Abstract
The promoter region of kappa-casein (κ-CN) gene in Indian native cattle and buffalo breeds was sequenced and analyzed for nucleotide variations. Sequence comparison across breeds of Indian cattle revealed a total of 7 variations in the promoter region, of which − 515 G/T, − 427 C/T, − 385 C/T, − 283 A/G and − 251 C/T were located within consensus binding sites for octamer-binding protein (OCT1)/pregnancy specific mammary nuclear factor (PMF), activator protein-2 (AP2), hepatocyte nuclear factor (HNF-1) and GAL4 transcription factors (TFs), respectively. These variations might be involved in gain or loss of potential transcription factor binding sites (TFBSs). Unlike the other 4 variants, the − 283 (A/G) variant located within HNF-1 TFBS was specific to Indian cattle as this change has not been observed in the Bos taurus sequence. Other TFBSs viz., MGF, TBP, NF-1, milk box and C/EBP were conserved across species. For the Indian native buffalo breeds, only 3 changes were identified in the promoter region; − 305 (A/C), − 160 (T/C) and − 141 (A/G) and most of the TFBSs were found to be conserved. However, deletion of two adjacent nucleotides located in and around binding site for C/EBP TF was identified in buffalo when compared with promoter sequence of bovine κ-CN. For κ-CN of Indian native cattle, a strong linkage disequilibrium (LD) was observed for variations 515 G/T, − 427 C/T and − 385 C/T in the promoter region; and for variations at codons 136 and 148 of exon-IV. Further, among intragenic haplotypes, variation − 427 C/T was found to be in LD with variations at codons 136 and 148. The information generated in the present work provides comprehensive characterization of κ-CN gene promoter and coding regions in Indian cattle and buffaloes and reported variations could become important candidates for carrying out further research in dairy traits.
Keywords: Kappa casein, Molecular characterization, Genetic variability, Promoter region, Coding sequences, Linkage disequilibrium
Graphical abstract
Variation analysis of regulatory and coding regions of kappa casein gene in Indian cattle and buffalo breeds of India
Highlights
-
•
The κ-casein gene (κ-CN) and its promoter region was sequence characterized in 15 Indian native cattle and 8 buffalo breeds.
-
•
Among the identified variations, four were located within TFBSs in Indian cattle, while all TFBSs were conserved in buffaloes.
-
•
The κ-CN CDS was highly conserved with only 3 and 2 non-synonymous changes for Indian cattle and buffaloes, respectively.
-
•
Across promoter and CDS, − 427 C/T was found to be in LD with variation at codons 136 and 148.
Introduction
Bovine milk is an important source of protein in several parts of the world including Asia, Africa and Europe. The major portion (80%) (Haug et al., 2007) of bovine milk protein is constituted by a casein (CN) cluster encoded by four tightly linked genes: αs1-CN (CSN1S1), β-CN (CSN2), αs2-CN (CSN1S2) and κ-CN (CSN3), located within a 250 kb segment of bovine autosome 6 (Caroli et al., 2009). Among these, calcium insensitive kappa-casein (κ-CN) is a 13 kb gene with 5 exons and 4 introns and is thought to be genetically divergent from other calcium sensitive caseins (αs1-, β- and αs2-CN) (Ginger et al., 1999). The κ-CN constitutes about 12% of casein protein. It stabilizes calcium sensitive caseins against precipitation, plays an important role in determining the size of casein micelles and upon cleavage by chymosin, and initiates micelle aggregation resulting in curd and cheese production (Threadgill et al., 1990). The κ-CN gene has been extensively studied in several species and eleven variants of this gene have been identified in cattle (Farrell et al., 2004). Among these, A and B are the most common protein variants that differ at codons 136 (A: Thr, ACC and B: Ile, ATC) and 148 (A: Asp, GAT and B: Ala, GCT) of exon-IV (Ron et al., 1994). Recent analysis had indicated the B variant to be significantly associated with content and yield of milk protein and fat, as well as having a role in curd and cheese making properties (Heck et al., 2009). Among the various dairy products, curd/cheese production occupies a huge market and hence it is important to study κ-CN and its variants that influence the manufacturing properties. In recent years, along with the coding region, variations in the promoter region of casein genes have also attracted research interest due to their influence on expression of milk protein genes. Variations in the flanking region individually or in combination with the coding region can affect the quality and quantity of milk (Caroli et al., 2009, Hoogendoorn et al., 2003, Martin et al., 2002, Pauciullo et al., 2013).
Sequence characterization of the promoter region of κ-CN in taurine cattle has revealed several variations involving putative transcription factor binding sites (TFBSs) (Adachi et al., 1996, Kaminski, 2000, Schild et al., 1994). These variations affect the gene transcript individually as well as in combination with other variations in the promoter or coding region of the casein gene in question (intragenic haplotypes) or other caseins (intragenic haplotypes) (Hobor et al., 2008, Kaminski, 2000, Keating et al., 2007, Schild et al., 1994). Such variations (intra/intergenic) and their association with milk traits have been well established in Bos taurus (Keating et al., 2007), however, such information is scanty in the Indian native cattle (Bos indicus) and buffalo (Bubalus bubalis) breeds. Only few reports are available on the polymorphism analysis in the Indian native cattle breeds and these also are restricted to the coding region (Sodhi et al., 2010). Moreover, no study has been undertaken to present a holistic profile of κ-CN promoter variants and their possible linkage with variants in the coding region in Indian cattle and buffaloes. The naturally evolved Indian native cattle and buffalo possess a unique gene pool and differ from their taurine counterparts in terms of adaptability, immunity and dairy traits. In terms of milk production, compared to exotic breeds, Indian cattle breeds have relatively low milk yield but are rich in fat and protein content while certain buffalo breeds (Murrah, Bhadawari) exhibit high milk yield as well as high fat percentage (Nivsarkar et al., 2000). Considering the significant role that the promoter region plays in influencing gene expression, the present study was undertaken to identify variations in the promoter as well as the coding region of κ-CN among different Indian native cattle and buffaloes. Analysis of linkage disequilibrium (LD) among coding and promoter variants was also carried out to identify intragenic haplotypes.
Materials and methods
Animal selection and DNA isolation
To determine the κ-CN gene diversity in Indian native cattle and buffalo breeds, blood samples of genetically unrelated animals were collected from the native breeding tracts. The only exception was the Red Sindhi breed which does not have any specific breeding tract owing to its low population size and blood samples were collected from organized farms located in the southern and central regions of India. A total of 122 animals representing 15 Indian native cattle breeds (sample size — 80) and 8 buffalo breeds (sample size — 42) were undertaken for the study. The breeds included in the study were of different aptitudes: dairy cattle (Gir, Tharparkar, Rathi, Red Sindhi, and Sahiwal); dual purpose cattle (Deoni, Gaolao, Hariana, Kankrej and Mewati); draft cattle (Amritmahal, Kangayam, Malnad Gidda, Red Kandhari and Umblachery) and dairy buffalo (Murrah, Jaffrabadi, Mehsana and Bhadawari), draft purpose buffalo (Assamese swamp, Chilika, Manipuri and Toda-semiwild). Genomic DNA was isolated by proteinase-K (Sigma, USA) digestion followed by standard phenol–chloroform extraction (Sambrook et al., 1989).
PCR amplification and sequence characterization of κ-CN 5′-flanking and coding regions
Genomic DNA from 80 Indian native cattle and 42 riverine buffaloes were amplified for sequence characterization of κ-CN 5′-flanking region (κ-CN5′) using two sets of overlapping primers P1 and P2 (Table 1). Moreover, the complete coding sequence (CDS) region including 5′-untranslated region (UTR), four exons and 3′-UTR of the buffalo κ-CN gene was amplified using five primer sets (C1–C5) (Table 1). These animals were also genotyped for non-synonymous (dN) mutations at exon-IV of the coding region using primer set G1.
Table 1.
Primer sets and their respective primer sequence for PCR amplification of κ-CN gene and its 5′-flanking region.
| Primers | Primer sequence | Annealing temp (°C) | Amplicon (bp) |
|---|---|---|---|
| Primer sets for amplification and sequencing of 5′-flanking region | |||
| P1 | F: 5′-CACTGTTGGTGGCAATGTAA-3′ R: 5′-AAGGGTGTTAGTGTCTTGCG-3′ |
59 | 567 |
| P2 | F: 5′-GGATCCCTA CTTTATATT-3′ R: 5′-CTTCCACTGTTAAGGAA-3′ |
46 | 687 |
| Primer sets for amplification and sequencing the coding region | |||
| C1 | For1: 5′-CCTCTGCATTCCATTAACCG-3′, Rev 1: 5′-GGGATCAATCTGGATTATGC-3′; |
60 | 600 |
| C2 | For 2: 5′-TCACATTGGCTATATCTCCC-3′, Rev 2: 5′-GAGACCAGAAGAATATCCAG-3′ |
58 | 370 |
| C3 | For 3: 5′-TGCCTTCTCTGTCACAGACT-3′, Rev 3: 5′-TCAACACGACTTGGCCTGTA-3′ |
60 | 444 |
| C4 | For 4: 5′-TTCACTCTGCTTCTGCTGCT-3′, Rev 4: 5′-ATTAGCCCATTTCGCCT TCT-3′ |
60 | 779 |
| C5 | For: 5′-AGGACTGTGTCTTTCTGTGA-3′, Rev: 5′-TCACGTTGATGTGTGGTA GA-3′ |
57 | 467 |
| Primer set for PCR-RFLP based genotyping (exon IV/HindIII–HaeIII loci) | |||
| G1 | For: 5′-AGCGCTGTGAGAAAGATG-3′ Rev: 5′-GTGCAACAACACTGGTAT-3′ |
55 | 935 |
PCR amplifications were performed in a 25 μl reaction mixture containing 100–200 ng of genomic DNA, 5 pmol of each primer, 200 μM of dNTP mix, 1.5 mM MgCl2 and 1.0 U Taq DNA polymerase (Invitrogen, Brazil). Amplification was performed in a thermal cycler (Eppendorf, Germany) for 30 cycles with specific annealing temperature for different primer sets (Table 1). PCR products were electrophoresed in 1.5% ethidium-bromide stained agarose gel and visualized under a UV-transilluminator. The PCR products of the promoter region and CDS including the untranslated regions (5′-UTR and 3′-UTR) were purified and sequenced using an ABI Prism® BigDye™ Terminator Cycle Sequencing Kit (version 3.1; Applied Biosystems, Foster City, CA). The resulting sequences were aligned using phrap assembler and phred basecaller program in CodonCode Aligner software (CodonCode Corporation, Dedham, MA, USA) and polymorphic sites were confirmed by visual inspection.
The transcription factor binding sites (TFBSs) were identified using TESS (http://www.cbil.upenn.edu/cgi-bin/tess/tess), MATCH (Kel et al., 2003), TRANSFAC (Matys et al., 2003) and AliBaba2.1 Search engine (Grabe, 2002). The haplotypic sequences were inferred using PHASE v2.1.1 software (http://www.stat.washington.edu/stephens/software.html). Molecular Evolutionary Genetic Analysis (MEGA) Software Version 5.0 (Tamura et al., 2011) was used for the comparative sequence analysis and phylogenetic sequence analyses employing the Neighbor-joining (NJ) method. This method does not require the assumption of a constant rate of evolution. Distances were estimated by the p-distance model and the standard errors of the estimates were obtained through 1000 bootstrap replicates.
Genotyping at exon-IV and its LD analysis with κ-CN5′ variants
For PCR-RFLP based genotyping of κ-CN at codons 136 and 148 of the exon-IV region, aliquots of 10 μl PCR products were digested separately each with 1.0 U of HindIII and HaeIII restriction enzymes and incubated at 37 °C for 4 h to differentiate the A and B alleles as described by Sodhi et al. (2010). Estimation of LD among variations identified in the promoter region (κ-CN5′) and the coding region at the HindIII/HaeIII locus in exon-IV was carried out using Haploview v 4.1 (Barrett et al., 2005).
For determination of phylogenetic relationship of κ-CN5′ among different mammalian species, nucleotide sequences from the GenBank and Ensemble databases include B. taurus (M755887), Ovis aries (L31372), Capra hircus (Z33882), Homo sapiens (ENSG00000171209), Pan troglodytes (ENSPTRG000000 16126), Macaca mulatta (ENSMMUG00000006031), Equus caballus (AY579426), Equus asinus (ZGEU429803), Equus zebra (EU429802), Mus musculus (AJ309571), Rattus norvegicus (ENSRNOG00000001951), Oryctolagus cuniculus (AJ309572), B. bubalis (AJ628346), Camelus dromedarius (AJ409280) and Sus scrofa (ENSSSCG0000000 9267) were used. Phylogeny for κ-CN5′ was constructed based on genetic distance (Fst values) using ‘MEGA5’ (Tamura et al., 2011).
Results and discussion
The analysis of κ-CN5′ is important to understand the role of factors involved in the regulation of milk protein gene expression and the transcriptional effects of polymorphisms located in such regions. In the present study, the − 565 bp (upstream to transcription start site) sequence of the promoter region followed by 5′-UTR (69 bp), CDS (573 bp) and 3′-UTR (207 bp) was sequence characterized for analysis of genetic variability at κ-CN gene among Indian native cattle and buffaloes.
In the κ-CN5′, the search for putative TFBSs revealed a total of 20 regulatory binding domains. Among these, TFBSs characteristic of the casein gene regulatory region such as, the binding site for TATA and CAAT box, binding sites for transcriptional activators [Glucocorticoid Receptor (GR), Nuclear Factor-1 (NF-1), Mammary Gland Factor (MGF) also known as a signal transducer and activator of transcription 5 (STAT5)] and repressors [Ying Yang factor (YY1) and pregnancy specific Mammary Nuclear Factor (PMF)] (Lenasi et al., 2005) were present. Besides these, other potential TFBSs viz., activator protein-2 (AP-2), CCAAT/Enhancer Binding Protein C/EBP), cAMP-Response Element Binding Protein (CREB), c-Jun, GAL4, GATA-binding factor 1 (GATA-1), Hepatocyte Nuclear Factor 1 (HNF1), Heat Shock transcription Factor 1 (HSF1), octamer-binding factor 1 (OCT-1), transcription factor 1 (PIT1, growth hormone factor 1) (POU1F1a), POU domain-class 2 transcription factor 1 (POU2F1), TATA-box binding protein (TBP) and transcription factor IID (TFIID) were identified. The conserved TATA box was located from − 25 to − 32 bp and consensus sequence of CAAT box was between − 75 and − 78 bp upstream from the transcriptional start site (Fig. 1a, Supplementary Tables S1a and S1b). The position of TATA box in cattle and buffalo was consistent with that of humans and murines.
Fig. 1.
a: Nucleotide sequence of κ-CN5′ and its putative TFBSs among Indian native cattle. Alphabets marked by arrow indicate site of variation, in comparison to B. taurus sequences (M75887). Variations in Indian native cattle are represented in nucleotide IUPAC codes. Variations in Indian native buffalo are shown in boxes, whereas the site of deletion in buffalo sequences is shaded. Positions are marked with respect to transcription start site.
b: Schematic representation for variations within κ-CN5′ between cattle and buffalo. The upper lane indicates variation between B. indicus and B. taurus (M75887), while the lower lane indicates variation between B. indicus and Indian buffalo (Bubalus bubalis).
Polymorphism analysis within the κ-CN5′-flanking and coding regions of Indian native cattle
Fifteen Indian native cattle breeds representing milk, dual-purpose and draft-type were evaluated for sequence variation in κ-CN5′. Clustal analysis of the sequences revealed seven polymorphic sites at positions − 559 (T/A), − 558 (C/T), − 515 (K: G/T), − 427 (Y: C/T), − 385 (Y: C/T); − 283 (R: A/G) and − 251 (Y: C/T) across the Indian native cattle samples (Table 2, Fig. 1a). The transition/transversion rate ratios were observed to be k1 = 0 for purines and k2 = 9.65 for pyrimidines with an overall transition/transversion bias of R = 1.596. These variations corresponded to a total of seven haplotypes in the κ-CN5′ (Supplementary Table S2). However, none of the variations or haplotypes were specific to a breed or a specific aptitude. This suggested that variations of κ-CN5′ with inter- and intragenic haplotypes of other milk protein genes might be responsible for different protein contents across various breeds and thus it may be used in combination with variations of other casein genes (interhaplotypic combination) along with their regulatory regions.
Table 2.
Variations within κ-CN5′ detected among the Indian native cattle in comparison to B. taurus (accession no. M75887) and their putative TFBSs along with their restriction site involved.
| Position | Allele | TFBSs | Restriction site | Reported by earlier workers |
|---|---|---|---|---|
| − 559 | T/A | – | – | – |
| − 558 | C/T | – | – | – |
| − 515 | T/G | OCT1 | – | Keating et al. (2007) and Schild et al. (1994) |
| − 427 | T/C | PMF | BclI | Keating et al. (2007) and Schild et al. (1994) |
| − 385 | T/C | AP2 | DdeI | Keating et al. (2007), Kaminski et al. (2000) and Schild et al. (1994) |
| − 283 | G/A | HNF1 | – | – |
| − 251 | T/C | GAL4 | – | Schild et al. (1994) |
The nucleotide variations observed in Indian cattle at positions − 515 G/T, − 427 C/T, − 385 C/T and − 251 C/T in zebu cattle have also been reported in B. taurus breeds (Kaminski, 2000, Keating et al., 2007, Schild et al., 1994). Mutations within the promoter region play a key role in alteration of transcriptional efficiency and thus influence gene expression and performance traits of animal (Kaminski, 2004). Further, functional characterization of bovine κ-CN5′ sequences (− 552 to + 18 bp) in mouse cell lines has suggested that polymorphism in the region − 439 to − 308 bp has pregnancy-specific TFBSs, whereas polymorphism in region − 307 to − 125 bp contains lactation specific TFBSs (Adachi et al., 1996). Therefore, nucleotide variations at positions − 283 A/G and − 251 C/T could possibly be involved in regulating the total protein/milk yield.
Moreover, variations at positions − 515 G/T, − 427 C/T and − 385 C/T have been reported (Adachi et al., 1996, Keating et al., 2007) to have a potential role in gain or loss of TFBSs since these are located within putative binding site for OCT1/PMF, PMF and AP-2 transcription factors (TFs), respectively. Besides, variations observed at positions − 283 (R: A/G) and − 251 (Y: C/T) were located within the binding site for HNF-1 and GAL4 TFs, respectively (Table 2). Among these, the variation − 283 (R: A/G) is specific to B. indicus and may possibly change specificity for binding of HNF-1, but its role in terms of difference in milk production from B. taurus is not clear. The other TFBSs such as MGF, TBP, NF-1, milk box and C/EBP remained conserved across the studied breeds of Indian native cattle (Fig. 1a).
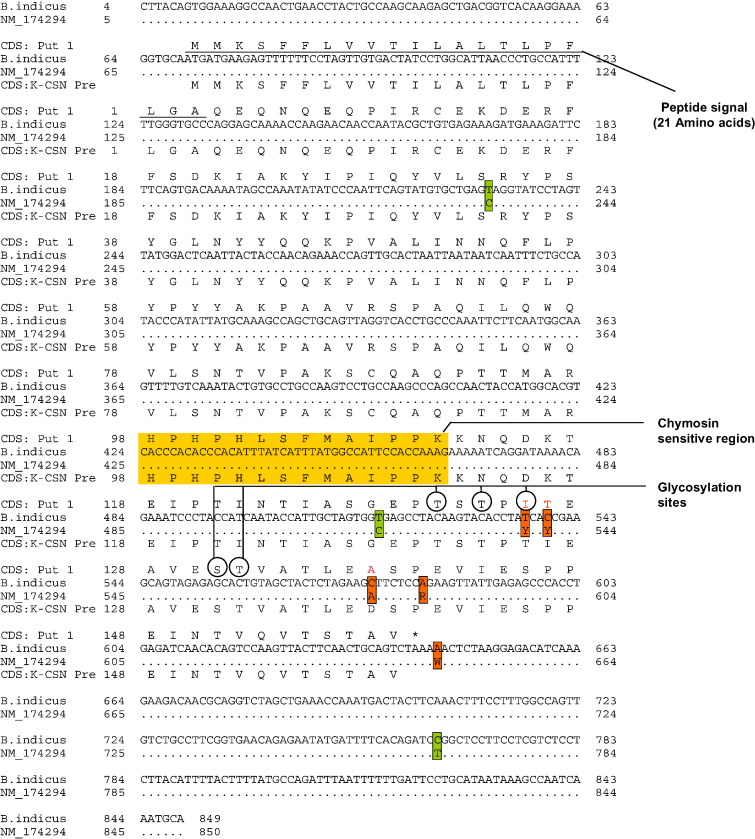
Clustal analysis for the 849 bp region comprising of 5′-UTR, CDS and 3′-UTR of κ-CN revealed five variations within Indian cattle breeds. Of these, four variations (536 T/C, 539 C/T, 575 C/A and 582 A/G) were present in the exon IV region and one (644 A/T) in the 3′-UTR region (Fig. 2). Variations at 536, 539 and 575 corresponded to non-synonymous (dN) change at codon 135 Ile/Thr, 136 Thr/Ile and 148 Ala/Asp, respectively. The point mutation at codons 135, 136 and 148 within exon-IV are considered to be important as they are located relatively closer to several glycosylation sites (amino acids at positions 131, 133 and 135 or 141, and 142) (Fig. 3) and could possibly affect the structure of protein and its glycosylation patterns (Creamer et al., 1998).
Fig. 2.
Variations in the κ-CN gene within Indian cattle and buffalo population.
Fig. 3.
Comparative sequence analysis of κ-CN gene among Indian cattle and taurine counterpart (NM_174294).
Moreover, sequence comparison of Indian cattle κ-CN CDS region with B. taurus sequence (GenBank accession: NM_174294) revealed a single deletion (A/−) (between 3 and 4 bp; not shown in Fig. 3) and eight variations: six in exon-IV (231 C/T, 516 C/T, 536 C/T, 539 T/C, 575 A/C and 582 G/A) and two in 3′-UTR (644 T/A and 764 C/T) in addition to the variations observed within Indian native cattle (Fig. 3). Out of these, the deletion and three variations at 231, 516 and 764 bp were found to be B. indicus specific. Genotype analysis of non-synonymous variations in exon-IV in Indian native cattle breeds (Sodhi et al., 2010) has indicated contrasting allelic pattern at the κ-CN locus in Indian native cattle in comparison to that reported for B. taurus. The allele A was predominant (0.908) in Indian cattle and mean frequency of allele B that has been reported as desirable allele for dairy trait (Stevanovic et al., 2000) was however substantially low (0.092).
Kaminski et al. (2000) have observed an intragenic haplotype of A/B alleles in the exon-IV with that of P/M allele in the regulatory region. The variation − 385 (Y: C/T) was genotyped across 124 dairy cattle with the DdeI restriction enzyme and three genotypes were observed (PP, PM and MM). The heterozygous genotype ‘PM’ of the regulatory region in association with the AA genotype of exon-IV (intragenic haplotype ‘PM/AA’) was observed to be associated with high protein percentage. Conversely, in studied Indian cattle breeds, 89% (71/80) animals had AA genotype but none of the individual revealed PM genotype (− 385 C/T) in the regulatory region. All these sequences might account for the different protein contents across the indicus and taurine breeds.
Analysis of LD between pairs of identified variations, varied from an almost lack of disequilibrium to complete disequilibrium, with a moderate degree of LD between the two regions. A pairwise calculation of LD using four gamete tests among 5 the variations observed in κ-CN5′ (− 515 G/T, − 427 C/T, − 385 C/T, − 283 A/G and − 251 C/T) and 2 variations in κ-CN exon-IV (539 C/T and 575 C/A) revealed two haploblocks (Fig. 4a and b). Block 1 consisted of 4 variants (− 515 G/T, − 427 C/T, − 385 C/T and − 283 A/G), while block 2 comprised of 3 variants (− 251 C/T, 539 C/T and 575 C/A). Overall three variations within κ-CN5′ (− 515 G/T, − 427 C/T, − 385 C/T) and two variations in κ-CN exon-IV (539 C/T and 575 C/A) exhibited high LD (Table 3, Fig. 4a and c). A strong LD was observed between the two variants of exon-IV with haplotype frequencies of 0.944 and 0.56 (Fig. 4d).
Fig. 4.
Pairwise measure of linkage disequilibrium (LD) relationship among the seven variations identified for haplotypes obtained for the promoter region and exon-IV/HindIII-HaeIII locus of κ-CN gene. (a) Two haploblocks were identified. Each diamond contains the level of LD measured by r2 between pairs of SNPs; darker tones correspond to increasing levels of r2; (b) haplotypes for the variations and their population frequency (light gray color). The transition from one block to the next block is displayed through frequency corresponding to the thickness of the line. Hedrick's multiallelic D′ (0.30) represents the degree of LD between the two blocks; (c) LD for variations from the promoter and exon-IV regions and (d) haplotypes for exon-IV region.
Table 3.
Linkage disequilibrium (LD) measured by r2 between pairs of SNPs observed for the promoter and coding regions of Indian native cattle κ-CN gene.
| Pair wise variations | LD values |
|---|---|
| − 515 G/T and − 427 C/T | 7.82 |
| − 515 G/T and − 385 C/T | 6.96 |
| − 427 C/T and − 385 C/T | 11.16 |
| 539 C/T and 575 C/A | 12.34 |
Polymorphism analysis within the κ-CN5′-flanking and coding regions of Indian native buffalo
Comparative sequence analysis of κ-CN5′ across indicine buffalo (riverine and swamp), indicine cattle, and B. taurus sequence (M75887) revealed a total of 19 variations (Fig. 1b). However, sequence analysis for κ-CN5′ (568 bp; − 563 to + 5 bp region) within Indian native buffaloes, revealed only three polymorphic sites at positions − 305 (M: A/C), − 160 (Y: T/C) and − 141 (R: A/G) (Fig. 1a). The transition/transversion rate ratios were k1 = 1 (purines) and k2 = 1 (pyrimidines) with an overall transition/transversion bias of R = 0.31. The observed variations corresponded to a total of six haplotypes (Supplementary Table S3). None of the variations detected in bubaline κ-CN5′ was found to be located within potential TFBSs. However, when compared with bovine (indicine as well as taurine) sequences, bubaline κ-CN5′ revealed a deletion of two adjacent nucleotides (at positions − 55 and − 54) located in and around the binding site for C/EBP TF. This might involve potential loss of binding of C/EBP TF, known to induce adipocyte differentiation. There was a 99.8% sequence homology between two water buffalo sub-species while it was 98.7% between cattle and buffalo.
Clustal analysis of the sequenced κ-CN UTRs and CDS region (849 bp) of buffalo breeds revealed three variations viz., 536 T/C, 540 T/C and 585 A/C, with all being located within exon-IV region (Fig. 2). In buffalo, variations at positions 536 and 585 bp were found to be non-synonymous causing a change at codons 135 Thr/Ile and 151 Glu/Asp, respectively. When Indian buffalo κ-CN CDS regions were compared with cattle as reference sequence, a total of 35 variations were observed, of which 68.27% were present in the exon-IV region. Out of 35 variations, 91.43% were found to be buffalo specific. The 3′-UTR region with 10 variations was comparatively more variable than 5′-UTR with a single variation (Supplementary Table S4).
Genotyping of the coding region at the HindIII/HaeIII loci within exon-IV using PCR-RFLP revealed all buffaloes as monomorphic with BB genotype. Similar to previous reports (Alipanah et al., 2008, Dayem et al., 2009, Mitra et al., 1998, Otaviano et al., 2005), protein variant B considered favorable for better milk and cheese quality was observed to be fixed in the present study. No LD was observed between the promoter and coding region variations.
There is a significant difference in the total protein, κ-CN and fat percent among cattle and buffalo with values 4.2, 3.6, 12.5 and 7.1, 4.6, 15.4% respectively. The identified variations in the regulatory and coding regions need to be studied further across large population sizes so as to pinpoint the variations responsible for the differing protein and fat contents. Sequence comparison for organization of κ-CN5′ among major dairy species (B. indicus, B. taurus, B. bubalis and C. hircus) indicated a highly conserved sequence (97.5%) (Fig. 5). Analysis for homology domain conservativeness among these species revealed differences at 9 potential TFBSs. Besides the divergence in ubiquitously expressed Oct-1 and CAAT box, differences were also observed among other important regulatory domains viz., PMF and GR TFBSs. Divergence at such important regulatory domains might reflect the possible distinctness at the functional level among different species. Analyzing the conserved nature of κ-CN5′, an approximately 400 bp region located about 800 bp upstream of the TATA box has been reported to be the most conserved across six different species (bovine, caprine, ovine, rabbit, mice and human) with two MGF/STAT5 sites to be highly conserved (Gerencser et al., 2002). Recently, the highly conserved nature of κ-CN5′ has been reported in camel (Pauciullo et al., 2013).
Fig. 5.
Homology for κ-CN5′ among the major dairy species (Bubalus bubalis, B. taurus, Capra hircus and B. indicus). Variations are highlighted and marked in boxes, whereas, gaps are represented by dashes. Only the putative TFBSs affected due to variations are marked in shaded regions.
Phylogenies drawn on the basis of nucleotide similarity between 15 different mammalian species revealed four major distinct groups. As expected, the ruminants were clustered together with B. taurus closely placed to the Indian native cattle, while species from the Equidae family being most distantly placed in the phylogenetic tree (Fig. 6). A similar finding across different livestock species has been reported by Mukesh et al. (2006) based on lineages of nucleotide and amino acid sequences.
Fig. 6.
N-J phylogenetic tree for the promoter region of kappa-casein among different mammalian species.
Conclusions
-
•
This is the first report for characterization and variation analysis in κ-CN coding and its promoter sequences among Indian native cattle and buffalo breeds. Characterization of the promoter region identified 20 putative TFBSs of different functional classes, including transcriptional activators (GR, NF-1) as well as repressors (YY1 and PMF).
-
•
For the κ-CN promoter region, among Indian cattle, a total of seven single nucleotide polymorphisms (SNPs) were found. The variations affected five putative TFBSs, of which the − 283 (A/G) variant located within binding site for HNF-1 was specific to Indian cattle. In buffalo, three SNPs were identified, however all the TFBSs remained conserved reflecting the highly conserved promoter region in buffalo than cattle and differential transcriptional regulation among the two species.
-
•
The novel SNPs in Indian native cattle at positions − 559 (T/C), − 558 (C/T) and − 283 (A/G); while in Indian buffalo at − 305 (A/C), − 160 (T/C) and − 141 (A/G) have been identified which could help to explore the underlying differences across bovine and bubaline κ-CN gene regulation.
-
•
Comparative analysis among Indian cattle and buffalo revealed a total of 15 SNPs and two insertions/deletions (INDELs). Thus, the observed mutations within the κ-CN promoter harboring the binding sites for mammary gland-specific as well as ubiquitous TFs might be responsible for differences in milk composition across cattle and buffalo breeds.
-
•
The untranslated regions (5′- and 3′-UTRs) also exhibited a high level of conservativeness in both cattle and buffalo. In Indian cattle, 5′-UTR was conserved while a single SNP in 3′-UTR was identified. However, in buffaloes, both 5′- and 3′-UTRs were conserved, suggesting high stability at the transcriptional level.
-
•
Within the CDS region, exon-IV was most polymorphic. Among the identified SNPs in exon-IV, three dN mutations in Indian cattle and two dN in buffalo was located in close proximity to glycosylation sites. This could possibly affect the structure of protein and its glycosylation patterns.
-
•
The presence of LD between coding (539 C/T and 575 C/A) and its promoter variations (− 427 C/T) may provide a basis for the simultaneous selection of desirable alleles at this important milk protein locus and improve the selection response of milk traits in Indian native cattle.
-
•
Further functional studies are required in zebu cattle and buffaloes to understand the influence of promoter variants on the regulation of κ-CN gene expression and/or its encoded products on milk trait performance.
Acknowledgments
The authors acknowledge the financial support to carry out the present research work by Department of Biotechnology (DBT) (BT/PR7467/AAQ/01/288/2006), New Delhi and Director, National Bureau of Animal Genetic Resources (NBAGR) and Indian Council of Agricultural Research (ICAR). Thanks are due to Mrs. P Kumari for the technical support.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mgene.2014.10.001.
Appendix A. Supplementary data
Supplementary tables.
References
- Adachi T., Ahn J.Y., Yamamoto K., Aoki N., Nakamura R., Matsuda T. Characterization of the bovine κ-casein gene promoter. Biosci. Biotechnol. Biochem. 1996;60:1937–1940. doi: 10.1271/bbb.60.1937. [DOI] [PubMed] [Google Scholar]
- Alipanah M., Kalashnikova L.A., Rodionov G.V. Kappa-casein and PRL-RSAI genotypic frequencies in two Russian cattle breeds. Arch. Zootech. 2008;57:131–138. [Google Scholar]
- Barrett J.C., Fry B., Maller J., Daly M.J. Haploview, analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Caroli A.M., Chessa S., Erhardt G.J. Milk protein polymorphisms in cattle: effect on animal breeding and human nutrition. J. Dairy Sci. 2009;92:5335–5352. doi: 10.3168/jds.2009-2461. [DOI] [PubMed] [Google Scholar]
- Creamer L.K., Plowman J.E., Liddell M.J., Smith M.H., Hill J.P. Micelle stability, k-casein structure and function. J. Dairy Sci. 1998;81:3004–3012. doi: 10.3168/jds.S0022-0302(98)75864-3. [DOI] [PubMed] [Google Scholar]
- Dayem A.M.H.A., Mahmoud K.G.M., Nawito M.F., Ayoub M.M., Darwish S.F. Genotyping of kappa-casein gene in Egyptian buffalo bulls. Livest. Sci. 2009;122:286–289. [Google Scholar]
- Farrell H.M.J., Jimenez-Flores R., Bleck G.T., Brown E.M., Butler J.E., Creamer L.K., Hicks C.L., Hollar C.M., Ng-Kwai-Hang K.F., Swaisgood H.E. Nomenclature of the proteins of cows' milk—sixth revision. J. Dairy Sci. 2004;87:1641–1674. doi: 10.3168/jds.S0022-0302(04)73319-6. [DOI] [PubMed] [Google Scholar]
- Gerencser A., Barta E., Boa S., Bosze Z., Whitelaw B. Comparative analysis on the structural features of the 5′ flanking region of kappa-casein genes from six different species. Genet. Sel. Evol. 2002;34:117–128. doi: 10.1186/1297-9686-34-1-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginger M.R., Grigor M.R. Comparative aspects of milk caseins. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1999;124:133–145. doi: 10.1016/s0305-0491(99)00110-8. [DOI] [PubMed] [Google Scholar]
- Grabe N. AliBaba2, context specific identification of transcription factor binding sites. In Silico. Biol. 2002;2:S1–S15. [PubMed] [Google Scholar]
- Haug A., Høstmark A.T., Harstad O.M. Bovine milk in human nutrition, a review. Lipids Health Dis. 2007;6:25-25. doi: 10.1186/1476-511X-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck J.M.L., Schennink A., Valenberg H.J.F.V., Bovenhuis H., Visker M.H.P.W., Arendonk J.A.M.V., Hooijdonk A.C.M.V. Effects of milk protein variants on the protein composition of bovine milk. J. Dairy Sci. 2009;92:1192–1202. doi: 10.3168/jds.2008-1208. [DOI] [PubMed] [Google Scholar]
- Hobor S., Kunej T., Dovc P. Polymorphisms in the kappa casein (CSN3) gene in horse and comparative analysis of its promoter and coding region. Anim. Genet. 2008;39:520–530. doi: 10.1111/j.1365-2052.2008.01764.x. [DOI] [PubMed] [Google Scholar]
- Hoogendoorn B., Coleman S.L., Guy C.A., Smith K., Bowen T., Buckland P.R., O'Donovan M.C. Functional analysis of human promoter polymorphisms. Hum. Mol. Genet. 2003;12:2249–2254. doi: 10.1093/hmg/ddg246. [DOI] [PubMed] [Google Scholar]
- Kaminski S. Association between polymorphism within regulatory and coding fragments of bovine kappa-casein gene and milk production traits. J. Anim. Feed Sci. 2000;9:435–446. [Google Scholar]
- Kaminski S. Polymorphism of milk protein genes in coding and regulatory regions and their effects on gene expression and milk performance traits. Anim. Sci. Paper. 2004;22:109–113. 41. [Google Scholar]
- Keating A.F., Davoren P., Smith T.J., Ross R.P., Cairns M.T. Bovine kappa-casein gene promoter haplotypes with potential implications for milk protein expression. Dairy Sci. 2007;90:4092–4099. doi: 10.3168/jds.2006-687. [DOI] [PubMed] [Google Scholar]
- Kel A.E., Gössling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E. MATCH, a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenasi T., Kokalj-Vokac N., Narat M., Baldi A., Dovc P. Functional study of the equine beta-casein and kappa-casein gene promoters. J. Dairy Res. 2005;72:34–43. doi: 10.1017/s0022029905001184. [DOI] [PubMed] [Google Scholar]
- Martin P., Szymanowska M., Zwierzchowski L., Leroux C. The impact of genetic polymorphisms on the protein composition of ruminant milks. Reprod. Nutr. Dev. 2002;42:433–459. doi: 10.1051/rnd:2002036. [DOI] [PubMed] [Google Scholar]
- Matys V., Fricke E., Geffers R., Gößling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Kloos D.U., Land S., Lewicki-Potapov B., Michael H., Münch R., Reuter I., Rotert S., Saxel H., Scheer M., Thiele S., Wingender E. TRANSFAC, transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A., Schlee P., Krause I., Blusch J., Werner T., Balakrishnan C.R., Pirchner F. Kappa-casein polymorphisms in Indian dairy cattle and buffalo, a new genetic variant in buffalo. Anim. Biotechnol. 1998;9:81–87. doi: 10.1080/10495399809525896. [DOI] [PubMed] [Google Scholar]
- Mukesh M., Mishra B.P., Kataria R.S., Sobti R.C., Ahlawat S.P. Sequence analysis of UTR and coding region of kappa-casein gene of Indian riverine buffalo (Bubalus bubalis) DNA Seq. 2006;17:94–98. doi: 10.1080/10425170600699950. [DOI] [PubMed] [Google Scholar]
- Nivsarkar A.E., Vij P.K., Tantia M.S. Directorate of Information and Publications of Agriculture Indian Council of Agricultural Research; New Delhi, India: 2000. Animal Genetic Resources of India, Cattle and Buffalo. [Google Scholar]
- Otaviano A.R., Tonhati H., Sena J.A.D., Muñoz M.F.C. Kappa-casein gene study with molecular variations in female buffaloes, Bubalus bubalis. Genet. Mol. Biol. 2005;28:237–241. [Google Scholar]
- Pauciullo A., Shuiep E.S., Cosenza G., Ramunno L., Erhardt G. Molecular characterization and genetic variability at κ-casein gene (CSN3) in camels. Gene. 2013;513:22–30. doi: 10.1016/j.gene.2012.10.083. [DOI] [PubMed] [Google Scholar]
- Ron M., Yoffe O., Ezra E., Medrano J.F., Weller J.I. Determination of effects of milk protein genotypes on production traits of Israeli Holsteins. J. Dairy Sci. 1994;77:1106–1113. doi: 10.3168/jds.S0022-0302(94)77046-6. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Cold Spring Harbour Lab. Press; Cold Spring Harbour, N.Y.: 1989. Molecular Cloning, A Laboratory Manual. [Google Scholar]
- Schild T.A., Wagner V., Geldermann H. Variants within the 5′-flanking regions of bovine milk protein genes. I. κ-casein-encoding gene. Theor. Appl. Genet. 1994;89:116–120. doi: 10.1007/BF00226992. [DOI] [PubMed] [Google Scholar]
- Sodhi M., Mishra B.P., Prakash B., Kaushik R., Singh K.P., Mukesh M. Distribution of major allelic variants at exon-IV of kappa casein gene in Indian native cattle. J. Appl. Anim. Res. 2010;38:117–121. [Google Scholar]
- Stevanovic M., Stanojčic S., Djurovic J., Verbic V. Molecular marker technologies and selection for the traits of economic interest. Biotechnol. Anim. Husb. 2000;16:25–34. [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5, Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill D.W., Womack J.E. Genomic analysis of the major bovine casein genes. Nucleic Acids Res. 1990;18:6935–6942. doi: 10.1093/nar/18.23.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.