Abstract
Background
Abnormal lipid levels are considered one of the most significant risk factors for atherosclerosis and coronary artery disease, two of the main causes of death worldwide. Apart from monogenic cases of hypercholesterolemia, most of the common dyslipidemias are caused by a number of low-impact polymorphisms. It has recently been reported that frequent polymorphisms at a large number of loci are significantly associated with one or more blood lipid parameters in many populations. Identifying these associations in different populations and estimating the possible interactions between genetic models are necessary to explain the underlying genetic architecture of the associated loci and their ultimate impact on lipid-associated traits.
Methods
We estimated the association between 144 common single-nucleotide polymorphisms (SNPs) from published genome-wide association studies and the levels of total cholesterol, low- and high-density lipoprotein–cholesterol, and triglycerides in 1273 individuals from the Genome Database of the Latvian Population. We analyzed a panel of 144 common SNPs with Illumina GoldenGate Genotyping Assays on the Illumina BeadXpress System.
Results
Ten SNPs at the CETP locus and two at the MLXIPL locus were associated with reduced high-density lipoprotein–cholesterol levels; one SNP at the TOMM40 locus was associated with increased low-density lipoprotein–cholesterol; and four SNPs at the MLXIPL locus were associated with increased log triglyceride levels. There was also a significant correlation between the number of risk alleles and all the lipid parameters, suggesting that the coexistence of many low-impact SNPs has a greater effect on the dyslipidemia phenotype than the individual effects of found SNPs.
Conclusion
We conclude that the CETP, MLXIPL, and TOMM40 loci are the strongest genetic factors underlying the variability in lipid traits in our population.
Keywords: Association study, Dyslipidemia, High-density lipoprotein–cholesterol, Total cholesterol, Low-density lipoprotein–cholesterol, Triglycerides
Highlights
-
•
Genotyping of 144 common SNPs associated with blood lipids in Latvian population
-
•
16 common SNPs are associated with blood lipid levels in Latvian population.
-
•
The number of risk alleles influences the level of blood lipids.
Introduction
Dyslipidemia is a disorder of lipid and lipoprotein metabolism, including overproductivity, deficiencies, or other changes in lipid metabolism, which results in abnormal amounts of lipids and lipoproteins in the blood. Dyslipidemia can manifest as an elevation of the total cholesterol (TC), low-density lipoprotein (LDL)–cholesterol, or triglyceride (TG) concentration, or a reduction in the high-density lipoprotein (HDL)–cholesterol concentration in the blood (Goldberg, 2013). Abnormal lipid levels are one of the essential causes of atherosclerosis and coronary artery disease (CAD) (Vance and Vance, 2008, World Health Organization, H.G.P., 1999). When lipoproteins in the plasma, and hence in the arterial wall, fall below a certain threshold level, atherogenesis does not occur. However, the absolute value of this threshold varies among individuals and may be very low in subjects with genetic and/or environmental factors that render their arteries highly susceptible to atherogenic processes (Vance and Vance, 2008, World Health Organization, H.G.P., 1999). Polygenic hypercholesterolemia is characterized by moderate elevations in LDL–cholesterol (3.63–7.77 mmol/L), with serum TG concentrations within the reference range. Like more extreme monogenic diseases, polygenic dyslipidemia is also associated with CAD. Although the main causes of dyslipidemia are high fat intake (particularly saturated fats) and obesity, genetic factors are also considered important, because many individuals vary in their responses to dietary cholesterol. However, there is no clear pattern of inheritance, and a combination of several genetic variants is generally required for this type of hypercholesterolemia (Durrington, 2003). Epidemiological studies have established that a reduction in plasma TC reduces the risk of coronary heart disease (CHD) (Bhagavan, 2002), indicating the importance of characterizing the most strongly associated SNPs with the main risk factors for elevated blood lipid levels. This characterization should facilitate the early diagnosis of these risk factors, before the development of CAD symptoms, and the administration of appropriate treatment. Recent genome-wide association studies (GWAS) and other human genetic studies have localized many common SNPs and many loci that influence the levels of different blood lipids, including previously known loci that are potentially involved in lipid metabolism (Aulchenko et al., 2009, Braun et al, Burkhardt et al., 2008, Chasman et al., 2008, Edmondson et al, Heid et al., 2008, Hiura et al., 2009, Igl et al, Johansen et al, Kathiresan et al., 2008, Kathiresan et al., 2009, Lanktree et al., 2009, Suchindran et al, Wallace et al., 2008, Waterworth et al, Willer et al., 2008). More than 40 loci have been associated with the levels of one or more blood lipid parameters, and contain many SNPs, but not all of these associations have been confirmed in other populations (Aulchenko et al., 2009, Burkhardt et al., 2008, Chasman et al., 2008, Heid et al., 2008, Hiura et al., 2009, Igl et al, Kathiresan et al., 2008, Kathiresan et al., 2009, Suchindran et al, Wallace et al., 2008, Waterworth et al, Willer et al., 2008, Ma et al, Pollin et al., 2008, Ridker et al., 2009, Kooner et al., 2008, Sabatti et al., 2009, Sandhu et al., 2008, Saxena et al., 2007, Shen et al). Frequent polymorphisms in ABCA1, ANGPTL3, APOA1, APOA5, APOB, APOE, CELSR2, CETP, CILP2, DOCK7, FADS2, GALNT2, GCKR, GPR109A/B, GPR81, HMGCR, KNTC1, LCAT, LDLR, LIPC, LIPG, LPL, MLXIPL, NCAN, NIACR1, NIACR2, PCSK9, PLTP, TOMM40, TRIB1, TTC39B, and other loci have been shown to be significantly associated with the blood levels of one or more lipid parameters in many populations (Aulchenko et al., 2009, Chasman et al., 2008, Edmondson et al, Hiura et al., 2009, Kathiresan et al., 2008, Ma et al, Ridker et al., 2009, Khovidhunkit et al). It has been demonstrated that a number of missense mutations at some of these loci can cause different types of monogenic hypercholesterolemia. For example, mutations in the ABCA1 gene cause familial HDL deficiency, or Tanger disease (Bhagavan, 2002). Similarly, mutations in the APOA5, APOB, and APOE genes cause different types of hyperlipoproteinemias or even familial hypercholesterolemia type B (Bhagavan, 2002, Marcais et al., 2005, Soria et al., 1989), but these are rare and usually more severe in their phenotypes. Confirmation of previously identified associations in different ethnic groups can give additional support to the underlying genetic architecture of the associated loci, especially when data from related populations are compared (Baba et al., 2009). Genetic structure studies of Europeans have shown that populations from Baltic countries (Estonia, Latvia, and Lithuania), together with Poland and the western part of Russia, form rather a homogeneous group, distinct from the rest of the Europe (Nelis et al., 2009). However, there is little information available on the SNPs associated with blood lipid levels in any of these countries.
Here, we report the associations between common SNPs and the plasma levels of different plasma lipids in a relatively large sample of the Latvian population. The main aims of this study were to investigate the associations between the most-informative SNPs from previous GWAS and four blood lipid parameters: TC, HDL–cholesterol, LDL–cholesterol, and TG in the Latvian population and to provide additional information to characterize the genetic factors that influence blood lipid levels.
Materials and methods
Subjects
We conducted this research using DNA samples from the Genome Database of the Latvian Population (LGDB), which included 18,888 participants in September 2011 when the study sample was selected (Ignatovica et al., 2011). We selected all individuals from this dataset for whom there was information on all four blood lipid parameters (TC, HDL, LDL, and TG), body mass index (BMI), glucose levels, sex, and age, resulting in 1581 samples. We then filtered out subjects with cardiovascular disease and those undergoing lipid-lowering therapies, resulting finally in 1345 samples. One sample was excluded as an outlier because of an extremely high TG level. A proportion (56.5%) of the samples matched those used in a previous study based on the same genotyping panel (Radovica et al., 2013). The genotypes of those samples were obtained from the database, and the remaining 585 samples were genotyped in this study. Written informed consent was acquired from all LGDB participants. The study protocol was approved by the Central Medical Ethics Committee of Latvia (protocol no 2007 A-7 and 01-29.1/25).
SNP data
We previously created a genotyping panel from GWAS, which contained 144 SNPs that were associated with one or more lipid traits (Aulchenko et al., 2009, Burkhardt et al., 2008, Chasman et al., 2008, Edmondson et al, Heid et al., 2008, Hiura et al., 2009, Kathiresan et al., 2008, Kathiresan et al., 2009, Wallace et al., 2008, Waterworth et al, Willer et al., 2008, Ma et al, Pollin et al., 2008, Ridker et al., 2009, Kooner et al., 2008, Sabatti et al., 2009, Sandhu et al., 2008, Saxena et al., 2007, Khovidhunkit et al). These SNPs occurred in more than 30 loci, including ABCA1, ANGPTL3, APOA1, APOA5, APOB, APOE, CELSR2, CETP, CILP2, DOCK7, FADS2, GALNT2, GCKR, GPR109A/B, HMGCR, LCAT, LDLR, LIPC, LIPG, LPL, MLXIPL, NCAN, NIACR1, NIACR1/KNTC1, NIACR2, NIACR2/GPR81, PCSK9, PLTP, TOMM40, TRIB1, and TTC39B. The SNP selection procedure is described in detail in our previous publication (Radovica et al., 2013).
Genotyping and quality control
All 144 SNPs were genotyped with the Illumina BeadXpress System (Illumina GoldenGate Genotyping Assay), according to the manufacturer's instructions. The quality control procedure applied to the raw data can be found in our previous article (Radovica et al., 2013). After quality control, the remaining sample consisted of 1273 individuals with 139 genotyped SNPs, with a successful genotyping rate of 99.76%.
Statistical analysis
The normal distributions of all quantitative variables were tested with the two most important parameters, the mean value and its standard deviation (SD) (Altman, 1991), and with the Shapiro–Wilk test. None of the lipid levels were normally distributed according to the Shapiro–Wilk test, even after various transformations. Therefore, to assess the influence of the covariates, we used linear regression, applying less-stringent normality criteria: the 68–95–99.7 rule or the three-sigma rule, according to which about 68% of values should fit within an interval of one SD, 95% in two SDs, and 99.7% in three SDs. Among all the variables tested, the TG levels were not normally distributed, so they were log transformed for further statistical analysis. We applied a linear regression analysis with and without the covariates (age, sex, BMI, and glucose levels), and tested epistasis and Hardy–Weinberg equilibrium with the PLINK v2.050 software (http://pngu.mgh.harvard.edu/purcell/plink/) (Purcell et al., 2007). The Bonferroni correction was used to calculate the significance level (0.05/139 = 3.5 × 10− 4). To calculate the joint effects, all SNPs in genes with more than one nominally associated SNP were divided into haploblocks using HapMap data and Haploview software v4.2 (Barrett, 2009, Barrett et al., 2005), and one SNP was chosen from each haploblock. The joint effect analyses were performed with the SPSS v13.0 software, using a one-sample t test. The association analysis of haplotypes was performed by PLINK toolset. A gene-by-gene interaction analysis was performed with the PLINK v2.050 software and GMDR software Beta 0.9 (http://sourceforge.net/projects/gmdr/) (Lou et al., 2007). Imputation was performed with the IMPUTE2 v2.2.2 software (https://mathgen.stats.ox.ac.uk/impute/impute_v2.html#home) (Howie et al., 2009, Marchini et al., 2007) for loci containing more than five SNPs. As reference haplotypes, we used the 1000 Genomes Phase I integrated variant set. The imputation region was set based on the coordinates of the first- and last-tested SNP at each locus. The SNPTEST v2.4.1 software was used to calculate the association between the imputed SNPs and the four lipid parameters (https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html#Download_and_Compilation) (Marchini et al., 2007). To accommodate uncertain genotypes, we used the –method threshold option, with a threshold value of 0.9. The additive model for association studies was chosen to calculate the P values. The Bonferroni threshold was calculated as follows: 0.05/number of our genotyped SNPs (n = 139) at all loci. Statistical power was calculated with the Quanto v1.2.4 software (Gauderman and Morrison, 2006). The minor allele frequencies (MAFs) of our genotyped SNPs ranged from 0.020 to 0.474. Taking into account this range of MAFs, our study had sufficient power (80%) to detect beta coefficients in the following range for each of the parameters: increased TC, 0.50–0.15; increased LDL–cholesterol, 0.45–0.15; reduced HDL–cholesterol, 0.200; and increased TG, 0.30–0.10.
Results
The clinical characteristics of the study subjects are listed in Table 1.
Table 1.
Sample characteristics.
| Variable | |
|---|---|
| n | 1273 |
| SNPs genotyped | 139 |
| Genotyping rate, % | 99.76% |
| Mean age, years ± SD (min–max) | 52.05 ± 13.73 (18–82) |
| Female gender, % | 67.0% |
| TC-level, mmol/L ± SD (min–max) | 5.78 ± 1.24 (1.95–11.7) |
| TC, median (Q1; Q3) | 5.70 (4.91; 6.55) |
| LDL-level, mmol/L ± SD (min–max) | 3.57 ± 1.07 (0.72–8.4) |
| LDL, median (Q1; Q3) | 3.48 (2.81; 4.24) |
| HDL-level, mmol/L ± SD (min–max) | 1.59 ± 0.44 (0.33–3.52) |
| HDL, median (Q1; Q3) | 1.53 (1.28; 1.87) |
| TG-level, mmol/L ± SD (min–max) | 1.40 ± 0.75 (0.29–5.87) |
| TG, median (Q1; Q3) | 1.22 (0.89; 1.74) |
| BMI, kg/m2 ± SD (min–max) | 27.02 ± 5.14 (15.64–59.73) |
SD, standard deviation; min, minimum value of variable; max, maximum value of variable; Q1, first quartile of the interquartile range; Q3, third quartile of the interquartile range; TC, total cholesterol; LDL, low-density lipoprotein; HDL, high-density lipoprotein; TG, triglyceride; BMI, body mass index.
SNP association tests
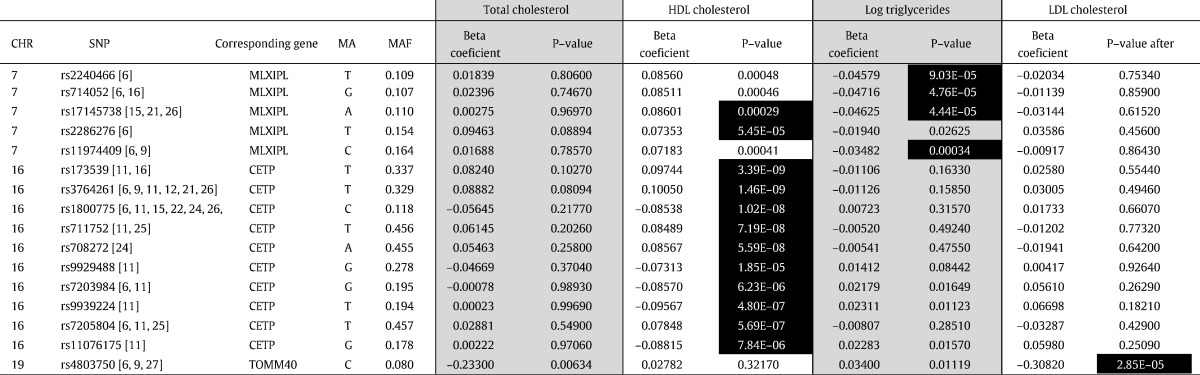
We performed linear regression analyses for all 139 SNPs with the four blood lipid parameters (TC, HDL, LDL, and logTG). In unadjusted analyses, 26 SNPs from nine loci were associated with TC, 22 SNPs from 10 loci with LDL, 58 SNPs from 13 loci with HDL, and 40 SNPs from 14 loci with logTG at the nominally significant level (P < 0.05). After age, sex, BMI, and glucose levels were added as covariates to the linear regression analysis, 24 SNPs at 11 loci were associated with TC, 23 SNPs at nine loci with LDL, 56 SNPs at 13 loci with HDL, and 40 SNPs at 14 loci with logTG, with the same significance level. Data for the associations of all SNPs are shown in Supplementary Table 1. In total, 16 SNPs remained significantly associated after the Bonferroni correction was applied (Table 2): 10 SNPs at the CETP (cholesteryl ester transfer protein) locus and two SNPs at the MLXIPL (MLX-integrating protein like) locus were associated with reduced HDL–cholesterol levels; one SNP at the TOMM40 (translocase of outer mitochondrial membrane 40) locus was associated with increased LDL–cholesterol; and four SNPs located at the MLXIPL locus were associated with increased logTG levels.
Table 2.
SNPs significantly associated with lipid parameters.
SNPs with P values < 0.05 are marked in bold; SNPs with P values < 0.05 after Bonferroni correction are black highlighted with white letters. All calculations were made with a linear regression analysis with covariates (age, sex, BMI, and glucose levels). CHR, chromosome; SNP, single-nucleotide polymorphism; MA, minor allele; MAF, minor allele frequency; HDL, high-density lipoprotein; LDL, low-density lipoprotein.
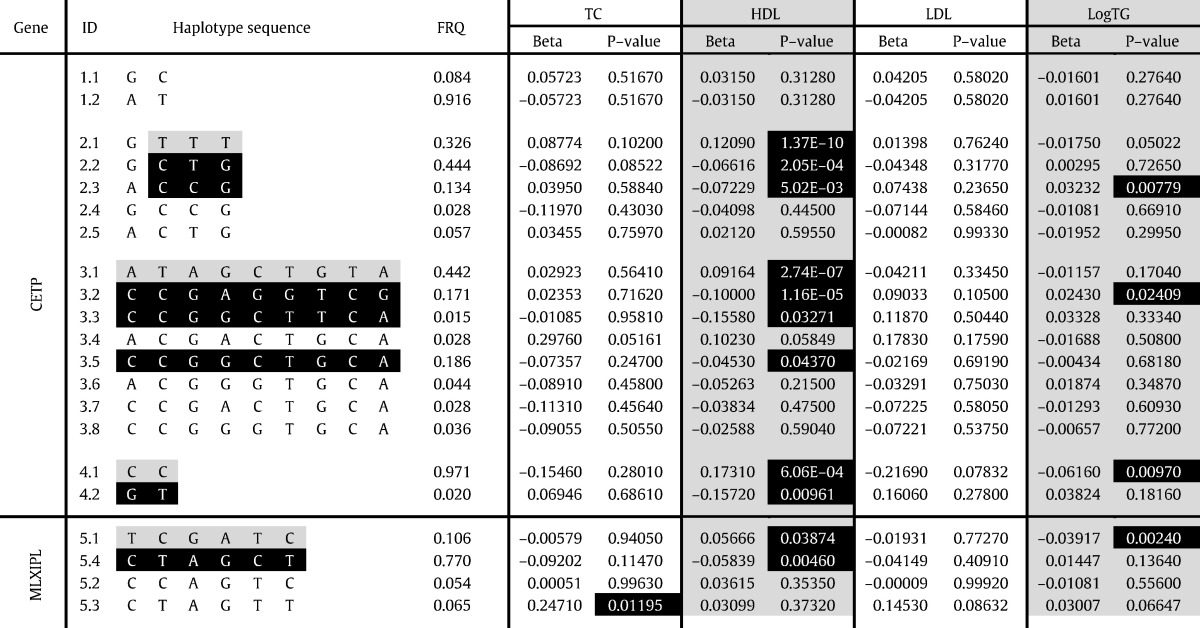
Haplotype association test
In order to explore the LD in association with changes in blood lipids detailed haplotype analysis was performed for both genes with more than one significantly associated SNP (MLXIPL and CETP). We thus selected all SNPs from our panel that were located within or in near proximity of CETP and MLXIPL gene and assigned them in haploblocks that were generated from the HapMap1/3 data. In total one haploblock was identified in the MLXIPL locus but four were found in CETP locus. Acquired haplotype data was further used for haplotype association test. Reconstructed haplotypes that exceeded the frequency of 0.01 and the results of association test are shown in Table 3. More detailed description of CETP haplotypes and their genomic localization can be found in our previous paper (Radovica et al., 2013).
Table 3.
Haplotypes in CETP and MLXIPL genes associated with blood lipid levels.
FRQ, frequency of haplotypes; TC, total cholesterol; HDL, high density lipoprotein cholesterol; LDL, low density lipoprotein cholesterol, logTG, logarithmic value of triglyceride level; ID, haplotype identification number (first digit—number of haploblock studied; second digit—number of individual haplotype); first haploblock of CETP gene consists of rs2241770 and rs16962767; second haploblock of CETP gene consists of rs12448528, rs173539, rs12708967 and rs3764261; third haploblock of CETP gene consists of rs1800775, rs711752, rs708272, rs1864163, rs9929488, rs7203984, rs9939224, rs7205804 and rs11076175; fourth haploblock of CETP gene consists of rs5880 and rs1800777; first haploblock of MLXIPL gene consists of rs2240466, rs1178979, rs714052, rs17145738, rs2286276 and rs11974409; haplotypes considered as “protective” are highlighted in grey; haplotypes considered as “risk” are highlighted in black with white letters; P-values lower than 0.05 are highlighted in black with white letters.
Gene–gene interaction test
Pair wise SNP–SNP interactions for all 139 SNPs were analyzed with the epistasis option implemented in the PLINK software package to determine whether interactions between two SNPs were associated with lipid levels. We found four pairs of SNPs with P values < 0.0001. One interacting SNP pair was associated with reduced HDL–cholesterol levels (rs1551894 in HMGCR, encoding 3-hydroxy-3-methylglutaryl-CoA reductase, interacting with rs6586891 in LPL, encoding lipoprotein lipase); two SNP pairs were associated with increased LDL–cholesterol levels (rs4803750 in TOMM40 interacting with rs157580 in APOE, encoding apolipoprotein E; and rs157580 in APOE interacting with rs405509 in TOMM40); and one pair of SNPs was associated with increased logTG (rs10889353 in ANGPTL3, encoding angioprotein-like 3, interacting with rs166358 in LIPC, encoding hepatic lipase). We also performed a gene–gene epistasis test with the GMDR software, but this analysis revealed no significant gene–gene interactions.
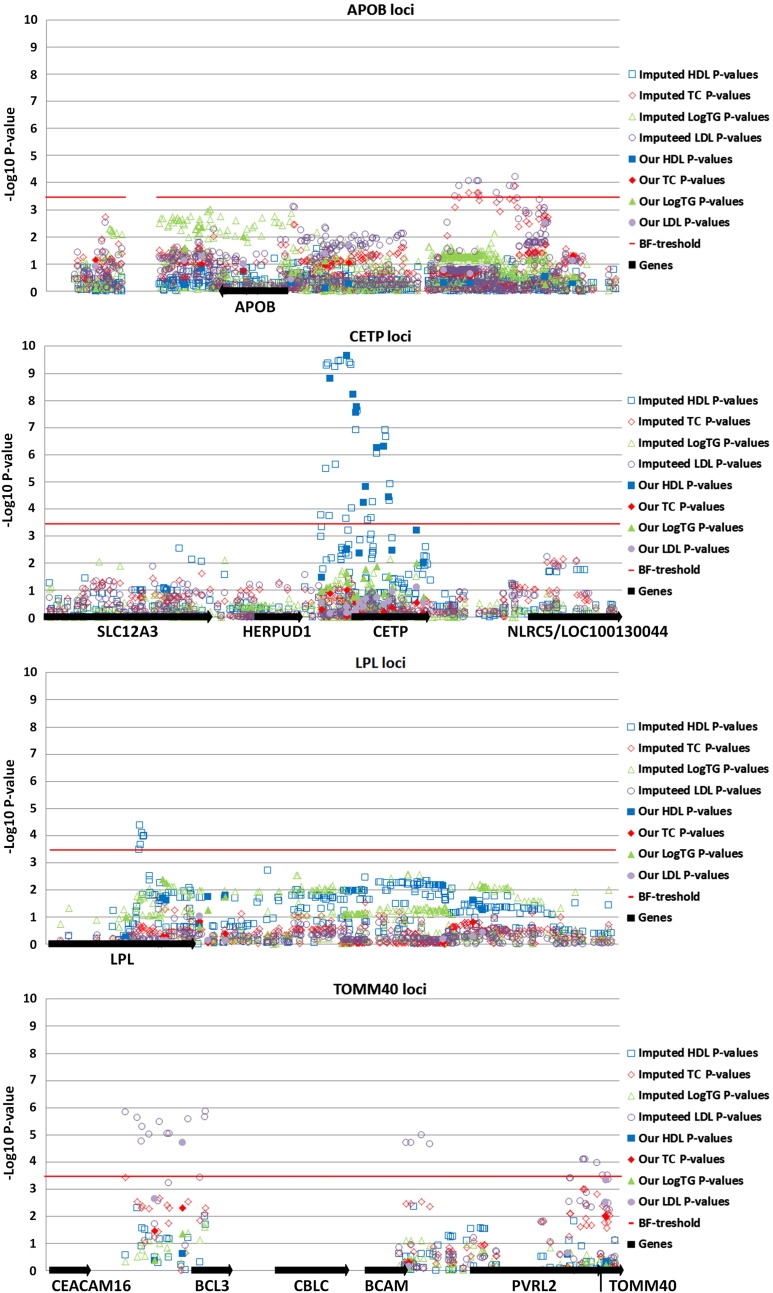
Imputation
We performed imputation and subsequent association analyses for 11 loci that contained five or more SNPs (CELSR2, APOB, HMGCR, MLXIPL, LPL, APOA1/5, KNTC1/NIACR1, LIPC, CETP, and TOMM40/APOE), using the 1000 Genomes Phase I integrated variant set of haplotypes as the reference and the coordinates of the outer SNPs from our 11 loci as the interval boundaries. At four loci, APOB (apolipoprotein B), CETP, LPL, and TOMM40, a number of SNPs displayed P values lower than the Bonferroni threshold for one or more lipid traits. To illustrate these results, we created Manhattan plots for these four loci (shown in Fig. 1). The P values and beta coefficients for the 71 corrected SNPs that were associated with one of the lipid traits after imputation and SNPTEST are shown in Supplementary Table 2. Thus, for APOB and LPL, only the imputed SNPs reached the appropriate significance level for association with LDL, TC, and HDL. One nucleotide deletion at chr2:21428937 (APOB locus) showed the strongest association with LDL cholesterol levels. rs289, located in the sixth intron of the LPL gene, displayed the strongest association with HDL–cholesterol. We found a number of imputed SNPs at the TOMM40 locus that reached the appropriate significance level for their associations with LDL–cholesterol levels, and had lower P values than our genotyped SNPs. The most strongly associated SNP was rs62117206, located in the intron of the BCL3 gene (B-cell leukemia/lymphoma 3). We found many SNPs with the appropriate significance level at the CETP locus for association with HDL–cholesterol; however none of these SNPs had a P value lower than that for rs3764261, which was genotyped with our panel. Most of the associated SNPs were found within introns, although some were located in 5′ or 3′ untranslated regions, but none occurred in a coding region.
Fig. 1.
Manhattan plots of imputed SNPs and the four lipid parameters for the four most strongly associated loci.
These data plots show the P values for SNPs associated with all four lipid traits: filled symbols indicate P values of the originally genotyped SNPs in our study; empty symbols indicate P values of the imputed SNPs; black arrows indicate genes located at the studied loci and their directions; red lines indicate the Bonferroni threshold of 0.05/139 = 3.60E − 04.
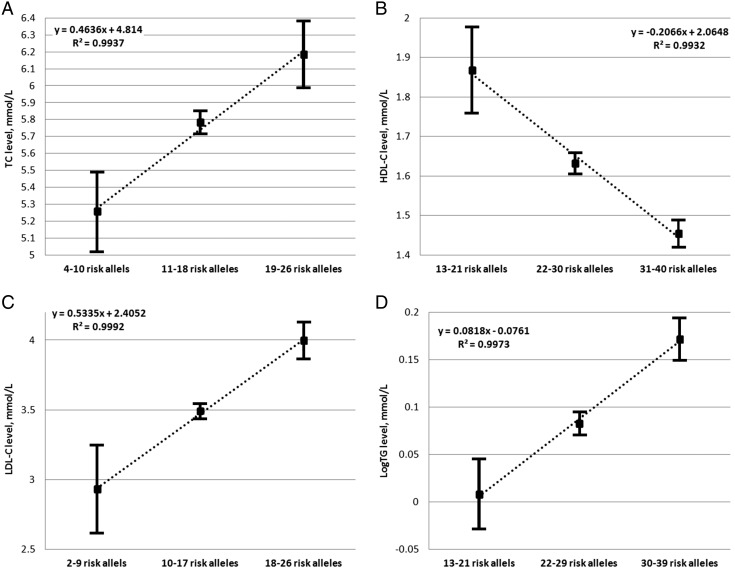
Joint effect analysis
Because the effects of individual SNPs are rather small, we decided to investigate the summary effects of associated SNPs on blood lipid parameters, using the risk allele dosage test and those SNPs that were nominally associated with different blood lipid levels before the Bonferroni correction, each of which represented one haploblock. Sixteen SNPs representing the ABCA1, ANGPTL3, APOB, APOE, DOCK7, HMGCR, LCAT, LIPC, and TOMM40 loci were included in case of TC; 24 SNPs representing the ABCA1, APOA1, CETP, HMGCR, NIACR1, LCAT, LIPC, LPL, MLXIPL, PLTP, and TRIB1 loci were included in cases of HDL cholesterol; 15 SNPs representing ABCA1, ANGPTL3, APOA1, APOB, APOE, DOCK7, HMGCR, LCAT, and TOMM40 loci were included in case of LDL–cholesterol; and 27 SNPs representing ABCA1, ANGPTL3, APOA1, APOA5, APOB, CETP, CILP2, DOCK7, GCKR, LIPC, LPL, MLXIPL, NCAN, and TOMM40 loci were included in case of logTG. The distribution of risk allele number within each group of lipid parameters analyzed was almost normally distributed and therefore we divided the sample into three equivalent groups based on the range of the risk allele distribution separately for each lipid parameter. We then performed a one-sample t test with the SPSS software to calculate the differences between these three groups. We found that the number of risk alleles correlated strongly with the blood lipid levels for all four parameters. The results of this analysis are shown in Fig. 2. In case of TC the group of individuals with 4–10 risk alleles had a mean TC level 0.53 mmol/L lower than that of the median group, represented by individuals with 11–18 risk alleles (P = 1.59E − 44). In the group with 19–26 risk alleles, the mean TC level was 0.40 mmol/L higher (P = 4.18E − 97) than that in the median group. The mean HDL–cholesterol level was 0.24 mmol/L higher (P = 1.59E − 44) in the group with 13–21 risk alleles and 0.18 mmol/L lower (P = 7.51E − 241) in the group with 31 − 40 risk alleles compared to the value for the median group (22–30 risk alleles). The mean LDL–cholesterol level was 0.56 mmol/L lower (P = 1.36E − 22) in the group with two to nine risk alleles and 0.51 mmol/L higher (P = 7.91E − 140) in the group with 10–17 risk alleles compared to the value in the median group (10–17 risk alleles). The mean logTG value was 0.07 log mmol/L lower (P = 1.06E − 31) in the group with 13–21 risk alleles and 0.09 log mmol/L higher (P = 1.76E − 34) in the group with 30–39 risk alleles compared to the median group (22–29 risk alleles).
Fig. 2.
Joint effect analyses for all blood lipid parameters.
Analysis of the joint risk allele effects on A, total cholesterol; B, high-density lipoprotein–cholesterol; C, low-density lipoprotein–cholesterol, and D, log triglycerides. The Y axis represents the level of the corresponding blood lipid (in mmol/L); the X axis represents three equal groups, based on the number of risk alleles; black squares indicate the mean values for the lipids in each group, with the 95% confidence interval of the mean difference. The dotted line indicates the linear correlation between the groups and the R2 value is shown for each lipid trait.
Discussion
The aim of this study was to identify associations between 144 SNPs selected from published GWAS and blood lipid levels in the Latvian population. This is the first report in which many genetic loci, involved in a number of lipid-related metabolic pathways, were simultaneously studied in a relatively large group representing the Latvian population (420 males, 853 females; for detailed data, see Table 1). Our study has convincingly identified associations between 16 of the 139 most strongly associated SNPs and one or more blood lipid parameters.
In our previous case–control study, we reported strong associations between 10 SNPs in CETP and reduced HDL–cholesterol (Radovica et al., 2013). In this study, we confirmed this strong evidence for this association between the CETP gene and HDL–cholesterol levels and discovered the new association of two SNPs at the MLXIPL locus (Table 2), using quantitative analyses. Moreover, haplotype analyses also confirmed the associations demonstrated in our previous study (Radovica et al., 2013). These results also agree well with those of other studies around the world (Chasman et al., 2008, Heid et al., 2008, Hiura et al., 2009, Kathiresan et al., 2008, Kathiresan et al., 2009, Willer et al., 2008, Ma et al, Ridker et al., 2009, Khovidhunkit et al, Thompson et al., 2005, Thompson et al., 2007) and are supported by the well-known function of CETP in lipid metabolism. CETP promotes the transfer of cholesteryl esters from HDL particles to apolipoprotein-B-containing particles in exchange for TG, allowing the receptor-mediated uptake of cholesterol esters by the liver and reducing HDL–cholesterol levels (Vance and Vance, 2008). In contrast, CETP deficiency increases HDL–cholesterol levels. Supporting the association between SNPs in the MLXIPL locus and HDL–cholesterol, several reports have linked MLXIPL and HDL–cholesterol. However, their relationship within the lipid metabolic pathways is not as direct as that of the SNP–CETP association. The protein encoded by the MLXIPL gene is a helix–loop–helix leucine zipper transcription factor, which forms a heterodimer with MLX to bind DNA (Cairo et al., 2001). This transcription factor complex is activated by high glucose and inhibited by cyclic adenosine monophosphate (cAMP) (Kawaguchi et al., 2001), and can repress E-box-dependent transcription (Cairo et al., 2001) or the carbohydrate response element motifs in the promoters of lipogenic enzymes. In adipose tissues, MLXIPL is a major determinant of adipose tissue fatty-acid synthesis and systemic insulin sensitivity (Herman et al.). Therefore, it is plausible that changes in MLXIPL expression indirectly influence HDL–cholesterol levels through various metabolic pathways.
We also found strong associations between four SNPs at the MLXIPL locus and increased logTG (Table 2). Because MLXIPL is a transcription factor that influences fatty-acid synthesis in adipose tissue, it is plausible that SNPs in this gene indirectly affect TG levels. We also found that a SNP in the TOMM40 locus is associated with increased LDL–cholesterol levels (Table 2). More precisely, this SNP is located in the first intron of the BCL3 gene (Fig. 1 TOMM40 locus). The protein encoded by the BCL3 gene is an inhibitor of subunit two of nuclear factor kappa-B (NF-κB) (Franzoso et al., 1992, Wulczyn et al., 1992). Because interactions between BCL3 and NF-κB have not yet been linked to lipid metabolism, we think that this SNP might be in linkage disequilibrium (LD) with functional SNPs in one of the genes encoding various apolipoproteins located downstream from the TOMM40 gene. For example, mutations in the APOE gene (located immediately downstream from TOMM40) can cause hyperlipoproteinemia type III (Utermann et al., 1979). Alternatively, the associated SNPs may be located within upstream regulatory elements of these apolipoprotein genes because the distance between the TOMM40 gene and, for example, the APOE gene is rather large (2.1 Mbp), arguing against a simple LD-based explanation.
Since the number of associated SNPs was found in CETP and MLXIPL gene, we estimated the association of individual haplotypes with all lipid levels. First it should be noted that in case of CETP the results of HDL associations corresponded exactly to our previous findings (Radovica et al., 2013). Most of the associated SNPs fall in second and third haploblock (Table 3). These haploblocks cover the region starting from ~ 2 kb upstream CETP gene, promoter region and include first ~ 10 kb of CETP gene. Interestingly, the strongest association was observed for two protective haplotypes with increased HDL levels: haplotype 2.1 from second haploblock and 3.1 from the third haploblock. For second haploblock it is clear that effect is explained by the presence of risk alleles from two SNPs rs173539 and rs3764261 that are both in strong LD with each other (R2 = 0.94). In case of third haploblock T allele of rs9939224 from the second intron of CETP gene is present only in two “risk” haplotypes and may be linked to the increased expression or functionality of CETP, thus decreasing the HDL-C levels as suggested by our previous observations (Radovica et al., 2013). The effect of “protective” haplotype may be explained by the presence of rs1800775 A allele that has been linked to changes in binding site Sp1/Sp3 in functional studies (Dachet et al., 2000, Thompson et al., 2004). In case of MLXIPL one protective haplotype was identified that was associated with increased HDL and decreased logTG levels and one risk haplotype associated with decreased HDL. Due to strong LD it is however less clear which of the tested SNPs may be responsible for these effects.
An epistasis analysis was performed to test whether there was any gene–gene interactions among the SNPs examined in this study. We found four pairs of SNPs that showed probable interactions in their associations with blood lipid parameters. Two pairs of SNPs, located again in the TOMM40 and APOE genes, showed functional or LD-based interactions that affected LDL–cholesterol levels. Another two interactions were associated with HDL–cholesterol (HMGCR and LPL) or logTG levels (ANGPTL3 and LIPC). The HMGCR gene encodes 3-hydroxy-3-methylglutaryl-CoA reductase, which catalyzes the crucial step in cholesterol biosynthesis (Vance and Vance, 2008). In contrast, the LPL gene encodes lipoprotein lipase, which hydrolyzes TGs derived from blood lipid particles. These are taken up into cells as fatty acids and are stored or delivered to the cholesterol or TG synthetic pathways (Vance and Vance, 2008). This interaction thus is supported by the known gene functions. ANGPTL3 encodes angiopoietin-like 3 protein, which belongs to the specific vascular endothelium growth factor protein family (Camenisch et al., 2002), and homozygous or compound heterozygous mutations in ANGPTL3 cause familial hypobetalipoproteinemia type II (Musunuru et al.). The LIPC gene encodes hepatic lipase, which is synthesized in the liver and acts in the uptake of lipoproteins and cholesteryl esters by hepatocytes, thus directly influencing plasma lipid levels (Cai et al., 1989). It is not known how these two proteins interact to influence lipid metabolism, but such interactions cannot be excluded. However, it should be noted that the majority of genes selected for this study have well-established functions in lipid metabolism. Therefore, all attempts to explain these interactions statistically, based on the relationships of these protein functions, are highly biased, and in all cases, further investigation is required in large well-characterized cohorts. Unfortunately, these findings were not supported by the GMDR software, perhaps because more-specific algorithms are used in GHMDR, taking into account, for example, covariates, “course of dimensionality”, and other criteria that are not included in the PLINK toolset and which therefore better exclude false positive results (Chen et al.).
For a more detailed analysis and to find other strongly associated SNPs at our test loci, we performed imputation for 11 loci that contained five or more SNPs (CELSR2, APOB, HMGCR, MLXIPL, LPL, APOA1/5, KNTC1/NIACR1, LIPC, CETP, and TOMM40/APOE). We found many imputed SNPs in the CETP gene that were associated with changes in HDL–cholesterol levels; although none of these associations was stronger than those of the originally genotyped SNPs (Fig. 1 CETP locus). These results again agree well with those of our previous study, in that CETP is the strongest and most common genetic factor to influence HDL–cholesterol levels in the Latvian population. Imputed SNPs at another three loci, APOB, LPL, and TOMM40, were significantly associated with one or more lipid trait (Fig. 1). All these SNPs were located in noncoding regions and many of them have already been associated with disease phenotypes, including CAD, myocardial infarction, and atherosclerosis. The APOB and LPL genes are known to be directly involved in lipid metabolism, and a number of functional mutations in these genes have already been reported to cause different types of monogenic familial hypercholesterolemia (http://www.hgmd.org/). Therefore, it is likely that common variants with smaller effects may play roles in milder forms of dyslipidemia. It is also possible that our cohort contained a number of patients with undiagnosed monogenic hypercholesterolemia. This may have influenced the association results either because of the LD between these mutations and the markers used in our study or simply by adding a stronger genetic modifier. A number of imputed SNPs at the TOMM40 locus reached the appropriate significance level in their associations with LDL–cholesterol levels. Again, the role of TOMM40 in the regulation of LDL metabolism is unclear (see discussion above).
To achieve our aim, we performed an allele dosage association test with nominally associated SNPs, each representing one haploblock, for all four blood lipid parameters to calculate the influence of the number of risk alleles on lipid levels. We divided our sample in three equivalent groups based on the number of risk alleles. There was a strong correlation between the number of risk alleles and the plasma lipid levels for all four lipid parameters (Fig. 2). Therefore, it is possible that the discovery and inclusion of additional lipid-influencing SNPs in similar analyses will lead to the development of prognostic tests for dyslipidemia and the prescription of appropriate drugs.
Conclusions
The results of this study confirm that of all the genes included in the analysis, CETP, MLXIPL, and TOMM40 are the genetic factors that most strongly influence the plasma lipid levels in our study population. We have also shown that these risk alleles have strong cumulative effects on all lipid parameters. It should be noted that this is the first replication study of a relatively large sample population from the Baltic States and Eastern Europe.
Abbreviations
- CAD
coronary artery disease
- TC
total cholesterol
- SNP
single-nucleotide polymorphism
- LDL
low-density lipoprotein
- HDL
high-density lipoprotein
- TG
triglyceride
- BMI
body mass index
- MAF
minor allele frequency
- CHR
chromosome
- MA
minor allele
- GWAS
genome-wide association studies
- cAMP
cyclic adenosine monophosphate
- LD
linkage disequilibrium
- SD
standard deviation
The following are the Supplementary data related to this article.
Association data for all 139 SNPs used in present study.
Imputed SNPs associated with one or more lipid traits.
Acknowledgments
The authors thank the Genome Database of the Latvian Population and the Latvian Biomedical Research and Study Centre for providing data and DNA samples.
This work was supported by the Latvian Research Program ”Biomedicine for health” and the European Social Fund within the project “Support for Doctoral Studies at the University of Latvia”, contract no. 2009/0138/1DP/1.1.2.1.2/09/IPIA/VIAA/004.
Contributor Information
I. Radovica, Email: ilze@biomed.lu.lv.
D. Fridmanis, Email: davids@biomed.lu.lv.
I. Silamikelis, Email: ivars.silamikelis@biomed.lu.lv.
L. Nikitina-Zake, Email: lienik@biomed.lu.lv.
J. Klovins, Email: klovins@biomed.lu.lv.
References
- Altman D.G. Practical Statistics for Medical Research. In: Altman D.G., editor. Chapman & Hall/CRC; London: 1991. [Google Scholar]
- Aulchenko Y.S. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009;41(1):47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K. Validating an alternate version of the chewing function questionnaire in partially dentate patients. BMC Oral Health. 2009;9:9. doi: 10.1186/1472-6831-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C. Haploview: visualization and analysis of SNP genotype data. Cold Spring Harb Protoc. 2009;2009(10) doi: 10.1101/pdb.ip71. (p. pdb ip71) [DOI] [PubMed] [Google Scholar]
- Barrett J.C. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bhagavan N.V. 4th ed. Harcourt/Academic Press; San Diego: 2002. Medical Biochemistry. (1016 pp.) [Google Scholar]
- Braun, T.R. et al., A replication study of GWAS-derived lipid genes in Asian Indians: the chromosomal region 11q23.3 harbors loci contributing to triglycerides. PLoS One. 7(5), e37056. [DOI] [PMC free article] [PubMed]
- Burkhardt R. Common SNPs in HMGCR in micronesians and whites associated with LDL-cholesterol levels affect alternative splicing of exon13. Arterioscler. Thromb. Vasc. Biol. 2008;28(11):2078–2084. doi: 10.1161/ATVBAHA.108.172288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S.J. Structure of the human hepatic triglyceride lipase gene. Biochemistry. 1989;28(23):8966–8971. doi: 10.1021/bi00449a002. [DOI] [PubMed] [Google Scholar]
- Cairo S. WBSCR14, a gene mapping to the Williams–Beuren syndrome deleted region, is a new member of the Mlx transcription factor network. Hum. Mol. Genet. 2001;10(6):617–627. doi: 10.1093/hmg/10.6.617. [DOI] [PubMed] [Google Scholar]
- Camenisch G. ANGPTL3 stimulates endothelial cell adhesion and migration via integrin alpha vbeta 3 and induces blood vessel formation in vivo. J. Biol. Chem. 2002;277(19):17281–17290. doi: 10.1074/jbc.M109768200. [DOI] [PubMed] [Google Scholar]
- Chasman D.I. Genetic loci associated with plasma concentration of low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, triglycerides, apolipoprotein A1, and apolipoprotein B among 6382 white women in genome-wide analysis with replication. Circ. Cardiovasc. Genet. 2008;1(1):21–30. doi: 10.1161/CIRCGENETICS.108.773168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G.B. et al., Practical and theoretical considerations in study design for detecting gene-gene interactions using MDR and GMDR approaches. PLoS One. 6(2), e16981. [DOI] [PMC free article] [PubMed]
- Dachet C. New functional promoter polymorphism, CETP/-629, in cholesteryl ester transfer protein (CETP) gene related to CETP mass and high density lipoprotein cholesterol levels: role of Sp1/Sp3 in transcriptional regulation. Arterioscler. Thromb. Vasc. Biol. 2000;20(2):507–515. doi: 10.1161/01.atv.20.2.507. [DOI] [PubMed] [Google Scholar]
- Durrington P. Dyslipidaemia. Lancet. 2003;362(9385):717–731. doi: 10.1016/S0140-6736(03)14234-1. [DOI] [PubMed] [Google Scholar]
- Edmondson, A.C. et al., Dense genotyping of candidate gene loci identifies variants associated with high-density lipoprotein cholesterol. Circ. Cardiovasc. Genet. 4(2), 145–155. [DOI] [PMC free article] [PubMed]
- Franzoso G. The candidate oncoprotein Bcl-3 is an antagonist of p50/NF-kappa B-mediated inhibition. Nature. 1992;359(6393):339–342. doi: 10.1038/359339a0. [DOI] [PubMed] [Google Scholar]
- Gauderman W., Morrison J. QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies. 2006. http://hydra.usc.edu/gxe
- Goldberg A.C. Dyslipidemia. 2013. http://www.merckmanuals.com/professional/endocrine_and_metabolic_disorders/lipid_disorders/dyslipidemia.html (September 2013 [cited 2014 31.01.2014]; Available from:)
- Heid I.M. Genome-wide association analysis of high-density lipoprotein cholesterol in the population-based KORA study sheds new light on intergenic regions. Circ. Cardiovasc. Genet. 2008;1(1):10–20. doi: 10.1161/CIRCGENETICS.108.776708. [DOI] [PubMed] [Google Scholar]
- Herman, M.A. et al., A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 484(7394), 333–338. [DOI] [PMC free article] [PubMed]
- Hiura Y. Identification of genetic markers associated with high-density lipoprotein-cholesterol by genome-wide screening in a Japanese population: the Suita study. Circ. J. 2009;73(6):1119–1126. doi: 10.1253/circj.cj-08-1101. [DOI] [PubMed] [Google Scholar]
- Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igl, W., et al., Modeling of environmental effects in genome-wide association studies identifies SLC2A2 and HP as novel loci influencing serum cholesterol levels. PLoS Genet. 6(1), e1000798. [DOI] [PMC free article] [PubMed]
- Ignatovica V. Single nucleotide polymorphisms of the purinergic 1 receptor are not associated with myocardial infarction in a Latvian population. Mol. Biol. Rep. 2011;39(2):1917–1925. doi: 10.1007/s11033-011-0938-4. [DOI] [PubMed] [Google Scholar]
- Johansen, C.T. et al., Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 42(8), 684–687. [DOI] [PMC free article] [PubMed]
- Kathiresan S. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008;40(2):189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009;41(1):56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T. Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc. Natl. Acad. Sci. U. S. A. 2001;98(24):13710–13715. doi: 10.1073/pnas.231370798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khovidhunkit, W., et al., Resequencing CETP, LIPC and LIPG Genes in Thai Subjects With Hyperalphalipoproteinemia. Am. J. Cardiol. [DOI] [PubMed]
- Kooner J.S. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat. Genet. 2008;40(2):149–151. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- Lanktree M.B. Replication of genetic associations with plasma lipoprotein traits in a multiethnic sample. J. Lipid Res. 2009;50(7):1487–1496. doi: 10.1194/jlr.P900008-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou X.Y. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am. J. Hum. Genet. 2007;80(6):1125–1137. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., et al., Genome-wide association analysis of total cholesterol and high-density lipoprotein cholesterol levels using the Framingham heart study data. BMC Med. Genet. 11, 55. [DOI] [PMC free article] [PubMed]
- Marcais C. Apoa5 Q139X truncation predisposes to late-onset hyperchylomicronemia due to lipoprotein lipase impairment. J. Clin. Invest. 2005;115(10):2862–2869. doi: 10.1172/JCI24471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Musunuru, K., et al., Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N. Engl. J. Med. 363(23), 2220–2227. [DOI] [PMC free article] [PubMed]
- Nelis M. Genetic structure of Europeans: a view from the North-East. PLoS One. 2009;4(5):e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollin T.I. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322(5908):1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radovica I. The association of common SNPs and haplotypes in CETP gene with HDL cholesterol levels in Latvian population. PLoS One. 2013;8(5):e64191. doi: 10.1371/journal.pone.0064191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker P.M. Polymorphism in the CETP gene region, HDL cholesterol, and risk of future myocardial infarction: genomewide analysis among 18 245 initially healthy women from the Women's Genome Health Study. Circ. Cardiovasc. Genet. 2009;2(1):26–33. doi: 10.1161/CIRCGENETICS.108.817304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatti C. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 2009;41(1):35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu M.S. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371(9611):483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Shen, H., et al., Familial defective apolipoprotein B-100 and increased low-density lipoprotein cholesterol and coronary artery calcification in the old order amish. Arch. Intern. Med. 170(20), 1850–1855. [DOI] [PMC free article] [PubMed]
- Soria L.F. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc. Natl. Acad. Sci. U. S. A. 1989;86(2):587–591. doi: 10.1073/pnas.86.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchindran, S., et al., Genome-wide association study of Lp-PLA(2) activity and mass in the Framingham Heart Study. PLoS Genet. 6(4), e1000928. [DOI] [PMC free article] [PubMed]
- Thompson J.F. Cholesteryl ester transfer protein promoter single-nucleotide polymorphisms in Sp1-binding sites affect transcription and are associated with high-density lipoprotein cholesterol. Clin. Genet. 2004;66(3):223–228. doi: 10.1111/j.1399-0004.2004.00289.x. [DOI] [PubMed] [Google Scholar]
- Thompson J.F. CETP polymorphisms associated with HDL cholesterol may differ from those associated with cardiovascular disease. Atherosclerosis. 2005;181(1):45–53. doi: 10.1016/j.atherosclerosis.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Thompson J.F. High-density genotyping and functional SNP localization in the CETP gene. J. Lipid Res. 2007;48(2):434–443. doi: 10.1194/jlr.M600372-JLR200. [DOI] [PubMed] [Google Scholar]
- Utermann G. Polymorphism of apolipoprotein E II. Genetics of hyperlipoproteinemia type III. Clin. Genet. 1979;15(1):37–62. [PubMed] [Google Scholar]
- Vance D.E., Vance J.E. 5th ed. Elsevier; Amsterdam, Boston: 2008. Biochemistry of Lipids, Lipoproteins and Membranes. (631 pp.) [Google Scholar]
- Wallace C. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am. J. Hum. Genet. 2008;82(1):139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth, D.M. et al., Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 30(11), 2264–2276. [DOI] [PMC free article] [PubMed]
- Willer C.J. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40(2):161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, H.G.P. WHO; Geneva: 1999. Familial hypercholesterolemia: report of a second WHO consultation. (WHO/HGN/FH/Cons/99.2) [Google Scholar]
- Wulczyn F.G., Naumann M., Scheidereit C. Candidate proto-oncogene bcl-3 encodes a subunit-specific inhibitor of transcription factor NF-kappa B. Nature. 1992;358(6387):597–599. doi: 10.1038/358597a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Association data for all 139 SNPs used in present study.
Imputed SNPs associated with one or more lipid traits.