val, 2II+ID vs. DD for 45-bp insertion/deletion, and 2AA + vs. GG for -866G/A), over-dominant (CT vs. CC+TT for Ala55Val, ID vs. II+DD for 45-bp insertion/deletion, and GA vs. AA+GG for -866G/A) and allelic (T vs. C for Ala55Val, I vs. D for 45-bp insertion/deletion, and A vs. G for -866G/A) models, respectively. Pooled ORs in subgroups stratified by ethnicity were also calculated. In addition, cumulative analyses were performed to track the evidence over sample size. Z test was used to determine the significance of pooled OR. The I2 was used to assess heterogeneity among studies (Higgins and Thompson, 2002). Heterogeneity was considered significant if I2 > 50%, when random-effect model (REM) was adopted as the pooling method; otherwise, fix-effect model (FEM) was used. Meta-analysis regression was performed to identify potential covariates, including ethnicity, publication year, sex, age and sample size, which may contribute to between-study heterogeneity. Sensitive analysis was performed to identify ‘outlying’ studies and decrease statistical heterogeneity if I2 > 50% using hetred module (Patsopoulos et al., 2008). Influence of each individual study on the pooled estimate was investigated with influence analysis. A study was suspected of excessive influence if the point estimate of its omitted meta-analysis was beyond the 95%CI of the full analysis. Publication bias was evaluated by funnel plot and Peters test (Peters et al., 2006). All statistical analyses were performed with STATA version 11.0 (Stata Corporation, College Station, TX, USA). P value < 0.05 was considered statistically significant (two-tailed).
Results
Characteristics of studies
A total of 37 published articles with 42 studies (Andersen et al., 2013, Dalgaard et al., 1999, Dalgaard et al., 2003, Esterbauer et al., 2001, Evans et al., 2000, Feng et al., 2004, Gu, 2005, Heidari et al., 2010, Hong et al., 2005, Kosuge et al., 2008, Kring et al., 2008, Kubota et al., 1998, Li, 2007, Li et al., 2007, Lin et al., 2009, Liu et al., 2012, Maestrini et al., 2003, Mancini et al., 2003, Marti et al., 2004, Mottagui-Tabar et al., 2008, Nieters et al., 2002, Ochoa et al., 2007, Oktavianthi et al., 2012, Papazoglou et al., 2012, Schauble et al., 2003, Shen, 2004, Shen, 2007, Srivastava et al., 2010, Sui et al., 2004, Urhammer et al., 1997, Wang et al., 2007, Wang et al., 2009, Xu, 2005, Yang, 2012, Yang et al., 2004, Yiew et al., 2010, Zou et al., 2011) were eligible for this meta-analysis, including 12 studies on Ala55Val (2011 cases and 2752 controls), 12 studies on 45-bp insertion/deletion (3416 cases and 3260 controls), and 18 studies on -886G/A (5780 cases and 10,196 controls). General characteristics and genotype distributions of the three polymorphisms were summarized in Table 1, Table 2, Table 3. In the Asian population, the average frequency of T allele in Ala55Val polymorphism was 38.5% in cases and 41.5% in controls, that of I allele in 45-bp insertion/deletion polymorphism was 12.3% in cases and 11.9% in controls, and that of A allele in -866G/A polymorphism was 41.0% in cases and 44.7% in controls. In the European population, the average frequency of T allele in Ala55Val polymorphism was 53.1% in cases and 51.7% in controls, that of I allele in 45-bp insertion/deletion polymorphism was 27.8% in cases and 27.0% in controls, and that of A allele in -866G/A polymorphism was 37.3% in cases and 39.3% in controls.
Table 1.
Characteristics of UCP2 gene Ala55Val (rs660339) polymorphism genotype distributions in studies included in this meta-analysis.
| First author | Year | Location | Ethnicity | Genotypes (CC/CT/TT) |
T allele frequency (%) |
% of male (case/control) | Mean age (case/control) | P for HWE | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | |||||||
| Urhammer SA | 1997 | Denmark | European | 41/67/36 | 56/86/40 | 48.3 | 45.6 | 100/100 | Na/Na | 0.5209 |
| Kubota T | 1998 | Japan | Asian | 15/13/14 | 64/97/57 | 48.8 | 48.4 | 28.6/56.0 | 47.1/58.2 | 0.1070 |
| Yang M | 2004 | China | Asian | 36/45/18 | 19/29/9 | 40.9 | 41.2 | Na/Na | Na/Na | 0.7066 |
| Sui Y | 2004 | China | Asian | 41/52/26 | 82/81/14 | 43.7 | 30.8 | 31.1/38.4 | 54.2/56.0 | 0.3266 |
| Shen LQ | 2004 | China | Asian | 96/34/14 | 37/9/2 | 21.5 | 13.5 | 50.0/50.0 | Na/Na | 0.1674 |
| Xu R | 2005 | China | Asian | 102/193/89 | 185/356/159 | 48.3 | 48.1 | 42.5/33.9 | 64.1/62.9 | 0.6236 |
| Mottagui-Tabar S | 2006 | Sweden | European | 57/136/88 | 90/227/127 | 55.5 | 54.2 | 0/0 | Na/Na | 0.5318 |
| Wang TN | 2007 | Taiwan | Asian | 211/104/9 | 90/24/0 | 18.8 | 10.5 | 38.0/51.9 | 55.86/58.95 | 0.2091 |
| Li Q | 2007 | China | Asian | 44/87/38 | 77/72/24 | 48.2 | 34.7 | 57.4/58.4 | 7.7/7.6 | 0.2842 |
| Kosuge K | 2008 | Japan | Asian | 56/80/46 | 91/199/79 | 47.3 | 48.4 | 70.9/64.2 | 50.1/50.7 | 0.1256 |
| Wang W | 2009 | China | Asian | 15/23/5 | 32/13/3 | 38.4 | 19.8 | 67.4/58.1 | Na/Na | 0.3086 |
| Yang RR | 2012 | China | Asian | 23/45/12 | 70/114/38 | 43.1 | 42.8 | Na/Na | Na/Na | 0.4670 |
Na: not available.
Table 2.
Characteristics of UCP2 gene 45-bp insertion/deletion polymorphism genotype distributions in studies included in this meta-analysis.
| First author | Year | Location | Ethnicity | Genotypes (DD/ID/II) |
I allele frequency (%) |
% of male (case/control) | Mean age (case/control) | P for HWE | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | |||||||
| Dalgaard LT | 1999 | Denmark | European | 371/293/80 | 177/149/28 | 30.4 | 29.0 | 100/100 | 19–20/19–20 | 0.6645 |
| Evans D | 2000 | Germany | European | 145/130/30 | 286/198/24 | 31.1 | 24.2 | Na/Na | Na/Na | 0.1621 |
| Esterbauer H | 2001 | Australia | European | 179/132/29 | 117/115/24 | 27.9 | 31.8 | 20/25 | 41.4/40.1 | 0.5751 |
| Nieters A | 2002 | Germany | European | 88/58/8 | 78/57/16 | 24.0 | 29.5 | 37.7/37.7 | 51.2/51.3 | 0.2585 |
| Maestrini S | 2003 | Italy | European | 211/124/25 | 51/42/10 | 24.2 | 30.1 | 29.2/Na | 45.0/Na | 0.7537 |
| Marti A | 2004 | Spain | European | 83/63/11 | 92/52/6 | 27.1 | 21.3 | Na/Na | 20–60/20–60 | 0.6875 |
| Feng QW | 2004 | China | Asian | 75/27/1 | 138/31/2 | 14.1 | 10.2 | Na/Na | Na/Na | 0.8618 |
| Hong QR | 2005 | China | Asian | 165/35/1 | 190/36/4 | 9.2 | 9.6 | 12.7/13.0 | 63.2/61.3 | 0.1485 |
| Ochoa MC | 2007 | Spain | European | 103/71/18 | 79/76/11 | 27.9 | 29.5 | 51.3/49.3 | 11.5/11.8 | 0.1963 |
| Yiew Sk | 2010 | Malaysia | Asian | 71/14/1 | 130/37/3 | 9.3 | 12.6 | Na/Na | Na/Na | 0.8454 |
| Papazoglou D | 2012 | Greece | European | 96/55/7 | 60/27/4 | 21.8 | 19.2 | 41.1/40.6 | 50.0/Na | 0.6684 |
| Liu XQ | 2012 | China | Asian | 463/141/12 | 696/199/15 | 13.4 | 12.6 | 51.8/49.8 | 45.4/44.6 | 0.8581 |
Na: not available.
Table 3.
Characteristics of UCP2 gene -866G/A (rs659366) polymorphism genotype distributions in studies included in this meta-analysis.
| First author | Year | Location | Ethnicity | Genotypes (GG/GA/AA) |
A allele frequency (%) |
% of male (case/control) | Mean age (case/control) | P for HWE | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | |||||||
| Esterbauer H | 2001 | Australia | European | 54/42/13 | 221/286/82 | 31.2 | 38.2 | 53.2/56.2 | 53.5/52.8 | 0.4905 |
| Esterbauer H | 2001 | Australia | European | 156/140/44 | 85/127/44 | 33.5 | 42.0 | 20.0/25.0 | 41.4/40.1 | 0.0642 |
| Dalgaard LT | 2003 | Denmark | European | 292/322/135 | 299/369/148 | 39.5 | 40.7 | 100/100 | 20.0/20.0 | 0.0696 |
| Schauble N | 2003 | Germany | European | 108/135/34 | 72/89/27 | 36.6 | 38.0 | 41.5/54.8 | 14.7/25.5 | 0.9524 |
| Mancini FP | 2003 | Italy | European | 96/82/20 | 183/165/26 | 30.8 | 29.0 | 66.7/69.0 | 36.8/45.0 | 0.1691 |
| Xu R | 2005 | China | Asian | 105/197/82 | 179/357/164 | 47.0 | 48.9 | 42.5/33.9 | 64.1/62.9 | 0.5881 |
| Gu GY | 2005 | China | Asian | 5/5/3 | 32/73/44 | 42.3 | 54.0 | 69.2/51.7 | 59.8/52.8 | 0.8668 |
| Ochoa MC | 2007 | Spain | European | 79/80/34 | 59/92/19 | 38.3 | 38.2 | 51.3/49.3 | 11.5/11.8 | 0.0573 |
| Wang TN | 2007 | Taiwan | Asian | 193/115/16 | 81/28/5 | 22.7 | 16.7 | 38.0/51.9 | 55.9/59.0 | 0.2163 |
| Li JN | 2007 | China | Asian | 10/16/13 | 21/28/13 | 53.8 | 43.5 | Na/Na | Na/Na | 0.5211 |
| Shen XJ | 2007 | China | Asian | 32/36/10 | 45/55/18 | 35.9 | 38.6 | Na/Na | Na/Na | 0.8595 |
| Kring SI | 2008 | Denmark | European | 88/96/41 | 114/131/49 | 39.6 | 38.9 | 100/100 | Na/Na | 0.2797 |
| Lin E | 2009 | Taiwan | Asian | 92/101/29 | 92/146/47 | 35.8 | 42.1 | 48.6/46.3 | 57.0/57.5 | 0.3915 |
| Srivastava N | 2010 | India | Asian | 73/86/41 | 106/113/21 | 42.0 | 32.3 | Na/Na | Na/Na | 0.2346 |
| Heidari J | 2010 | Iran | Asian | 16/48/11 | 27/41/7 | 46.7 | 36.7 | 24.0/45.3 | 44.3/35.9 | 0.1252 |
| Zou HY | 2011 | China | Asian | 176/281/133 | 623/1115/489 | 46.4 | 47.0 | 23.7/40.8 | 55.7/55.8 | 0.8140 |
| Oktavianthi S | 2012 | Indonesia | Asian | 60/120/37 | 127/195/64 | 44.7 | 41.8 | 64.5/51.8 | 47.3/50.1 | 0.4552 |
| Andersen G | 2012 | Denmark | European | 583/754/210 | 1133/1499/521 | 37.9 | 40.3 | Na/Na | Na/Na | 0.5028 |
Na: not available.
Quantitative synthesis
The results of pooled analyses were summarized in Table 4.
Table 4.
Pooled measures for the associations of the UCP2 Ala55Val, 45-bp insertion/deletion, and -866G/A polymorphisms with overweight and obesity.
| Population | Inherited model | Before sensitive analysis |
After sensitive analysis |
Excluded studies in sensitive analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Numbers of cases/controls | Pooled OR (95% CI) |
I2 (%) |
Numbers of cases/controls | Pooled OR (95% CI) |
I2 (%) |
|||||
| FEM | REM | FEM | REM | |||||||
| Ala55Val | ||||||||||
| Overall | Dominant | 2011/2752 | 1.19 (1.04–1.36)⁎ | 1.27 (0.99–1.62) | 65.4 | 1799/2531 | 1.08 (0.94–1.25) | 1.11 (0.91–1.36) | 42.6 | Li (2007), Wang et al. (2009) |
| Recessive | 1687/2638 | 1.24 (1.06–1.45)⁎ | 1.29 (1.06–1.56)⁎ | 24.7 | – | – | – | – | – | |
| Co-dominant 1 | 2011/2752 | 1.12 (0.97–1.29) | 1.17 (0.91–1.50) | 62.0 | 1799/2531 | 1.01 (0.87–1.18) | 1.02 (0.84–1.25) | 35.1 | Li (2007), Wang et al. (2009) | |
| Co-dominant 2 | 1687/2638 | 1.26 (1.05–1.52)⁎ | 1.38 (1.03–1.84)⁎ | 51.3 | 1568/2461 | 1.18 (0.97–1.42) | 1.22 (0.96–1.54) | 25.4 | Sui et al. (2004) | |
| Additive | 2011/2752 | 1.24 (1.09–1.41)⁎ | 1.34 (1.04–1.72)⁎ | 69.5 | 1475/2417 | 1.07 (0.92–1.23) | 1.09 (0.90–1.31) | 33.9 | Li (2007), Wang et al., 2007, Wang et al., 2009 | |
| Over-dominant | 2011/2752 | 1.00 (0.89–1.13) | 1.04 (0.85–1.27) | 54.6 | 1968/2704 | 0.98 (0.86–1.11) | 0.99 (0.83–1.19) | 43.5 | Wang et al. (2009) | |
| Allelic | 2011/2752 | 1.17 (1.07–1.27)⁎ | 1.25 (1.07–1.48)⁎ | 64.6 | 1723/2402 | 1.08 (0.98–1.19) | 1.14 (0.98–1.32) | 46.2 | Li (2007), Sui et al. (2004) | |
| Asian | Dominant | 1586/2126 | 1.24 (1.06–1.44)⁎ | 1.34 (0.99–1.81) | 70.7 | 1050/1791 | 1.02 (0.85–1.21) | 1.03 (0.82–1.31) | 35.0 | Li (2007), Wang et al., 2007, Wang et al., 2009 |
| Recessive | 1262/2012 | 1.28 (1.06–1.55)⁎ | 1.39 (1.06–1.82)⁎ | 37.8 | – | – | – | – | – | |
| Co-dominant 1 | 1586/2126 | 1.15 (0.98–1.36) | 1.22 (0.90–1.67) | 68.0 | 1192/1536 | 1.12 (0.92–1.36) | 1.13 (0.88–1.45) | 29.9 | Kosuge et al. (2008), Li (2007), Wang et al. (2009) | |
| Co-dominant 2 | 1262/2012 | 1.31 (1.06–1.63)⁎ | 1.50 (1.01–2.20)⁎ | 59.9 | 1143/1835 | 1.19 (0.95–1.50) | 1.28 (0.92–1.80) | 41.2 | Sui et al. (2004) | |
| Additive | 1586/2126 | 1.29 (1.11–1.50)⁎ | 1.41 (1.04–1.93)⁎ | 74.0 | 1050/1791 | 1.07 (0.90–1.26) | 1.10 (0.85–1.43) | 49.9 | Li (2007), Wang et al., 2007, Wang et al., 2009 | |
| Over-dominant | 1586/2126 | 1.03 (0.89–1.19) | 1.09 (0.84–1.40) | 61.8 | 1361/1709 | 1.08 (0.92–1.27) | 1.10 (0.88–1.37) | 39.2 | Kosuge et al. (2008), Wang et al. (2009) | |
| Allelic | 1586/2126 | 1.20 (1.08–1.33)⁎ | 1.32 (1.07–1.63)⁎ | 64.6 | 974/1662 | 1.04 (0.93–1.18) | 1.09 (0.91–1.30) | 39.7 | Li (2007), Sui et al. (2004), Wang et al. (2007) | |
| European | Dominant | 425/626 | 1.04 (0.78–1.40) | 1.04 (0.78–1.40) | 0.0 | – | – | – | – | – |
| Recessive | 425/626 | 1.15 (0.87–1.52) | 1.15 (0.87–1.52) | 0.0 | – | – | – | – | – | |
| Co-dominant 1 | 425/626 | 0.99 (0.72–1.35) | 0.99 (0.72–1.35) | 0.0 | – | – | – | – | – | |
| Co-dominant 2 | 425/626 | 1.14 (0.80–1.61) | 1.14 (0.80–1.61) | 0.0 | – | – | – | – | – | |
| Additive | 425/626 | 1.07 (0.80–1.42) | 1.07 (0.80–1.42) | 0.0 | – | – | – | – | – | |
| Over-dominant | 425/626 | 0.92 (0.72–1.18) | 0.92 (0.72–1.18) | 0.0 | – | – | – | – | – | |
| Allelic | 425/626 | 1.07 (0.90–1.28) | 1.07 (0.90–1.28) | 0.0 | – | – | – | – | – | |
| 45-bp insertion/deletion | ||||||||||
| Overall | Dominant | 3416/3260 | 1.02 (0.92–1.13) | 1.01 (0.86–1.17) | 45.7 | – | – | – | – | – |
| Recessive | 3416/3260 | 1.19 (0.95–1.49) | 1.13 (0.85–1.51) | 28.6 | – | – | – | – | – | |
| Co-dominant 1 | 3416/3260 | 1.00 (0.89–1.12) | 0.99 (0.86–1.15) | 33.4 | – | – | – | – | – | |
| Co-dominant 2 | 3416/3260 | 1.16 (0.92–1.46) | 1.08 (0.78–1.51) | 41.4 | – | – | – | – | – | |
| Additive | 3416/3260 | 1.04 (0.93–1.15) | 1.01 (0.86–1.19) | 56.2 | 3111/2752 | 0.97 (0.87–1.09) | 0.96 (0.83–1.12) | 38.7 | Evans et al. (2000) | |
| Over-dominant | 3416/3260 | 0.98 (0.88–1.09) | 0.98 (0.86–1.11) | 20.4 | – | |||||
| Allelic | 3416/3260 | 1.04 (0.95–1.13) | 1.02 (0.89–1.17) | 53.0 | 3111/2752 | 0.98 (0.90–1.08) | 0.98 (0.87–1.10) | 32.2 | Evans et al. (2000) | |
| Asian | Dominant | 1006/1481 | 1.07 (0.88–1.30) | 1.07 (0.86–1.34) | 11.8 | – | – | – | – | – |
| Recessive | 1006/1481 | 0.96 (0.50–1.87) | 0.96 (0.50–1.87) | 0.0 | – | – | – | – | – | |
| Co-dominant 1 | 1006/1481 | 1.08 (0.89–1.32) | 1.09 (0.87–1.36) | 11.8 | – | – | – | – | – | |
| Co-dominant 2 | 1006/1481 | 0.98 (0.50–1.90) | 0.98 (0.50–1.90) | 0.0 | – | – | – | – | – | |
| Additive | 1006/1481 | 1.06 (0.88–1.28) | 1.06 (0.84–1.32) | 15.8 | – | – | – | – | – | |
| Over-dominant | 1006/1481 | 1.08 (0.89–1.32) | 1.09 (0.87–1.36) | 11.4 | – | – | – | – | – | |
| Allelic | 1006/1481 | 1.05 (0.89–1.26) | 1.05 (0.87–1.27) | 5.8 | – | – | – | – | – | |
| European | Dominant | 2410/1779 | 1.00 (0.88–1.13) | 0.98 (0.80–1.20) | 57.5 | 2105/1271 | 0.91 (0.79–1.05) | 0.91 (0.76–1.09) | 33.7 | Evans et al. (2000) |
| Recessive | 2410/1779 | 1.22 (0.96–1.55) | 1.16 (0.82–1.65) | 47.6 | – | – | – | – | – | |
| Co-dominant 1 | 2410/1779 | 0.96 (0.84–1.10) | 0.96 (0.80–1.15) | 42.5 | – | – | – | – | – | |
| Co-dominant 2 | 2410/1779 | 1.18 (0.92–1.51) | 1.12 (0.74–1.68) | 58.5 | 2105/1271 | 1.00 (0.76–1.32) | 0.97 (0.68–1.39) | 34.1 | Evans et al. (2000) | |
| Additive | 2410/1779 | 1.02 (0.91–1.16) | 1.00 (0.80–1.25) | 67.3 | 2105/1271 | 0.93 (0.81–1.06) | 0.92 (0.76–1.12) | 47.4 | Evans et al. (2000) | |
| Over-dominant | 2410/1779 | 0.94 (0.82–1.07) | 0.94 (0.81–1.09) | 22.3 | – | – | – | – | ||
| Allelic | 2410/1779 | 1.03 (0.94–1.14) | 1.01 (0.85–1.21) | 65.3 | 2105/1271 | 0.96 (0.86–1.07) | 0.95 (0.81–1.11) | 44.1 | Evans et al. (2000) | |
| -866G/A | ||||||||||
| Overall | Dominant | 5780/10,196 | 0.93 (0.86–0.99)⁎ | 0.94 (0.83–1.06) | 52.7 | 5440/9940 | 0.95 (0.88–1.02) | 0.97 (0.86–1.02) | 43.9 | Esterbauer et al. (2001) |
| Recessive | 5780/10,196 | 0.96 (0.88–1.05) | 1.02 (0.88–1.18) | 44.9 | – | – | – | – | ||
| Co-dominant 1 | 5780/10,196 | 0.93 (0.86–1.00) | 0.92 (0.82–1.03) | 44.3 | – | – | – | – | ||
| Co-dominant 2 | 5780/10,196 | 0.91 (0.83–1.01) | 0.98 (0.82–1.17) | 52.9 | 5580/9956 | 0.88 (0.80–0.98)⁎ | 0.91 (0.79–1.05) | 28.2 | Srivastava et al. (2010) | |
| Additive | 5780/10,196 | 0.93 (0.86–0.99)⁎ | 0.95 (0.84–1.08) | 60.7 | 5240/9700 | 0.93 (0.86–1.00) | 0.95 (0.85–1.06) | 44.7 | Esterbauer et al. (2001), Srivastava et al. (2010) | |
| Over-dominant | 5780/10,196 | 0.95 (0.89–1.02) | 0.93 (0.85–1.03) | 32.6 | – | – | – | – | – | |
| Allelic | 5780/10,196 | 0.95 (0.91–1.00) | 0.98 (0.90–1.07) | 57.6 | 5580/9956 | 0.94 (0.89–0.99)⁎ | 0.95 (0.88–1.03) | 44.4 | Srivastava et al. (2010) | |
| Asian | Dominant | 2142/4356 | 1.01 (0.90–1.14) | 1.07 (0.87–1.32) | 58.1 | 1596/3957 | 1.03 (0.90–1.17) | 1.08 (0.88–1.32) | 42.4 | Lin et al. (2009), Wang et al. (2007) |
| Recessive | 2142/4356 | 1.05 (0.91–1.21) | 1.11 (0.88–1.40) | 45.8 | – | – | – | – | – | |
| Co-dominant 1 | 2142/4356 | 1.00 (0.88–1.13) | 1.04 (0.86–1.26) | 45.7 | – | – | – | – | – | |
| Co-dominant 2 | 2142/4356 | 1.03 (0.88–1.22) | 1.13 (0.83–1.54) | 59.4 | 1942/4116 | 0.95 (0.80–1.13) | 0.97 (0.77–1.21) | 24.6 | Srivastava et al. (2010) | |
| Additive | 2142/4356 | 1.03 (0.91–1.15) | 1.10 (0.87–1.37) | 66.1 | 1396/3717 | 0.99 (0.87–1.13) | 1.04 (0.83–1.30) | 45.4 | Lin et al. (2009), Srivastava et al. (2010), Wang et al. (2007) | |
| Over-dominant | 2142/4356 | 0.98 (0.88–1.10) | 1.00 (0.87–1.14) | 19.8 | – | – | – | – | – | |
| Allelic | 2142/4356 | 1.02 (0.95–1.11) | 1.08 (0.92–1.26) | 63.6 | 1720/3831 | 1.02 (0.93–1.11) | 1.06 (0.92–1.22) | 41.5 | Lin et al. (2009), Srivastava et al. (2010) | |
| European | Dominant | 3638/5840 | 0.88 (0.81–0.96)⁎ | 0.86 (0.75–0.97)⁎ | 36.8 | – | – | – | – | – |
| Recessive | 3638/5840 | 0.90 (0.80–1.01) | 0.95 (0.79–1.14) | 39.1 | – | – | – | – | – | |
| Co-dominant 1 | 3638/5840 | 0.89 (0.81–0.98)⁎ | 0.85 (0.73–0.98)⁎ | 42.2 | – | – | – | – | – | |
| Co-dominant 2 | 3638/5840 | 0.85 (0.74–0.94)⁎ | 0.87 (0.72–1.05) | 33.3 | – | – | – | – | – | |
| Additive | 3638/5840 | 0.88 (0.80–0.95)⁎ | 0.86 (0.76–0.98)⁎ | 41.5 | – | – | – | – | – | |
| Over-dominant | 3638/5840 | 0.94 (0.86–1.02) | 0.88 (0.76–1.01) | 48.5 | – | – | – | – | – | |
| Allelic | 3638/5840 | 0.91 (0.86–0.97)⁎ | 0.91 (0.84–0.99)⁎ | 31.5 | – | – | – | – | – | |
Dominant model: CT+TT vs. CC for Ala55Val, ID+II vs. DD for 45-bp insertion/deletion, and GA+AA vs. GG for -866G/A.
Recessive model: TT vs. CT+CC for Ala55Val, II vs. ID+DD for 45-bp insertion/deletion, and AA vs. GA+GG for -866G/A.
Co-dominant 1 model: CT vs. CC for Ala55Val, ID vs. DD for 45-bp insertion/deletion, and GA vs. GG for -866G/A.
Co-dominant 2 model: TT vs. CC for Ala55Val, II vs. DD for 45-bp insertion/deletion, and AA vs. GG for -866G/A.
Additive model: 2TT+CT vs. CC for Ala55Val, 2II+ID vs. DD for 45-bp insertion/deletion, and 2AA+GA vs. GG for -866G/A.
Over-dominant model: CT vs. CC+TT for Ala55Val, ID vs. II+DD for 45-bp insertion/deletion, and GA vs. AA+GG for -866G/A.
Allelic model: T vs. C for Ala55Val, I vs. D for 45-bp insertion/deletion, and A vs. G for -866G/A.
DHWE: deviating from Hardy–Weinberg equilibrium.
FEM: fixed effect model; REM: random effect model.
P < 0.05.
UCP2 Ala55Val polymorphism
In the overall population, T allele was found to be significantly associated with an increased risk of overweight and obesity in the recessive (FEM: OR = 1.24, 95%CI = 1.06–1.45), co-dominant 2 (REM: OR = 1.38, 95%CI = 1.03–1.84), additive (REM: OR = 1.34, 95%CI = 1.04–1.72), and allelic (REM: OR = 1.25, 95%CI = 1.07–1.48) models.
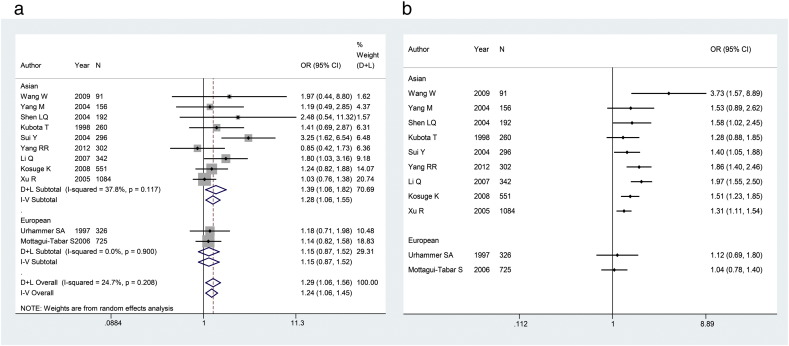
Subgroup analysis stratified by ethnicity revealed a significant association in recessive (FEM: OR = 1.28, 95%CI = 1.06–1.55), co-dominant 2 (REM: OR = 1.50, 95%CI = 1.01–2.20), additive (REM: OR = 1.41, 95%CI = 1.04–1.93) and allelic (REM: OR = 1.32, 95%CI = 1.07–1.63) models for Asians. No significant association between Ala55Val polymorphism and risk of overweight and obesity was found in any of the models for Europeans. The pooled ORs for the association of Ala55Val polymorphism with overweight and obesity under recessive model in the main and cumulative meta-analysis were presented in Fig. 1.
Fig. 1.
Stratified analysis of the association of Ala55Val polymorphism with overweight and obesity by ethnicity. (a) Meta-analysis for the association of the Ala55Val polymorphism with overweight and obesity under recessive model (TT vs. CT+CC). (b) Cumulative meta-analysis for the association of Ala55Val polymorphism with overweight and obesity under recessive model.
UCP2 45-bp insertion/deletion polymorphism
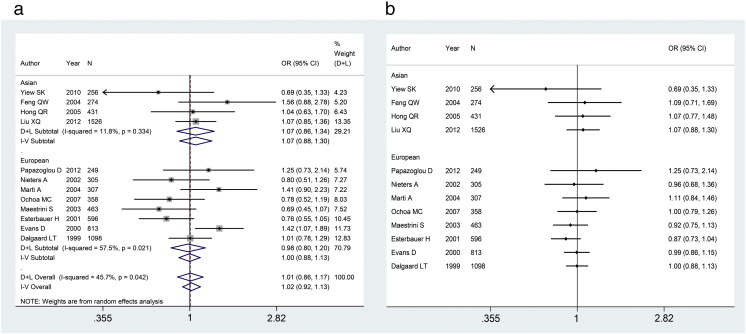
The meta-analysis showed no significant association between the 45-bp insertion/deletion polymorphism and the risk of overweight and obesity in dominant (FEM: OR = 1.02, 95%CI = 0.92–1.13), recessive (FEM: OR = 1.19, 95%CI = 0.95–1.49), co-dominant 1 (FEM: OR = 1.00, 95%CI = 0.89–1.12), co-dominant 2 (FEM: OR = 1.16, 95%CI = 0.92–1.46), additive (REM: OR = 1.04, 95%CI = 0.93–1.15), over-dominant (FEM: OR = 0.98, 95%CI = 0.88–1.09) and allelic (REM: OR = 1.02, 95%CI = 0.89–1.17) models. The results remained non-significant after stratification by ethnicity. The pooled ORs for the association of 45-bp insertion/deletion polymorphism with overweight and obesity under dominant model in the main and cumulative meta-analysis were presented in Fig. 2.
Fig. 2.
Stratified analysis of the association of 45-bp insertion/deletion polymorphism with overweight and obesity by ethnicity. (a) Meta-analysis for the association of the 45-bp insertion/deletion polymorphism with overweight and obesity under dominant model (ID+II vs. DD). (b) Cumulative meta-analysis for the association of 45-bp insertion/deletion polymorphism with overweight and obesity under dominant model.
UCP2 -866G/A polymorphism
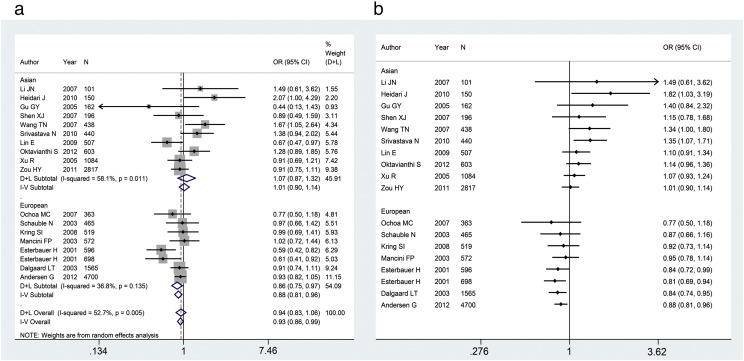
In the overall population, no significant association between -866G/A polymorphism and risk of overweight and obesity was found in any of the models. Subgroup analysis stratified by ethnicity revealed significant associations in dominant (FEM: OR = 0.88, 95%CI = 0.81–0.96), co-dominant 1 (FEM: OR = 0.89, 95%CI = 0.81–0.98), co-dominant 2 (FEM: OR = 0.85, 95%CI = 0.74–0.94), additive (FEM: OR = 0.88, 95%CI = 0.80–0.95), and allelic (FEM: OR = 0.91, 95%CI = 0.86–0.97) models for Europeans. However, no significant association was found in any inherited models in the population of Asian descent. The pooled ORs for the association of -866G/A polymorphism with overweight and obesity under dominant model in the main and cumulative meta-analysis were presented in Fig. 3.
Fig. 3.
Stratified analysis of the association of -866G/A polymorphism with overweight and obesity by ethnicity. (a) Meta-analysis for the association of the -866G/A polymorphism with overweight and obesity under dominant model (GA+AA vs. GG). (b) Cumulative meta-analysis for the association of -866G/A polymorphism with overweight and obesity under dominant model.
Source of heterogeneity and sensitive analysis
As shown in Table 4, significant heterogeneity (I2 > 50%) was observed among studies of the Ala55Val, 45-bp insertion/deletion, and -866G/A polymorphisms in some inherited models. Univariate meta-regression with the covariates of ethnicity, publication year, sex (ratio of male in case group to that in control group) and age (ratio of mean age in case group to that in control group) revealed that no covariates had excessive influence on between-study heterogeneity regarding the association of each polymorphism with overweight and obesity.
Significant heterogeneity was observed among studies of the Ala55Val polymorphism under all the inherited models except for the recessive one in the overall population. When stratified by ethnicity, the heterogeneity still existed in participants of Asian descent. Studies that were key contributors to between-study heterogeneity were indicated in Table 4. After sensitive analysis, the associations between T allele and risk of overweight and obesity were no longer significant in the co-dominant 2, additive, and allelic inherited models among the overall population.
Significant heterogeneity was observed among studies regarding the associations of the 45-bp insertion/deletion polymorphism with overweight and obesity in additive and allelic models among the overall population. When stratified by ethnicity, the heterogeneity was significantly decreased in the population of Asian descent, but still existed in that of European descent. Results of sensitive analysis indicated that one study (Evans et al., 2000) was mainly responsible for the between-study heterogeneity in additive and allelic models for the overall population, and that in dominant, co-dominant 2, additive and allelic models for the population of European descent, considering the association of the 45-bp insertion/deletion polymorphism with overweight and obesity. After exclusion of the study, the associations still remained non-significant in all the inherited models.
Significant heterogeneity was observed among studies regarding the associations of the -866G/A polymorphism with overweight and obesity in dominant, co-dominant 2, additive, and allelic models among the overall population. When stratified by ethnicity, the heterogeneity was significantly decreased in the population of European descent, but still existed in that of Asian descent. After sensitive analysis, the associations between A allele and risk of overweight and obesity were significant in the co-dominant 2 (FEM: OR = 0.99, 95%CI = 0.80–0.98) and allelic (FEM: OR = 0.94, 95%CI = 0.89–0.99) models in the overall population. However, the associations remained non-significant in all the aforementioned inherited models among the population of Asian descent.
Influence analysis
With articles that were key contributors to heterogeneity being excluded, no individual study was found to have excessive influence on the pooled effect under all the mentioned inherited models for any of the polymorphisms (data not shown).
Publication bias evaluation
After exclusion of studies that were key contributors to between-study heterogeneity, no publication bias was detected in all the aforementioned inherited models using funnel plot and Peters test regarding the associations between the three polymorphisms with overweight and obesity (data not shown).
Discussion
UCP2 gene was first identified as a candidate gene for obesity by Fleury et al. (1997). Since then, the associations of UCP2 gene polymorphisms (Aal55Val, 45-bp insertion/deletion, and -866G/A) with overweight and obesity have been widely studied; however, the results were conflicting. Considering the small effect of a genetic variant, a large sample size is certainly required in the primary genetic association study with adequate power. In order to obtain a robust conclusion regarding the role of UCP2 gene polymorphisms in the risk of overweight and obesity based on a larger scale of population, we performed the meta-analyses consisting 42 studies to evaluate the associations. The pooled ORs from our meta-analyses revealed a risk effect of T allele in Ala55Val polymorphism in populations of Asian descent, a protective effect of A allele in -866G/A polymorphism in populations of European descent and no effect of 45-bp insertion/deletion polymorphism on overweight and obesity. Cumulative meta-analyses showed an increased accuracy in the estimate of effects with larger samples aggregated for the three polymorphisms in UCP2 gene.
Between-study heterogeneity is a usual concern in meta-analyses (Munafo and Flint, 2004), which was also observed in our analysis under most of the inherited models regarding the association of UCP2 polymorphisms with overweight and obesity. After stratification analysis by ethnicity, the heterogeneity in some comparison groups remained significant. To further explore the source of heterogeneity, we performed a meta-regression analysis involving covariates such as publication year, sex, age, and sample size. However, none of the covariates was found to contribute to the between-study heterogeneity regarding the associations of aforementioned polymorphisms with overweight and obesity. It suggested that some other confounding factors, such as design quality, representativeness and general characteristics of the participants, environmental exposures, and non-uniform genotyping methods may be responsible for the heterogeneity. However, we failed to evaluate the role of these factors in the source of heterogeneity in our analysis due to inadequate information in the original articles.
No significant between-study heterogeneity (I2 < 50) was found in the recessive model regarding the association of Ala55Val polymorphism with overweight and obesity. Available data suggested a stable association of T allele of Ala55Val polymorphism with an increased risk of overweight and obesity in the recessive model for the Asian population. Studies indicated that the Ala55Val polymorphism had effects on metabolic rate and nocturnal physical activity (Astrup et al., 1999, Klannemark et al., 1998, Walder et al., 1998). T allele of the polymorphism was reported to be associated with lower energy expenditure and lower fat oxidation (Astrup et al., 1999), which may be implicated in the pathogenesis of overweight and obesity. In this meta-analysis, we failed to fully explore the source of between-study heterogeneity observed in most of the inherited models. Although significant associations were revealed in the co-dominant 2, additive and allelic models in the meta-analysis, the conclusions were limited because of the unsolved heterogeneity.
Significant between-study heterogeneity was found in the additive and allelic models regarding the association of 45-bp insertion/deletion polymorphism with overweight and obesity in the overall population. One study (Evans et al., 2000) that was a key contributor to the heterogeneity was identified using sensitive analysis. The between-study heterogeneity largely decreased after the key contributor was excluded, and the conclusions were consistent with those before sensitive analysis. In our meta-analysis, no evidence was observed on the association of UCP2 45-bp insertion/deletion polymorphism with overweight and obesity both in European and Asian populations.
Associations of -866G/A polymorphism with the risk of obesity were evaluated using meta-analysis by Andersen et al. (2013) and Liu et al. (2013) before. In this study, we performed a meta-analysis based on 15,976 individuals from 18 studies with strict eligible criteria to provide robust evidence. Available data in our meta-analysis suggested a stable association of -866G/A polymorphism with overweight and obesity in the population of European descent. The pooled ORs for GA+AA vs. GG, GA vs. GG, AA vs. GG and A vs. G for Europeans were 0.88, 0.89, 0.85 and 0.91, respectively, which were consistent with previous researches (Andersen et al., 2013, Liu et al., 2013). Besides, we identified a significant association of -866G/A polymorphism with overweight and obesity under additive model (2AA+GA vs GG) for the population of European descent. However, no evidence was observed in all the aforementioned models for the population of Asian descent. Oberkofler et al. indicated a reduced UCP2 mRNA level in intraperitoneal adipose tissue in obese individuals (Oberkofler et al., 1998). A allele of -866G/A polymorphism was reported to be associated with enhanced UCP2 mRNA expression in adipose tissue in European population (Esterbauer et al., 2001), which may explain its protective effect against overweight and obesity.
Obesity is a heterogeneous disorder caused by complex gene × gene and gene × environment interactions. The effects of Ala55Val and -866G/A polymorphisms on overweight and obesity differed in Europeans and Asians, which may be attributed to different genetic backgrounds. Significant linkage disequilibrium has been observed among Ala55Val, 45-bp insertion/deletion, and -866G/A polymorphisms in overweight and obese populations (Esterbauer et al., 2001, Ochoa et al., 2007, Oktavianthi et al., 2012, Wang et al., 2007). Lack of consideration of the effects of adjacent loci may partially explain the discrepancies among individual studies. Since limited information was provided in the included studies, effects of haplotypes on overweight and obesity could not be evaluated in our meta-analysis, which require further investigation.
The conclusions of our meta-analysis should be considered under the following limitations. First, the outcome of interest was not uniform in all the included studies. Cases were defined as morbidly obese population, obese population, or combined population of overweight and obesity in different studies. The cutoff used for overweight or obesity differed in studies with the same target population. Second, subgroup analysis was performed to decrease potential between-study heterogeneity; however, limited studies and small sample size in some subgroups may lead to an underpowered result. Third, since insufficient information was provided in the original studies, we failed to fully explore the source of between-study heterogeneity under some circumstances. Last, language was restricted to English and Chinese in the search strategy, which may limit the result.
Conclusions
In conclusion, meta-analyses of currently available studies supported that T allele of Ala55Val polymorphism was associated with an increased risk of overweight and obesity under recessive model, especially for populations of Asian descent; and A allele of -866G/A polymorphism had a protective effect on overweight and obesity, especially for European populations. However, 45-bp insertion/deletion polymorphism is not a susceptibility locus to overweight and obesity. Additional association studies with large sample size and sufficient information allowing powerful stratification analyses are needed to provide more robust evidence to the conclusions. The roles of haplotypes of UCP2 gene in the pathogenesis of overweight and obesity require to be clarified in future researches.
Funding
The study was sponsored by a grant from the Ministry of Science and Technology of People's Republic of China (2006BA105A01).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- Andersen G. The frequent UCP2-866G>A polymorphism protects against insulin resistance and is associated with obesity: a study of obesity and related metabolic traits among 17 636 Danes. Int. J. Obes. (Lond.) 2013;37:175–181. doi: 10.1038/ijo.2012.22. [DOI] [PubMed] [Google Scholar]
- Astrup A., Toubro S., Dalgaard L.T., Urhammer S.A., Sorensen T.I., Pedersen O. Impact of the v/v 55 polymorphism of the uncoupling protein 2 gene on 24-h energy expenditure and substrate oxidation. Int. J. Obes. Relat. Metab. Disord. 1999;23:1030–1034. doi: 10.1038/sj.ijo.0801040. [DOI] [PubMed] [Google Scholar]
- Dalgaard L.T., Pedersen O. Uncoupling proteins: functional characteristics and role in the pathogenesis of obesity and Type II diabetes. Diabetologia. 2001;44:946–965. doi: 10.1007/s001250100596. [DOI] [PubMed] [Google Scholar]
- Dalgaard L.T., Sorensen T.I., Andersen T., Hansen T., Pedersen O. An untranslated insertion variant in the uncoupling protein 2 gene is not related to body mass index and changes in body weight during a 26-year follow-up in Danish Caucasian men. Diabetologia. 1999;42:1413–1416. doi: 10.1007/s001250051312. [DOI] [PubMed] [Google Scholar]
- Dalgaard L.T. Mutational analysis of the UCP2 core promoter and relationships of variants with obesity. Obes. Res. 2003;11:1420–1427. doi: 10.1038/oby.2003.191. [DOI] [PubMed] [Google Scholar]
- Diano S., Horvath T.L. Mitochondrial uncoupling protein 2 (UCP2) in glucose and lipid metabolism. Trends Mol. Med. 2012;18:52–58. doi: 10.1016/j.molmed.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Esterbauer H. A common polymorphism in the promoter of UCP2 is associated with decreased risk of obesity in middle-aged humans. Nat. Genet. 2001;28:178–183. doi: 10.1038/88911. [DOI] [PubMed] [Google Scholar]
- Evans D. Frequency of and interaction between polymorphisms in the beta3-adrenergic receptor and in uncoupling proteins 1 and 2 and obesity in Germans. Int. J. Obes. Relat. Metab. Disord. 2000;24:1239–1245. doi: 10.1038/sj.ijo.0801402. [DOI] [PubMed] [Google Scholar]
- Feng Q.W., Dong Y., Su Q., Xing H.L., Zuo J.N. Association between polymorphism of uncoupling protein 2 gene and type 2 diabetes in Chinese. Acad. J. Shanghai Second Med. Univ. 2004;24:108–110. [Google Scholar]
- Fleury C. Uncoupling protein-2: a novel gene linked to obesity and hyperinsulinemia. Nat. Genet. 1997;15:269–272. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- Gu G.Y. 2005. Relationships between the functional polymorphism in the promoter of uncoupling protein 2 gene and obesity and type 2 diabetes. (D) [Google Scholar]
- Heidari J., Akrami S.M., Heshmat R., Amiri P., Fakhrzadeh H., Pajouhi M. Association study of the -866G/A UCP2 gene promoter polymorphism with type 2 diabetes and obesity in a Tehran population: a case control study. Arch. Iran. Med. 2010;13:384–390. [PubMed] [Google Scholar]
- Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Hong Q.R. Relationship between uncoupling protein 2 gene polymorphism and obesity in children and adolescent. Chin. J. Endocrinol. Metab. 2005;21:545–546. [Google Scholar]
- Klannemark M., Orho M., Groop L. No relationship between identified variants in the uncoupling protein 2 gene and energy expenditure. Eur. J. Endocrinol. 1998;139:217–223. doi: 10.1530/eje.0.1390217. [DOI] [PubMed] [Google Scholar]
- Kosuge K. Human uncoupling protein 2 and 3 genes are associated with obesity in Japanese. Endocrine. 2008;34:87–95. doi: 10.1007/s12020-008-9111-9. [DOI] [PubMed] [Google Scholar]
- Krempler F. A functional polymorphism in the promoter of UCP2 enhances obesity risk but reduces type 2 diabetes risk in obese middle-aged humans. Diabetes. 2002;51:3331–3335. doi: 10.2337/diabetes.51.11.3331. [DOI] [PubMed] [Google Scholar]
- Kring S.I. Genotype–phenotype associations in obesity dependent on definition of the obesity phenotype. Obes. Facts. 2008;1:138–145. doi: 10.1159/000137665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T. Molecular screening of uncoupling protein 2 gene in patients with noninsulin-dependent diabetes mellitus or obesity. J. Clin. Endocrinol. Metab. 1998;83:2800–2804. doi: 10.1210/jcem.83.8.4994. [DOI] [PubMed] [Google Scholar]
- Li Q. 2007. Study of relevant risk factor and molecular epidemiology on polymorphism of UCP2 gene in children simple obesity. (D) [Google Scholar]
- Li J.N., He L., Zhou L.L., Ye F., Dong C.P. Relationship of uncoupling protein 2-866G/A polymorphism with obesity in northern Chinese. J. Pract. Med. 2007;23:1992–1994. [Google Scholar]
- Lin E., Pei D., Huang Y.J., Hsieh C.H., Wu L.S. Gene–gene interactions among genetic variants from obesity candidate genes for nonobese and obese populations in type 2 diabetes. Genet. Test. Mol. Biomarkers. 2009;13:485–493. doi: 10.1089/gtmb.2008.0145. [DOI] [PubMed] [Google Scholar]
- Liu X.Q. A 45-bp insertion/deletion polymorphism in uncoupling protein 2 is not associated with obesity in a Chinese population. Biochem. Genet. 2012;50:784–796. doi: 10.1007/s10528-012-9520-6. [DOI] [PubMed] [Google Scholar]
- Liu L., Zhao X., Kang S., Zhang D. An association between -866G/A polymorphism in the promoter of UCP2 and obesity: a meta-analysis. Gene. 2013;514:41–47. doi: 10.1016/j.gene.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Maestrini S. Lack of association between UCP2 gene polymorphisms and obesity phenotype in Italian Caucasians. J. Endocrinol. Invest. 2003;26:985–990. doi: 10.1007/BF03348196. [DOI] [PubMed] [Google Scholar]
- Mancini F.P. Variants of uncoupling protein-2 gene and obesity: interaction with peroxisome proliferator-activated receptorgamma2. Clin. Endocrinol. (Oxf.) 2003;59:817–822. doi: 10.1046/j.1365-2265.2003.01926.x. [DOI] [PubMed] [Google Scholar]
- Marti A., Corbalan M.S., Forga L., Martinez-Gonzalez M.A., Martinez J.A. Higher obesity risk associated with the exon-8 insertion of the UCP2 gene in a Spanish case–control study. Nutrition. 2004;20:498–501. doi: 10.1016/j.nut.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar S., Hoffstedt J., Brookes A.J., Jiao H., Arner P., Dahlman I. Association of ADRB1 and UCP3 gene polymorphisms with insulin sensitivity but not obesity. Horm. Res. 2008;69:31–36. doi: 10.1159/000111793. [DOI] [PubMed] [Google Scholar]
- Munafo M.R., Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20:439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Nieters A., Becker N., Linseisen J. Polymorphisms in candidate obesity genes and their interaction with dietary intake of n-6 polyunsaturated fatty acids affect obesity risk in a sub-sample of the EPIC-Heidelberg cohort. Eur. J. Nutr. 2002;41:210–221. doi: 10.1007/s00394-002-0378-y. [DOI] [PubMed] [Google Scholar]
- Oberkofler H., Liu Y.M., Esterbauer H., Hell E., Krempler F., Patsch W. Uncoupling protein-2 gene: reduced mRNA expression in intraperitoneal adipose tissue of obese humans. Diabetologia. 1998;41:940–946. doi: 10.1007/s001250051011. [DOI] [PubMed] [Google Scholar]
- Ochoa M.C. Association between obesity and insulin resistance with UCP2–UCP3 gene variants in Spanish children and adolescents. Mol. Genet. Metab. 2007;92:351–358. doi: 10.1016/j.ymgme.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Oktavianthi S. Uncoupling protein 2 gene polymorphisms are associated with obesity. Cardiovasc. Diabetol. 2012;11:41. doi: 10.1186/1475-2840-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazoglou D. Uncoupling protein-2 45-base pair insertion/deletion polymorphism: is there an association with severe obesity and weight loss in morbidly obese subjects? Metab. Syndr. Relat. Disord. 2012;10:307–311. doi: 10.1089/met.2012.0003. [DOI] [PubMed] [Google Scholar]
- Patsopoulos N.A., Evangelou E., Ioannidis J.P. Sensitivity of between-study heterogeneity in meta-analysis: proposed metrics and empirical evaluation. Int. J. Epidemiol. 2008;37:1148–1157. doi: 10.1093/ije/dyn065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.L., Sutton A.J., Jones D.R., Abrams K.R., Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- Schauble N. No evidence for involvement of the promoter polymorphism -866 G/A of the UCP2 gene in childhood-onset obesity in humans. Exp. Clin. Endocrinol. Diabetes. 2003;111:73–76. doi: 10.1055/s-2003-39232. [DOI] [PubMed] [Google Scholar]
- Shen L.Q. 2004. Effects of the related factors of environment and mutation of the uncoupling protein 2 gene on the simple obesity in children. (D) [Google Scholar]
- Shen X.J. 2007. Relationships of uncoupling protein 2 gene -866G/A polymorphism with metabolic syndrome in Chinese Han. (D) [Google Scholar]
- Srivastava N., Prakash J., Lakhan R., Agarwal C.G., Pant D.C., Mittal B. A common polymorphism in the promoter of UCP2 is associated with obesity and hyperinsulenemia in northern Indians. Mol. Cell. Biochem. 2010;337:293–298. doi: 10.1007/s11010-009-0311-2. [DOI] [PubMed] [Google Scholar]
- Sui Y., Weng J.P., Xiu L.L., Wang J., Yan J.H. Additive effects of the variants in the beta(3)-adrenergic receptor and uncoupling protein-2 genes on obesity in Chinese. Chin. J. Epidemiol. 2004;21:229–232. [PubMed] [Google Scholar]
- Urhammer S.A. Mutational analysis of the coding region of the uncoupling protein 2 gene in obese NIDDM patients: impact of a common amino acid polymorphism on juvenile and maturity onset forms of obesity and insulin resistance. Diabetologia. 1997;40:1227–1230. doi: 10.1007/s001250050811. [DOI] [PubMed] [Google Scholar]
- Walder K. Association between uncoupling protein polymorphisms (UCP2–UCP3) and energy metabolism/obesity in Pima indians. Hum. Mol. Genet. 1998;7:1431–1435. doi: 10.1093/hmg/7.9.1431. [DOI] [PubMed] [Google Scholar]
- Wang T.N. UCP2 A55V variant is associated with obesity and related phenotypes in an aboriginal community in Taiwan. Int. J. Obes. (Lond.) 2007;31:1746–1752. doi: 10.1038/sj.ijo.0803648. [DOI] [PubMed] [Google Scholar]
- Wang W., Zhang W.W., Xie Z.M., Li Y.R., Pei X.F. A pioneer study on relationship of peroxisome proliferator-activated receptor-γ2 and uncoupling protein 2 gene polymorphism with simple obesity in Chengdu. Mod. Prev. Med. 2009;36:938–941. [Google Scholar]
- Xu R. 2005. Association between uncoupling protein 2 gene polymorphisms and obesity in Japanese. (D) [Google Scholar]
- Yang R.R. 2012. Relationship between polymorphism of height and obesity-associated gene and growth and development in middle school in Chongqing. (D) [Google Scholar]
- Yang M. An association study of resting energy expenditure, total and regional body fat with the polymorphism of UCP2 gene. Shanghai Med. J. 2004;27:237–240. [Google Scholar]
- Yiew S.K. No association between peroxisome proliferator-activated receptor and uncoupling protein gene polymorphisms and obesity in Malaysian university students. Obes. Res. Clin. Pract. 2010;4:E325–E331. doi: 10.1016/j.orcp.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Zhang C.Y. Uncoupling protein-2 negatively regulates insulin secretion and is a major link between obesity, beta cell dysfunction, and type 2 diabetes. Cell. 2001;105:745–755. doi: 10.1016/s0092-8674(01)00378-6. [DOI] [PubMed] [Google Scholar]
- Zou H.Y. The association of uncoupling proteins 2-866G/A (UCP2-866G/A) polymorphism with obesity. Mol. Cardiol. China. 2011;11:158–163. [Google Scholar]