Abstract
Metabolic disorders including type 2 diabetes, obesity and hypertension have growing prevalence globally every year. Genome-wide association studies have successfully identified many genetic markers associated to these diseases, but few studied their interaction effects. In this study, twenty candidate SNPs from sixteen genes are selected, and a lasso-multiple regression approach is implemented to consider the SNP–SNP interactions among them in an Asian population. It is found out that the main effects of the markers are weak but the interactions among the candidates showed a significant association to diseases. SNPs from genes CDKN2BAS and KCNJ11 are significantly associated to risk for developing diabetes, and SNPs from FTO and APOA5 might interact to play an important role for the onset of hypertension.
Keywords: Metabolic disorder, Candidate genes, Interaction effects
Introduction
Diabetes mellitus has reached the scale of a global epidemic with 347 million affected people in 2012. During the past year, its prevalence has been over 8.5% worldwide and 9.4% in Hong Kong (Li et al., 2012). It produces various complications that may cause damage to the eyes, nerves, kidneys and heart, causing a significant public health care burden. Diabetes is a metabolic disorder that is due to defects in insulin secretion and action. Its etiology has a complex nature, and involves multiple genetic markers and environmental factors. Previous genome-wide association studies on diabetes, obesity and hypertension have identified many genetic markers that have significant associations (Ng et al., 2008, Scheffold et al., 2011, Vaxillaire et al., 2008). However, most studies reported only a marginal effect of the SNPs, and very few reported interaction effects (Neuman et al., 2010), either among genes or between genes and environmental factors. Some studies concerned interactions, but nothing significant was discovered (Wu et al., 2008). It is likely that the lack of reports about the interaction effects is due to methodology rather than dataset. Furthermore, using traditional statistical methods alone such as linear regression may not identify the important markers even if the sample size is very large. Noise variable correlates with the true signal may reduce the power of detecting real features. In this study, twenty candidate markers that were previously reported to be marginally associated to metabolic diseases were selected, and a lasso-multiple regression (LMR) method was used to evaluate interactions among them in Chinese subjects. It was found out that although some markers have only a moderate marginal effect, the SNP–SNP interactions showed a significant influence.
Material and method
Sample subjects
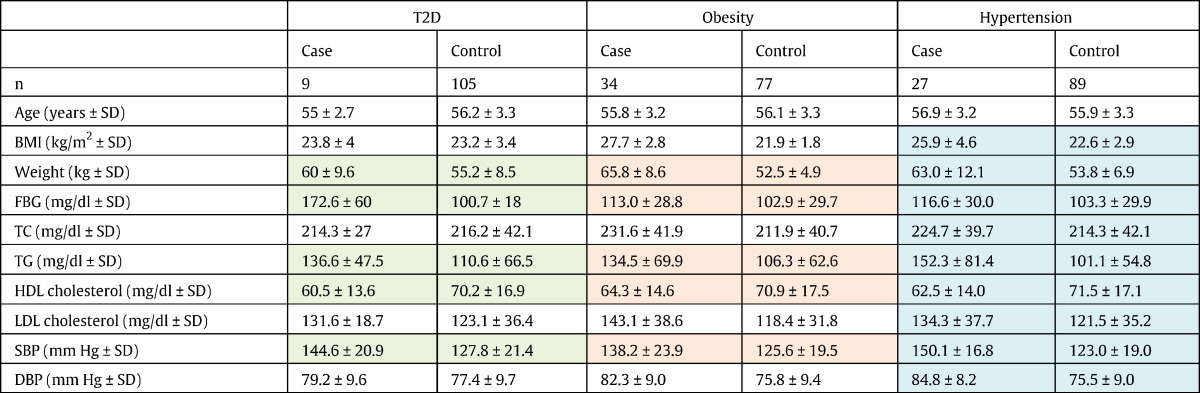
The subjects were recruited through random telephone dialing based on the residential telephone directory. Written informed consents for DNA testing were obtained, and the study was approved by the Clinical Research Ethics Committee of the Chinese University of Hong Kong. In total 115 subjects were recruited. All of them were female, of Chinese Han ethnicity and aged between 50 and 64 years. Body mass index, fasting blood glucose, systolic blood pressure, diastolic blood pressure, hypertension history, total cholesterol, triglycerides, high and low density lipoprotein, and type 2 diabetes history were collected. Genomic DNA was extracted by ROCHE Kit, and amplification was done by polymerase chain reaction. The DNA templates with high quality were sent to the McGill University & Genome Quebec Innovation Centre for sequencing on Sequenom Platform. Obesity was defined as having a body-mass-index (BMI) of > 25 kg/m2, adjusted for Asians (Barba et al., 2004, Chiu et al., 2011, Force, 2000). Hypertension was defined as having a systolic blood pressure of > 140 mm Hg and/or a diastolic blood pressure of > 90 mm Hg. Diabetes was defined as having fasting blood glucose of at least 7.0 mg/dl and/or pharmacologic treatment. The sample characteristics for cases and controls of the three phenotypes were summarized in Table 1.
Table 1.
Clinical characteristics of cases and controls for T2DM, obesity and hypertension.
The clinical characteristics (rows) are tabled for the three metabolic disorders by case and control groups. The features that have significant differences by two sample t-test in case and control groups are highlighted by colored cells.
Abbreviations: T2DM: type 2 diabetes mellitus, BMI: body mass index, FBG: fast blood glucose, TC: total cholesterol, TG: total triglyceride, HDL: high density lipoprotein, LDL: low density lipoprotein, SBP: systolic blood pressure, DBP: diastolic blood pressure.
Candidate gene selection
Twenty SNPs from 16 genes were selected as the candidate markers. These include eight genes predisposing type 2 diabetes (CDKN2A/B, CDKAL1, HHEX, IGF2BP2, KCNJ11, PPARG, TCF7L2, and WFS1), four genes predisposing obesity (FTO, APOA5, INSIG2, and MC4R), ATGR1 for non-alcoholic fatty liver disease (NAFLD), two genes predisposing hypertension (SLC2A9, STK39), and CDKN2BAS for heart attack. The complete references were in Table 2.
Table 2.
The list of candidate SNPs/genes.
This table lists the 20 candidate SNPs used in search for pair-wise interactions. The Gene name column gives the gene in which the SNP is located; the Disease column gives which disorder the SNP is previously identified to have associations, and the literature source is given in the Reference column. The MAF column contains the minor allele frequency of the SNP calculated by the samples of this study; the OR (T2DM, obesity, hypertension) columns give the odds ratio calculated using current samples by a two-way contingency table, which provides a direct evaluation on the SNP relevant disease risk.
| No | SNP name | Gene name | Disease | MAF | OR |
OR |
OR |
Reference |
|---|---|---|---|---|---|---|---|---|
| T2DM | Obesity | Hypertension | ||||||
| 1 | rs2383208 | CDKN2A/B | T2D | 0.40 | 4.1 | 4.0 | 2.6 | Saxena, Voight et al. (2007) |
| 2 | rs13266634 | CDKAL1 | T2D | 0.45 | 1.2 | 2.1 | 1.8 | Chimienti, Devergnas et al. (2004) |
| 3 | rs4712523 | CDKAL1 | T2D | 0.38 | 2.1 | 0.8 | 2.0 | Steinthorsdottir, Thorleifsson et al. (2007) |
| 4 | rs1111875 | HHEX | T2D | 0.35 | 2.9 | 1.1 | 1.2 | van Vliet-Ostaptchouk, Onland-Moret et al. (2008) |
| 5 | rs4402960 | IGF2BP2 | T2D | 0.21 | 1.0 | 1.4 | 1.9 | Nielson, Czech et al. (1999) |
| 6 | rs5219 | KCNJ11 | T2D | 0.38 | 1.9 | 0.8 | 0.8 | Vaxillaire et al. (2008) |
| 7 | rs1801282 | PPARG | T2D | 0.07 | 1.0 | 5.1 | 0.5 | Altshuler, Hirschhorn et al. (2000) |
| 8 | rs7903146 | TCF7L2 | T2D | 0.04 | 2.7 | 2.4 | 3.1 | Grant, Thorleifsson et al. (2006) |
| 9 | rs10012946 | WFS1 | T2D | 0.08 | 3.0 | 2.8 | 3.6 | Sandhu, Weedon et al. (2007) |
| 10 | rs3751812 | FTO | Obesity | 0.15 | 1.1 | 1.9 | 4.4 | Frayling, Timpson et al. (2007); Hennig, Fulford et al. (2009) |
| 11 | rs662799 | APOA5 | Obesity | 0.32 | 2.0 | 1.9 | 2.6 | Corella, Lai et al. (2007) |
| 12 | rs766605 | INSIG2 | Obesity | 0.03 | 1.8 | 1.7 | 2.4 | Dina, Meyre et al. (2007) |
| 13 | rs10871777 | MC4R | Obesity | 0.12 | 0.6 | 2.4 | 1.1 | Loos, Lindgren et al. (2008) |
| 14 | rs12970134 | MC4R | Obesity | 0.14 | 0.5 | 6.1 | 1.1 | Chambers, Elliott et al. (2008) |
| 15 | rs3772622 | ATGR1 | NAFLD | 0.46 | 0.3 | 1.4 | 0.8 | Yoneda, Hotta et al. (2009) |
| 16 | rs1014290 | SLC2A9 | Hypertension | 0.37 | 3.1 | 3.1 | 3.3 | Vitart, Rudan et al. (2008) |
| 17 | rs6449213 | SLC2A9 | Hypertension | 0.02 | 1.4 | 5.2 | 7.3 | Vitart, Rudan et al. (2008) |
| 18 | rs737267 | SLC2A9 | Hypertension | 0.01 | 0.0 | 5.1 | 7.0 | Vitart, Rudan et al. (2008) |
| 19 | rs3754777 | STK39 | Hypertension | 0.24 | 1.7 | 1.7 | 0.7 | Wang, O'Connell et al. (2009) |
| 20 | rs2383207 | CDKN2BAS | Heart attack | 0.40 | 1.0 | 0.9 | 1.7 | Scheffold et al. (2011) |
MAF is minor allele frequency.
OR is odds ratio.
NAFLD is non-alcoholic fatty liver disease.
Lasso-multiple regression (LMR) method for selecting variables
Lasso-multiple regression method is composed of two steps: Step 1: The lasso (Tibshirani, 1996) is used to select a subset of variables including all pair-wise interaction terms. Lasso is a linear regression model with one additional penalization term on the coefficients. The result of the penalization is that some coefficients can be shrunk to absolute zero, achieving a feature selection effect. It puts a coefficient to zero if the corresponding variable does not affect the dependent variable, and vice versa, thus produces a small subset of markers in which numbers can be handled by traditional methods (Fan and Li, 2001, Zou, 2006). The lasso is implemented by R package glmnet (Friedman et al., 2010). Step 2: Multiple linear regression is then performed on the lasso-selected variables to obtain variable significance. Stepwise model selection by AIC is applied to achieve an optimal combination of variables using R package stepAIC.
Sample size calculation and adjusting for multiple testing issue
The sample size is estimated by the standard formula for case and control study:
where r is the ratio of number of controls to cases, which is 2.7 in this study; σ2 is the variance of the response, which is 0.20. Assume that power is 80%, and type I error is 5%; the difference in the denominator is taken to be one standard error. The number of cases needed is 24, and the number of controls needed is 64. Thus the sample size of this study satisfies the specified power and type I error. Multiple testing is adjusted by Bonferroni correction, and an adjusted p-value less than 0.05 is considered to be statistically significant.
Results
Linkage disequilibrium
The pairwise linkage disequilibrium was calculated. Among the 190 combinations of 20 SNPs, only two pairs had an LD coefficient of correlation (r) greater than 0.3, and six pairs with 0.2 < |r| < 0.3. Most of the SNP pairs were in linkage equilibrium and this ensured the randomness of the data. The high LD SNP pairs are not among the significant markers we identified. Thus the result is not confounded by LD.
Hardy–Weinberg equilibrium (HWE) test
HWE tests on 20 SNPs were performed and permutation was done 1000 times to obtain p-values. All SNPs were in Hardy–Weinberg equilibrium except rs6449213 and rs737267. However, they were not in the final markers selected, and did not affect the conclusion.
Interactions for type 2 diabetes mellitus
Lasso selected six variables including one main effect and five interactions, which were passed to multiple linear regressions (Supplemental Table S1). After adjusting for multiple comparisons, the pair (rs2383207, rs5219) remained to be significant with p-value 0.029 (Table 3). The power of this significant variable is 69.5% and the type I error is 20% based on the current sample size, using the method suggested by Hsieh et al. (1998). The SNP rs2383207 was from gene CDKN2BAS on chromosome 9p21.3, which was reported to be associated with myocardial infarction in American Caucasians (Center for Health Protection, Hong Kong, 2012), Europeans (Scheffold et al., 2011), and Koreans (Shen et al., 2008). rs5219 representing KCNJ11 was previously reported to be associated with type 2 diabetes (Vaxillaire et al., 2008). Their marginal p-values were 0.027 and 0.11, respectively, before adjusting for multiple comparisons. Both SNPs had only a moderate main effect but their interaction was significantly associated with T2DM. The lasso selected markers explained 33.47% (R-square) of the variation in the T2D phenotype. This finding reflected the complex nature of diabetes and showed the importance of including interaction effects in genetic analysis.
Table 3.
Significant SNPs found for metabolic disorders.
This table summarizes the SNP sets that are identified in this study to have a significant interaction effect on metabolic disorders. The first column is the associated disorder. The second column gives the SNP sets. The “Genes” column shows in which genes the SNPs are located. The fourth column gives the disorder the genes are previously reported to have associations. The fifth column gives the SNP set's odds ratio. The last column shows the p-values of the SNPs after adjusting by multiple testing.
| Metabolic disorder | SNP sets | Genes | Previous identified associated disorders | Odds ratio of identified sets | Adjusted p-value |
|---|---|---|---|---|---|
| T2D | (rs2383207, rs5219) | (CDKN2BAS, KCNJ11) | (Heart attack, T2D) | 8.5 | 0.029 |
| Obesity | rs4402960 | IGF2BP2 | T2D | 2.4 | 0.045 |
| (rs1014290, rs4402960) | (SLC2A9, IGF2BP2) | (Hypertension, T2D) | 6.4 | 0.003 | |
| (rs3751812, rs662799) | (FTO, APOA5) | (Obesity, obesity) | 11.7 | 0.007 | |
| Hypertension | (rs12970134, rs4402960) | (MC4R, IGF2BP2) | (Obesity, T2D) | 3.0 | 0.002 |
SNP: Single nucleotide polymorphism; T2D: type 2 diabetes.
Interactions for obesity
For this phenotype, LMR selected 16 markers including main effects, SNP–SNP interactions and SNP–age interactions (Supplemental Table S2). rs4402960 was found to be marginally significant with an adjusted p-value of 0.045 (Table 3). rs1014290 and rs4402960 from genes SLC2A9 and IGF2BP2 were found to be significantly associated to obesity with adjusted p-value 0.003, rs3751812 and rs662799 from FTO and APOA5 were significant with an adjusted p-value of 0.007 (Table 3). For the given sample size and type I error 30%, the power of rs1014290 and rs4402960 is 64.5% for rs3751812 and rs662799 is 70%, respectively. SLC2A9 and IGF2BP2 were previously reported to be related to hypertension and type 2 diabetes (Haupt et al., 2009, Vasan et al., 2001); here they were found to be also associated with the classification of obesity.
Interactions for hypertension
LMR selected 21 markers, which could explain 36% (adjusted R-squared statistic) of the variation in hypertension. Logistic regression gave a prediction error rate of 13.9% on hypertension, with 93.26% specificity (true negative/negative) and 61.5% sensitivity (true positive/positive). Thirteen markers had a p-value of < 0.05 before adjusting for multiple testing, in which four pairs were SNP–age interactions. This indicated that age played an important role in hypertension onset risk (Vasan et al., 2001). After adjusting for multiple comparisons, one SNP pair rs12970134 and rs4402960 from genes MC4R and IGF2BP2 remained to be significant with an adjusted p-value of 0.002 (Table 3). With the existing sample size, this interaction set has type I error 20% and power 71%. rs3751812, a SNP from FTO, paired with age was associated to hypertension with an adjusted p-value of 0.083 (Supplemental Table S3).
Discussion
The candidate genes had a weak main effect but strong interaction effects
Different from the reports concerning Western populations, the main effects of the candidate genes in this Asian cohort were much weaker; however, strong interaction effects were found among the SNPs. In French Caucasians, rs5219 (KCNJ11) was reported to have a strong association with T2DM (p-value = 0.009) (Vaxillaire et al., 2008). Here its main effect was not significant, but combined with rs2383207 (CDKN2BAS) it showed strong interaction effects (p-value = 0.029, odds ratio = 8.5). rs3751812 (FTO) and rs662799 (APOA5) both had strong main effects in Caucasian population (Schmitz et al., 2010, Shen et al., 2008), but in this study they exhibited negligible main effect but strong interaction effect on obesity (p-value = 0.007, odds ratio = 11.7). For hypertension, none of the candidate hypertension SNPs was found to be significant marginally, but the joint effect of rs12970134 (MC4R) and rs4402960 (IGF2BP2) showed strong risk for hypertension with a p-value of 0.002 and an odds ratio of 3.0.
Validation of identified significant pairs by generalized multifactor dimension reduction (GMDR)
We use the popular interaction method GMDR to cross-check the identified pairs (Lou et al., 2007). For the T2D phenotype, the final reported pair (rs2383207, rs5219) is ranked the 3rd best model by GMDR. For the obesity phenotype, the top reported marker by LMR is also identified by GMDR as the top one marker with the highest accuracy. For the hypertension phenotype, the final reported two pairs (rs12970134, rs4402960 and rs3751812, rs13266634) are also the top first and second pairs by GMDR, respectively. Thus the validation shows that the identified interaction sets have high consistency, while the proposed LMR eliminates most markers in the first step and may have smaller noise in the second step of significance evaluation.
Biological implications of the interactive pairs
Glucose-stimulated insulin secretion from the pancreatic β cell is dependent upon the ability of the KATP channel to alter the voltage polarization of the cell membrane (Ashcroft and Rorsman, 1990), and mutations in KCNJ11 have been reported to cause pediatric diabetes (Ioannou et al., 2011). CDKN2B–CDKN2A is the strongest genetic susceptibility locus for cardiovascular diseases, and it is also linked to several cancers, intracranial aneurysm, type 2 diabetes, periodontitis, Alzheimer's disease, endometriosis, frailty in the elderly, and glaucoma. In this study, two SNP pairs were found to be associated with metabolic syndrome. One such SNP pair (CDKN2BAS, KCNJ11) interaction was found to be significant in type 2 diabetes. CDKN2BAS (previously known as CDKN2B antisense RNA) is an RNA gene (a non-protein coding gene). Multiple alternatively spliced transcript variants have been generated from this gene, and all of them are long non-coding RNAs. It has been proposed that the recruitment of DNA methyltransferase DNMT3b by DNA:RNA triplexes may be a common and generally used pathway in epigenetic regulation, as indicated by research data from this gene (Schmitz et al., 2010). Thus the CDKN2BAS and KCNJ11 interaction may be explained by the recruitment of DNA methyltransferase DNMT3b by the non-coding RNA encoded by CDKN2BAS acting on KCNJ11. Although the target gene of the non-coding RNA encoded by CDKN2BAS is not known at present, and whether it might act in concert with KCNJ11 is not entirely clear, it has been reported that the non-coding RNA coded by ANRIL (an alias of CDKN2BAS) is associated with type 2 diabetes. ANRIL is also involved in the recruitment of both polycomb complexes, PRC1 (Yap et al., 2010) and PRC2 (Schmitz et al., 2010), which are histone methyltransferase complexes. Deregulation of ANRIL by risk-associated single-nucleotide polymorphisms is thought to change the expression level of a closely linked gene p15INK4B and/or other target genes (which might be on a different chromosomal location such as KCNJ11), thereby leading to the development of type II diabetes or coronary heart disease in humans (Schmitz et al., 2010). Although no direct action of ANRIL non-coding RNA for the regulation of KCNJ11 has been reported, transcriptome profiling and select chromatin modifications have revealed that the removal of polycomb group (PcG)-mediated repression on stage-specific genes is a key mechanism for the induction of developmental regulators during pancreatic endocrine differentiation (Xie et al., 2013). The same study also revealed that KCNJ11 is one of the in-vivo-differentiated functional endocrine (FE) signature genes that are activated and associated with distinct patterns of chromatin modifications. Alternatively, the putative explanation underlying the association between ANRIL and miscellaneous unrelated diseases has been hypothesized due to the role of ANRIL in the replicative function of the stem cell with advancing age (Sharpless and Depinho, 2007); hence diabetes could be due to the interactive and the additive effect of the slower replicative function of the pancreatic cells and the lower activity of the KCNJ11.
The finding of two SNPs' (FTO, APOA5) interaction was observed in the obese subjects in our study. Fat mass and obesity-associated protein is a homolog in the AlkB family proteins, and it is the first mRNA demethylase that has been identified, and, also, it has been hypothesized that a nucleic acid methylation status is linked to increased fat mass (Gerken et al., 2007). The finding of a risk allele of FTO having an effect on food intake or satiety has been independently replicated in five subsequent studies (Cecil et al., 2008, Haupt et al., 2009, Timpson et al., 2008, Wardle et al., 2008, Wardle et al., 2009). FTO has efficient oxidative demethylation activity targeting the abundant N6-methyladenosine (m6A) residues in RNA in vitro, and N6-methyladenosine (m6A) is the most prevalent internal modification in messenger RNA (Jia et al., 2012). Over 12,000 m6A sites have been characterized in the transcripts of more than 7000 human genes (Dominissini et al., 2012). From a search in identified m6A peaks in the HepG2 cell line and the normal human brain, no significant peakscore for m6A peaks in the APOA5 gene could be found; however, there is a high peakscore (5.08) for the m6A peak in the USF1 gene near the 3′UTR. The USF1 gene has been found to have a possible interaction with the APOA5 gene on the risk for the metabolic syndrome (Singmann et al., 2009). USF1 is a ubiquitous transcription factor governing the expression of numerous genes of lipid and glucose metabolism and insulin stimuli have been shown to reduce the association of the USF1 transcription factor to APOA5 E-box (Singmann et al., 2009). Thus the gene–gene interaction we observe between FTO and APOA5 might be mediated through USF1 as a transcription factor of APOA5.
In summary, this study examined the risk of 20 candidate genes in Chinese subjects using a Lasso multiple regression method. We found out that the candidate SNPs had a weak main effect but strong interaction effects on type 2 diabetes mellitus, obesity and hypertension. Furthermore, the two interaction pairs we observed as being associated with metabolic syndrome could be explained by known gene regulators. For CDKN2BAS and KCNJ11 interaction in diabetes subjects, long non-coding RNA regulation of KCNJ11 via polycomb group (PcG) regulation might be involved; whereas for the FTO and APOA5 interaction, the demethylation activity of FTO affecting APOA5 via its transcription factor USF1 could be involved. Metabolic disorders have important etiology in environmental factors, and this study is limited by that only age–gene interactions are considered; more interplay of environment and genetic variables would be included in future study.
Acknowledgments
This research study was partly supported by donations from the Croucher Foundation, CUHK Direct Grant 4054077, internal funding from the Chinese University of Hong Kong and RGC-GRF grant project number 476013.
Footnotes
The supplemental data contain the multiple regression output using lasso-selected variables for T2DM, obesity and hypertension, respectively. Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.mgene.2014.04.010.
Appendix A. Supplementary data
Supplementary tables.
References
- Ashcroft F.M., Rorsman P. Atp-sensitive K + channels—a link between B-cell metabolism and insulin-secretion. Biochem. Soc. Trans. 1990;18(1):109–111. doi: 10.1042/bst0180109. (Feb) [DOI] [PubMed] [Google Scholar]
- Barba C., Cavalli-Sforza T., Cutter J., Darnton-Hill I., Deurenberg P., Deurenberg-Yap M. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet. 2004;363(9403):157–163. doi: 10.1016/S0140-6736(03)15268-3. (Jan 10) [DOI] [PubMed] [Google Scholar]
- Cecil J.E., Tavendale R., Watt P., Hetherington M.M., Palmer C.N. An obesity-associated FTO gene variant and increased energy intake in children. N. Engl. J. Med. 2008;359(24):2558–2566. doi: 10.1056/NEJMoa0803839. (Research Support, Non-U.S. Gov't, Dec 11) [DOI] [PubMed] [Google Scholar]
- Center for Health Protection, Hong Kong 2012. http://www.chp.gov.hk/files/pdf/ncd_watch_oct2012_chin.pdf Available from.
- Chiu M., Austin P.C., Manuel D.G., Shah B.R., Tu J.V. Deriving ethnic-specific BMI cutoff points for assessing diabetes risk. Diabetes Care. 2011;34(8):1741–1748. doi: 10.2337/dc10-2300. (Aug) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S. Topology of the human and mouse m(6)A RNA methylomes revealed by m(6)A-seq. Nature. 2012;485(7397):201-U84. doi: 10.1038/nature11112. (May 10) [DOI] [PubMed] [Google Scholar]
- Fan J.Q., Li R.Z. Variable selection via nonconcave penalized likelihood and its oracle properties. J. Am. Stat. Assoc. 2001;96(456):1348–1360. (Dec) [Google Scholar]
- Force I.O.T. World Health Organization; Sydney, Australia: 2000. The Asia-Pacific Perspective: Redefining Obesity and Its Treatment. [Google Scholar]
- Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010;33(1):1–22. (Feb) [PMC free article] [PubMed] [Google Scholar]
- Gerken T., Girard C.A., Tung Y.C.L., Webby C.J., Saudek V., Hewitson K.S. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–1472. doi: 10.1126/science.1151710. (Nov 30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt A., Thamer C., Staiger H., Tschritter O., Kirchhoff K., Machicao F. Variation in the FTO gene influences food intake but not energy expenditure. Exp. Clin. Endocrinol. Diabetes. 2009;117(4):194–197. doi: 10.1055/s-0028-1087176. (Research Support, Non-U.S. Gov't, Apr) [DOI] [PubMed] [Google Scholar]
- Hsieh F.Y., Bloch D.A., Larsen M.D. A simple method of sample size calculation for linear and logistic regression. Stat. Med. 1998;17(14):1623–1634. doi: 10.1002/(sici)1097-0258(19980730)17:14<1623::aid-sim871>3.0.co;2-s. (Jul 30) [DOI] [PubMed] [Google Scholar]
- Ioannou Y.S., Ellard S., Hattersley A., Skordis N. KCNJ11 activating mutations cause both transient and permanent neonatal diabetes mellitus in Cypriot patients. Pediatr. Diabetes. 2011;12(2):133–137. doi: 10.1111/j.1399-5448.2010.00743.x. (Mar) [DOI] [PubMed] [Google Scholar]
- Jia G.F., Fu Y., Zhao X., Dai Q., Zheng G.Q., Yang Y. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2012;8(12):1008-. doi: 10.1038/nchembio.687. (vol 7, pg 885, 2011) (Dec) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Oldenburg B., Chamberlain C., O'Neil A., Xue B., Jolley D. Diabetes prevalence and determinants in adults in China mainland from 2000 to 2010: a systematic review. Diabetes Res. Clin. Pract. 2012;98(2):226–235. doi: 10.1016/j.diabres.2012.05.010. (Nov) [DOI] [PubMed] [Google Scholar]
- Lou X.Y., Chen G.B., Yan L., Ma J.Z., Zhu J., Elston R.C. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am. J. Hum. Genet. 2007;80(6):1125–1137. doi: 10.1086/518312. (Jun) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman R.J., Wasson J., Atzmon G., Wainstein J., Yerushalmi Y., Cohen J. Gene-gene interactions lead to higher risk for development of type 2 diabetes in an Ashkenazi Jewish population. PLoS One. 2010;5(3) doi: 10.1371/journal.pone.0009903. (Mar 26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M.C.Y., Park K.S., Oh B., Tam C.H.T., Cho Y.M., Shin H.D. Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes. 2008;57(8):2226–2233. doi: 10.2337/db07-1583. (Aug) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffold T., Kullmann S., Huge A., Binner P., Ochs H.R., Schols W. Six sequence variants on chromosome 9p21.3 are associated with a positive family history of myocardial infarction: a multicenter registry. BMC Cardiovasc. Disord. 2011;7:11. doi: 10.1186/1471-2261-11-9. (Mar) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz K.M., Mayer C., Postepska A., Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24(20):2264–2269. doi: 10.1101/gad.590910. (Oct 15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless N.E., Depinho R.A. How stem cells age and why this makes us grow old. Nat. Rev. Mol. Cell Biol. 2007;8(9):703–713. doi: 10.1038/nrm2241. (Sep) [DOI] [PubMed] [Google Scholar]
- Shen G.Q., Li L., Rao S., Abdullah K.G., Ban J.M., Lee B.S. Four SNPs on chromosome 9p21 in a South Korean population implicate a genetic locus that confers high cross-race risk for development of coronary artery disease. Arterioscler. Thromb. Vasc. 2008;28(2):360–365. doi: 10.1161/ATVBAHA.107.157248. (Feb 1) [DOI] [PubMed] [Google Scholar]
- Singmann P., Baumert J., Herder C., Meisinger C., Holzapfel C., Klopp N. Gene—gene Interaction between APOA5 and USF1: two candidate genes for the metabolic syndrome. Obes. Facts. 2009;2(4):235–242. doi: 10.1159/000227288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. B Methodol. 1996;58(1):267–288. [Google Scholar]
- Timpson N.J., Emmett P.M., Frayling T.M., Rogers I., Hattersley A.T., McCarthy M.I. The fat mass- and obesity-associated locus and dietary intake in children. Am. J. Clin. Nutr. 2008;88(4):971–978. doi: 10.1093/ajcn/88.4.971. (Oct 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan R.S., Larson M.G., Leip E.P., Kannel W.B., Levy D. Assessment of frequency of progression to hypertension in nonhypertensive participants in the Framingham Heart Study: a cohort study. Lancet. 2001;358(9294):1682–1686. doi: 10.1016/S0140-6736(01)06710-1. (Nov 17) [DOI] [PubMed] [Google Scholar]
- Vaxillaire M., Veslot J., Dina C., Proenca C., Cauchi S., Charpentier G. Impact of common type 2 diabetes risk polymorphisms in the DESIR prospective study. Diabetes. 2008;57(1):244–254. doi: 10.2337/db07-0615. (Multicenter Study Research Support, Non-U.S. Gov't, Jan) [DOI] [PubMed] [Google Scholar]
- Wardle J., Carnell S., Haworth C.M.A., Farooqi I.S., O'Rahilly S., Plomin R. Obesity associated genetic variation in FTO is associated with diminished satiety. J. Clin. Endocrinol. Metab. 2008;93(9):3640–3643. doi: 10.1210/jc.2008-0472. (Sep) [DOI] [PubMed] [Google Scholar]
- Wardle J., Llewellyn C., Sanderson S., Plomin R. The FTO gene and measured food intake in children. Int. J. Obes. 2009;33(1):42–45. doi: 10.1038/ijo.2008.174. (Jan) [DOI] [PubMed] [Google Scholar]
- Wu Y., Li H.X., Loos R.J.F., Yu Z.J., Ye X.W., Chen L.H. Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Diabetes. 2008;57(10):2834–2842. doi: 10.2337/db08-0047. (Oct) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R., Everett L.J., Lim H.W., Patel N.A., Schug J., Kroon E. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2012.11.023. (Jan 9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K.L., Li S.D., Munoz-Cabello A.M., Raguz S., Zeng L., Mujtaba S. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. (Jun 11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H. The adaptive lasso and its oracle properties. J. Am. Stat. Assoc. 2006;101(476):1418–1429. (Dec) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.