Abstract
White sponge nevus (WSN) is an autosomal dominant hereditary disease. Keratin 4 (KRT4) and Keratin 13 (KRT13) gene mutations were involved in the WSN. We recruited two WSN Chinese families, and oral lesion biopsy with hematoxylin and eosin staining showed that patients had significant pathological characteristics. The mutations of KRT4 and KRT13 gene were detected by PCR and direct sequencing. The multiple alignments of KRT13 from 23 diverse species homology analyses were performed by the ClustalW program. The KRT13 expression was measured by Real-Time RT-PCR and Western blot analysis. Sequencing analysis revealed two mutations of KRT13 gene: one mutation was 332T>C and amino acid change was Leu111Pro. Another mutation was 340C>T and amino acid change was Arg114Cys. The sequence of KRT13 was highly conserved. Real-Time RT-PCR and Western blot analysis results show that KRT13 expression level is lower in patient but keep almost no change in mRNA level. When cells were treated with MG132, KRT13 protein level was increased and kept almost the same in normal and patient cells. We identified two heritable mutations in the KRT13 gene, which were associated with the development of WSN. The abnormal degradation of KRT13 protein of WSN may probably associate with the abnormal ubiquitination process.
Keywords: Gene mutation, Keratin 13 (KRT13), Oral disease, Ubiquitination, White sponge nevus (WSN)
Introduction
WSN is an uncommon hereditary disease firstly described by Hyde in 1909 and coined by Canon in 1935 (Cannon, 1935). It is characterized by white, thickened, folded and spongy lesions of the oral mucosa, although the esophageal, laryngeal, nasal and anogenital mucosa might also be affected (Jorgenson and Levin, 1981). The plaques of WSN are considered to be benign since the lesions are asymptomatic and painless in many cases although they may undergo alternate periods of remission and exacerbation due to infections. This disorder is often manifested in early childhood and showed no gender preference.
WSN occurred on the surface of the mucosa, the disorder of epithelium, as well as epithelial cells. Electron microscope assay of oral mucosa from WSN patient showed cellular keratinization in the mucosa. What's more, McGininis observed keratinizing epithelium in those tissues by focusing on the ultrastructural features of the tissues (McGinnis and Turner, 1975). Epithelia could express KRTs in well-defined type I/type II pairs in tissue and differentiation-specific manners. KRTs, which played an important role in the constitution of the cellular cytoskeleton, were the largest subgroup of the intermediate filament proteins and they were usually found in skins and other epithelial tissues. Till now, KRTs had been shown to play a regulatory role in electrolyte transport, post-translational modifications and protection against degradation (Majumdar et al., 2012). KRTs could provide epithelia with mechanical support. Moreover, abnormal KRT mutations, like KRT4 and KRT13 had been shown to be closely linked to WSN (Smith, 2003). Both oral and anogenital mucosae were able to express type II KRT4 and its type I partner, KRT13. What's more, KRT4 and KRT13 gene mutations were shown to represent the underlying cause of WSN recently (Kimura et al., 2013).
In our previous work, we investigated two causative mutations for WSN in two Chinese families, which could affect a functionally important segment of the KRT13 protein. In addition, homology analysis of KRT13 showed that a portion of the amino acid sequence of KRT13 was highly conserved in 23 diverse species. Real-Time RT-PCR and Western blot analysis results show that KRT13 protein level is lower in WSN patient but keep almost no change in mRNA level. Further analysis suggests that the abnormal degradation of KRT13 protein may probably associate with abnormal ubiquitination process.
Materials and methods
Ethical approval
Blood samples and oral epidermis were obtained from the patients who were well informed of all the purposes that might be used in the research and all the issues were approved by Tongji University's ethical committee.
Case report
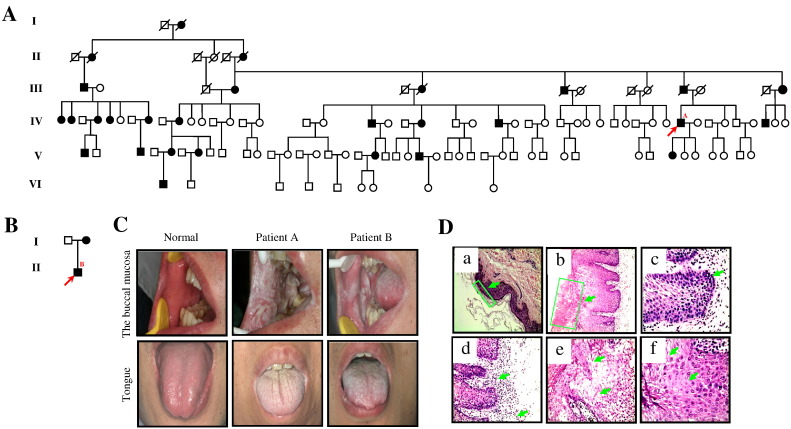
The proband in WSN-A family was a 43-year-old male Chinese patient from Hunan province, who was affected by white asymptomatic oral plaques and was clinically diagnosed as WSN. 23.3% of the six-generation-family members were recruited in the study. With pedigree analysis, 28 patients with genetic mode of the disease in this family have autosomal dominant disorder. In addition, the major position lesions of these patients were white plaques of the tongue and the buccal mucosa on both sides (Fig. 1A). The diagnosis of WSN in the two families was supported by the family history and the clinical and histopathological findings. Another proband was a 29-year-old male Chinese patient from Zhejiang province, and this family (named WSN-B) had two generations (Fig. 1B). He had a significant white spongy patch on the bilateral buccal mucosa and on the surface of the tongue.
Fig. 1.
Two WSN pedigrees from China and clinicopathological report.
(A) Pedigree of proband's family from Hunan Province, China (WSN-A). There are 28 cases in this family. Arrow indicates proband A, the 43-year-old male. (B) Pedigree of proband's family from Zhejiang Province, China (WSN-B). Arrow indicates proband B, the 29-year-old male. (C) White spongy plaques on the tongue, the buccal mucosa of normal, proband A from WSN-A family and proband B from WSN-B family. (D) Histopathology of the proband A lesion from WSN-A family. Thickened epidermis and transparent cytoplasm in keratinocytes, the epithelium is generally thickened, showing both hyper parakeratosis and acanthosis with the intact basal layer expression. Hematoxylin–eosin for all straining, original magnification 100 × or 400 ×. (a) Normal epidermis (100 ×). (b) Epithelial hyperplasia, parakeratosis, acanthosis (100 ×). (c) Stratum basale has 1–2 layers of cells, similar to the normal epidermis (400 ×). (d) The up arrow shows an infiltrate of inflammatory cells, the down arrow indicates that the small vessels of infiltrates have extravasated blood (400 ×). (e) The up arrow shows that spinous cells are loosening, the down arrow indicates intracellular edema with a characteristic basket-weave appearance (400 ×). (f) The up arrow shows vacuolization of the spinous cells, the down arrow shows that spinous cells have cytoplasmic basophilic mucoid secretions (400 ×).
Clinical data
Pathological features of WSN were determined by clinical data, pictures and oral cell structure of WSN lesions which was observed with the optical microscope by paraffin method.
Mutation detection
After an informed consent and as approved by the Medical Ethics Committee, Tongji University School of Stomatology, venous blood samples were obtained from the affected patients of both families and a number of unaffected members from WSN-A family, in which four unaffected members were chosen as controls. Genomic DNA was extracted from peripheral blood lymphocytes of the family members (QIAGEN DNA Blood Mini Kit, Germany). KRT4 and KRT13 genes were amplified by PCR and sequenced using the primers of KRT4 and KRT13 designed by us (Table 1, Table 2).
Table 1.
The primers of KRT4.
| Primer name | Primer (5′ to 3′) | Sizes (bp) |
|---|---|---|
| K4F1 | TCTATCCTGAGAAGTGGCAAGGCAA | 1086 |
| K4R1 | GGACTCAGGACCCCTCTCTTCTAAC | |
| K4F2 | TTGGCTGGTGCCTTTATCAACACAG | 580 |
| K4R2 | TAGTAAGTGAGGACTCAGTATGCAA | |
| K4F3 | CATGTATCTGAATAAACCCTGGCCT | 296 |
| K4R3 | CTCCCTACCATCCTTCGTCTCTTAC | |
| K4F4 | TCTTATCCTCCTTACTCCTCTGCAG | 454 |
| K4R4 | AAGCCACTCTAATGCAGTGAGTGCC | |
| K4F5 | GCCTGTATCAGGTCAGTGAAGGCTT | 253 |
| K4R5 | ATCTCCTGAGAGATCCATACCACC | |
| K4F6 | TTCTCTGTGTCTTCTTGAGTCTTAG | 184 |
| K4R6 | CTTGTGCTCCATCTGGAAGGGAGT | |
| K4F7 | GGCACTGCTGACCACCTATCTAATG | 289 |
| K4R7 | CTCATGTTCACACCCTGCCTAACC | |
| K4F8 | ATTCTGTGCTCCCTGAGGCTCACCG | 104 |
| K4R8 | TACAGCAGGCGTGGAAGGTACTTAC | |
| K4F9 | GCTGGCAGCATTGGAATAACTGTGT | 799 |
| K4R9 | TATACAGGATTCTAGAGACCTCTCC |
Table 2.
The primers of KRT13.
| Primer name | Primer (5′ to 3′) | Sizes (bp) |
|---|---|---|
| K13F1 | AAGTGTAGTTGGCACCAAGTCAGAG | 758 |
| K13R1 | TTGTGAATTCCGTCCACACCTAGTC | |
| K13F2 | CCTAGAGGTTACCTTCTTCTGCATT | 163 |
| K13R2 | ACTAAGAGACTCCTGGGAAAGGAGA | |
| K13F3 | TGTCTCATATCATTTATGTCTTGCT | 269 |
| K13R3 | TCCTTCTCACTGGAGGTTGTTGAGC | |
| K13F4 | ATGAGTAGCCTCACACATCTTCCTT | 245 |
| K13R4 | CTGGCTATATGGGATGGGCTATGTG | |
| K13F5 | CACCTTCTTCCTGCTTGCATTTCAG | 197 |
| K13R5 | GGAAACCTAGAGGTGGTCGCCACCG | |
| K13F6 | AACTTCTTGTCTGTCGCTCCACCA | 475 |
| K13R6 | ATCAGATGTTCCCTCCATGGTCTCT | |
| K13F7 | GACAGACAGGAAAGACAGACAGTCC | 986 |
| K13R7 | CACCTAAGAAACCAGGCCAGAATCA |
Multiple alignment of the KRT13 protein sequence from 23 species was created using the ClustalW program (http://www.genome.jp/tools/clustalw/). The 23 species consisted of human, chimpanzee, northern white-cheeked gibbon, olive baboon, gorilla, black-capped chickadee, common marmoset, cow, sheep, horse, elephant, dog, northern greater galago, rabbit, rat, mouse, pig, Tasmanian devil, cat, guinea pig, platypus, xenopus and zebrafish. The result was collected and sorted to construct the similarity comparison table, both of which gave a clear image of evolutionary relationship through the protein sequences.
KRT13 RNA expression by Real-Time RT-PCR
The oral epithelial cells from normal and WSN patient were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in 5% CO2. Real-time RT-PCR was used to determine KRT13 gene expression. Total cellular RNA was extracted using Trizol Reagent (Invitrogen). RNA was reverse transcribed using the Reverse Transcription Kit (Tiangen) and PCR was conducted with SuperReal PreMix Plus (SYBR Green) Kit (Tiangen). Reactions were analyzed using 7500 Real-time PCR system (Applied Biosystems). The samples were normalized to β-actin, and the relative abundance of transcripts was calculated by the cycle threshold (Ct) models.
Western blot and reagent
The oral epithelial cells from normal and WSN patient were lysed in RIPA buffer (50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 1% Protein Inhibitor). After boiling at 100 °C for 10 min, assay for KRT13 protein by SDS-PAGE and Western blot. Then we analyzed the protein expression in oral mucosal epithelial cells which were treated with/without MG132 (10 ng/ml) for proteasome inhibition for 2 h.
Results
Clinical phenotypes and evolution
After the oral clinical examination, the oral mucosal lesions of the proband and his affected family members were detected as a symptomatic, soft, white, spongy mucosal plaques with thick, folded surfaces. Probands A and B had white lesions bilaterally on the lips, on the lateral margin of the tongue and on the bilateral buccal mucosa (Fig. 1C).
Histopathology of the white plaque regions from the mucosa of the patients revealed that the epithelium was usually thickened, being observed with hyperparakeratosis and acanthosis, as well as the intact basal layer. The epithelial tissue of the whole spinous layer exhibited intracellular edema with a so-called basket-weave appearance, in which the atypical nuclei were absent. Nucleoli and perinuclear bodies were also noted and the intercellular spaces between vacuolated cells were narrow. Besides, the intercellular bridges were gloomy while basal cell layers had no rare appearance (Fig. 1D).
Identification of mutation in KRT13
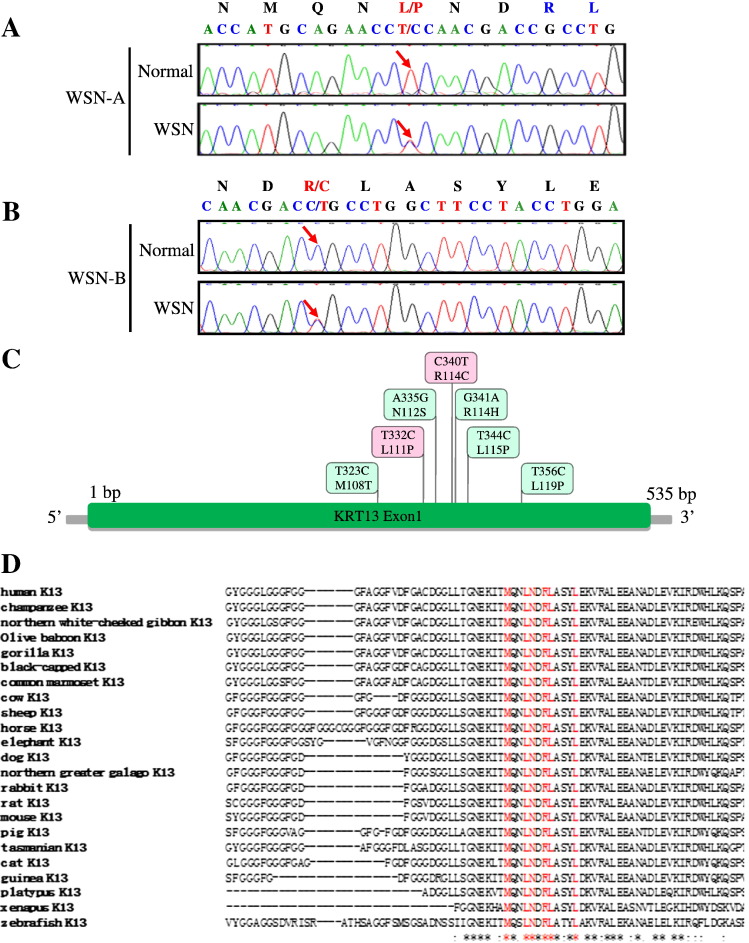
The mutation was confirmed by PCR and sequencing. We detected the direct sequencings of PCR products which revealed two mutations in the KRT13 gene by comparing with the Single Nucleotide Polymorphism (SNP) Database in NCBI (http://www.ncbi.nlm.nih.gov/snp/) and the Human Gene Mutation Database (http://www.hgmd.org/). In WSN-A family, the arrow indicates the position of the mutation 332T>C, which predicts the amino acid change Leu111Pro in the KRT13 coding sequence (Fig. 2A). In WSN-B family, the arrow indicates the position of the mutation 340C>T, which predicts the amino acid change Arg114Cys in the KRT13 polypeptide (Fig. 2B). All the reported mutations till now in the exon 1A of the KRT13 gene were listed (Fig. 2C). The protein homology analysis of KRT13 protein showed that a portion of the amino acid sequence of KRT13 from various species is highly conserved and it shared high similarity in human, chimpanzee, northern white-cheeked gibbon, olive baboon, gorilla, black-capped chickadee, common marmoset, cow, sheep, horse, elephant, dog, northern greater galago, rabbit, rat, mouse, pig, Tasmanian devil, cat, guinea pig, platypus, xenopus and zebrafish (Fig. 2D).
Fig. 2.
Two KRT13 mutations in two Chinese families with WSN.
(A) Partial DNA sequences of exon 1A of the KRT13 gene from the WSN-A family. The arrow indicates the position of the mutation 332T>C, the mutation predicts the amino acid change L111P in the KRT13 polypeptide from the WSN patient. (B) Partial DNA sequences of exon 1A of the KRT13 gene from the WSN-B family. The arrow indicates the position of the mutation 340C>T. The mutation predicts the amino acid change R114C in the KRT13 polypeptide. (C) The exon 1A of the KRT13 gene and the reported mutations being discovered till now. (D) A portion of the amino acid sequence of KRT13 is shown from 23 diverse species. Alignments of amino acid sequences for human, chimpanzee, northern white-cheeked gibbon, olive baboon, gorilla, black-capped chickadee, common marmoset, cow, sheep, horse, elephant, dog, northern greater galago, rabbit, rat, mouse, pig, Tasmanian devil, cat, guinea pig, platypus, xenopus and zebrafish KRT13 proteins. Red means published associated mutations in KRT13 gene; “*” means that the residues or nucleotides in that column are identical in all sequences in the alignment; “:” means that conserved substitutions have been observed; “.” means that semi-conserved substitutions are observed.
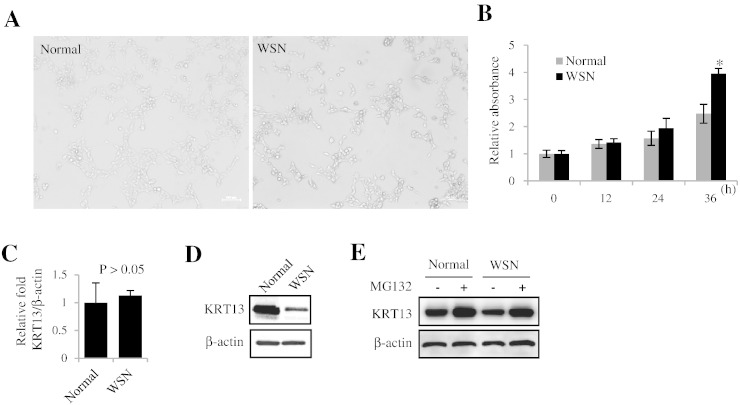
Protein expression of KRT13 is decreased in the WSN patient cells
To further investigate the pathogenesis mechanism of WSN, we take the tissues from oral mucosa of WSN patient and normal as control cell from the same family for cell culture. All these two cell lines are attached cells showing typical morphology of oral mucosa cells (Fig. 3A). But the growth curve shows that WSN patient cell line grows faster than normal control cell line (Fig. 3B). We check the KRT13 gene expression level between normal and WSN on transcriptional level by Real-time RT-PCR. The results show that the KRT13 gene in normal expression has no significant difference between WSN patient cells and normal control (Fig. 3C). Then we check the KRT13 protein expression level by Western blot analysis. The results show that the KRT13 protein expression level in normal was much higher than that in the WSN, which suggest that WSN patient has lower KRT13 protein level that may be associated with the protein degradation process but not in the transcriptional level (Fig. 3D). To test this hypothesis, we treat the cell with protease inhibitor MG132. The KRT13 protein expression levels in WSN patient cell are getting back at almost the same as normal control (Fig. 3E). Thus we concluded that KRT13 gene point mutation at a very shorten key position of alpha helix will lead to the abnormal degradation of KRT13 protein which is probably associated with the abnormal ubiquitination process.
Fig. 3.
Protein expression of KRT13 is decreased in WSN patient cells.
(A) Observation of oral mucosal epithelial cell morphology from normal and WSN patient.
(B) The growth curve of WSN patient cell line and normal control cell line (P < 0.05). (C) Quantification of KRT mRNA (P < 0.05). All images shown are representative and data are mean ± s.e.m. of n ≥ 3 independent experimental groups. Results are representative of three independent experiments. All experiments were performed at least three times with similar results. (D) Analysis of the protein expression between normal and WSN. (E) Analysis of the protein expression in oral mucosal epithelial cells with/without MG132 treatment.
Discussion
In our study, we report two causative mutations for WSN in two Chinese families, which could affect a functionally important segment of the KRT13 protein.
Whitten firstly did the electron microscopic examination of the focus tissues (Whitten, 1970). Because associated symptoms of the skin are missing, it is classified as a distinctive disease among the congenital leukokeratosis. Ordinarily, the mucosal lesions symptoms were detected by clinic examination when they were birth or in the early age (Metz and Metz, 1979). The cause of the disease may originally be hereditary. However, the pathogenesis of WSN remains unclear. Though the disease is called the mole, the disorder does not have a mole feature. Furthermore, the symptoms were caused by normal keratinization (Rugg et al., 1995). Researchers found that the abnormal expressions of several KRTs, mainly KRT4 or KRT13, played an important role in the generation of WSN and the KRT expression is not always parallel with each other (Su et al., 1993). More specifically, a mutation in the mucosal KRT4 was associated with oral WSN. In mouse, KRT4 was necessary for internal epithelial integrity (Ness et al., 1998). Another mutation in the KRT4 gene revealed a heterozygous missense mutation 1345G>A in KRT4, predicting an amino acid change of Glu449Lys, which caused WSN (Chao et al., 2003). Later on several other mutants of the KRTs had been found to have a close link to WSN. The KRT4 gene mutation 1558G>A in the 2B region is predicted to change a glutamic acid to a lysine at amino acid Glu520Lys (Shimizu et al., 2012). Kimura detected a deletion mutation of the KRT4 gene (N160Del) (Kimura et al., 2013). Richard found a point mutation in KRT13 of WSN that was the hereditary mucosal epithelial and reported a missense mutation 356T>C in 1A region of the helical rod domain of KRT13 that leads to Leu115Pro (Richard et al., 1995). Rugg identified two novel KRT13 mutations, a family mutation of 344T>C with an amino acid change of Leu115Pro, and another family mutation of 323T>C with an amino acid change of Met118Thr (Rugg et al., 1999). A heterozygous missense mutation 335A>G was detected in exon 1 of the KRT13 gene, predicting the amino acid change Asn112Ser in the 1A domain of the KRT13 polypeptide (Terrinoni et al., 2001). Shibuya identified a heterozygous missense mutation at 332T>C, the amino acid change was Leu111Pro (Shibuya et al., 2003). This was correspondent with WSN-A family's mutation. Nishizawa analyzed KRT13 mutation G341A, which predicts the amino acid change Arg114His in the KRT13 polypeptide (Nishizawa et al., 2008). Liu report mutational analysis in familial and sporadic patients with WSN, a novel mutation was 340C>T and the amino acid change was Arg114Cys (Liu et al., 2011).
We have presented two cases of WSN in two Chinese families with typical clinical and pathological features and found two mutations in KRT13 gene. Although WSN patients had no significant physical pain, they often complained of an altered texture of the mucosa or the bad looking created by the lesions. A case of WSN which showed a definite improvement following penicillin administration was reported (Alinovi et al., 1982). Another report said that oral tetracycline rinse improved symptoms of WSN (Lim and Ng, 1992). Victoria A acid can also inhibit keratosis formation (Irvine and McLean, 1999). Many WSN patients made therapy treatment medication with nystatin, antihistamines, vitamins and mouth rinses. Azithromycin, tetracycline and penicillin had succeeded during the clinical progress (Songu et al., 2012). However, there is no standard treatment protocol for WSN. McDonagh first reported that the tetracycline medicine will be effective for four WSN patients (McDonagh et al., 1990). After that, white sponge nevus could be successfully treated with tetracycline mouth rinse (Becker et al., 1997). Four cases also improved with tetracycline mouth rinse (Otobe et al., 2007). However, long-term low dose treatment of systemic antibiotic therapy maintained the remission of WSN (Lamey et al., 1998). Till now, systemic antibiotics or local applications of retinoic acid could provide limited benefits but both are poorly effective. Dufrasne found a surgical resection which proved to be effective (Dufrasne et al., 2011), as the patient was free of recurrence two years later. These methods together with the iPS technology may point a way to the treatment of the rare disease. To lead to the proper diagnosis and treatment of this rare disease, it is of great importance to collaborate anamnesis, clinical examination and pathologic findings.
In addition, homology analysis of KRT13 showed that a portion of the amino acid sequence of KRT13 was highly conserved in 23 diverse species. Further analysis suggests that the abnormal degradation of KRT13 protein may probably associate with abnormal ubiquitination. Real-time RT-PCR and Western blot analysis results show that KRT13 protein level is lower in WSN patient but keep almost no change in mRNA level. The exact pathogenesis of WSN is still unclear. We found that when treating the cell with protease inhibitor MG132, the KRT13 protein expression levels are getting back at almost the same. Thus we concluded that the abnormal degradation of KRT13 protein may probably be associated with abnormal ubiquitination. The KRT13 gene point mutation at a very shorten key position of alpha helix will lead to the abnormal degradation of KRT13 protein. Jaitovich demonstrated that ubiquitin was associated with KRT proteins' immunoprecipitation from shear-stressed cells and that the MG132 prevented the degradation and ubiquitination of the KRT8 intermediate filament (IF) network (Jaitovich et al., 2008). In Micah's study, they found that KRT IF network is known to colocalize with ubiquitin in response to stress and explain the role of the ubiquitin proteasome pathway in KRT intermediate filament protein degradation, especially in IF-related disease (Micah et al., 2010). Here the relationship between ubiquitination and KRT13 is reported for the first time.
In summary, we detected two different mutations of KRT13 from two Chinese families, which may be related to the development of WSN. We would like to perform the immunoprecipitation coupled with mass spectrometry (Co-IP-MS) assay and RNA sequence, trying to find the downstream regulated genes of KRT13 and clarifying the pathogenesis of WSN. At the same time, we induced pluripotent stem cell (iPS) technology and we are able to transform human WSN-fibroblast cells and wild type fibroblasts into iPS, and the iPS cells could be transformed into human epidermal cells to search for a series of effective compounds to treat WSN by drug screening. All the efforts may contribute to the molecular therapy for WSN. This research is a fundamental work in the fields of the KRT13 mutation analysis. Our results from WSN have a vital significance to reveal pathogenic mechanism of WSN. It may play an important part in prenatal gene diagnosis, gene therapy, drug screening and clinical treatment for WSN in the future. Gene-based diagnosis and gene therapy for WSN may become available in the near future while it provides reference and instruction for treating other KRT related diseases.
Acknowledgments
This work was supported, in whole or in part, by the National Basic Research Program (973 Program), no. 2013CB967501, the Natural Science Foundation of Shanghai, no. 12ZR1434200, the Fundamental Research Funds for the Central Universities, no. 20120072110016, the Key Program of Shanghai, no. 074119614, and the Shanghai Science and Technology Committee Program, no. 13JC1405400.
We are grateful to the participating individuals and their families. We thank Y. E. Sun (UCLA) for the critical review of the manuscript.
Contributor Information
Shangfeng Liu, Email: liusf23@gmail.com.
Xiaoping Wang, Email: wxp990@yahoo.com.cn.
References
- Alinovi A., Benoldi D., Pezzarossa E. White sponge nevus: successful treatment with penicillin. Acta Derm. Venereol. 1982;63:83–85. [PubMed] [Google Scholar]
- Becker L., Lutz C., Erbard H., Bröcker E., Hamm H. White sponge naevus successfully treated with tetracycline mouth rinse. Acta Derm. Venereol. 1997;77:413. doi: 10.2340/0001555577413. [DOI] [PubMed] [Google Scholar]
- Cannon A. White sponge nevus of the mucosa (naevus spongiosus albus mucosae) Arc. Dermatol. 1935;31:365. [Google Scholar]
- Chao S., Tsai Y., Yang M., Lee J. A novel mutation in the keratin 4 gene causing white sponge naevus. Br. J. Dermatol. 2003;148:1125–1128. doi: 10.1046/j.1365-2133.2003.05337.x. [DOI] [PubMed] [Google Scholar]
- Dufrasne L., Magremanne M., Parent D., Evrard L. Current therapeutic approach of the white sponge naevus of the oral cavity. Bull. Group. Int. Rech. Sci. Stomatol. Odontol. 2011;50:1–5. [PubMed] [Google Scholar]
- Irvine A., McLean W. Human keratin diseases: the increasing spectrum of disease and subtlety of the phenotype–genotype correlation. Br. J. Dermatol. 1999;140:815–828. doi: 10.1046/j.1365-2133.1999.02810.x. [DOI] [PubMed] [Google Scholar]
- Jaitovich A., Mehta S., Na N., Ciechanover A., Goldman R.D., Ridge K.M. Ubiquitin-proteasome-mediated degradation of keratin intermediate filaments in mechanically stimulated A549 cells. J. Biol. Chem. 2008;283:25348–25355. doi: 10.1074/jbc.M801635200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson R.J., Levin L.S. White sponge nevus. Arc. Dermatol. 1981;117:73. [PubMed] [Google Scholar]
- Kimura M., Nagao T., Machida J., Warnakulasuriya S. Mutation of keratin 4 gene causing white sponge nevus in a Japanese family. Int. J. Oral Maxillofac. Surg. 2013;42:615–618. doi: 10.1016/j.ijom.2012.10.030. [DOI] [PubMed] [Google Scholar]
- Lamey P.J., Bolas A., Napier S.S., Darwazeh A.M., Macdonald D.G. Oral white sponge naevus: response to antibiotic therapy. Clin. Exp. Dermatol. 1998;23:59–63. doi: 10.1046/j.1365-2230.1998.00325.x. [DOI] [PubMed] [Google Scholar]
- Lim J., Ng S.K. Oral tetracycline rinse improves symptoms of white sponge nevus. J. Am. Acad. Dermatol. 1992;26:1003–1005. doi: 10.1016/s0190-9622(08)80340-4. [DOI] [PubMed] [Google Scholar]
- Liu X., Li Q., Gao Y., Song S., Hua H. Mutational analysis in familial and sporadic patients with white sponge naevus. Br. J. Dermatol. 2011;165:448–451. doi: 10.1111/j.1365-2133.2011.10404.x. [DOI] [PubMed] [Google Scholar]
- Majumdar D., Tiernan J.P., Lobo A.J., Evans C.A., Corfe B.M. Keratins in colorectal epithelial function and disease. Int. J. Exp. Pathol. 2012;93:305–318. doi: 10.1111/j.1365-2613.2012.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonagh A., Gawkrodger D., Walker A. White sponge naevus successfully treated with topical tetracycline. Clin. Exp. Dermatol. 1990;15:152–153. doi: 10.1111/j.1365-2230.1990.tb02056.x. [DOI] [PubMed] [Google Scholar]
- McGinnis J.P., Turner J.E. Ultrastructure of the white sponge nevus. Oral Surg. Oral Med. Oral Pathol. 1975;40:644–651. doi: 10.1016/0030-4220(75)90374-6. [DOI] [PubMed] [Google Scholar]
- Metz J., Metz G. Naevus spongiosus albus mucosae. Review and personal observations. Z. Hautkr. 1979;54:604–612. [PubMed] [Google Scholar]
- Ness S.L., Edelmann W., Jenkins T.D., Liedtke W., Rustgi A.K., Kucherlapati R. Mouse keratin 4 is necessary for internal epithelial integrity. J. Biol. Chem. 1998;273:23904–23911. doi: 10.1074/jbc.273.37.23904. [DOI] [PubMed] [Google Scholar]
- Nishizawa A., Nakajima R., Nakano H., Sawamura D., Takayama K., Satoh T., Yokozeki H. A de novo missense mutation in the keratin 13 gene in oral white sponge naevus. Br. J. Dermatol. 2008;159:974–975. doi: 10.1111/j.1365-2133.2008.08716.x. [DOI] [PubMed] [Google Scholar]
- Otobe I., De Sousa S., Matthews R., Migliari D. White sponge naevus: improvement with tetracycline mouth rinse: report of four cases. Clin. Exp. Dermatol. 2007;32:749–751. doi: 10.1111/j.1365-2230.2007.02538.x. [DOI] [PubMed] [Google Scholar]
- Richard G., Delaurenzi V., Didona B., Bale S.J., Compton J.G. Keratin-13 point mutation underlies the hereditary mucosal epithelia disorder white sponge nevus. Nat. Genet. 1995;11:453–455. doi: 10.1038/ng1295-453. [DOI] [PubMed] [Google Scholar]
- Micah R., Rogel A.J., Ridge Karen M. Vol. 7. 2010. The role of the ubiquitin proteasome pathway in keratin intermediate filament protein degradation; pp. 71–76. (Proc. Am. Thorac. Soc.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugg E., McLean W., Allison W., Lunny D., Macleod R., Felix D., Lane E., Munro C. A mutation in the mucosal keratin K4 is associated with oral white sponge nevus. Nat. Genet. 1995;11:450–452. doi: 10.1038/ng1295-450. [DOI] [PubMed] [Google Scholar]
- Rugg E., Magee G., Wilson N., Brandrup F., Hamburger J., Lane E. Identification of two novel mutations in keratin 13 as the cause of white sponge naevus. Oral Dis. 1999;5:321–324. doi: 10.1111/j.1601-0825.1999.tb00097.x. [DOI] [PubMed] [Google Scholar]
- Shibuya Y., Zhang J., Yokoo S., Umeda M., Komori T. Constitutional mutation of keratin 13 gene in familial white sponge nevus. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2003;96:561–565. doi: 10.1016/s1079-2104(03)00372-x. [DOI] [PubMed] [Google Scholar]
- Shimizu A., Yokoyama Y., Shimomura Y., Ishikawa O. White sponge nevus caused by a missense mutation in the keratin 4 gene. Eur. J. Dermatol. 2012;22:571–572. doi: 10.1684/ejd.2012.1765. [DOI] [PubMed] [Google Scholar]
- Smith F.J. The molecular genetics of keratin disorders. Am. J. Clin. Dermatol. 2003;4:347–364. doi: 10.2165/00128071-200304050-00005. [DOI] [PubMed] [Google Scholar]
- Songu M., Adibelli H., Diniz G. White sponge nevus: clinical suspicion and diagnosis. Pediatr. Dermatol. 2012;29:495–497. doi: 10.1111/j.1525-1470.2011.01414.x. [DOI] [PubMed] [Google Scholar]
- Su L., Morgan P., Thomas J., Lane E. Expression of keratin 14 and 19 mRNA and protein in normal oral epithelia, hairy leukoplakia, tongue biting and white sponge nevus. J. Oral Pathol. Med. 1993;22:183–189. doi: 10.1111/j.1600-0714.1993.tb01054.x. [DOI] [PubMed] [Google Scholar]
- Terrinoni A., Rugg E., Lane E., Melino G., Felix D., Munro C., McLean W. A novel mutation in the keratin 13 gene causing oral white sponge nevus. J. Dent. Res. 2001;80:919–923. doi: 10.1177/00220345010800031401. [DOI] [PubMed] [Google Scholar]
- Whitten J. The electron microscopic examination of congenital keratoses of the oral mucous membranes: I. White sponge nevus. Oral Surg, Oral Med, Oral Pathol. 1970;29:69–84. doi: 10.1016/0030-4220(70)90413-5. [DOI] [PubMed] [Google Scholar]