Abstract
Introduction
Interleukin 1 beta (IL- 1β), a key proinflammatory cytokine encoded by the interleukin 1 beta gene, has been associated with chronic inflammation and plays an important role in lung inflammatory diseases including lung cancer. Elevated levels of Interleukin 1proteins, in particular interleukin 1 beta greatly enhance the intensity of the inflammatory response.
Aim
To study the role of interleukin 1 beta-31C > T and -511 T > C polymorphism in the pathogenesis of non small cell lung cancer (NSCLC).
Materials and methods
One hundred and ninety non small cell lung cancer patients and 200 healthy age, sex, smoking and dwelling matched controls were used for polymorphic analysis by polymerase chain reaction—restriction fragment length polymorphism (PCR-RFLP) followed by sequencing. Normal tissues of 48 histopathologically confirmed non small cell lung cancer patients were taken for mRNA expression analysis. Quantitation of interleukin 1 beta was carried out by quantitative real time PCR.
Result
The T/T genotype of interleukin 1 beta-31 gene was significantly associated with increased risk of NSCLC [(P = 0.001, OR – 2.8 (95%CI 1.52–5.26)]. The interleukin 1 beta − 511 T > C does not show any difference between the NSCLC and control group (P = 0.3, OR – 0.72 (95%CI 0.41–1.28). Quantitative analysis of mRNA showed significant association with interleukin 1 beta T allele as compared to the interleukin 1 beta-31C allele (P = 0.006).
Conclusion
We conclude that lung cancer risk genotype interleukin 1 beta-31TT results in increased expression of interleukin 1 beta mRNA in lung cancer patients. Our data suggest that this genotype (IL1β -31TT) in the interleukin 1 beta regulatory region provide a microenvironment with elevated inflammatory stimuli and thus increasing the risk for lung cancer.
Keywords: Polymorphism, Non small cell lung cancer, Inflammation, Restriction fragment length polymorphism (RFLP)
Introduction
Lung cancer kills more people annually than the next four cancer types combined, and as many as one billion people may die worldwide from lung cancer this century (Jha, 2009). Tobacco smoking is considered to be the leading cause of lung cancer, with approximately 85% of deaths directly linked to smoking. It has been observed that all smokers did not develop lung cancer but other factors such as individual genetic susceptibility and inflammation may play a role in the development of lung cancer. Chronic inflammation has long been deemed to be an important factor in the pathogenesis of many human cancers, including lung cancer. Many lung cancer pathogens can cause inflammation in the respiratory tract and lung parenchyma. Elevated numbers of inflammatory cells and higher levels of pro-inflammatory cytokines are seen in lung cancer tumor microenvironments and their surrounding stromal tissue (Balkwill and Mantovani, 2001, Takizawa et al., 2000). Many pro inflammatory cytokines have been shown to be involved in the pathogenesis of lung cancer. However, the cellular and molecular mechanisms underlying this relationship are not well elucidated.
Interleukin 1 beta is an important cytokine in regulating the expression of several genes involved in various inflammatory processes (Dinarello, 1996, Dinarello, 2002a). Interleukin 1 beta plays an important role in various inflammatory diseases including lung cancer (Azad et al., 2008, Mayne et al., 1999). Sustained induction of interleukin 1 beta enhances the intensity of the inflammatory response and creates an inflammatory micro-environment in advantage of tumor initiation and/or promotion (Dinarello, 2006). The amount of interleukin 1 beta secreted to the microenvironment is regulated at the messenger RNA (mRNA) level and protein level. The processing of the pro-interleukin 1 beta precursor into active interleukin 1 beta molecules is mediated by the caspase-1 enzyme (Dinarello, 1996, Apte et al., 2006b). Different cell types including lung epithelial cells upon exposure to various chemicals and other environmental agents produce and secrete interleukin 1 beta (Apte et al., 2006a, Dinarello, 1996). It has also been shown that the interleukin 1beta gene may be induced by cigarette smoke in human cells in vitro (Hellermann et al., 2002). The crucial role of interleukin 1 beta in susceptibility to 3-methylcholanthrene-induced carcinogenesis has been recently investigated in interleukin 1 beta knock-out mice. In the mice deficient in interleukin 1 beta, tumours developed slower and in fewer animals than in wild-type mice (Shirakawa et al., 1993, Apte et al., 2006a). Several single-nucleotide polymorphisms (SNPs) such interleukin 1 beta-31C > T (rs1143627), interleukin 1 beta-511 T > C (rs16944), in the promoter region of the gene have been identified. The role of interleukin 1 beta in increasing the risk of lung cancer has been observed. In particular, the interleukin 1 beta-31 C > T polymorphism has been has been examined in several association studies. Some studies have reported an association between this SNP and risk for cancer and inflammatory diseases (Lind et al., 2005, Zienolddiny et al., 2004, Chen et al., 2006a, Haukim et al., 2002, Bidwell et al., 1999). Several studies have shown that both -31C/T and -511 T > C are significantly associated with the increased risk of non small cell lung cancer (Zienolddiny et al., 2004). The interleukin 1 beta-31 C > T is being considered as a potential candidate in regulating the expression of interleukin 1 beta due to the shift from T to C at position -31 that mediates a change from TATAAA to CATA AA and a potential disruption of the TATA box (Wobbe and Struhl, 1990). A study conducted in A549 cells showed that the expression of interleukin 1 beta was higher in cells carrying -31 T allele as compared to the normal wild -31 C allele (Lind et al., 2007) while studies have so far failed to show any functional relevance of -511 SNP to alter gene transcription (Chen et al., 2006b, El Omar et al., 2000).
In view of the above findings, we conduct a case control study to investigate the polymorphic association of interleukin 1 beta-31C > T and interleukin 1 beta-511 T > C with non small cell lung cancer in our population. Also mRNA expression of interleukin 1beta was analysed in normal tissues of lung cancer patients by quantitative real time PCR.
Materials and methods
Materials
This study included 190 lung cancer patients and 200 age, sex, dwelling and smoking matched controls. All the lung cancer cases selected for study were histologically confirmed non small lung cancer cases. Patients were recruited from Out Patients Department, Department of Medical Oncology and Department of Cardiovascular and thoracic surgery, Sher-i-Kashmir Institute of Medical Sciences, Srinagar (SKIMS) between April 2010 and March 2012. All those patients who had prior history of cancer other than lung cancer and patients who had received any chemotherapy/radiotherapy were excluded from our study. All participants of the control group were selected from individuals receiving routine medical examinations in the same hospital, with no history of any malignancy, or any symptoms of other acute or chronic inflammatory lung diseases (i.e. COPD and asthma), peritonitis, rheumatoid arthritis, or late stage kidney diseases. Ethical approval was taken from the ethical committee of Sheri Kashmir institute of medical sciences Srinagar and World Medical Association (Declaration of Helsinki) protocol was followed. All the participants were informed and written consent was taken for participation in the study.
Peripheral blood samples (5 ml) were collected in EDTA vials from lung cancer patients and healthy controls and later stored at − 80 °C till further use. Tissue samples were stored in RNA later (Sigma Aldrich) and kept at − 80 °C till further use. A detailed questionnaire was completed for each patient and control. The questionnaire included information on the number of cigarettes smoked daily/quantity of tobacco smoked every day and the number of years the subject had been smoking. For smoking status, a person who had smoked (cigarette/Hokka) at least once a day for > 1year was regarded as smoker. The cases included 163 (85.79%) males and 27 (14.71%) females with age distribution of ≤ 50 years 59 (31.05%) and > 50 years of age were 131 (68.95%) with mean age of 57.80 ± 10.97. The control group had 170 (85%) males and 30 (15%) females with age distribution ≤ 50 years 65 (32.5%) and > 50years 135 (67.5%) with mean age of 56.69 ± 12.22. Smokers in cases and controls were 114 (60%), 116 (58%) and non smokers were 76 (40%) and 84 (42%) respectively. Non small cell lung cancer included 130 (68.42%) cases who had stage I and stage II and 60 (3158%) cases who had stage III and IV. Squamous cell carcinoma patients were 140 (73.69%) while others which included large cell carcinoma, bronchogenic carcinoma and adenocarcinoma were 50 (26.32%). Out of 190 patients 71 had developed grade 1, 76 had developed grade 2 and 43 presented with grade 3 non small cell lung cancer (Table 1).
Table 1.
Selected demographic variables and risk factors of non small cell lung cancer cases and controls.
| Variables |
Cases N = 190 (%) |
Controls N = 200 (%) |
P value |
|---|---|---|---|
| All subjects | |||
| Age in years | |||
| ≤ 50 | 59 (31.05) | 65 (32.5) | 0.8 |
| > 50 | 131 (68.95) | 135 (67.5) | |
| Sex | |||
| Male | 163 (85.79) | 170 (85) | 0.88 |
| Female | 27 (14.21 | 30 (15) | |
| Smoking status | |||
| Nonsmoker | 76 (40.0) | 84 (42) | |
| Smoker | 114 (60.0) | 116 (58) | 0.76 |
| Dweller | 127 (66.80) | 129 (64.5) | |
| Rural | 63 (33.15) | 71 (35.5) | |
| Urban | 0.67 | ||
| Stage | |||
| I & II | 130 (68.42) | – | – |
| III & IV | 60 (31.58) | ||
| Histology | |||
| SCCa | 140 (73.69) | ||
| Othersb | 50 (26.32) | – | – |
| Grade 1 | 71 (37.36) | ||
| Grade 2 | 76 (40.0) | ||
| Grade 3 | 43 (22.6) | ||
Adjusted for age, smoking, gender and dwelling.
G1 - well differentiated, G 2- moderately differentiated, G3- poorly differentiated.
Squamous cell carcinoma.
Others include adenocarcinoma, large cell carcinoma, brancogenic carcinoma.
Interleukin 1 beta genotyping
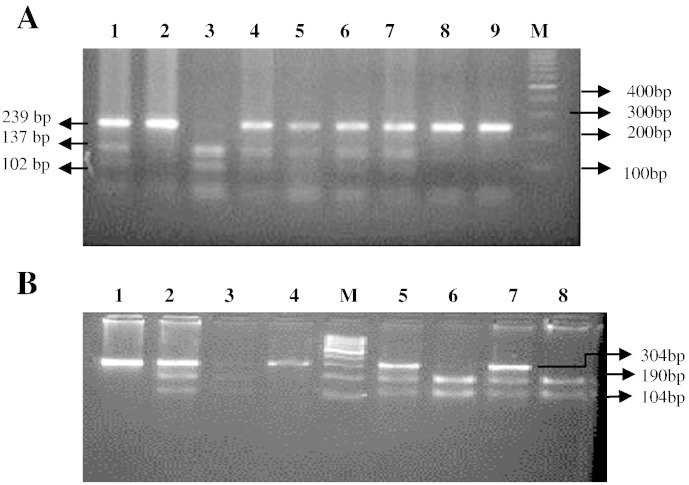
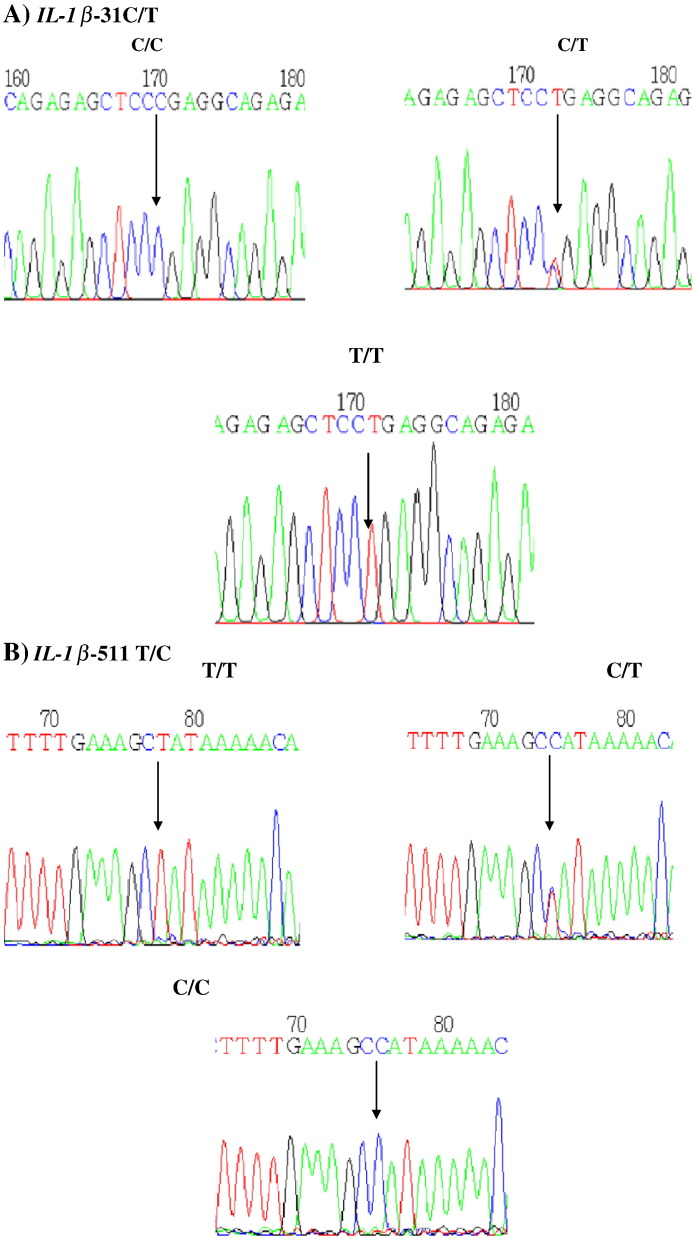
DNA extraction was performed according to the manufacturer's protocol for Qiagen DNA extraction kits (Qiagen, Hilden, NRW, Germany). DNA content was quantified by spectrophotometric absorption (Nanodrop Spectrophotometer, BioLab, Scoresby, VIC, Australia). Polymerase chain reaction (PCR) was performed using an iCycler Thermal Cycler (Bio- Rad, Hercules, CA, USA). Interleukin 1 beta-31C > T (rs1143627) and interleukin 1 beta-511 T > C (rs16944) genotypes were determined using polymerase chain reaction–restriction fragement length polymorphism (PCR-RFLP) method followed by DNA sequencing. Amplification of the target region was carried out by polymerase chain reaction using the specific forward and reverse primers. Primers were designed and selected using Primer3, version 0.4.0 software. For interleukin 1 beta-31 C > T, the primers were forward 5′-AGAAGCTTCCACCAATACTC-3′ and reverse 5′-AGCACCTAGTTGTAAGGAAG-3′. For interleukin 1 beta-511 T > C, the primers were forward 5′-TGG CAT TGA TCT GGT TCA TC-3′and reverse 5′-GTT TAG GAA TCT TCC CAC TT-3′. The PCR reaction mixture consisted of Taq 1.5 U (Ferments), sense and antisense primers (0.5 μmol/l), MgCl2 (50 mmol/l), dNTP (0.2 mmol/l), and DNA template (1 μg) and was subjected to an initial denaturing step of 4 min at 95 °C, then 35 cycles of denaturing for 30 s at 95 °C, annealing for 30 s at 56 °C, extension for 30 s at 72 °C, and a final extension step of 10 min at 72 °C. Digestion of the amplified products of interleukin 1 beta-31C > T and interleukin 1 beta-511 T > C was done by using 10 units restriction endonucleases Alu1 (New England Biolabs) and Ava1 (New England Biolabs) respectively and incubated at 37 °C for 16 h. The digested products were checked on 3% agaroses gel, the RFLP picture for iinterleukin 1beta − 31 genotype was identified as (C/C - 239 bp), (T/T- 137/102 bp), (T/C- 239/137/102 bp) and interleukin 1 beta-511 was identified as (T/T- 304 bp), (C/C -190/114), (T/C- 304/190/114 bp) Fig. 1. RFLP results were later confirmed by DNA sequencing Fig. 2.
Fig. 1.
A: Three percent electrophoresis results for the IL-1β − 31 genotype. Lane M represents the 100 bp DNA marker; lane 3 represents the IL-1β − 31 T/T genotype; lanes 1, 4, 5,6,7 represent the heterogeneous T/C genotypes; and lane 2,8,9 represents the C/C genotype.
B: Three percent electrophoresis results of the IL-1β − 511 genotype. Lane M represents the 100 bp DNA marker; lane 1,4 represents the IL-1β − 511 T/T genotype; lanes 2, 3, 5,7 represent the heterogeneous C/T genotype; and lane 1,4 represents C/C genotype.
Fig. 2.
A. IL-1β − 31 sequencing results. Arrow indicates the SNP location.
B. IL-1β − 511 sequencing results. The arrow indicates the SNP location.
Interleukin 1 beta (IL-1β) mRNA expression
Total RNA was extracted by using TRIZOL (Sigma Aldrich, USA) from normal tissues of lung cancer tissue. Integrity of the mRNA was checked on 1% agarose gel and quantified at 260/280 ratio. RNA was converted to cDNA by using first strand cDNA synthesis kit according to manufactures protocol (Fermentas, USA). Dilution of the cDNA was performed to get uniform quantity of cDNA in all samples. PCR primers were designed for interleukin 1 beta and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) over introns to avoid the amplification of possible traces of genomic DNA contamination by using Primer Express 3.0 software, Applied Biosystems and glyceraldehyde 3-phosphate dehydrogenase mRNA (GenBank accession number NM-002046). Glyceraldehyde 3-phosphate dehydrogenase was used as an internal control. For interleukin 1 beta primer sequence were as follows- forward 5'-GCACGATGCACCTGTACGAT and reverse 5'-CACCAAGCTTTTTTGCTGTGAGT-3' and for glyceraldehyde 3-phosphate dehydrogenase, the primers were as follows: forward 5′-GATCCGCATAATCTGCATGGT-3′ and reverse 5′-GATCCGCATAATCTGCATGGT-3′. Quantitative Real time PCR (Agilent Biotechnologies, Germany) was performed for the detection of interleukin 1 beta mRNA by recruiting Appllied Biosystems Inc StepOne software v2.0. PCR was performed containing Maxima® SYBR Green qPCR Master Mix (2X) and all the samples (unknown and standards) were run in triplicates and accompanied by non template control (NTC). Thermal cycling conditions included 40 cycles of 30s at 55 °C. The melting curves of all final real time PCR products were analysed for determination of genuine products and contamination of non specific products and primer dimer. All amplified products of real time polymerase chain reaction were subjected for separation on 2% agarose gel electrophoresis for ensuring the correct amplification products. Delta CT (ΔCT) method was used to check the interleukin 1 beta mRNA expression in normal lung tissue of lung cancer patients by normalization against glyceraldehyde 3-phosphate dehydrogenase used as a reference gene.
Statistical analysis
The distribution of the genotypes in controls was compared with that expected from Hardy-Weinberg equilibrium (HWE) by the chi square (χ2) test. Odds ratios (ORs) and their 95% confidence intervals (CIs), with adjustments for age, sex dwelling and smoking were calculated by Fisher's exact test/Chi square test as appropriate. Students unpaired t test was used to compare the means and standard deviation. All reported P values were based on two-sided tests. Significance level was taken at p ≤ 0.05. Statistical tests were performed using the software SPSS 16.0 (SPSS Inc., Chicago, Illinois).
Results
Interleukin 1 beta SNPs
A total of 190 Lung cancer cases and 200 healthy controls were successfully evaluated for genotyping of interleukin 1 beta-31C > T and interleukin 1 beta-511 T > C. The frequencies of tested genotypes in cases and controls are given in (Table 2). The observed genotypes for the controls population was in complete accordance with the Hardy Weinberg equilibrium (P > 0.05). The distribution of genotypes of interleukin 1 beta-31 C/T in controls were -31C/C(29.5%), -31C/T (55.5%), -31TT (15%) while in cases the distribution was -31 C/C (17.8%), -31C/T (56.3%) and -31 T/T (25.7%). The frequency of interleukin 1 beta genotypes was higher in cases with odds ratio of 1.6 (95% CI 1.01-2.7) in -31 CT and 2.8 (95% CI- 1.52-5.26) in -31TT respectively (Table 2). The distribution of both -31CT and -31TT observed in cases showed a strong significant association with non small cell lung cancer (P < 0.05) (Table 2). The interleukin 1 beta − 31 T allele occurred more significantly in the non small cell lung cancer group than in the control group (P = 0.002) (Table 3). The distribution of genotypes of interleukin 1 beta-511 T/C in controls was -511 T/T(23.5%), -511 T/C(45%),-511C/C(31%) and in cases genotype distribution was -511 T/T(22.63%), -511 C/T(55.26%) and -511 C/C (22.10%). We found odds ratio of 1.27(95% CI 0.77–2.10) and 0.72 (95% CI 0.41–1.28) in -511 CT and TT genotypes respectively (P > 0.5) (Table 2). Also in our study group, we could not find any significant association of -511C allele with non small cell lung cancer (P = 0.2) (Table 3). Interleukin 1 beta-31TT genotype did not show any significant association with various clinical parameters such as age and gender (P > 0.05) while smoking, histology, dwelling and stage showed significant association with -31TT genotype (P < 0.05) (Table 4). Interleukin 1 beta-511CC genotype showed significant association with gender and dwelling (P < 0.05) while other clinical parameters such as smoking, pathological stage and histology did not show any association with -511 C/C genotype (P > 0.05) (Table 4).
Table 2.
Association between Interleukin1 beta (IL-1β) genotypes and non small cell lung cancer.
| Genotype | Cases N = 190(%) |
Control N = 200(%) |
P value OR(CI 95%) |
|---|---|---|---|
| IL1β 31 C > T | |||
| CC | 34 (17.8) | 59 (29.5) | Reference |
| CT | 107 (56.3) | 111 (55.5) | 0.04, 1.6 (1.01-2.7) |
| TT | 49 (25.7) | 30 (15) | 0.001, 2.8 (1.52-5.26) |
| Allele | |||
| C | 175 | 229 | 0.002, 1.57 (1.18–2.08) |
| T | 205 | 171 | ref. |
| IL1β 511 T > C | |||
| TT | 43 (22.63) | 47 (23.5) | 0.37, 1.27 (0.77-2.10) |
| CT | 105 (55.26) | 90 (45.0) | 0.3, 0.72 (0.41-1.28) |
| CC | 42 (22.10) | 63 (31) | 0.25, 0.84 (0.64- 1.12) |
| Allele | |||
| T | 191 | 184 | |
| C | 189 | 216 |
IL1 β -31 -CC homozygous wild, CT heterozygous, TT homozygous variant.
IL1 β -511 TT homozygous wild, CT heterozygous, CC homozygous variant.
Adjusted for age, smoking, gender, and dwelling.
Table 3.
Clinicopathological relevance of IL-1β genotypes in non small cell lung cancer.
| Parameters | Genotype (IL-1β 31 C > T) N = 190 |
P value | Genotype (IL-1β 511 T > C) N = 190 |
P value | ||||
|---|---|---|---|---|---|---|---|---|
| CC | TC | TT | TT | TC | CC | |||
| Age | ||||||||
| ≤ 50 years | 09 | 30 | 20 | 0.22 | 13 | 34 | 12 | 0.8 |
| > 50 years | 25 | 77 | 29 | 30 | 71 | 30 | ||
| Gender | ||||||||
| Male | 31 | 93 | 39 | 0.30 | 43 | 83 | 37 | 0.00 |
| Female | 03 | 14 | 10 | 00 | 05 | 22 | ||
| Smoking satus | ||||||||
| Nonsmoker | 11 | 38 | 27 | 0.04 | 19 | 43 | 14 | 0.5 |
| Smoker | 23 | 69 | 22 | 23 | 61 | 28 | ||
| Histology | ||||||||
| Sq.CC | 29 | 82 | 29 | 0.01 | 33 | 75 | 32 | 0.7 |
| Othersa | 05 | 25 | 20 | 10 | 30 | 10 | ||
| Dweller | ||||||||
| Rural | 27 | 22 | 28 | 0.01 | 26 | 74 | 27 | 0.01 |
| Urban | 07 | 35 | 21 | 17 | 31 | 30 | ||
| Pathological stage | ||||||||
| I & II | 18 | 81 | 31 | 0.03 | 25 | 72 | 33 | 0.12 |
| III & IV | 16 | 26 | 18 | 18 | 33 | 09 | ||
| Pathological grade | ||||||||
| GI | 17 | 35 | 19 | 0.47 | 20 | 39 | 12 | 0.17 |
| G2 | 11 | 45 | 20 | 12 | 47 | 17 | ||
| G3 | 06 | 27 | 10 | 11 | 19 | 13 | ||
G1- well differentiated, G 2- moderately differentiated, G3- poorly differentiated
Others include adenocarcinoma, large cell carcinoma, brancogenic carcinoma.
Table 4.
IL-1β mRNA expression in normal lung tissue samples of non small cell lung cancer cases in relation to IL-1 β 31 T > C genotypes.
| Genotypes |
IL1β mRNA Expression (mean ± SD) |
Number | P value |
|---|---|---|---|
| CC | 2.89 ± 2.21 | 10 | 0.006 |
| CT + TT | 6.18 ± 3.48 | 38 |
IL1 β -31 -CC homozygous wild, CT heterozygous, TT homozygous variant.
Interleukin 1 beta mRNA expression in non small cell lung cancer
We included 48 lung cancer patients for detection interleukin 1 beta mRNA expression in adjacent normal lung tissues of lung cancer patients. We analysed the effect of interleukin 1 beta-31 T SNP which is located in the core promoter of interleukin 1 beta gene with the mRNA expression. The mRNA expression was checked by ΔCT method with expression ratio of (amount of interleukin 1 beta mRNA/amount of glyceraldehyde 3-phosphate dehydrogenase mRNA) by applying the formula ΔCT = (CT GAPDH- CT IL1B mRNA). The ΔCT values of normal tissues of lung cancer patients was significantly higher in cases carrying interleukin 1 beta-31 T allele with a mean ± standard deviation (SD) of 6.16 ± 3.48 as compared to cases carrying interleukin 1 beta-31C allele with a mean ± standard deviation (SD) of 2.89 ± 2.21 (P = 0.006) (Table 5). However, we could not find any significant association between various clinicopathological characteristics and interleukin 1 beta mRNA expression in normal lung tissues of non small cell lung cancer patients.
Table 5.
The relationship between clinicopathological variables and IL-1β mRNA expression in lung cancer.
| Clinical parameters |
IL1β mRNA Expression (mean ± SD) |
Number | P value |
|---|---|---|---|
| Age | |||
| ≤ 50 years | 5.59 ± 4.32 | 16 | 0.8 |
| > 50 years | 5.42 ± 3.11 | 32 | |
| Sex | |||
| Male | 5.32 ± 3.38 | 38 | 0.5 |
| Female | 6.08 ± 4.13 | 10 | |
| Smoking status | |||
| Nonsmoker | 5.90 ± 3.81 | 15 | 0.58 |
| Smoker | 5.29 ± 3.42 | 33 | |
| Histology | |||
| SCCa | 5.41 ± 3.68 | 35 | 0.8 |
| Othersb | 5.66 ± 3.17 | 13 | |
| Dwelling | |||
| Rural | 5.76 ± 3.77 | 30 | 0.48 |
| Urban | 5.02 ± 3.09 | 18 | |
| Pathological stage | |||
| I &II | 6.16 ± 3.64 | 31 | 0.06 |
| III&IV | 4.20 ± 2.95 | 17 | |
| Pathological gradec | |||
| G1 | 5.23 ± 3.93 | 21 | 0.67 |
| G2 + G3 | 5.67 ± 3.22 | 27 | |
Squamous cell carcinoma.
Others include adenocarcinoma, large cell carcinoma, brancogenic carcinoma.
G1- well differentiated, G 2- moderately differentiated, G3- poorly differentiated.
Discussion
Epidemiological studies have supported the concept that chronic inflammation frequently precedes the development of lung cancer in adults (Tockman et al., 1987). Interleukin-1beta (IL-1β), a key proinflammatory cytokine encoded by the interleukin 1 beta gene, has been associated with chronic inflammation and plays an important role in lung inflammatory diseases, including lung cancer (Coussens and Werb, 2002, Ballaz and Mulshine, 2003, Smith et al., 2006). Together with tumor necrosis factor alpha (TNFα), IL-1α and interleukin 1 beta are defined as “alarm cytokines” that are secreted by macrophages and initiate inflammation (Dinarello, 1996, Dinarello, 2002b, Apte and Voronov, 2002). Interleukin 1 alpha and Interleukin 1beta cause inflammation but more importantly they induce the expression of pro-inflammatory genes which particularly include cyclooxygenase type 2 (COX-2), inducible nitric oxide synthase (iNOS), interleukin- 6 (IL-6) and other chemokines/cytokines (Apte et al., 2006b).
Interleukin 1 beta is expressed in 90% of invasive breast carcinomas and to a lesser extent in ductal in situ carcinomas (DISC) and benign lesions (Jin et al., 1997). In advanced breast carcinomas, high interleukin 1 beta content correlated well with other parameters of aggressive tumors (estrogen receptor negativity, high tumor grade, presence of p53, absence of bcl-2 and expression of pro-inflammatory cytokines, i.e., interleukin 1 alpha and interleukin-8) (Jin et al., 1997, Miller et al., 2000). The elevated expression of interleukin 1 beta has been reported in various cancers and patients with increased expression were related with bad prognosis (Lewis et al., 2006). Inflammation in the lung may be modulated by host genetic factors such as polymorphisms in inflammatory genes. Identification of polymorphisms in inflammatory genes may help understanding interindividual differences in susceptibility to lung cancer. Polymorphism of a specific gene can have effects on gene transcription, the stability of the mRNA, or the quantity and activity of the resulting protein. Thus, screening and risk evaluation is important for purposes of cancer prevention or detection. A specific interleukin 1 beta haplotype -3893 G, -1464 G, -511 C and -31 T in the promoter region of interleukin 1 beta was found to be associated with increased interleukin 1 beta mRNA and increased risk of risk of non small cell lung cancer (Landvik et al., 2009). The two functionally important polymorphisms in the promoter region of interleukin 1 beta-31 T/C (rs1143627) and interleukin 1 beta-511C/T (rs16944) have been observed to be associated with the risk of lung cancer (Kai-song et al., 2010).
In this study we examined the risk of non small cell lung cancer association with two polymorphisms interleukin 1 beta-31C > T (rs1143627) and interleukin 1 beta-511 T > C (rs16944) and effect of interleukin 1 beta –31TT on its mRNA expression. We found both heterozygous (-31CT) and homozygous (-31TT) carriers of the interleukin 1 beta − 31 T/C had a significantly higher risk of lung cancer (P < 0.05). Our results were in accordance with the previous studies which have shown association of interleukin -31C > T and interleukin 1 beta-31 T/T with the non small cell lung cancer (Zienolddiny et al., 2004, Haukim et al., 2002, Asada et al., 2006). We also observed that -31 T allele is significantly associated with non small cell lung cancer as compared to controls (P = 0.002). The heterozygous and homozygous subjects were particularly at higher risk of lung cancer with -31TC and -31TT genotype having 1.6 and 2.8 fold increased risk for the non small cell lung cancer respectively (Table 2). Thus our report supports the role of interleukin 1 beta-31C > T in predisposing the risk of lung cancer in our population. However we did not observe any significant role of interleukin 1 beta-511 T > C polymorphism in risk of non small cell lung cancer as has been previously reported (Bidwell et al., 1999, Kai-song et al., 2010, Asada et al., 2006). Thus we found that in this group, the risk was more closely related to polymorphisms of interleukin 1 beta-31 T rather than interleukin 1 beta-511C. The discrepancy in our report with the previous studies that have shown association of both the SNP's (IL-1β -31 C > T and IL-1β -511 T > C) could be due to the ethnic variation. When clinicopathological characteristics were correlated with interleukin 1 beta-31 C > T and interleukin 1 beta-511 T > C polymorphism, interleukin 1 beta-31TT genotype showed significant association with smoking, histology, dwelling, pathological stage while as interleukin 1 beta-511 showed significant association with gender and dwelling only (P < 0.05).
We investigated interleukin 1 beta mRNA expression in normal lung tissues of non small cell lung cancer patients using qRT PCR. We observed an increasing trend of interleukin 1 beta mRNA expression in normal tissues of non small cell lung cancer patients with genotypes interleukin 1 beta-31CT and interleukin 1 beta-31TT as compared to -31CC genotypes (P = 0.006). We did not find any clinicopathological factor affecting interleukin 1 beta mRNA expression and thus our study may help in understanding the role of -31 T allele in affecting the expression of interleukin 1 beta mRNA. Functional studies of the SNP have also shown an increasing trend of mRNA expression in normal tissues of lung cancer patients. The decreased expression of the C allele observed in our report is already confirmed by previous study (Landvik et al., 2009). Accordingly the high expression of T allele as found in our study has been reported by Helge Lind et al in A549 cells in A549 cells (Lind et al., 2007).
Similarly Kent Hart, observed a higher expression of the interleukin 1 beta mRNA in lungs of the patients carrying the SEPS1-105G/G- IL1β - 31 T/T genotype (Hart et al., 2008). This is an interesting observation since inflammatory cytokines such as interleukin 1 beta have been shown to induce DNA damage and inhibit DNA repair in vitro (Jaiswal et al., 2000). The decreased expression found in our study with interleukin 1 beta-31C allele has been already postulated due to differential binding patterns of nuclear proteins to oligonucleotide probes containing the interleukin 1 beta-31C allele compared to those with the interleukin 1 beat -31 T allele as a result of specific binding of the transcription factor Yin Yang 1 (YY1) (Landvik et al., 2012).
It has already been shown that the down-regulation of YY1 by the use of siRNA results in increased tumor necrosis factor alpha (TNF-α) induced interleukin 1 beta expression. Further YY1 has been shown to have inhibitory effects on transcription (Shi et al., 1997), thus YY1 when binds to the interleukin 1 beta core promoter (TAATA) promoter could inhibit gene expression, this may explain the previously observed lower promoter activity in reporter vectors with the interleukin 1 beta-31C promoter compared to-31 T promoter (Lind et al., 2007).
In conclusion, our work has indicated that the molecular status of interleukin 1 beta may play a significant role in the pathogenesis of non small cell lung carcinoma. Over expression of interleukin 1 beta mRNA was noted in non small cell lung cancer and more so in lung cancer with interleukin 1 beta-31 T allele. These findings imply that continued research into interleukin 1 beta mRNA expression and polymorphisms will be an important source of information in understanding the role of inflammation in the pathogenesis of lung cancer.
Acknowledgement
The authors gratefully acknowledge the financial support provided by the Sher-i-Kashmir Institute of Medical Sciences, Kashmir for this work. Our thanks are also due to the Technical Staff, particularly Mr Fayaz Ahmad and Mr Rafiq Ahmad of the operation theatre of the Department of Cardiovascular Thoracic Surgery, SKIMS Srinagar who helped us in procuring the tissue samples.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- Apte R.N., Voronov E. Interleukin-1-a major pleiotropic cytokine in tumor–host interactions. Semin. Cancer Biol. 2002;12:277–290. doi: 10.1016/s1044-579x(02)00014-7. [DOI] [PubMed] [Google Scholar]
- Apte R.N., Krelin Y., Song X., Dotan S., Recih E., Elkabets M., Carmi Y., Dvorkin T., White R.M., Gayvoronsky L., Segal S., Voronov E. Effects of micro-environmentand malignant cell-derived interleukin-1 in carcinogenesis, tumour invasiveness and tumour–host interactions. Eur. J. Cancer. 2006;42:751–759. doi: 10.1016/j.ejca.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Apte R.N., Dotan S., Elkabets M., White M.R., Reich E., Carmi Y., Song X., Dvozkin T., Krelin Y., Voronov E. The involvement of IL-1 in tumorigenesis, tumor invasiveness, metastasis and tumor-host interactions. Cancer Metastasis Rev. 2006 Sep;25(3):387–408. doi: 10.1007/s10555-006-9004-4. [DOI] [PubMed] [Google Scholar]
- Asada M., Yasuda H., Ebihara S., Tomita N., Suzuki S., Sato M., Kubo H., Yamaya M. Interleukin-1beta gene polymorphisms associated with risk of lung cancer in Japanese. Lung Cancer. 2006;54:261–263. doi: 10.1016/j.lungcan.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Azad N., Rojanasakul Y., Vallyathan V. Inflammation and lung cancer: Roles of reactive oxygen/nitrogen species. J. Toxicol. Environ. Health B Crit. Rev. 2008;11:1–15. doi: 10.1080/10937400701436460. [DOI] [PubMed] [Google Scholar]
- Balkwill F., Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- Ballaz S., Mulshine J.L. The potential contributions of chronic inflammation to lung carcinogenesis. Clin. Lung Cancer. 2003;5:46–62. doi: 10.3816/CLC.2003.n.021. [DOI] [PubMed] [Google Scholar]
- Bidwell J., Keen L., Gallagher G., Kimberly R., Huizinga T., McDermott M.F., Oksenberg J., McNicholl J., Pociot F., Hardt C., D'Alfonso S. Cytokine gene polymorphism in human disease: on-line databases. Genes Immun. 1999;1:3–19. doi: 10.1038/sj.gene.6363645. [DOI] [PubMed] [Google Scholar]
- Chen H., Wilkins L.M., Aziz N., Cannings C., Wyllie D.H., Bingle C., Rogus J., Beck J.D., Offenbacher S., Cork M.J., Rafie-Kolpin M., Hsieh C.M., Kornman K.S. Duff GW Single nucleotide polymorphisms in the human interleukin-1B gene affect transcription according to haplotype context. Hum. Mol. Genet. 2006;15:519–529. doi: 10.1093/hmg/ddi469. [DOI] [PubMed] [Google Scholar]
- Chen H., Wilkins L.M., Aziz N., Cannings C., Wyllie D.H., Bingle C., Rogus J., Beck J.D., Offenbacher S., Cork M.J., Rafie-Kolpin M., Hsieh C.M., Kornman K.S., Duff G.W. Single nucleotide polymorphisms in the human interleukin-1B gene affect transcription according to haplotype context. Hum. Mol. Genet. 2006;15:519–529. doi: 10.1093/hmg/ddi469. [DOI] [PubMed] [Google Scholar]
- Coussens L.M., Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello C.A. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–2147. [PubMed] [Google Scholar]
- Dinarello C.A. The IL-1 family and inflammatory diseases. Clin. Exp. Rheumatol. 2002;20(5 Suppl. 27):S1–S13. (Sep-Oct) [PubMed] [Google Scholar]
- Dinarello Charles. A primer for clinicians. Third edition. Amgen; Thousand Oaks, CA: 2002. Proinflammatory and anti-inflammatory cytokines in rheumatoid arthritis; pp. 21–51. [Google Scholar]
- Dinarello C.A. The paradox of pro-inflammatory cytokines in cancer. Cancer Metastasis Rev. 2006;25:307–313. doi: 10.1007/s10555-006-9000-8. [DOI] [PubMed] [Google Scholar]
- El Omar E.M., Carrington M., Chow W.H., McColl K.E., Bream J.H., Young H.A., Herrera J., Lissowska J., Yuan C.C., Rothman N., Lanyon G., Martin M., Fraumeni J.F., Jr., Rabkin C.S. Interleukin 1polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- Hart Kent, Haugen Aage, Zienolddiny Shanbeh. Allele-specific induction of IL1B–31 T/C promoter polymorphism by lung carcinogens. Mutat. Res. 2008;656:14–18. doi: 10.1016/j.mrgentox.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Haukim N., Bidwell J.L., Smith A.J., Keen L.J., Gallagher G., Kimberly R., Huizinga T., McDermott M.F., Oksenberg J., McNicholl J., Pociot F., Hardt C., D'Alfonso S. Cytokine gene polymorphism in human disease: on-line databases, supplement 2. Genes Immunol. 2002;3:313–330. doi: 10.1038/sj.gene.6363881. [DOI] [PubMed] [Google Scholar]
- Hellermann G.R., Nagy S.B., Kong X., Lockey R.F., Mohapatra S.S. Mechanism of cigarette smoke condensate-induced acute inflammatory response in human bronchial epithelial cells. Respir. Res. 2002;3(1):2. doi: 10.1186/rr172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal M., LaRusso N.F., Burgart L.J., Gores G.J. Inflammatory cytokines induce DNA damage and inhibit DNA repair in cholangiocarcinoma cells by a nitric oxide-dependent mechanism. Cancer Res. 2000;60:184–190. [PubMed] [Google Scholar]
- Jha P. Avoidable global cancer deaths and total deaths from smoking. Nat. Rev. Cancer. 2009;9:655–664. doi: 10.1038/nrc2703. [DOI] [PubMed] [Google Scholar]
- Jin L., Yuan R.Q., Fuchs A., Yao Y., Joseph A., Schwall R., Schnitt S.J., Guida A., Hastings H.M., Andres J., Turkel G., Polverini P.J., Goldberg I.D., Rosen E.M. Expression of interleukin-1beta in human breast carcinoma. Cancer. 1997;80:421–434. doi: 10.1002/(sici)1097-0142(19970801)80:3<421::aid-cncr10>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Kai-song Wu., Zhou Xin, Zheng Fang, Xian-qun Xu., Lin Yu-hui, Yang Jiong. Influence of interleukin-1 beta genetic polymorphism, smoking and alcohol drinking on the risk of non-small cell lung cancer. Clin. Chim. Acta. 2010;411:1441–1446. doi: 10.1016/j.cca.2010.05.035. [DOI] [PubMed] [Google Scholar]
- Landvik Nina E., Hart Kent, Skaug Vidar, Stangeland Lodve B., Haugen Aage, Zienolddiny Shanbeh. A specific interleukin-1beta haplotype correlates with high levels of IL1B mRNA in the lung and increased risk of non-small cell lung cancer. Carcinogenesis. 2009;30:1186–1192. doi: 10.1093/carcin/bgp122. [DOI] [PubMed] [Google Scholar]
- Landvik X. Tekpli, Anmarkrud K.H., Haugen A., Zienolddiny S. Molecular Characterization of a Cancer-Related Single Nucleotide Polymorphism in the Pro-Inflammatory Interleukin-1B. Gene. 2012;51:168–175. doi: 10.1002/mc.21910. [DOI] [PubMed] [Google Scholar]
- Lewis A.M., Varghese S., Xu H., Alexander H.R. Interleukin 1and cancer progression: the emerging role of interleukin-1 receptor antagonist as a novel therapeutic agent in cancer treatment. J. Transl. Med. 2006;4:48. doi: 10.1186/1479-5876-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind Helge, Zienolddiny Shanbeh, Ryberg David, Skaug Vidar, Phillips David H., Haugen Aage. Interleukin 1receptor antagonist gene polymorphism and risk of lung cancer: a possible interaction with polymorphisms in the interleukin 1 beta gene. Lung Cancer. 2005;50:285–290. doi: 10.1016/j.lungcan.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Lind Helge, Haugen Aage, Zienolddiny Shanbeh. Differential binding of proteins to the IL1B–31 T/C polymorphism in lung epithelial cells. Cytokine. 2007;38:43–48. doi: 10.1016/j.cyto.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Mayne S.T., Buenconsejo J., Janerich D.T. Previous lung disease and risk of lung cancer among men and women nonsmokers. Am. J. Epidemiol. 1999;149:13–20. doi: 10.1093/oxfordjournals.aje.a009722. [DOI] [PubMed] [Google Scholar]
- Miller Lauri J., Kurtzman Scott H., Anderson Kathleen, Wang Yanping, Stankus Marra, Renna Maria, Lindquist Richard, Barrows George, Kreutzer Donald L. Interleukin 1family expression in human breast cancer: interleukin-1 receptor antagonist. Cancer Investig. 2000;18:293–302. doi: 10.3109/07357900009012171. [DOI] [PubMed] [Google Scholar]
- Shi Y., Lee J.S., Galvin K.M. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta. 1997;1332:49–66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- Shirakawa F., Saito K., Bonagura C.A., Galson D.L., Fenton M.J., Webb A.C. Auron PE The human prointerleukin 1 beta gene requires DNA sequences both proximal and distal to the transcription start site for tissue-specific induction. Mol. Cell. Biol. 1993;13:1332–1344. doi: 10.1128/mcb.13.3.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.J., Perfetti T.A., King J.A. Perspectives on pulmonary inflammation and lung cancer risk in cigarette smokers. Inhal. Toxicol. 2006;18:667–677. doi: 10.1080/08958370600742821. [DOI] [PubMed] [Google Scholar]
- Takizawa H., Tanaka M., Takami K., Ohtoshi T., Ito K., Satoh M., Okada Y., Yamasawa F., Umeda A. Increased expression of inflammatory mediators in small-airway epithelium from tobacco smokers. Am. J. Physiol. Lung Cell. Mol. Physiol. 2000;278:906–913. doi: 10.1152/ajplung.2000.278.5.L906. [DOI] [PubMed] [Google Scholar]
- Tockman M.S., Anthonisen N.R., Wright E.C., Donithan M.G. Airways obstruction and the risk for lung cancer. Ann. Intern. Med. 1987;106:512–518. doi: 10.7326/0003-4819-106-4-512. [DOI] [PubMed] [Google Scholar]
- Wobbe C.R., Struhl K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol. Cell. Biol. 1990;10:3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zienolddiny S., Ryberg D., Maggini V., Skaug V., Canzian F. Haugen A Polymorphisms of the interleukin-1 beta gene are associated with increased risk of non-small cell lung cancer. Int. J. Cancer. 2004;109:353–356. doi: 10.1002/ijc.11695. [DOI] [PubMed] [Google Scholar]