Abstract
The Nuclear receptor 1 family I member 3 (NR1I3), also known as the Constitutive Androstane Receptor (CAR), was initially characterized as a key regulator of xenobiotic metabolism. However, recent biochemical and structural data suggest that NR1I3 is activated in response to metabolic and nutritional stress in a ligand-independent manner. Thus, we prospected the Bovine NR1I3 gene for polymorphisms and studied their association with feed efficiency traits in Nellore cattle. First, 155 purebred Nellore bulls were individually measured for Residual Feed Intake (RFI) and the 25 best (High Feed Efficiency group, HFE) and the 25 worst animals (Low Feed Efficiency group, LFE) were selected for DNA extraction. The entire Bovine NR1I3 gene was amplified and polymorphisms were identified by sequencing. Then, one SNP different between HFE and LFE groups was genotyped in all the 155 animals and in another 288 animals totalizing 443 Nellore bulls genotyped for association of NR1I3 SNPs with feed efficiency traits. We found 24 SNPs in the NR1I3 gene and choose a statistically different SNP between HFE and LFE groups for further analysis. Genotyping of the 155 animals showed a significant association within SNP and RFI (p = 0.04), Residual Intake and BW Gain (p = 0.04) and Dry Matter Intake (p = 0.01). This SNP is located in the 5′flanking promoter region of NR1I3 gene and different alleles alter the binding site for predicted transcriptional factors as HNF4alpha, CREM and c-MYB, leading us to conclude that NR1I3 expression and regulation might be important to feed efficiency.
Keywords: NR1I3, CAR, Residual feed intake, Nellore, Beef cattle
Introduction
Feed efficiency in beef cattle is a phenotypic trait where several cellular processes and gene networks are important as cellular growth and proliferation, cell signaling, drug metabolism, protein synthesis, lipid metabolism and carbohydrate metabolism (Chen et al., 2011). As a consequence, no major gene has been found to be related to feed efficiency yet (Moore et al., 2009). Therefore, it is rational to consider genes that are implicated in more than one of the processes described above. The Nuclear receptor 1 family I member 3 (NR1I3), previously known as Constitutive Androstane Receptor (NR1I3), was initially characterized as a key regulator of xenobiotic metabolism (Wei et al., 2000, Zhang et al., 2002). NR1I3 was considered a “xenobiotic sensor” of cells together with the closely related orphan nuclear hormone receptor – the pregnane X receptor (PXR, NR1I2) – regulating the transcription of several genes as cytochrome p450 enzymes (CYPs), ABC transporters and other genes which protect the body from a diverse array of harmful chemicals (Willson and Kliewer, 2002). However, recent biochemical and structural data suggest that, as opposed to merely sensing xenobiotic stress, NR1I3 is activated in response to metabolic or nutritional stress in a ligand-independent manner (Ding et al., 2006). NR1I3 has also been implicated to play a role in various physiological pathways such as energy metabolism (Konno et al., 2008) and homeostasis of lipids (Roth et al., 2008), triglycerides (Maglich et al., 2009), cholesterol, biliary acids, bilirubin and other endogenous hydrophobic molecules (Handschin and Meyer, 2005).
Regarding energy metabolism, it was shown that administration of NR1I3 agonists increased serum triglycerides only in wild-type mice (NR1I3+/+), but not in NR1I3−/− knockout mice, demonstrating the importance of NR1I3 being physically present for carbohydrate metabolism. In another experiment, it was shown that administration of NR1I3 agonists decreased hepatic lipin-1 expression, reducing the activation of oxidative pathways of fatty acids which reduced lipid blood levels (Finck et al., 2006). These effects mostly occurred because NR1I3 interacts with a number of hormonal-responsive transcription factors to regulate energetic metabolism altering gluconeogenesis and lipidogenesis. These factors include FoxO1 (Forkhead Box O1), FoxA2 (Forkhead Box A2), CREB (c-AMP response element binding protein) and PGC1alpha (Peroxisome proliferator activated gamma coactivator 1alpha) (Konno et al., 2008). Recently it was shown that NR1I3 activation by TCPOBOP (an agonist ligand) prevented obesity in mice receiving a high-fat diet. This outcome was credited to the inhibition of lipidogenesis due to decreased VLDL secretion and triglyceride export along with increased energetic expenditure and peripheral fat mobilization (Gao et al., 2009).
Greger and coworkers (Greger et al., 2006) demonstrated the ontogeny of NR1I3 mRNA expression and some of its targets in neonatal calves revealing that NR1I3 expression is variably expressed depending on age. They concluded that NR1I3 and their target genes exhibited ontogenetic changes that are likely of importance for handling of xeno-and endobiotics with increasing age and this may be important especially during periods of potential metabolic stress, such as during the weaning process (Greger et al., 2006). To our knowledge, there was no other work relating NR1I3 gene in cattle. Thus, based on the importance of NR1I3 on physiological pathways related to energy and lipid metabolism in mice and humans, we seek to prospect new polymorphisms in NR1I3 gene from Bos taurus indicus, and evaluate their possible association with feed efficiency in Nellore cattle.
Materials and methods
All animal protocols were approved by the Institutional Animal care and Use Committee of Faculdade de Zootecnia e Engenharia de Alimentos, Universidade de São Paulo (FZEA-USP).
Experiment 1: prospection of polymorphisms in the bovine NR1I3 gene
Cattle population
In this experiment, 155 Nellore bulls (18 to 22 month-old and 396 ± 43 kg BW) were evaluated in feeding trials carried out at FZEA-USP and at Universidade Federal do Paraná (UFPR), located in Curitiba, PR, Brazil. Cattle were raised under grazing (primarily Brachiaria spp) conditions until around 18 months of age, and they were then enrolled in feeding trials. The feeding trial period varied from 56 to 84 days, after an adaptation period of 21 to 28 days, established to adapt the cattle to the feedlot diet and to the place. Cattle were housed in individual pens or in group pens composed of Calan Broadbent feeding doors (American Calan Inc., Northwood, NH, USA), with 25 m2 as a minimum space per animal. Individual and group pens were soil-surfaced and contained automatic water fountains. Dry matter intake (DMI) was measured daily and average daily gain (ADG) was computed as the slope of the linear regression of body weight (BW) on feeding days. Cattle were weighed after 18-hour fasting period in the beginning and the end of the test and at 21 to 28-d intervals during the test period. Residual feed intake (RFI) was calculated as the difference between the observed and expected DMI of the animal, predicted by regression equation as a function of average metabolic weight and ADG (Koch et al., 1963). Residual intake and BW gain (RIG) and Residual BW Gain (RG) were calculated as recently proposed by Berry and Crowley (Berry and Crowley, 2012). All 155 animals were ranked by Residual Feed Intake and then, two groups were formed: “group HFE” (High Feed Efficiency group with the 25 highest RIF animals) and “group LFE” (Low Feed Efficiency group with the 25 lowest RIF animals).
DNA collection, extraction, PCR amplification and sequencing
Total DNA from these 50 animals was isolated from blood samples by the salting out method (Paneto et al., 2008). DNA concentration and purity were evaluated in a spectrophotometer (Nanodrop 2000c, Thermo Scientific, USA) and DNA quality was checked by 1.0% agarose gel electrophoresis before PCR amplification. The NR1I3 gene (Genbank accession NM_001079768) was amplified by PCR using eight pairs of specific primers (Supplemental Table 1) which were designed to cover the entire length of the gene, starting at -399nt position above 5′UTR until 416nt position at the end of 3′UTR (nt + 5309) with products around 800 bp with 100 bp overlaps (Robin Hiesinger, 2002). Each PCR reaction were performed following manufacturer's instructions (Taq 2X Master Mix, New England BioLabs, USA) with 100 μg of genomic DNA in convencional thermocycler (Mastercycler®, Eppendorf, EUA) with the following parameters [initial denaturation at 95 °C for 5 min; 35 cycles at 95 °C for 30 s, 60 °C for 30 s and 68 °C for 60 s; final extension at 68 °C for 5 min]. Then, 40 μL of each sample were concentrated in a vacuum concentrator (Concentrator 5301, Eppendorf, USA) to a final volume of 15 μL. The final concentration was checked by running the samples in a 1% agarose gel with Low DNA Mass Ladder (#822878, Invitrogen, USA). All samples with concentration of 20 ng/μL were considered appropriate for sequencing and were purified following manufacturer's instructions (ExoSAP-IT® #78201, USB, USA) in which 10 μL of sample were supplemented of 5 μL of ExoSAP-IT and taken to the thermocycler under the following conditions [37 °C for 15 min and 80 °C for 15 min]. The sequencing of PCR products was performed in the DNA Sequencing Service of the Center for Human Genome Studies (Instituto de Biociências, Universidade de São Paulo, IB-USP). Two complementary strands of DNA were sequenced for each sample with the same specific primers used for PCR (Table 1). The reverse sequence was inverted using BioEdit Sequence Alignment Editor® (Hall, 1999) and compared with forward sequence using Data Analysis in Molecular Biology and Evolution® (Xia and Xie, 2001).
Table 1.
SNPs of bovine NR1I3 gene found in Nellore cattle, their genotypic frequency and MAF.
| SNPs | Nomenclature | NCBI ref. n°. | Genotypes |
MAF | N° of DNA sequences | ||
|---|---|---|---|---|---|---|---|
| A1A1 | A1A2 | A2A2 | |||||
| SNP01 | c.-81-176G > A | rs207913429 | 39 | 3 | 0.036 | 42 | |
| SNP02 | c.-78G > A | ss492952008 | 10 | 22 | 10 | 0.500 | 42 |
| SNP03 | c.107G + 141T > C | ss492952011 | 14 | 23 | 12 | 0.480 | 49 |
| SNP04 | c.107G + 192C > A | ss492952014 | 48 | 1 | 0.020 | 49 | |
| SNP05 | c.107G + 243T > C | ss492952017 | 44 | 5 | 0.051 | 49 | |
| SNP06 | c.107G + 321C > T | ss492952020 | 14 | 23 | 12 | 0.480 | 49 |
| SNP07 | c.239 T-400G > A | ss492952023 | 15 | 20 | 11 | 0.457 | 46 |
| SNP08 | c.239 T-344 T > C | rs109470451 | 41 | 5 | 0.054 | 46 | |
| SNP09 | c.239 T-261 T > C | ss492952028 | 22 | 19 | 5 | 0.315 | 46 |
| SNP10 | c. 265 T > C | ss492952030 | 29 | 13 | 4 | 0.228 | 46 |
| SNP11 | c.309G > A | ss492952033 | 29 | 13 | 4 | 0.239 | 46 |
| SNP12 | c.409G-35A > T | ss492952036 | 28 | 14 | 4 | 0.290 | 46 |
| SNP13 | c.448G > A | ss492952039 | 43 | 3 | 0.033 | 46 | |
| SNP14 | c.489 T > C | rs211651336 | 43 | 3 | 0.033 | 46 | |
| SNP15 | c.549G-105G > A | ss492952045 | 44 | 3 | 0.032 | 47 | |
| SNP16 | c.549G-25C > T | ss492952048 | 30 | 7 | 10 | 0.287 | 47 |
| SNP17 | c.695C-29C > A | ss492952051 | 44 | 3 | 0.032 | 47 | |
| SNP18 | c.696G > A | ss492952054 | 44 | 3 | 0.032 | 47 | |
| SNP19 | c.738 T > C | ss492952057 | 29 | 7 | 10 | 0.293 | 46 |
| SNP20 | c.798C > T | rs134389810 | 44 | 3 | 0.032 | 47 | |
| SNP21 | c.917G + 20G > A | rs132968864 | 28 | 14 | 3 | 0.222 | 45 |
| SNP22 | c.917G + 294C > T | ss492952066 | 25 | 17 | 3 | 0.256 | 45 |
| SNP23 | c.*136C + 61G > C | rs384288081 | 41 | 3 | 0.033 | 44 | |
| SNP24 | c.*136C + 123C > T | ss492952072 | 42 | 3 | 0.033 | 45 | |
Polymorphism identification and haplotype analysis
The NR1I3 gene sequences from all animals were aligned and compared using ClustalW2 software (Larkin et al., 2007) and SNPs were identified. Animals were considered heterozygous for a specific SNP when both sequences (forward and reverse) presented at the same position a “R” (when it detects adenine and guanidine), or a “Y” (when it detects thymine and cytosine), or a “M” (when it detects adenine and cytosine), an “S” (when it detects guanidine and cytosine) or a “W” (when it detects adenine and thymine). The terminology used to describe the SNPs followed Den_Dunnen and Antorinakis (den Dunnen and Antonarakis, 2000). All detected SNPs in these samples were compared with the ones described in the dbSNP Short Genetic Variations from Entrez Database (NCBI, USA, Table 3). Linkage disequilibrium and inferred haplotypes across the NR1I3 SNPs in Nellore cattle were estimated by the Haploview software (Barrett, 2009). SNPs, whose minor allele frequencies (MAFs) < 0.001 and those that failed to meet Hardy-Weinberg equilibrium (HWE; P < 0.0001) where excluded from the haplotype structure.
Table 3.
Comparison of alellic frequency of bovine NR1I3 SNPs between HFE and LFE groups.
| SNP | Alelles | HFE | LFE | P value |
|---|---|---|---|---|
| SNP01 | A | 3 | 0 | 0.0643 |
| G | 37 | 44 | ||
| SNP02 | A | 16 | 26 | 0.1884 |
| G | 22 | 20 | ||
| SNP03 | C | 25 | 22 | 0.2338 |
| T | 21 | 30 | ||
| SNP04 | A | 2 | 0 | 0.1447 |
| C | 46 | 50 | ||
| SNP05 | C | 2 | 3 | 0.6801 |
| T | 46 | 47 | ||
| SNP06 | T | 27 | 20 | 0.1075 |
| C | 21 | 30 | ||
| SNP07 | A | 17 | 25 | 0.1958 |
| G | 27 | 23 | ||
| SNP08 | C | 2 | 3 | 0.7187 |
| T | 42 | 45 | ||
| SNP09 | C | 14 | 16 | 0.8769 |
| T | 30 | 32 | ||
| SNP10 | C | 13 | 8 | 0.1415 |
| T | 31 | 40 | ||
| SNP11 | A | 13 | 8 | 0.1415 |
| G | 31 | 40 | ||
| SNP12 | T | 14 | 8 | 0.1425 |
| A | 32 | 38 | ||
| SNP13 | C | 1 | 2 | 0.5572 |
| G | 45 | 44 | ||
| SNP14 | C | 3 | 0 | 0.0782 |
| T | 43 | 46 | ||
| SNP15 | A | 3 | 0 | 0.0606 |
| G | 41 | 50 | ||
| SNP16 | T | 15 | 10 | 0.1229 |
| C | 29 | 40 | ||
| SNP17 | A | 1 | 2 | 0.6345 |
| C | 43 | 48 | ||
| SNP18 | A | 1 | 2 | 0.6345 |
| G | 43 | 48 | ||
| SNP19 | C | 15 | 12 | 0.3388 |
| T | 29 | 36 | ||
| SNP20 | T | 1 | 2 | 0.6345 |
| C | 43 | 48 | ||
| SNP21 | A | 10 | 10 | 0.9103 |
| G | 36 | 34 | ||
| SNP22 | T | 10 | 13 | 0.3960 |
| C | 36 | 31 | ||
| SNP23 | C | 2 | 1 | 0.6115 |
| G | 44 | 41 | ||
| SNP24 | T | 1 | 2 | 0.5310 |
| C | 45 | 42 |
Experiment 2: validation of NR1I3 gene polymorphism
Animals and DNA sampling
For validation of the results found in the previous experiment, DNA samples from the 105 animals left from the first analysis (total of 155 minus the 50 already sequenced) and another set of 288 Nellore bulls tested for feed efficiency in similar conditions as already described were used.
Genotyping using high resolution melting (HRM)
A primer pair for the SNP01 (G > A) was designed as already described to amplify the small fragment containing the target SNP of c.-81-176G > A, which was further subjected to HRM analysis (Supplemental Table 1). HRM was performed on sequencing-verified samples in order to determine the Tm and characterize of the melting curve profiles of different genotypes (GG and AG). PCR reactions were performed on StepOne real-time PCR detection system (Applied Biosystems®, USA). All samples were amplified in duplicate, and each run contained a non-template control (NTC) and two type individuals as the heterozygous and homozygous genotypes. The 20 μL reaction volume included 1 × Fast EvaGreen® Master Mix (Biotium Inc., USA), 20 ng DNA, 0,1 μM each primer and sterile water. The cycling conditions were an initial denaturation at 95 °C for 10 min, 40 cycles of 15 s at 95 °C, 1 min at 60 °C. For HRM analysis, the PCR product was denatured by rising temperature to 95 °C at 4.8 °C/s and was then cool down to 55 °C at 2.5 °C/s for hybridization. The melting curve was acquired by increasing the temperature from 55 °C to 95 °C at a ramp rate of 4.8 °C/s with 25 acquisitions per degree of temperature. The HRM Software v2.0.1 (Applied Biosystems®, USA) was used to auto group the melt curve, and each genotype produced easily discriminated melt curves (amplitude and/or shape of the curves).
Prediction of functional consequences of NR1I3 gene polymorphism
We used PROMO software for detecting known transcription regulatory elements using species-tailored searches (Farre et al., 2003, Messeguer et al., 2002) to predict whether the SNP in the promoter region of NR1I3 affects transcription factor (TF) binding. DNA sequences of 10 nucleotides at each side of the SNP01 (5′-CATGAAACTC(G/A)GCAGTCGGGA-3′) were used as inputs, and all available TF and TF sites were tested.
Statistical analysis
Experiment 1: All data were presented as mean plus standard deviation. The alellic and genotypic frequencies for each SNP were estimated by simply analyzing the genotypes. The comparisons between the phenotypic traits from the two groups (HFE × LFE) were analyzed by the unpaired T-test. Analysis of the allelic frequencies between both groups was performed with the χ2 test. P values ≤ 0.05 were considered significant. When the p value was between 0.05 and 0.10 it was considered as tendency for statistical significance. These analyses were performed with the GraphPad Prism 5 software (GraphPad, USA).
- Experiment 2: For the association analysis between SNP and phenotypic traits, the PROC MIXED (SAS Enterprise Guide 4.2, USA), using the following model were used:
where Yijk = the phenotypic value of trait; μ = the general mean of the trait; CGi = the fixed effect of contemporary group; α1 = the regression coefficient of covariate of age at the measurement; α2 = the regression coefficient of covariate of days on feeding; α3 = the regression coefficient associated with the number of favorable alleles for each marker; sik = the random effect of sire (sik ~ N(0, σs2)); and eijk = the random residual error (eijk ~ N(0, σe2).
Days on feed was not significant and was excluded from the model. The average allele substitution effect was estimated by regressing the phenotype on the number of copies of one allele of a single SNP using the cited mixed model in SAS.
Results
SNP prospection and genotyping
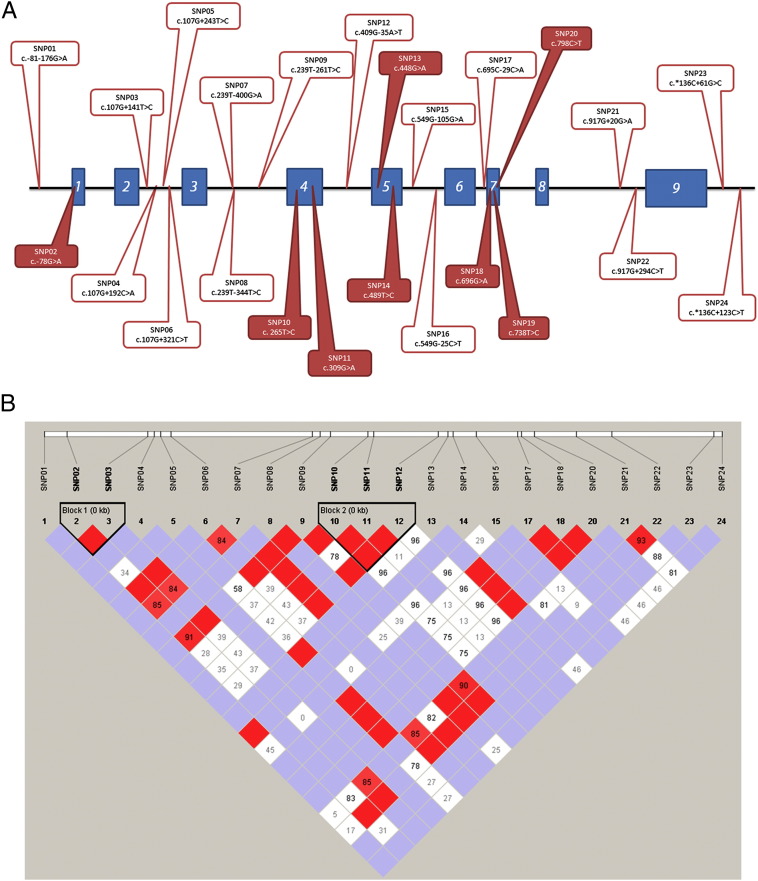
Initially, we selected 50 out of the 155 animals phenotypically measured for residual feed intake (RFI). From these 50 animals, one was removed from the analysis due to low quality DNA turning impossible the amplification by PCR. Thus, DNA from 49 Nellore bulls was used to perform 8 PCRs per animal. Some sequences were not used due to low quality as indicated in Table 1. Of all reliable DNA sequences obtained (more than 94%), we found 24 SNPs thoroughly the NR1I3 gene (Fig. 1), which presented different genotypic and allelic frequencies (Table 1). From the 24 SNPs obtained, six were already described at the dbSNP Short Genetic Variations from NCBI at the time this article was finished (08/26/2013) namely: SNP01 (rs207913429), SNP08 (rs109470451), SNP14 (rs211651336), SNP20 (rs134389810), SNP21 (rs132968864) and SNP23 (rs384288081).
Fig. 1.
Spatial localization of SNPs found in bovine NR1I3 gene of Nellore cattle and their haplotype analysis. (A) NR1I3 gene is composed of 9 exons (represented here by blue boxes) and 8 introns (represented here by black lines between two blue boxes). Red balloons are SNPs in exons of the NR1I3 gene. Clear balloons are SNP above 5′UTR, SNPs in introns or SNPs after the 3′UTR. (B) Linkage disequilibrium (LD) plot of NR1I3 SNPs from Nellore cattle. Color scheme is according to Haploview r2 scheme. Numbers in each cell stand for pairwise r2-values (%) between corresponding SNPs.
Linkage disequilibrium and haplotype analysis
After excluding SNP16 and SNP19 from the haplotype analysis due to the HWE criteria for exclusion, we performed the pairwise linkage disequilibrium analysis that showed the presence of two blocks namely Block 1 (SNP02 4and SNP03) and Block 2 (SNP10, SNP11 and SNP12; Fig. 1).
Analysis of NR1I3 gene polymorphisms between high and low feed efficiency groups
Next, we compared the HFE and LFE groups and determined that the HFE group presented decreased RFI (p < 0.0001), Dry Matter Intake (DMI; p < 0.0001) and Feed Conversion Ratio (FCR; p = 0.0232, Table 2) when compared to LFE group. On the other hand, increased RIG (p < 0.0001) and Feed Efficiency (FE; p = 0.0232) was found in the HFE group. No difference was detected for ADG (p = 0.7188) and Residual BW Gain (RG) between both groups (Table 2).
Table 2.
Phenotypic comparison from selected animals with high (HFE) and low RFI (LFE).
| Phenotype (n = 49) | Group HFE (n = 24) |
Group LFE (n = 25) |
p value |
|---|---|---|---|
| RFI (kg DM/day) | − 0.81 ± 0.42 | 0.94 ± 0.47 | p < 0.0001 |
| RIG | 1.78 ± 1.74 | − 2.02 ± 1.12 | p < 0.0001 |
| RG | 0.08 ± 0.38 | − 0.07 ± 0.31 | p = 0.2980 |
| Initial BW (kg) | 399.35 ± 46.38 | 392.46 ± 40.69 | p = 0.7565 |
| Final BW (kg) | 502.12 ± 60.77 | 492.62 ± 45.50 | p = 0.7338 |
| Mean BW (kg) | 450.74 ± 52.23 | 442.54 ± 41.71 | p = 0.7188 |
| ADG (kg/day) | 1.56 ± 0.41 | 1.57 ± 0.33 | p = 0.6455 |
| DMI (kg/day) | 8.61 ± 0.97 | 10.34 ± 1.11 | p < 0.0001 |
| FCR (kg DM/kg gain) | 5,87 ± 1,58 | 6,89 ± 1,65 | p = 0.0232 |
| FE (kg gain/kg DM) | 0.18 ± 0.04 | 0.15 ± 0.03 | p = 0.0232 |
Groups HFE and LFE where statistically compared by unpaired T test with a p value set to p < 0.05.
We further compared both groups for the allelic frequency of each of the 24 SNPs found in the bovine NR1I3 gene. Only three SNPs presented tendency for significant differences between both groups: SNP01 (p = 0.0643), SNP14 (p = 0.0782) and SNP15 (p = 0.0606; Table 3). In the next lines, we choose to characterize the importance of SNP01 for feed efficiency, since the SNP01, SNP14 and SNP15 were apparently in linkage (Fig. 1).
Association of SNP01 with Feed Efficiency Traits in different Nellore herds
To validate our results, we further evaluated the frequency of the SNP01 in all the 155 animals (which consisted of four different populations) for association analysis with feed efficiency traits. The MAF of SNP01 in this population was 2.58% (allele A). Interestingly, we found statistical association between SNP01 and RFI (p = 0.04) and RIG (p = 0.04) with an estimated effect of allelic substitution (G → A) of − 0.68 kg DM/day and + 0.85, respectively (Table 4).
Table 4.
Association between SNP01 in different populations of Nellore cattle.
| Populations | Number of animals | MAF | Association study p value (allelic substitution G > A effect) |
||
|---|---|---|---|---|---|
| RFI | RIG | DMI | |||
| Experiment 1 | 155 | 0.0258 | 0.04 (− 0.68) | 0.04 (+ 0.85) | 0.09 (− 0.67) |
| Pop. A | 44 | 0.0909 | 0.0059 (− 0.49) | 0.0124 (+ 0.56) | 0.01 (− 0.59) |
| Pop. B | 44 | 0 | |||
| Pop. C | 30 | 0 | |||
| Pop. D | 37 | 0 | |||
| Other populations | |||||
| Pop. E | 48 | 0.0417 | 0.73 | 0.74 | 0.62 |
| Pop. F | 15 | 0.0333 | 0.45 | 0.37 | 0.99 |
| Pop. G | 113 | 0.0044 | 0.14 | 0.14 | 0.18 |
| Pop. H | 69 | 0.0145 | 0.58 | 0.5 | 0.9 |
| Pop. I | 43 | 0 | |||
| All together | 443 | 0.0181 | 0.17 | 0.17 | 0.32 |
The allelic substitution effect was only calculated for the associations of SNP01 with phenotypic traits that presented p < 0.05. RIF and DMI units are the same presented in the text and in Table 2.
Next, for validation purposes, we evaluate the effect of SNP01 in another five different populations assessed for Feed Efficiency Traits, totalizing 443 genotyped Nellore animals. Considering all the nine populations, four of them presented the allele G fixed (Table 3). The overall MAF when considering all animals genotyped was only 1.81% (16/886). However, when we evaluate the association between SNP01 and feed efficiency traits in each population alone, only one population (with MAF = 9.09%) presented a statistically association of SNP01 with feed efficiency traits as the RFI (p = 0.0059), RIG (p = 0.0124) and DMI (p = 0.01). In this population, the effect of the allelic substitution of the allele G for allele A of SNP01 decreased the RFI (− 0.49 kg DM/day) and DMI (− 0.59 kg/day). On the other hand, the same allelic substitution increased the RIG (+ 0.56).
Pedigree analysis
From all the 16 heterozygote (AG) animals found in all animals genotyped (16/443), we were able to perform pedigree analysis of 12 of them. Three animals were half-sibs from one sire. The other 9 animals were or great-grandsons (6 animals) or four-greats-grandson (3) from another sire.
In silico prediction of the biological effect of SNP01
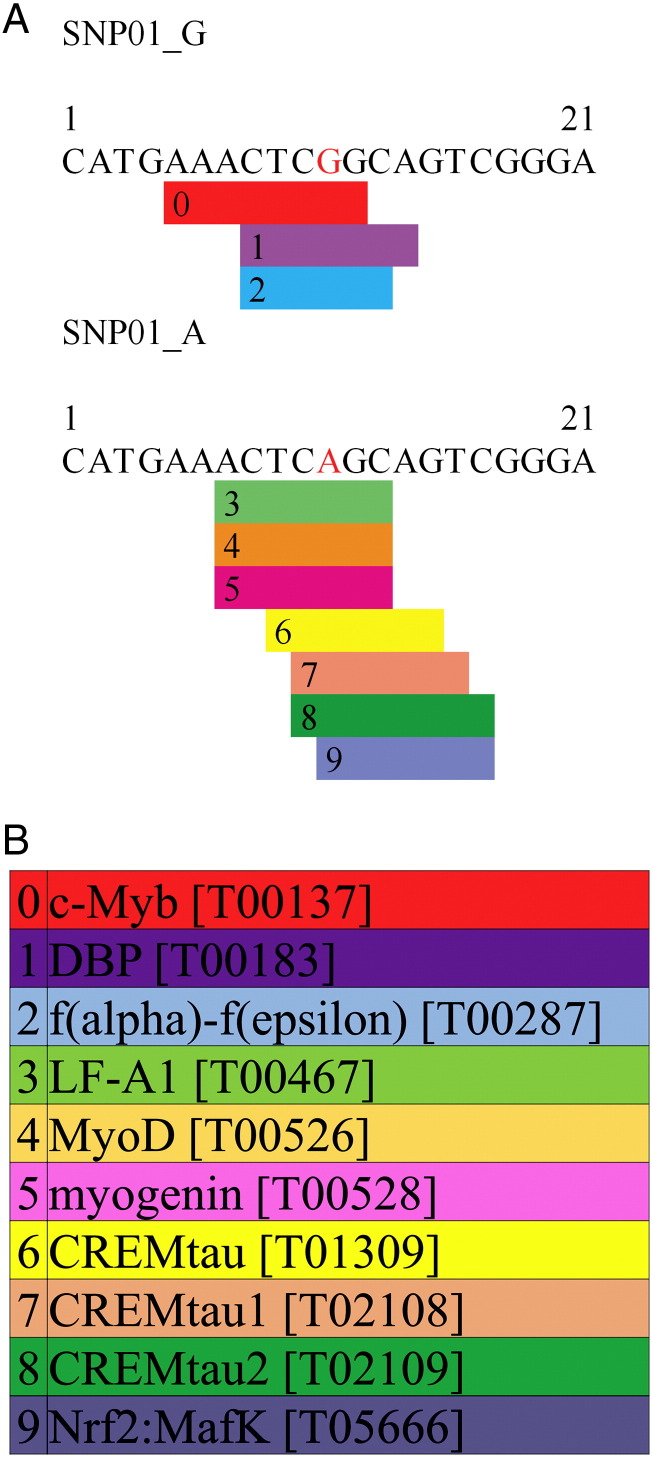
The SNP01 is located within the 5′flanking region of the bovine NR1I3 gene (Fig. 1). Thus, we evaluated this SNP as a potential part of the promoter region of the NR1I3 gene. Both alleles of SNP01 (G/A) were evaluated in silico for being potential transcription factor binding sites. We found different TF binding sites that are altered by the SNP01 (G > A). When G is the allele, three TFs could bind this promoter region of bovine NR1I3. However, when A substitutes G, these three TFs lost their binding sites in the bovine NR1I3 gene and another set of seven TFs could now bind the promoter region of the bovine NR1I3 (Fig. 2).
Fig. 2.
Transcription factor binding sites for the 5′flanking promoter region of bovine NR1I3 gene with the SNP01 (G > A). (A) Different TFs binding between DNA sequence with the allele G and allele A. (B) description of the TFs and their code according to Promo software (Farre et al., 2003, Messeguer et al., 2002).
Discussion
Here, we prospected polymorphisms in the bovine NR1I3 gene and found 24 new SNPs in Nellore cattle, being six already described for Bos taurus. In addition, we were able to significantly associate one of them to feed efficiency traits as RFI, DMI and a new index trait known as Residual Intake and BW Gain (RIG), a recently described measure for feed efficiency which also considers growth rate contrasting to the more commonly used, the Residual Feed Intake (Berry and Crowley, 2012). This SNP is located in the promoter region of NR1I3 gene and the change of G to A, being the A the favorable allele, alters the binding site for predicted transcriptional factors, leading us believe that NR1I3 gene expression and regulation might be important for feed efficiency.
In this work, our approach was first to prospect NR1I3 gene polymorphisms in the two groups of animals classified by their RFI in a population of 155 animals (HFE x LFE groups containing 25 animals each) and statistically analyzed the allelic frequency in these two groups. Later, we selected one SNP for validation process, which consisted of genotyping all the 155 animals and another 288 different animals evaluated for feed efficiency traits. The method for selecting a SNP based on groups of animals from extreme RFI in a population was proven as a good option since after genotyping the 155 animals, a statistically significant association between SNP01 and RIF, RIG and DMI were demonstrated. On the other hand, when we seek for this polymorphism in another set of animals totalizing 443 genotyped animals, the MAF of this SNP was too low (MAF = 0.0181), which might compromised the association analysis with the traits. This emerged two questions: 1) are SNPs in bovine NR1I3 gene important for feed efficiency? and 2) the low MAF of this SNP in Bos indicus is good or bad for selection purposes? We believe that discover SNPs in the bovine NR1I3 gene and associate them with feed efficiency traits, even in a small population of animals (155), is important since it opens opportunities to add these gene variations to a set of molecular targets for cattle selection. The low MAF of SNPs should not be viewed as bad and must be regarded as a great opportunity to use these markers for rapidly increase the net effect of a chosen phenotype, in this case, feed efficiency. It is important to be aware that regular Genome Wide Association Studies (GWAS) for feed efficiency in beef and dairy cattle discovered genomic regions that respond for great part of a phenotype, but not all of it (Lu et al., 2013, Rolf et al., 2012, Yao et al., 2013). Another set of significant genes and their variation that are also responsible for the phenotype will be discovered with approaches like the one that we performed here.
Prior work in feed efficiency molecular genetics has shown that this phenotype is a polygenic characteristic with no major gene affecting it (Moore et al., 2009). However, as in cancer research, studies should emphasize in determining the key regulators of molecular pathways responsible for the phenotype (Jones, 2008). As such, a recently published study from Chen et al. (2011) on differential global gene expression between high and low RFI beef cattle animals have demonstrated the importance of seven pathways: cellular growth and proliferation, cellular assembly and organization, cell signaling, drug metabolism, protein synthesis, lipid metabolism and carbohydrate metabolism. The NR1I3 gene was already shown to be a key regulator of the following mechanisms: cellular growth and proliferation (Costa et al., 2005, Huang et al., 2005), cell signaling (Braeuning et al., 2011), drug metabolism (Wei et al., 2000), protein synthesis (Ross et al., 2010) and metabolism of lipids and carbohydrates (Gao et al., 2009, Rezen et al., 2011).
In our experiment, high and low-RFI groups presented similar ADG, RG and final BW (Table 2) differing drastically in daily DMI (p < 0.0001). Thus, it is plausible to affirm that more efficient animals measured by RFI eat less to produce the same amount of beef, most probably because they are more energetically competent then the less efficient animals. It is usual consider that ruminants exist in a constant state of potential glucose deficiency with a continuous level of gluconeogenesis in the liver (Cunningham and Klein, 2007). Hence, animals that are more prone to deal with this stressful physiological situation should be more energetically efficient as well. Studies on NR1I3 function confirmed that NR1I3 activation inhibits gluconeogenesis and lipidogenesis with a net anti-obesity effect in mice (Gao et al., 2009). The in vivo significance of NR1I3-mediated suppression of gluconeogenesis was supported by two reports that activation of NR1I3 ameliorated hyperglycemia and improved insulin sensitivity (Dong et al., 2009, Gao et al., 2009). Therefore, we could hypothesize that SNP01 somehow is responsible for reduced transcriptional NR1I3 mRNA expression, resulting in diminished NR1I3 protein and consequently decreasing NR1I3 activation and its pleiotropic effects, leading ultimately to a decrease in the NR1I3 induced inhibition of gluconeogenesis and lipogenesis. Supporting our hypothesis, low RFI animals (more efficient ones) presented decreased expression of several genes related to drug metabolism in comparison to the high RFI animals (Chen et al., 2011), some of them known to be under NR1I3 transcriptional control such as ABCC4, CYP1A2 and CYP2C18 (Chen and Goldstein, 2009, Lee et al., 2007, Renga et al., 2011).
Accordingly, it is plausible that a genetic polymorphism in NR1I3 5′ flanking promoter region is associated with such a polygenic characteristic as feed efficiency. Interestingly, the allele substitution of G to A in the SNP01 hindered the binding sites for c-MYB and LF-A1/HNF-4α whereas create sites for binding of CREM agonistic spliced variant isoforms (CREMtau, CREMtau1 and CREMtau2), NRF2 and two muscle related TFs, myogenin and MyoD in the NR1I3 promoter region (Fig. 2). From all these possibilities, loss of HNF4α transcriptional regulation of NR1I3 seems to be important since HNF4α is a critical component of PGC-1α-mediated gluconeogenesis (Rhee et al., 2003). HNF4α and NR1I3 receptors compete for binding to the DR1 motif in the promoter of gluconeogenic genes and could compete for common coactivators such as GRIP-1 and PGC-1α (Miao et al., 2006). This could be a way for NR1I3 down-regulate key genes in hepatic lipid and glucose metabolism (Miao et al., 2006). Therefore, if HNF4α is not able to induce the transcription of NR1I3 expression, less NR1I3 protein will compete for the DR1 motif and coactivators leading to a positive balance for HNF4α-mediated gluconeogenesis in more efficient animals.
In conclusion, we showed that genetic polymorphism of NR1I3 gene is significantly associated with feed efficiency in Nellore cattle, in a small population (155 animals) but not in the other set of 288 animals. This could be in part explained by the low MAF of the associated SNP, even being fixed in some populations of Nellore cattle. Our studies now focuses in characterize the expression and the transcriptional activity of the bovine NR1I3 gene in efficient and inefficient animals.
The following is the supplementary data related to this article.
Primer pair sequence, melting temperature (Tm) and expected PCR product size in base pairs for NR1I3 gene amplification and sequencing or HRM analysis.
Acknowledgements
The authors acknowledge Soraya Silva for her kind help during the experiment. This work was supported by FAPESP grants (2010/05650-5; 2007/04445-6 and 2008/11363-9) and a fellowship from CNPq/PIBIC for P.A. Alexandre.
References
- Barrett J.C. Cold Spring Harbor protocols 2009. 2009. Haploview: visualization and analysis of SNP genotype data. (pdb ip71) [DOI] [PubMed] [Google Scholar]
- Berry D.P., Crowley J.J. Residual intake and body weight gain: a new measure of efficiency in growing cattle. J. Anim. Sci. 2012;90:109–115. doi: 10.2527/jas.2011-4245. [DOI] [PubMed] [Google Scholar]
- Braeuning A., Heubach Y., Knorpp T., Kowalik M.A., Templin M., Columbano A., Schwarz M. Gender-specific interplay of signaling through beta-catenin and CAR in the regulation of xenobiotic-induced hepatocyte proliferation. Toxicol. Sci. 2011;123:113–122. doi: 10.1093/toxsci/kfr166. [DOI] [PubMed] [Google Scholar]
- Chen Y., Goldstein J.A. The transcriptional regulation of the human CYP2C genes. Curr. Drug Metab. 2009;10:567–578. doi: 10.2174/138920009789375397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Gondro C., Quinn K., Herd R.M., Parnell P.F., Vanselow B. Global gene expression profiling reveals genes expressed differentially in cattle with high and low residual feed intake. Anim. Genet. 2011;42:475–490. doi: 10.1111/j.1365-2052.2011.02182.x. [DOI] [PubMed] [Google Scholar]
- Costa R.H., Kalinichenko V.V., Tan Y., Wang I.C. The CAR nuclear receptor and hepatocyte proliferation. Hepatology. 2005;42:1004–1008. doi: 10.1002/hep.20953. [DOI] [PubMed] [Google Scholar]
- Cunningham J.G., Klein B.G. Sauders Elsevier; St. Louis: 2007. Veterinary Phisiology. [Google Scholar]
- den Dunnen J.T., Antonarakis S.E. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Ding X., Lichti K., Kim I., Gonzalez F.J., Staudinger J.L. Regulation of constitutive androstane receptor and its target genes by fasting, cAMP, hepatocyte nuclear factor alpha, and the coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha. J. Biol. Chem. 2006;281:26540–26551. doi: 10.1074/jbc.M600931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Saha P.K., Huang W., Chen W., Abu-Elheiga L.A., Wakil S.J., Stevens R.D., Ilkayeva O., Newgard C.B., Chan L., Moore D.D. Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18831–18836. doi: 10.1073/pnas.0909731106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre D., Roset R., Huerta M., Adsuara J.E., Rosello L., Alba M.M., Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck B.N., Gropler M.C., Chen Z., Leone T.C., Croce M.A., Harris T.E., Lawrence J.C., Jr., Kelly D.P. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Gao J., He J., Zhai Y., Wada T., Xie W. The constitutive androstane receptor is an anti-obesity nuclear receptor that improves insulin sensitivity. J. Biol. Chem. 2009;284:25984–25992. doi: 10.1074/jbc.M109.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger D.L., Philipona C., Blum J.W. Ontogeny of mRNA abundance of nuclear receptors and nuclear receptor target genes in young cattle. Domest. Anim. Endocrinol. 2006;31:76–87. doi: 10.1016/j.domaniend.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999:95–98. [Google Scholar]
- Handschin C., Meyer U.A. Regulatory network of lipid-sensing nuclear receptors: roles for CAR, PXR, LXR, and FXR. Arch. Biochem. Biophys. 2005;433:387–396. doi: 10.1016/j.abb.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Huang W., Zhang J., Washington M., Liu J., Parant J.M., Lozano G., Moore D.D. Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol. Endocrinol. 2005;19:1646–1653. doi: 10.1210/me.2004-0520. [DOI] [PubMed] [Google Scholar]
- Jones D. Pathways to cancer therapy. Nat. Rev. Drug Discov. 2008;7:875–876. doi: 10.1038/nrd2748. [DOI] [PubMed] [Google Scholar]
- Koch R.M., Swiger L.A., Chambers D., Gregory K.E. Effciency of feed use in beef cattle. J. Anim. Sci. 1963;22:486–494. [Google Scholar]
- Konno Y., Negishi M., Kodama S. The roles of nuclear receptors CAR and PXR in hepatic energy metabolism. Drug Metab. Pharmacokinet. 2008;23:8–13. doi: 10.2133/dmpk.23.8. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee C.H., Ito Y., Yanagiba Y., Yamanoshita O., Kim H., Zhang S.Y., Kamijima M., Gonzalez F.J., Nakajima T. Pyrene-induced CYP1A2 and SULT1A1 may be regulated by CAR and not by AhR. Toxicology. 2007;238:147–156. doi: 10.1016/j.tox.2007.05.026. [DOI] [PubMed] [Google Scholar]
- Lu D., Miller S., Sargolzaei M., Kelly M., Vander Voort G., Caldwell T., Wang Z., Plastow G., Moore S. Genome-wide association analyses for growth and feed efficiency traits in beef cattle. J. Anim. Sci. 2013;91:3612–3633. doi: 10.2527/jas.2012-5716. [DOI] [PubMed] [Google Scholar]
- Maglich J.M., Lobe D.C., Moore J.T. The nuclear receptor CAR (NR1I3) regulates serum triglyceride levels under conditions of metabolic stress. J. Lipid Res. 2009;50:439–445. doi: 10.1194/jlr.M800226-JLR200. [DOI] [PubMed] [Google Scholar]
- Messeguer X., Escudero R., Farre D., Nunez O., Martinez J., Alba M.M. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- Miao J., Fang S., Bae Y., Kemper J.K. Functional inhibitory cross-talk between constitutive androstane receptor and hepatic nuclear factor-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1alpha. J. Biol. Chem. 2006;281:14537–14546. doi: 10.1074/jbc.M510713200. [DOI] [PubMed] [Google Scholar]
- Moore S.S., Mujibi F.D., Sherman E.L. Molecular basis for residual feed intake in beef cattle. J. Anim. Sci. 2009;87:E41–E47. doi: 10.2527/jas.2008-1418. [DOI] [PubMed] [Google Scholar]
- Paneto J.C., Ferraz J.B., Balieiro J.C., Bittar J.F., Ferreira M.B., Leite M.B., Merighe G.K., Meirelles F.V. Bos indicus or Bos taurus mitochondrial DNA - comparison of productive and reproductive breeding values in a Guzerat dairy herd. Genet. Mol. Res. 2008;7:592–602. doi: 10.4238/vol7-3gmr449. [DOI] [PubMed] [Google Scholar]
- Renga B., Migliorati M., Mencarelli A., Cipriani S., D'Amore C., Distrutti E., Fiorucci S. Farnesoid X receptor suppresses constitutive androstane receptor activity at the multidrug resistance protein-4 promoter. Biochim. Biophys. Acta. 2011;1809:157–165. doi: 10.1016/j.bbagrm.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Rezen T., Rozman D., Pascussi J.M., Monostory K. Interplay between cholesterol and drug metabolism. Biochim. Biophys. Acta. 2011;1814:146–160. doi: 10.1016/j.bbapap.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Rhee J., Inoue Y., Yoon J.C., Puigserver P., Fan M., Gonzalez F.J., Spiegelman B.M. Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin Hiesinger P. 2002. Multiple Primer Design with Primer 3. [Google Scholar]
- Rolf M.M., Taylor J.F., Schnabel R.D., McKay S.D., McClure M.C., Northcutt S.L., Kerley M.S., Weaber R.L. Genome-wide association analysis for feed efficiency in Angus cattle. Anim. Genet. 2012;43:367–374. doi: 10.1111/j.1365-2052.2011.02273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J., Plummer S.M., Rode A., Scheer N., Bower C.C., Vogel O., Henderson C.J., Wolf C.R., Elcombe C.R. Human constitutive androstane receptor (CAR) and pregnane X receptor (PXR) support the hypertrophic but not the hyperplastic response to the murine nongenotoxic hepatocarcinogens phenobarbital and chlordane in vivo. Toxicol. Sci. 2010;116:452–466. doi: 10.1093/toxsci/kfq118. [DOI] [PubMed] [Google Scholar]
- Roth A., Looser R., Kaufmann M., Blattler S.M., Rencurel F., Huang W., Moore D.D., Meyer U.A. Regulatory cross-talk between drug metabolism and lipid homeostasis: constitutive androstane receptor and pregnane X receptor increase Insig-1 expression. Mol. Pharmacol. 2008;73:1282–1289. doi: 10.1124/mol.107.041012. [DOI] [PubMed] [Google Scholar]
- Wei P., Zhang J., Egan-Hafley M., Liang S., Moore D.D. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- Willson T.M., Kliewer S.A. PXR, CAR and drug metabolism. Nat. Rev. Drug Discov. 2002;1:259–266. doi: 10.1038/nrd753. [DOI] [PubMed] [Google Scholar]
- Xia X., Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Yao C., Spurlock D.M., Armentano L.E., Page C.D., Jr., Vandehaar M.J., Bickhart D.M., Weigel K.A. Random Forests approach for identifying additive and epistatic single nucleotide polymorphisms associated with residual feed intake in dairy cattle. J. Dairy Sci. 2013;96(10):6716–6729. doi: 10.3168/jds.2012-6237. [DOI] [PubMed] [Google Scholar]
- Zhang J., Huang W., Chua S.S., Wei P., Moore D.D. Modulation of acetaminophen-induced hepatotoxicity by the xenobiotic receptor CAR. Science. 2002;298:422–424. doi: 10.1126/science.1073502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer pair sequence, melting temperature (Tm) and expected PCR product size in base pairs for NR1I3 gene amplification and sequencing or HRM analysis.