Abstract
Objectives
Liver cirrhosis is a condition in which the liver slowly deteriorates and malfunctions due to chronic injury. HCV is one of the major causes of liver fibrosis and ultimate progression to cirrhosis. Transforming growth factor-beta1 (TGF-β1), one of the three isoforms of TGF-β, is a pleiotrophic cytokine that regulates the proliferation and differentiation of cells, embryonic development, wound healing and angiogenesis. This study aimed to evaluate the role of serum TGF-β1 and − 509 C>T promoter gene polymorphism in the development of liver cirrhosis.
Design and methods
Besides routine liver profiles, serum TGF-β1 was measured in 40 liver cirrhosis patients and 40 controls using ELISA technique. TGF-β1 − 509 C>T promoter gene polymorphism was detected using PCR-RFLP technique.
Results
TGF-β1 − 509 CT and TT genotype frequencies were significantly higher in the cirrhotic group (52.5%, 25%; respectively) than control group (10%, 7.5%; respectively); OR = 16.238 (95% CI 5.391–48.914, p < 0.05). The − 509 T allele carriers are more prone to develop liver cirrhosis than − 509 C allele carriers; OR = 7.359 (95% CI 3.325–16.288, p < 0.05). Serum TGF-β1 was significantly higher in cirrhotic group (11.79 ± 1.45 ng/ml) than control group (8.67 ± 1.23 ng/ml); p < 0.05. Also serum TGF-β1 was significantly higher in TT genotype than CT and CC genotypes (p < 0.05). A significant positive correlation was observed between serum TGF-β1 and alkaline phosphatase (r = 0.559, p < 0.05); AST (r = 0.573, p < 0.05). A significant negative correlation was observed between serum TGF-β1 and albumin (r = − 0.331, p < 0.05).
Conclusion
There is an association between serum TGF-β1, − 509 CT and TT genotypes of TGF-β1 gene and the higher risk for liver cirrhosis development of liver cirrhosis.
Keywords: Transforming growth factor-β1 (TGF-β1), Liver cirrhosis, PCR-RFLP
Introduction
Chronic liver diseases (CLDs) are the most important health problems according to the current gastroenterology literature. CLDs are defined by the continuity of the clinical and biochemical evidence of hepatic dysfunction for longer than six months (Nawar et al., 2011).
Chronic hepatitis C is one of the major causes of liver fibrosis, with distortion of the hepatic architecture and ultimate progression to cirrhosis resulting in nodule formation. Approximately more than 3% of the total world population is chronically infected with HCV and due to gradual increase in the prevalence of HCV; Chronic HCV is predicted to raise at least 3 folds by the year 2020 (Waqar et al., 2011).
Clinical features of cirrhosis are derived from the morphologic alterations and often reflect the severity of the hepatic damage rather than the etiology of the underlying liver disease. Loss of functioning hepatocellular mass may lead to jaundice, edema, coagulopathy and a variety of metabolic abnormalities; finally leading to hepatocellular carcinoma (HCC) (Gines et al., 2004).
Transforming growth factor-β (TGF-β) is a multifunctional cytokine involved in the regulation of growth and differentiation of a variety of normal cells and organs. It also plays an important role in the regulation of wound healing, angiogenesis, extracellular matrix formation, immunosuppression and cancer development (Dong et al., 2008).
Changes in the production of TGF-β have been linked to numerous disease states, including atherosclerosis and fibrous diseases of the kidney, liver and lung. In liver diseases, the persistence of chronic inflammation, as observed in chronic viral hepatitis, plays a major role in determining the shift in the TGF-β signaling pathway from tumor suppression to fibrogenesis accelerating liver fibrosis and increasing the risk of HCC (Matsuzaki et al., 2007).
TGF-β1 is one of the three isoforms of TGF-β that arrests the cell cycle in the G1-phase, thereby inhibiting cell proliferation and triggering apoptosis (Dong et al., 2008). TGF-β1 is a central regulator in chronic liver disease contributing to all stages of disease progression from initial liver injury through inflammation and fibrosis to cirrhosis and HCC. The role of TGF-β1 in modulating liver fibrosis progression has been extensively investigated. This cytokine favors the transition of hepatic stellate cells to myofibroblast-like cells, stimulates the synthesis of extracellular matrix proteins and inhibits their degradation (Bissell, 2001).
Several single nucleotide polymorphisms (SNPs) have been described in the human TGF-β1 locus, on chromosome 19q13. Two SNPs are located in the promoter region, at the positions − 800 G>A and − 509 C>T from the first transcribed nucleotide. Two further SNPs are located in the codons 10 (CTG>CCG) and 25 (CGG>CCG) of exon 1, leading to amino acid substitutions Leu10Pro and Arg25Pro. These allelic variants result in significant differences with regard to TGF-β1 expression and plasma concentration, therefore, they may predispose to, or protect against cirrhosis and HCC (Gewaltig et al., 2002, Falleti et al., 2008).
The present study was conducted to evaluate the role of TGF-β1 − 509 C>T promoter gene polymorphism and serum TGF-β1 level in the development of liver cirrhosis.
Subjects and methods
This study was performed on 40 patients having liver cirrhosis (Child Pugh Score A) age ranged between 40 and 60 years (25 males and 15 females) and 40 age and sex matched healthy individuals taken as control group with no history of liver disease, free on clinical examination and normal liver function tests. The patients were attending the Outpatient Clinic of Tropical Department, Kasr El-Aini Hospital, Cairo University. Informed oral consents were taken from all participants and the study was approved by the local ethical committee.
All patients were subjected to full history taking, complete clinical examination, abdominal ultrasound, while laboratory investigations were done to all subjects (Table 1).
Table 1.
Laboratory data of the studied groups.
| Cirrhotic group (n = 40) | Control group (n = 40) | p-Value | |
|---|---|---|---|
| T. bilirubin (mg/dl)a | 1.27 (0.85–3.20) | 0.35 (0.25–0.53) | < 0.05 |
| AST (U/L)a | 61 (46–101) | 17 (14–21) | < 0.05 |
| ALT (U/L)a | 49.5 (40.75–68.75) | 12.0 (10–13.75) | < 0.05 |
| ALP (U/L)a | 100 (82–118) | 61 (52–70) | < 0.05 |
| GGT (U/L)a | 36 (24–53) | 18 (13–21) | < 0.05 |
| Albumin (g/dl)b | 2.50 ± 0.40 | 4.1 ± 0.40 | < 0.05 |
| TGF-β1 (ng/ml)b | 11.79 ± 1.45 | 8.67 ± 1.23 | < 0.05 |
| PT (second)b | 18.18 ± 3.44 | 11.66 ± 0.37 | < 0.05 |
| PC (%)b | 52.70 ± 16.49 | 94.85 ± 4.23 | < 0.05 |
| INRa | 1.6 (1.4–2.0) | 1.03 (1–1.07) | < 0.05 |
Data presented as median (25th–75th percentile).
Data presented as mean ± SD.
Specimen collection
Eight milliliters of venous blood was withdrawn from each subject; 3 ml was collected on plain tubes, sera were separated for assay of TGF-β1 and liver functions, 1.8 ml was collected in sodium citrated tubes for assay of prothrombin time (PT) and the remaining part was collected on sterile EDTA tubes for detection of TGF-β1 − 509 C>T promoter gene polymorphism using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).
Routine liver functions were assayed using dedicated kits on Dimension RxL chemistry auto-analyzer (Siemens Health Care Diagnostics, Inc. Newark, DE 19714, USA).
Serum TGF-β1 was determined using human TGF-β1 ELISA kit (quantitative sandwich enzyme immunoassay technique), provided by DRG International, Inc. (USA).
DNA isolation and TGF-β1 genotyping
Genomic DNA was isolated from peripheral blood leucocytes using Biospin Whole Blood Genomic DNA Extraction kit (Bioflux Corporation, Arrow Plaza, Tokyo, Japan).
Enzymatic amplification was performed by PCR using Master Taq polymerase enzyme supplied by Fermentas (LT-02241 Vilnius, Lithuania) and Hybaid thermal cycler (Promega Corporation, 2800 Woods Hollow Road, Madison, USA).
Amplification of the promoter region (− 509 C>T) of the TGF-β1 gene (rs 1800469) as proposed by Falleti et al. (Falleti et al., 2008) using forward primer: 5′- GGAGAGCAATTCTTACAGGTG -3′ and reverse primer: 5′-TAGGAGAAGGAGGGTCTGTC -3′ provided by Operon Biotechnologies GmbH (Nattermannallee 1, 50674, Cologne, Germany).
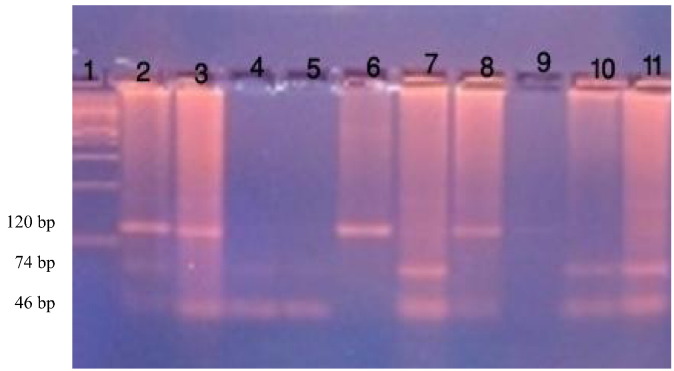
The PCR reaction mixture (25 μl) contained 12.5 μl 2 × PCR Master Mix {10 × PCR buffer, 4 mM MgCl2, 0.5 Taq DNA polymerase/μl, 0.4 mM dNTPs (dATP, dCTP, dGTP, dTTP)} 1 μl of each primer (25 pmol), 2.5 μl of genomic DNA and 8 μl sterilized nuclease-free water. The reaction was carried out with the following cycles; 95 °C for 5 min; 35 cycles of 95 °C for 1 min, 58 °C for 1 min and 72 °C for 1 min; and 72 °C for 10 min for final extension. Amplified bands (120 bp) were detected by electrophoresis on 1.5% agarose gel containing ethidium bromide and visualized by UV transillumination (Promega, USA) (Fig. 1).
Fig. 1.
Electrophoresis in 3% agarose gel after staining with ethidium bromide showing PCR-RFLP analysis of the TGF-β1 − 509 C/T polymorphism. Lane 1: 100 bp ladder, Lanes 2, 3: CT genotype, Lanes 4, 5, 7, 10, 11: CC genotype, Lanes 6, 8: TT genotype, Lane 9: no product.
The amplified products (8 μl) were digested with 5 units Fast Digest DdeI restriction enzyme (supplied by Fermentas, Lithuania, www.thermoscientificbio.com/fermentas) at 37 °C for 15 min. The digested products were then visualized by 3.5% agarose gel electrophoresis for identification of TGF-β1 (− 509 C>T) promoter gene polymorphism. Two bands at 74 bp and 46 bp identified CC homozygous wild pattern, three bands at 120 bp, 74 bp, and 46 bp identified CT heterozygous mutant pattern, one band at 120 bp identified TT homozygous mutant pattern, as shown in Fig. (1).
Statistical analysis
Data obtained from the study was coded and entered using SPSS software package version 15.0 (Echo soft Corporation, USA). Qualitative data was expressed as frequency and percentage (%). Comparison between qualitative data was done by Chi squared test. Parametric data was summarized using mean and standard deviation, while non-parametric data was summarized as median and percentiles for quantitative variables. t-Test and non-parametric Mann–Whitney test were used to compare two groups, while ANOVA and nonparametric test (Kruskal Wallis test) were used to compare multiple groups. Pearson correlation was used to correlate quantitative variables. Risk estimate was calculated by Odds Ratio (OR). The p-value was considered significant at < 0.05.
Results
A significant positive correlation was observed between serum TGF-β1 and alkaline phosphatase (r = 0.559, p < 0.05), AST (r = 0.573, p < 0.05). A significant negative correlation was observed between serum TGF-β1 and albumin (r = − 0.331, p < 0.05).
Discussion
In this present study, the mean serum level of TGF-β1 was significantly higher in liver cirrhosis group than in control group (p < 0.05). These results were in agreement with Okumoto et al. (2004) who found that plasma TGF-β1 concentrations in patients with cirrhosis (median, 3.5 ng/ml; range, 0.9–9.5 ng/ml) were significantly higher than in healthy subjects (median, 2.0 ng/ml; range, 0.6–3.2 ng/ml; p < 0.05).
Also in the current study it was found that mean serum level of TGF-β1 was significantly higher in subjects with TGF-β1 − 509 TT than CT and CC genotypes; p < 0.05 (Table 2).
Table 2.
Distribution of TGF-β1 − 509 genotypes among studied groups.
| TGF-β1 − 509 genotypes | Cirrhotic group (n = 40) | Control group (n = 40) | p-Value |
|---|---|---|---|
| CC | 9 (22.5%) | 33 (82.5%) | < 0.05 |
| CT | 21 (52.5%) | 4 (10%) | |
| TT | 10 (25%) | 3 (7.5%) |
Strategies aimed at disrupting TGF-β1 synthesis and/or signaling pathways markedly decreased fibrosis in experimental models (Bataller and Brenner, 2005). Accordingly, tissue and serum levels of TGF-β1 are elevated in fibrosis and over-expression of TGF-β1 in transgenic mice and application of exogenous TGF-β1 can induce organ fibrosis (Kopp et al., 1996). In patients with chronic HCV infection it has been shown that the viral core protein up-regulates TGF-β1 transcription, thus exacerbating liver fibrosis progression (Falleti et al., 2008, Taniguchi et al., 2004).
Our study showed that TGF-β1 − 509 CT and TT genotype frequencies were significantly higher in cirrhotic group (52.5%, 25%; respectively) than control group (10%, 7.5%, respectively); p < 0.05.
Logistic regression analysis revealed that − 509 CT and TT genotypes are more likely to be associated with high risk of developing liver cirrhosis; OR = 16.238 (95% CI 5.391–48.914, p < 0.05). The − 509 T allele carriers are 7 times more prone to develop liver cirrhosis than − 509 C allele carriers; OR = 7.359 (95% CI 3.325–16.288, p < 0.05).
This was in agreement with the study done by Falleti et al. (2008), which was conducted on 188 Italian patients transplanted for liver cirrhosis and a 140 healthy control subjects, they found that CT and TT genotypes were significantly higher in cirrhotic group (45.2%, 28.2%; respectively) than control group (32.4%, 11.7%; respectively); p < 0.05. Also, they reported that T allele frequency was significantly higher in cirrhotic group (51%) than in control group (37%). Thus it indicates that the mutant risky T allele was found to be a significant risk factor for the development of liver cirrhosis; OR = 1.721 (95% CI 1.256–2.358, p < 0.05) (Table 3).
Table 3.
Comparative study of laboratory data between TGF-β1 − 509 genotypes.
| CC (n = 42) | CT (n = 25) | TT (n = 13) | p-Value | |
|---|---|---|---|---|
| T. bilirubin (mg/dl)a | 0.4 | 0.99 | 1.35 | < 0.05 |
| (0.27–0.68) | (0.43–1.6) | (0.67–3.18) | ||
| a | b | c | ||
| AST (U/L)a | 14 | 27 | 12 | > 0.05 |
| (12–18.5) | (16.5–51) | (10.5–25.5) | ||
| a | a | a | ||
| ALT (U/L)a | 13 | 46 | 34 | > 0.05 |
| (11–18) | (24.5–61) | (14.5–62.5) | ||
| a | a | a | ||
| ALP (U/L)a | 51 | 77 | 67 | > 0.05 |
| (50–66) | (59–134) | (52–99) | ||
| a | a | a | ||
| Albumin (g/dl)b | 3.87 ± 0.91 | 3.15 ± 0.89 | 2.74 ± 0.91 | < 0.05 |
| a | b | b | ||
| TGF-β1 (ng/ml)b | 7.9 ± 0.62 | 9.27 ± 0.65 | 11.9 ± 1.19 | < 0.05 |
| a | b | c | ||
| PT (second)a | 12 | 16.7 | 16.9 | < 0.05 |
| (11.5–12) | (13–19.15) | (12.5–19.3) | ||
| a | b | b | ||
| PC (%)a | 94 | 63 | 59 | < 0.05 |
| (89.8–98) | (47.6–80) | (43.2–84.5) | ||
| a | b | b | ||
| INRb | 1.14 ± 0.28 | 1.53 ± 0.46 | 1.42 ± 0.32 | < 0.05 |
| a | b | b |
Groups bearing different initials are significantly different from each other at p = 0.05.
Data presented as median (25th–75th percentile).
Data presented as mean ± SD. Groups bearing same initials are not significantly different from each other at p = 0.05.
In accordance with the current study, an Egyptian study done by Hanafy and Abdo (2011), who found that the genotype and allele distributions of TGF-β1 − 509 C>T SNPs; were significantly different between the chronic HCV patients and the control group, they reported that the proportions of TT (35.5%) and CC (18.3%) among patient group are different from control group (22.1%, 29.4% respectively). With regard to allelic variance, the frequency of T allele was significantly higher in HCV patients (58.3%) than control group (46%) p = 0.05. Dai et al.'s (2008) study which was conducted on Taiwanese HCV patients, showed similar results.
In Germany, Gewaltig et al. (2002) carried a study on the association of SNPs in the TGF-β1 gene − 509 C>T and the rate of progression of HCV-induced fibrosis. They concluded that the homozygous TT genotype of − 509 C>T showed a slow progression of fibrosis (Table 4).
Table 4.
Risk estimate of TT and CT genotypes.
| TGF-β1 − 509 genotypes | Cirrhotic group (n = 40) | Control group (n = 40) | OR | 95% CI | p-Value |
|---|---|---|---|---|---|
| CC | 9 (22.5%) | 33 (82.5%) | 16.238 | (5.391–48.914) | < 0.05 |
| CT + TT | 31 (77.5%) | 7 (17.5%) |
Falleti et al. (2008) reported that the TGF-β1 high producers − 509 T and Arg25Pro C were found to be more represented in cirrhotic patients rather than in control subjects. These observations suggest a role of TGF-β1 SNPs, present both in the promoter and in the coding regions, in causing accelerated liver fibrogenesis in patients with different types of liver disease, not only of viral origin. Specific genotypes at the codons 10 and 25 have been associated with faster fibrosis progression in patients with chronic hepatitis C and hereditary hemochromatosis (Gewaltig et al., 2002, Osterreicher et al., 2005).
The results of Li et al. (2007) were in contrast to ours and revealed that the frequency of TGF-β1 − 509 CC genotype was higher in liver cirrhosis (73.5%) than control group (37.7%), p < 0.05, but there was no significant difference in allele frequency among studied groups. Also other Chinese studies done by Yang et al. (2005) and Wang et al. (2008) were contradictory to the current results. They reported that the C allele at − 509 may play an important role in the progression of liver cirrhosis than the T allele. Romani et al. (2011) found no significant difference in the allelic frequency distribution of SNPs at − 509 C/T between HCV patients and the healthy controls (Table 5).
Table 5.
Risk estimate of T allele.
| Allele | Cirrhotic group (n = 80) | Control group (n = 80) | OR | 95% CI | p-Value |
|---|---|---|---|---|---|
| C (protective allele) | 39 (48.7%) | 70 (87.5%) | 7.359 | (3.325–16.288) | < 0.05 |
| T (risky allele) | 41 (51.3%) | 10 (12.5%) |
Conclusion
There is an association between serum TGF-β1, − 509 CT and TT genotypes of TGF-β1 gene and the higher risk for liver cirrhosis development of liver cirrhosis. Further studies are, however, needed to confirm and extend these results.
References
- Bataller R., Brenner D.A. Liver fibrosis. J. Clin. Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell D.M. Chronic liver injury, TGF-β, and cancer. Exp. Mol. Med. 2001;33:179–190. doi: 10.1038/emm.2001.31. [DOI] [PubMed] [Google Scholar]
- Dai C.Y., Chuang L.P., Lee W.C., Pan J.F., Huang M.Y., Hsieh N.J., Hou Z.Y., Lin S.C., Chen M.Y., Hsieh L.Y., Wang W.Y., Chang M.L. Association between transforming growth factor-beta 1 polymorphism and virologic characteristics of chronic hepatitis C. Transl. Res. 2008;152(4):151–156. doi: 10.1016/j.trsl.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Dong Z.Z., Yao D.F., Tao M., Qiu L.W., Zong L., Wu W., Wu X.H., Yao D.B., Meng X.Y. Clinical impact of plasma TGF-β1 and circulating TGF-β1 mRNA in diagnosis of hepatocellular carcinoma. Hepatobiliary Pancreat Dis. Int. 2008;7(3):288–295. [PubMed] [Google Scholar]
- Falleti E., Fabris C., Toniutto P., Fontanini E., Cussigh A., Bitetto D., Fornasiere E., Aveilini C., Minisini R., Pirisi M. TGF-β1 genotypes in cirrhosis: relationship with the occurrence of liver cancer. Cytokine. 2008;44:256–261. doi: 10.1016/j.cyto.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Gewaltig J., Mangasser-Stephan K., Gartung C., Biesterfeld S., Gressner A.M. Association of polymorphisms of the transforming growth factor-beta1 gene with the rate of progression of HCV-induced liver fibrosis. Clin. Chim. Acta. 2002;316:83–94. doi: 10.1016/s0009-8981(01)00738-0. [DOI] [PubMed] [Google Scholar]
- Gines P., Cardenas A., Arroyo V., Rodes J. Management of cirrhosis and ascites. N. Engl. J. Med. 2004;350:1646–1654. doi: 10.1056/NEJMra035021. [DOI] [PubMed] [Google Scholar]
- Hanafy S.M., Abdo A. Impact of single nucleotide polymorphism of TGF-β1 gene (SNP-codon 10) on hepatocellular carcinoma risk in Egyptian patients following HCV infection. Aust. J. Basic Appl. Sci. 2011;5(9):1814–1821. [Google Scholar]
- Kopp J.B., Factor V.M., Mozes M., Nagy P., Sanderson N., Bottinger E.P. Transgenic mice with increased plasma levels of TGF-β1 develop progressive renal disease. Lab. Invest. 1996;74:991–1003. [PubMed] [Google Scholar]
- Li H., Wu H.L., Lv H., Wei H.S., Wang H.B., Wang P.L. The association of TGF-β1 and AT1R gene polymorphisms with hereditary susceptibility and clinical phenotype of HBV-induced liver cirrhosis. Zhonghua Yi Xue Za Zhi. 2007;24(3):298–301. [PubMed] [Google Scholar]
- Matsuzaki K., Murata M., Yoshida K., Sekimoto G., Uemura Y., Sakaida N. Chronic inflammation associated with hepatitis C virus infection perturbs hepatic transforming growth factor beta signaling, promoting cirrhosis and hepatocellular carcinoma. Hepatology. 2007;46:48–57. doi: 10.1002/hep.21672. [DOI] [PubMed] [Google Scholar]
- Nawar E.A., Abul-Fadl A.M., Hassanin B.D., Abdel Haie O.M., El-Tokhy M. Clinical value of transforming growth factor beta as a marker of fibrosis in adolescents with chronic liver diseases. J. Am. Sci. 2011;7(3):464–472. [Google Scholar]
- Okumoto K., Hattori E., Tamura K., Kiso S., Watanabe H., Saito K. Possible contribution of circulating transforming growth factor-beta1 to immunity and prognosis of unresectable hepatocellular carcinoma. Liver Int. 2004;24:21–28. doi: 10.1111/j.1478-3231.2004.00882.x. [DOI] [PubMed] [Google Scholar]
- Osterreicher C.H., Dhtz C., Stickel F., Hellerbrand C., Penz M., Hofer H. TGF-β1 codon 25 gene polymorphism is associated with cirrhosis in patients with hereditary hemochromatosis. Cytokine. 2005;31:142–148. doi: 10.1016/j.cyto.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Romani S., Azimzadeh P., Mohebbi S.R., Kazemian S., Almasi S., Naghoosi H., Derakhshan F., Zali M.R. Investigation of transforming growth factor-beta 1 gene polymorphisms among Iranian patients with chronic hepatitis C. Hepat. 2011;11(11):901–906. doi: 10.5812/kowsar.1735143X.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H., Kato N., Otsuka M., Goto T., Yoshida H., Shiratori Y. Hepatitis C virus core protein upregulates transforming growth factor-beta 1 transcription. J. Med. Virol. 2004;72:52–59. doi: 10.1002/jmv.10545. [DOI] [PubMed] [Google Scholar]
- Wang H., Zhao Y.P., Gao C.F., Ji Q., Gressner A.M., Yang A.X. Transforming growth factor-beta 1 gene variants increase transcription and are associated with liver cirrhosis in Chinese. Cytokine. 2008;43(1):20–25. doi: 10.1016/j.cyto.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Waqar A., Ijaz B., Gull S., Asad S., Khalid S., Sarwar M.T., Sumrin A., Shahid I., Hassan S. Molecular, genetic and imaging techniques for HCV fibrosis evaluation. Virol. J. 2011;8:53–57. doi: 10.1186/1743-422X-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.X., Wang H., Gao C.F., Xu L.L., Zhao W.J. Effect of polymorphism of transforming growth factor-beta 1 gene on HBV-induced liver cirrhosis. Zhonghua Yi Xue Za Zhi. 2005;85(15):1021–1026. [PubMed] [Google Scholar]