Abstract
Molecular genetic fingerprints of eleven Hedychium species from Northeast India were developed using PCR based markers. Fifteen inter-simple sequence repeats (ISSRs) and five amplified fragment length polymorphism (AFLP) primers produced 547 polymorphic fragments. Positive correlation (r = 0.46) was observed between the mean genetic similarity and genetic diversity parameters at the inter-species level. AFLP and ISSR markers were able to group the species according to its altitude and intensity of flower aroma. Cophenetic correlation coefficients between the dendrogram and the original similarity matrix were significant for ISSR (r = 0.89) compared to AFLP (r = 0.83) markers. This genetic characterization of Hedychium from Northeast India contributes to the knowledge of genetic structure of the species and can be used to define strategies for their conservation and management.
Abbreviations: AFLP, amplified fragment length polymorphism; DNA, deoxyribonucleic Acid; GUBH, Gauhati university botanical herbarium; HCA, hierarchical clustering; ISSR, inter- simple sequence repeat; MI, marker index; NE, North east; NTSYS, numerical taxonomy system; PCA, principal component analysis; PCR, polymerase chain reaction; PIC, polymorphic information content; UPGMA, unweighted pair-group arithmetic average.
Keywords: AFLP, Genetic diversity, Hedychium, PCA, Northeast India
Highlights
-
•
Molecular genetic fingerprints of eleven Hedychium species from Northeast India were developed using PCR based markers.
-
•
Fifteen ISSRs and five AFLP primers produced 547 polymorphic fragments.
-
•
AFLP and ISSR markers were able to group the species according to its altitude and intensity of flower aroma.
-
•
Genetic characterization of Hedychium from Northeast India contributes to the knowledge of genetic structure of the species and can be used to define strategies for their conservation and management.
Introduction
Hedychium J. Köenig (Zingiberaceae) is an economically important genus, consisting of 65 species worldwide of which Northeast (NE) India has the highest species concentration (24 out of 65) (Wood et al., 2000). Hedychium species are widely cultivated for their perfume, as a useful raw material for manufacturing paper, medicinal and for its horticultural significance. The medicinal efficacies of the essential oil extracted from leaves, flowers and rhizomes of these plants including cercaricidal properties (Warren and Peters, 1968), molluscicidal activity (Saleh et al., 1982), antimicrobial activities (Gopanraj et al., 2005), and anti-inflammatory and analgesic effects (Shrotriya et al., 2007) are well established.
In spite of huge ornamental and medicinal significance, limited knowledge is available for many members of Hedychium. In addition, high intra and interpopulation variation has led to a debate concerning species concepts and boundaries. Also, the genus is facing the loss of genetic diversity, due to uncontrolled uprooting of the rhizome and the whole plant in huge quantities for preparation of traditional herbal medicine. Species level taxonomy of Zingiberaceae propagating vegetatively is unresolved because morphological differences are inconclusive due to the short lived flowers and owning to the fact that plants flower in monsoon season. So there is an urgent need for sustainable management to conserve the germplasm; however sustainable management has so far not succeeded and further aggravating the extinction. For any future analysis using Hedychium, it may be practical to have information regarding the genetic relatedness of all species. The data on genetic similarity is a mandatory requirement to study the evolutionary history of a species, as well as for other studies, such as intraspecific variations, genetic resources conservation, etc. In the traditional taxonomy of Hedychium, the taxonomic characters are in bract arrangement, number of flowers per bract and petal colors. Such characters simply offered each species a relative, but arbitrary location in the genus, without any quantification of similarity. The quantification of similarity, however, can be easily realized by using molecular techniques.
Molecular markers are used as the taxonomic aid because of advantages of high polymorphism, non-pleiotropy and clear identifying alleles, etc. The major challenge associated with any molecular method is to determine the appropriate taxonomic level at which it is most informative and to correlate it with morphologically definable taxonomic groupings. Inter-simple sequence repeat (ISSR) markers are useful in detecting genetic polymorphisms between and within the species by generating a large number of markers that target multiple microsatellite loci distributed across the genome (Das et al., 2011). Among many researchers, amplified fragment length polymorphism (AFLP) is the marker technology of choice since it combines the reliability of classical restriction-based fingerprinting with the speed and convenience of polymerase chain reaction (PCR)-based marker techniques (Das et al., 2011, Lu et al., 2002, Anderson et al., 1993). The AFLP technique rapidly generates hundreds of highly replicable DNA markers, thus allowing high resolution genotyping. These DNA fingerprinting techniques have been used successfully for confirmation of relationship of Boesenbergia (Vanijajivaa et al., 2005); phylogenetic analysis in Amomum (Kaewsri et al., 2007) and genetic relationship in Curcuma (Das et al., 2011). There is only one report on the phylogenetic analysis and genetic mapping of Chinese Hedychium using sequence related amplified polymorphism (SRAP: Gao et al., 2008). Therefore, the present study was undertaken to investigate the intra- as well as inter-species genetic relationship using principal component analysis (PCA) and hierarchical clustering (HCA) among the species of the Hedychium occurring in NE India by PCR based molecular markers.
Materials and methods
Plant material
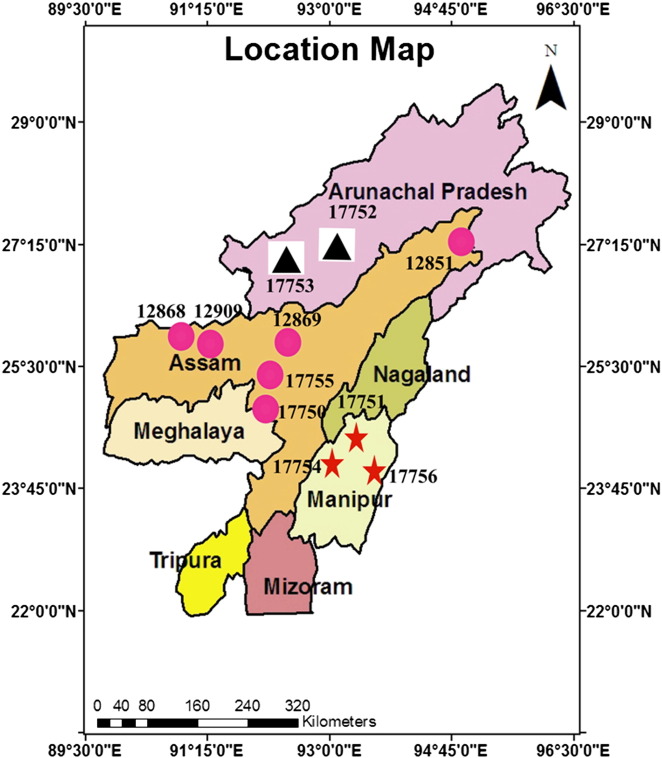
The materials for the present study consisted of 11 species of the genus Hedychium, collected from different locations of NE India (Fig. 1, Table 1). Two individuals of Hedychium coronarium, Hedychium chrysoleucum, Hedychium stenopetalum, Hedychium spicatum, Hedychium gardnerianum and Hedychium flavescens were collected and only a single individual of the remaining five species were collected. Plantlets were maintained in a greenhouse under semi-shade and high humidity (RH 80%) with a 16 h photoperiod at 28 ± 2 °C for hardening. Intermittent mist was supplied for 30 s at 15 min intervals. Herbarium vouchers for the seventeen individuals were deposited in the Gauhati University Botanical Herbarium (GUBH, recognized by American Botanical Society).
Fig. 1.
Geographical distribution of the Hedychium species studied. The numbering of the dots (Assam collections) and triangles (Arunachal Pradesh) and stars (Manipur collections) corresponds to the Hedychium species according to the collection places. Samples have been shown in the figures by the voucher numbers. H. coronarium J. Koenig (17755.1, 17755.2); H. dekianum A.S.Rao & Verma (12868); H. flavum Roxb. (12909); H. stenopetalum Lodd (12869.1, 12869.2); H. spicatum Lodd (17750.1, 17750.2); H. chrysoleucum Hook (12851.1, 12851.2); H. gardnerianum Wall. ex Spreng (17753.1, 17753.2); H. marginatum Clarke (17754); H. flavescens Lodd (17752.1, 17752.2); H. aurantiacum Wall (17751); 11. H. coccineum Wall (17756).
Table 1.
List of Hedychium species used in the study with their geographical origin, morphological information and habitat description.
| S. no. | Species name | Voucher no | Morphological description | Habitat |
|---|---|---|---|---|
| 1 | Hedychium coronarium J. Koenig | 17755.1 17755.2 |
Leaves: ablong or ablong-lanceolate; bracts: large ablong imbricate; flowers: white | Swamps and wet meadows |
| 2 | H. dekianum A.S.Rao & Verma | 12868 | Leaves: ablong or ablong-lanceolate; bracts: large ablong; flowers: white | Lowlands shaded areas |
| 3 | H. flavum Roxb. | 12909 | Leaves: ablong or ablong-lanceolate; bracts: large ablong; flowers: yellow | Humid and shaded areas |
| 4 | H. stenopetalum Lodd. | 12869.1 12869.2 |
Leaves: ablong or ablong-lanceolate; Bracts: ablong; flowers:light orange | Humid and shaded areas |
| 5 | H. spicatum Lodd. | 17750.1 17750.2 |
Leaves: ablong or ablong-lanceolate; bracts: large ablong; flowers: white | From moderately to highly wet areas |
| 6 | H. chrysoleucum Hook. | 12851.1 12851.2 |
Leaves: ablong or ablong-lanceolate; bracts: large ablong; flowers: white tinged with yellow | Humid and shaded areas |
| 7 | H. gardnerianum Wall. ex Spreng. | 17753.1 17753.2 |
Leaves: ablong, white pulverulent beneath; bracts: large ablong; flowers: bright lemon yellow | Windward hilly slopes |
| 8 | H. marginatum Clarke | 17754 | Leaves: ablong or ablong-lanceolate; bracts: ablong; flowers: yellow flower with reddish-brown inflorescence | Windward moist hilly slope |
| 9 | H. flavescens Lodd. | 17752.1 17752.2 |
Leaves: ablong or ablong-lanceolate; bracts: large ablong imbricate; flowers: sulfur yellow | Windward hilly slope |
| 10 | H. aurantiacum Wall. | 17751 | Leaves: long ablong; bracts: ablong; flowers: bright orange yellow | Foot hills receiving heavy rainfall |
| 11 | H. coccineum Wall. | 17756 | Leaves: lanceolate; bracts: ablong; flowers: small bright crimson | Foot hills and moist grassland |
DNA extraction
Total genomic DNA was extracted from fresh tender leaves using a DNeasy Plant mini kit. For DNA extraction two different leaves from each individual of Hedychium were used and subsequent experimental analysis was also carried out in duplicates. The quality and quantity of the extracted DNA were confirmed to be consistent by running the extracted DNA on 0.8% agarose gel, stained with ethidium bromide (0.5 μg/μL). The DNA samples having the A260/280 ratio in between 1.6 and 1.8 and A260/230 ratio in between 2.0 and 2.2 (Wilfinger et al. 1997) were taken into consideration for subsequent marker analysis.
ISSR and AFLP analyses
The experimental procedures for ISSR and AFLP analyses were as described previously (Das et al., 2011). Fifteen primers were used for ISSR analysis and five sets of primers were used for AFLP fingerprinting (Table 3).
Table 3.
Degree of polymorphism and polymorphic information content for inter-species genetic relationship in Hedychium.
| Primer code no | TNB | NPB | % P | PIC | MI |
|---|---|---|---|---|---|
| ISSR primer | |||||
| 17898B | 5 | 4 | 80.0 | 0.39 | 31.20 |
| 826 | 11 | 11 | 100.0 | 0.35 | 35.00 |
| HB12 | 13 | 13 | 100.0 | 0.39 | 39.00 |
| HB13 | 8 | 7 | 87.5 | 0.45 | 39.37 |
| 17899A | 11 | 11 | 100.0 | 0.34 | 34.00 |
| 816 | 9 | 9 | 100.0 | 0.38 | 38.00 |
| 817 | 9 | 8 | 88.8 | 0.40 | 35.52 |
| 811 | 10 | 6 | 60.0 | 0.29 | 17.40 |
| 825 | 12 | 12 | 100.0 | 0.33 | 33.00 |
| 807 | 9 | 9 | 100.0 | 0.45 | 45.00 |
| 824 | 10 | 9 | 90.0 | 0.33 | 29.70 |
| HB15 | 7 | 6 | 85.7 | 0.33 | 28.28 |
| 814 | 12 | 12 | 100.0 | 0.39 | 39.00 |
| 17899A | 6 | 5 | 83.3 | 0.42 | 34.98 |
| 809 | 9 | 8 | 88.8 | 0.39 | 34.63 |
| Total | 141 | 131 | 1364.10 | 5.24 | 514.08 |
| Mean | 9.71 | 9.00 | 90.94 | 0.37 | 34.27 |
| AFLP primer | |||||
| MseI-CAC/EcoRI-ACT | 111 | 111 | 100 | 0.27 | 27 |
| MseI-CAG/EcoRI-ACT | 46 | 46 | 100 | 0.32 | 32 |
| MseI-CAT/EcoRI-ACT | 110 | 110 | 100 | 0.30 | 30 |
| MseI-CTC/EcoRI-ACT | 51 | 51 | 100 | 0.29 | 29 |
| MseI-CTG/EcoRI-ACT | 98 | 98 | 100 | 0.28 | 28 |
| Total | 416 | 416 | 500 | 1.46 | 146 |
| Mean | 83.2 | 83.2 | 100 | 0.292 | 29.2 |
TNB total number of bands, NPB number of polymorphic bands, % P percentage of polymorphism, PIC polymorphic information content, MI marker index.
Data analysis
For scoring and analysis of data from two molecular marker systems, duplicate samples from each individual were tested. The total number of monomorphic and polymorphic bands which were clear, unambiguous and reproducible was scored for all the tested primers. Data counting/scoring was carried out by using a binary number system for the presence or absence of each fragment in each sample. To avoid taxonomic ambiguities, all bands were taken into consideration, only the presence of a band was taken as an indicative. To compare the efficiency of each primer polymorphic information content (PIC); as a marker discrimination power, was computed using the formula PIC = 1 − ∑ pi2, where pi is the frequency of ith allele at a given locus and also a marker index (MI) was calculated (Powell et al., 1996). MI defined as the product of the polymorphism percentage and PIC, is used to estimate the overall utility of each marker system and was calculated according to Kesari et al. (2010). Measure of degree of similarity among 11 species was established as a percentage of polymorphic bands, and a matrix of genetic similarity compiled using Dice's coefficient (Dice, 1945). A dendrogram representing the genetic relationship among all Hedychium species was generated for each marker system (ISSR and AFLP) by applying unweighted pair-group arithmetic average (UPGMA) method (Sneath and Sokal, 1973) using the SHAN subroutine through the NTSYS-pc (numerical taxonomy system, 2.2 version) (numerical taxonomy system, Applied Biostatistics, NY) (Rohlf, 2005). To know the goodness of fit for 11 species of Hedychium to a specific cluster in the UPGMA algorithm, the relationships between the original similarity indices and cophenetic values were evaluated, and the Mantel's test (Mantel, 1967) was executed for it. PCA was also performed with modules STAND, CORR, and EIGEN of NTSYS-pc using the Euclidean distances derived from the standardized values using the NTSYS-pc-2.2 for only polymorphic bands. The filtration of data was carried out by STAND module before PCA analysis (Milligan and Cooper, 1987). Genetic diversity was estimated for intra and inter-species variation using POPGENE, version 1.32 (Yeh et al., 1997). The levels of genetic variability within species were estimated using five variables: the observed number of alleles per locus (na), the effective number of alleles per locus (ne), proportion of polymorphic loci (P) per species, Shannon information index (I), Nei's gene diversity (h) (McDermott and McDonald, 1993). HCA was further created for Hedychium, where each profile is a string with 11 entries (number of species analyzed). HCA was performed with Cluster (v. 3.0) and visualized with TreeView (Eisen et al., 1998). Pair wise linkage was used as the routine for hierarchical clustering (de Hoon et al., 2004). Species in the study were clustered according to the similarity of their phylogenetic profiles and if there was a similarity of one species to that of the other species, the value 1 was assigned at that position (red), if not, 0 was assigned (black).
Results
ISSR and AFLP polymorphisms
The intra-species genetic diversity was studied among 12 individuals of six species of Hedychium. Out of the 20 ISSR primers, 5 gave successful amplification with a total of 28 bands, of which 25 were polymorphic with polymorphism of 89.28%. The highest number of polymorphic fragments was observed for 826 (7) and the lowest number of polymorphic fragments was observed for 17898A (3), with an average of 5.0 per primer. The PIC varied from 0.31 (807) to 0.41 (17898A) with an average PIC of 0.37 (Table 2).
Table 2.
Degree of polymorphism and polymorphic information content for intra-species genetic relationship in Hedychium.
| Primer code no | TNB | NPB | % P | PIC | MI |
|---|---|---|---|---|---|
| 807 | 6 | 5 | 83.33 | 0.31 | 25.46 |
| 826 | 7 | 7 | 100.00 | 0.37 | 36.80 |
| 17898A | 5 | 3 | 60.00 | 0.41 | 25.0 |
| 17899A | 6 | 6 | 100.00 | 0.40 | 40.04 |
| HB12 | 4 | 4 | 100.00 | 0.38 | 37.84 |
| Total | 28 | 25 | |||
| Mean | 5.6 | 5.0 | 89.28 | 0.374 | 33.03 |
The inter-species genetic diversity was studied for the 11 Hedychium species of NE India. Of the 20 ISSR primers, 15 gave the successful amplification with a total of 141 bands, of which 131 were polymorphic with polymorphism of 91%. The highest number of polymorphic fragments obtained was for HB 13 (13) and the lowest for 17898B (4) with an average of 9.00 per primer. The polymorphic information content (PIC) varied from 0.29 (811) to 0.45 (HB13) with an average PIC of 0.37. Of the 8 AFLP primer combinations five gave the successful amplification with a total of 416 polymorphic fragments with 100% polymorphism. The PIC varied from 0.27 (MseI-CAC/EcoRI-ACT) to 0.32 (MseI-CAG/EcoRI-ACT) with an average value of 0.29. The MI varied from 27 (MseI-CAC/EcoRI-ACT primer combination) to 32 (MseI-CAG/EcoRI-ACT primer combination: Table 3).
Gene diversity
Intra-species diversity among the 12 individuals of 6 species was found to be low as revealed by ISSR markers. The average value of na, ne, h, I and % P varied from 1.01 ± 0.14 to 1.33 ± 0.48, 1.01 ± 0.09 to 1.24 ± 0.34, 0.01 ± 0.00 to 0.14 ± 0.01, 0.01 ± 0.00 to 0.20 ± 0.02, and 1.85 to 33.33 respectively. But the inter-species gene diversity was found to be high among the 11 species of Hedychium as revealed by ISSR and AFLP markers. The average value of na and ne, varied from 1.99 to 2.00, and 1.51 to 1.53, respectively. The average values of h and I were found to be 0.31 and 0.48, respectively (Table 4).
Table 4.
Genetic diversity parameters for Hedychium.
| Population | Sample size (n) |
Observed no of alleles (na) |
Effective no of alleles (ne) |
Nei's gene diversity (h) | Shannon's information index (I) | % P | |
|---|---|---|---|---|---|---|---|
| Intra-species | ISSR | ||||||
| H. coronarium | 2 | 1.33 ± 0.48 | 1.24 ± 0.34 | 0.14 ± 0.01 | 0.20 ± 0.02 | 33.33 | |
| H. chrysoleucum | 2 | 1.01 ± 0.14 | 1.01 ± 0.09 | 0.01 ± 0.00 | 0.01 ± 0.00 | 1.85 | |
| H. stenopetalum | 2 | 1.05 ± 0.23 | 1.03 ± 0.16 | 0.02 ± 0.01 | 0.03 ± 0.00 | 5.56 | |
| H. spicatum | 2 | 1.05 ± 0.23 | 1.03 ± 0.16 | 0.02 ± 0.01 | 0.03 ± 0.01 | 5.56 | |
| H. gardnerianum | 2 | 1.05 ± 0.23 | 1.03 ± 0.16 | 0.02 ± 0.01 | 0.03 ± 0.01 | 5.56 | |
| H. flavescens | 2 | 1.05 ± 0.23 | 1.03 ± 0.16 | 0.02 ± 0.01 | 0.03 ± 0.01 | 5.56 | |
| Total gene diversity | 12 | 1.92 ± 0.26 | 1.60 ± 0.30 | 0.19 ± 0.14 | 0.09 ± 0.19 | 40.00 | |
| Inter-species | Total gene diversity (ISSR) | 11 | 1.99 ± 0.08 | 1.53 ± 0.32 | 0.32 ± 0.15 | 0.48 ± 0.18 | 90.94 |
| Total gene diversity (AFLP) | 11 | 2.00 ± 0.08 | 1.51 ± 0.32 | 0.31 ± 0.15 | 0.48 ± 0.18 | 100.00 |
Genetic similarity analysis
Cluster analysis and PCA are valuable for determining relationships among individuals of the same and different species (Crawford, 1990). The PCA plot and UPGMA dendrogram of the intra-species genetic diversity study by ISSR markers showed two clusters for 12 individuals of Hedychium. Cluster I consisted of the individuals from H. coronarium, H. chrysoleucum, H. flavescens, H. stenopetalum and H. spicatum. Cluster II consisted of the individuals from H. gardnerianum. Dendrogram showed the highest genetic similarity between the individuals of H. chrysoleucum and the lowest genetic similarity between the individuals of H. coronarium. Mantel test of cophenetic correlation matrix and the similarity matrix revealed a very good degree of confidence (r = 0.96) in the association obtained for the 12 individuals of Hedychium (Supplementary Fig. 1)
For inter-species genetic relationship among 11 species of Hedychium, PCA of 15 ISSRs and 5 AFLP primer combinations displayed differential grouping patterns. The highest cumulative contribution of the first three principal components to total variation was found to be by ISSR markers (57.24%) compared to AFLP (42.94%) marker. The highest percent contribution of Eigenvalues of ISSR marker system is implicating better information of relatedness for the species studied. PCA analysis of ISSR and AFLP data-set for inter-species genetic relationship showed two discrete clusters (Fig. 2).
Fig. 2.
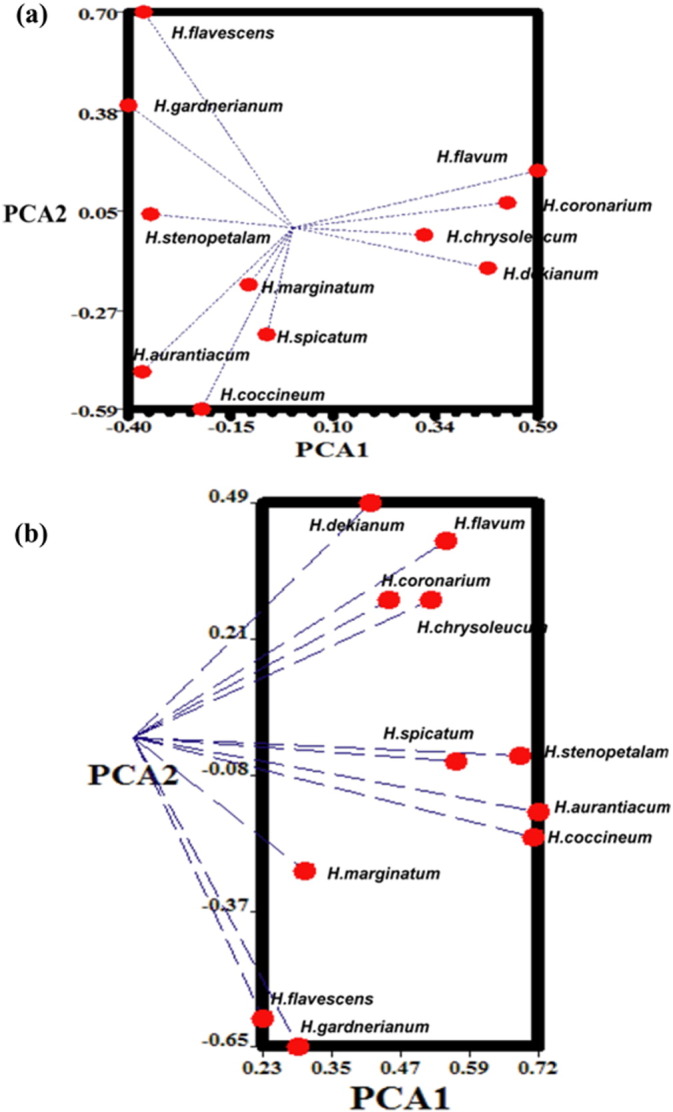
PCA plots of the 11 Hedychium species of Northeast India based on molecular markers. (a). ISSR (b). AFLP. The first three components explained 57.24% and 42.94%, of the variation. PCA analysis of ISSR and AFLP dataset segregates species collected from different altitudes and flower aroma into two groups represented by dotted lines.
The DICE genetic similarity values derived from the ISSR data ranged from 0.58 (H. flavescens and Hedychium dekianum) to 0.79 (Hedychium coccineum and Hedychium aurantiacum). Similarly AFLP showed the lowest genetic similarity coefficient between H. chrysoleucum and H. stenopetalum (0.63) while the highest value was calculated between H. coccineum and H. aurantiacum (0.79). This result showed that the inter-species genetic similarity of the genus Hedychiumis almost the same using two different PCR based molecular markers. Dendrogram constructed based on AFLP similarity matrix, showed two distinct groups for the 11 species of Hedychium at the level of similarity of 0.60, placing H. flavescens and H. edychium marginatum in cluster II and the rest of the species in the cluster I. H. flavescens and H. marginatum had 63.2% similarity between them. Cluster I formed two subgroups. Subgroup I consisted of H. edychium; flavum, H. coronarium, H. dekianum and H. chrysoleucum. Subgroup II of cluster I consisted of H. spicatum, H. stenopetalum, H. coccineum, H. aurantiacum and H. gardnerianum. The level of similarity between H. coccineum and H. aurantiacum was found to be 0.790, which is of similar magnitude to that of the ISSR (0.787) dendrogram. The species clustering of ISSR dendrogram was similar to AFLP dendrogram in the subgroup I of cluster I (Supplementary Fig. 2a & b). The high amount of red area in ISSR and AFLP HCA compared to the other dendrograms signifies better informativeness (Supplementary Fig. 2c & d).
The separation approach as revealed by the Mantel test comparing the results of ISSR and AFLP indicated a significant correlation among the 11 different Hedychium species. Mantel test yielded results ranging from good fit to very good fit of cophenetic values (0.83 < r < 0.86) (Table 5).
Table 5.
Comparison of correlation coefficient between similarity matrices and co-phenetic matrices derived from different markers.
| Similarity matrix values | |||
| ISSR | AFLP | ||
| Cophenetic correlation | ISSR | 0.86 | 0.06 |
| AFLP | − 0.02 | 0.83 | |
Note: Above diagonal values represent correlation coefficients between similarity matrices and below diagonal values represent correlation coefficients between co-phenetic matrices. Values on the diagonal represent co-phenetic correlation for markers.
Discussion
Hedychium commonly known as ‘ginger lily’ or ‘butterfly lily’is well known for its medicinal and ornamental importance. Classification of genus has been contentious and no report on genetic relationships of Hedychium species of NE India exists. Relying much on the morphological characters alone in species delineation has its own limitations since they are not always complete representative of the genetic structure. Molecular markers assume great significance, as these methods detect polymorphism by assaying subsets of the total amount of DNA sequence variation in a genome.
The intra-species variation in terms of percentage of polymorphism, observed number of alleles, effective number of alleles, Nei's gene diversity and Shannon's information index was found to be highest in H. coronarium and to be lowest in H. chrysoleucum. This might be the result of cross pollination in H. coronarium and vegetative propagation in other individuals of the species.
Marker studies separated Hedychium into 2 main clusters based on different altitudes and intensities of flower aroma. In the ISSR dendrogram, species of the moist lowlands with strong fragrant flower (H. coronarium, H. dekianum, H. flavum, H. chrysoleucum, H. stenopetalum, H. spicatum, H. marginatum, H. coccineum, H. aurantiacum) were combined in cluster I. The strong aroma of the flowers may be the evolutionary derived trait among the species. A small cluster (cluster II) consisted of species of H. flavescens and H. gardnerianum that possessed geographic similarity of high altitude and has yellow and mildly fragrant flowers. AFLP marker analysis too closely follows the altitude based clustering of ISSR marker analysis. However, our study of PCR based molecular markers deviates from the DNA analysis by other researchers who split Hedychium into three groups in terms of the species distribution according to altitude; each group correlated to a separate climatic zone (Wood et al., 2000). The discrepancy of our study from the earlier reported study by SPAR based marker was the positioning of H. spicatum which was found to cluster with H. stenopetalum (Gao et al., 2008).
The PCA and cluster analyses of a binary matrix (ISSR and AFLP) showed a unique genetic structure in Hedychium. These results suggest that the manner of polymorphism differs because of marker specificity. Modest correlation between marker systems was also reported in soybean (Powell et al., 1996), maize (Pejic et al., 1998), and safflower (Sehgal and Raina, 2005). In addition, the relation is assumed to depend on the genome coverage and sequence type recognized by each marker system (Pejic et al., 1998, Powell et al., 1996, Sehgal and Raina, 2005). Therefore, ISSR and AFLP are reliable methods for calculating genetic relationships reflecting coding and non-coding regions of the genome and they could well be used in aiding identification as well as classification of the Zingiberaceae using more species in each genus. Given the correlation of similarity matrix data from different markers, we therefore prefer to combine them in analyses, because this approach generally helps to overcome errors or introgression at one locus, and to increase explanatory power (Edwards and Beerli, 2000, Nixon and Carpenter, 2005).
This is the first report on genetic relationships for Hedychium species of NE India producing trees and PCA plot with high topological support for most nodes using ISSR and AFLP markers. The UPGMA, hierarchical clustering and PCA analysis of ISSR and AFLP datasets represent our best hypothesis of genetic relationship among Hedychium. We recommend the use of ISSR and AFLP topology as the most complete framework for future studies of phylogenetic comparative analyses, tests of bio-geographic hypotheses and models of trait evolution. This study confirmed that two marker utilizations (ISSR and AFLP) can be crucial for estimation of the relatedness of Hedychium at the inter-species level.
The following are the Supplementary data related to this article.
Intra-species genetic relationship study of six species of Hedychium. (a). PCA (ISSR). The first three components explained 69.04% of the variation. (b). The UPGMA dendrogram for inter-species genetic relationship study of six Hedychium species based on the ISSR data set. (c). Correlation coefficient of the similarity matrix and the cophenetic matrix of the ISSR dataset (r = 0.96) highlighting a high level of confidence of dendrogram formed for the intra-species study of six species of Hedychium.
Phylogenetics and hierarchical clustering.UPGMA dendrogram of (a) ISSR; (b) AFLP maker system; (c) HCA of all of the samples by ISSR; (d) HCA of all of the samples by AFLP.
Acknowledgments
Thanks to the Department of Biotechnology, IIT Guwahati, Tezpur University and M S Swaminathan Research Foundation for extending all infrastructural facilities. Thanks to Dr. Sarma, Gauhati University for the kind supply of study material. AP, SM and LR thank the Department of Biotechnology (DBT) Government of India for funding the project by way of DBT Twinning Programme for NE (BT/33/NE/TBP/2010).
Contributor Information
Supriyo Basak, Email: supriyo@iitg.ac.in.
Aadi Moolam Ramesh, Email: aadi@iitg.ac.in.
Vigya Kesari, Email: vigyakesari@gmail.com.
Ajay Parida, Email: ajay@mssrf.res.in.
Sudip Mitra, Email: sudipmitra@yahoo.com.
Latha Rangan, Email: latha_rangan@yahoo.com, lrangan@iitg.ernet.in.
References
- Anderson J.A., Churchill G.A., Autrique J.E., Tanksley S.D., Sorrells M.E. Optimizing parental selection for genetic linkage maps. Genome. 1993;36:181–186. doi: 10.1139/g93-024. [DOI] [PubMed] [Google Scholar]
- Crawford D.J. Wiley; New York: 1990. Plant Molecular Systematics: Macromolecular Approaches. [Google Scholar]
- Das A., Kesari V., Satyanarayan V.M., Parida A., Rangan L. Genetic relationship of Curcuma species from Northeast India using PCR-based markers. Mol. Biotechnol. 2011;49:65–76. doi: 10.1007/s12033-011-9379-5. [DOI] [PubMed] [Google Scholar]
- de Hoon M.J.L., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Dice L.R. Measures of the Amount of Ecologic Association Between Species. Ecology. 1945;26:297–302. [Google Scholar]
- Edwards S.V., Beerli P. Perspective: gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution. 2000;54:1839–1854. doi: 10.1111/j.0014-3820.2000.tb01231.x. [DOI] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Liu N., Huang B., Hu X. Phylogenetic analysis and genetic mapping of Chinese Hedychium using SRAP markers. Sci. Hortic. 2008;117:369–377. [Google Scholar]
- Gopanraj G., Dan M., Shiburaj S., Sethuraman M.G., George V. Chemical composition and antibacterial activity of the rhizome oil of Hedychium larsenii. Acta Pharma. 2005;55(3):315–320. [PubMed] [Google Scholar]
- Kaewsri W., Paisooksantivatana Y., Veesommai U., Eiadthong W., Vajrodaya S. Phylogenetic analysis of Thai Amomum (Alpinioideae: Zingiberaceae) using AFLP markers. Kasetsart J. Nat. Sci. 2007;41:213–226. [Google Scholar]
- Kesari V., Sathyanarayana V.M., Parida A., Rangan L. Molecular marker-based characterization in candidate plus trees of Pongamia pinnata, a potential biodiesel legume. AoB Plants. 2010 doi: 10.1093/aobpla/plq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- Lu G., Cao J.S., Chen H. Genetic linkage map of Brassica campestris L. using AFLP and RAPD markers. J. Zhejiang Univ. Sci. A. 2002;3(5):600–605. [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27(2):209–220. [PubMed] [Google Scholar]
- McDermott J.M., McDonald B.A. Gene flow in plant pathosystems. Annu. Rev. Phytopathol. 1993;31:353–373. [Google Scholar]
- Milligan G.W., Cooper M.C. The Ohio State University; Columbus, OH: 1987. A Study of Variable Standardization, College of Administrative Science Working Paper Series, 87–63. [Google Scholar]
- Nixon K.C., Carpenter J.M. On simultaneous analysis. Cladistics. 2005;12:221–241. doi: 10.1111/j.1096-0031.1996.tb00010.x. [DOI] [PubMed] [Google Scholar]
- Pejic I., Ajmone-Marsan P., Morgante M., Kozumplick V., Castiglioni P., Taramino G., Motto M. Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs and AFLPs. Theor. Appl. Genet. 1998;97:1248–1255. [Google Scholar]
- Powell W., Morgante M., Andre C., Hanafey M., Vogel J., Tingey S., Rafalski A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 1996;2:225–238. [Google Scholar]
- Rohlf F.J. Exeter Software; Setauket, NY: 2005. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, Version 2.2. [Google Scholar]
- Saleh M.M., Kamal R.M., Abdulla W.A. A contribution to the molluscicidal activity of Hedychium gardinarum. Planta Med. 1982;45(7):166. doi: 10.1055/s-2007-971366. [DOI] [PubMed] [Google Scholar]
- Sehgal D., Raina S.N. Genotyping safflower (Carthamus tinctorius) cultivars by DNA fingerprints. Euphytica. 2005;146:67–76. [Google Scholar]
- Shrotriya S., Ali M.S., Saha A., Bachar S.C., Islam M.S. Anti-inflammatory and analgesic effects of Hedychium coronarium Koen. Pak. J. Pharm. Sci. 2007;20(1):47–51. [PubMed] [Google Scholar]
- Sneath P.H.A., Sokal R.R. Freeman; San Francisco: 1973. Numerical Taxonomy. [Google Scholar]
- Vanijajivaa O., Sirirugsab P., Suvachittanont W. Confirmation of relationships among Boesenbergia (Zingiberaceae) and related genera by RAPD. Biochem. Syst. Ecol. 2005;33:159–170. [Google Scholar]
- Warren K.S., Peters P.A. Cercariae of Schistosoma mansoni and plants: attempt to penetrate Phaseolus vulgaris and Hedychium coronarium produces a cercaricide. Nature. 1968;217(5129):647–648. doi: 10.1038/217647a0. [DOI] [PubMed] [Google Scholar]
- Wilfinger W.W., Mackey K., Chomczynski P. Effect of pH and ionic strength on the spectrophotometric assessment of nucleic acid purity. Biotechniques. 1997;22(3):474–476. doi: 10.2144/97223st01. (478–81) [DOI] [PubMed] [Google Scholar]
- Wood T.H., Whitten W.M., Williams N.H. Phylogeny of Hedychium and related genera (Zingiberaceae) based on ITS sequence data. Edinb. J. Bot. 2000;57(2):261–2170. [Google Scholar]
- Yeh F.C., Yang R.C., Boyle T.B.J., Ye Z.H., Mao J.X. Molecular Biology and Biotechnology Centre, University of Alberta; Alberta: 1997. POPGENE, the User Friendly Shareware for Population Genetic Analysis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Intra-species genetic relationship study of six species of Hedychium. (a). PCA (ISSR). The first three components explained 69.04% of the variation. (b). The UPGMA dendrogram for inter-species genetic relationship study of six Hedychium species based on the ISSR data set. (c). Correlation coefficient of the similarity matrix and the cophenetic matrix of the ISSR dataset (r = 0.96) highlighting a high level of confidence of dendrogram formed for the intra-species study of six species of Hedychium.
Phylogenetics and hierarchical clustering.UPGMA dendrogram of (a) ISSR; (b) AFLP maker system; (c) HCA of all of the samples by ISSR; (d) HCA of all of the samples by AFLP.