Abstract
Recent studies from East Asia and Canadian National Collection of Insects have established the utility of DNA barcoding technique in identification of true bugs. The present study is an expansion of the database by adding mitochondrial cytochrome c oxidase I (mtCOI) sequences from forty three species of indigenous true bugs of India. mtCOI gene analysis of infraorder Pentatomomorpha covering a total of seventy three species that belong to five superfamilies; Pentatomoidea, Coreoidea, Pyrrhocoroidea, Lygaeoidea and Aradoidea revealed more than 3% interspecific distances in all the taxa studied except for two cases which showed barcode sharing. Less than 2% intra-specific divergence was observed in 97% of the taxa analysed and the average interspecies genetic distance was about 29 times higher than the average intraspecies genetic divergence. Distinct sequence divergence pattern at generic level and NJ clustering analysis suggests that COI barcode is an excellent molecular marker for species level identification of unknown taxa; however it may not be useful for resolving deep levels of divergence. Species identification even at nymphal stage could be achieved confirming the efficacy of this technique.
Abbreviations: COI, Cytochrome c oxidase I; CBOL, Consortium for the Barcode of Life; iBOL, International Barcode of Life, dNTPs, deoxynucleotide triphosphates; EtBr, Ethidium bromide; PEG, Polyethylene glycol; K2P, Kimura-2-parameter; NCBI, National Centre for Biological Information; BLAST, Basic local alignment search tool, MEGA, Molecular evolutionary genetics analysis; NJ, Neighbour joining; MP, Maximum parsimony
Keywords: DNA barcoding, mtCOI, Heteroptera, Pentatomomorpha, Western Ghats, India
Graphical abstract
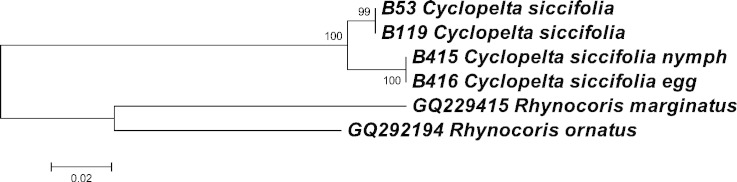
Fig. 1. NJ tree based on mtCOI from Cyclopelta siccifolia in three different life stages; egg, nymph and adult. Two Cimicomorpha taxa Rhynocoris marginatus and Rhynocoris ornatus are used as outgroups.
Highlights
-
•
mtCOI barcoding of true bugs from Western Ghats of India is reported.
-
•
Study includes 9 families and 73 species; 43 from present work and 30 from GenBank.
-
•
mtCOI barcodes can be used for species level identification of unknown bug taxa.
-
•
Species identification even at nymphal stage could be achieved with this technique.
-
•
Present study adds 41 new mtCOI sequences to the existing global database.
Introduction
Pentatomomorpha is a large group of many phytophagus as well as seed-eating insects, commonly called as plant bugs, with more than 10,000 species worldwide (Dolling, 1991, Schuh and Slater, 1995). Morphology-based study of Pentatomomorpha has a long history and several workers have recognized different superfamilies in this infraorder (Schuh and Slater, 1995) but there is little agreement on their relationships. Recently Yao et al. (2012) reported phylogeny of this infraorder based on fossil and extant species morphology. Molecular work to facilitate identification and to seek relationship amongst the various families is necessary in this vast group.
Initial work on COI-based identification of some Heteropteran groups (Damgaard, 2008, Memon et al., 2006) reported limited utility of this technique but recent work from Jung et al. (2011) and Park et al. (2011) have confirmed the efficacy of DNA barcoding for identification of true bug species. Whilst Damgaard (2008) considered COI sequences from the 3′ end, the species in Memon et al. (2006) work were from taxonomically disputed groups. Jung et al. (2011) presented COI variation in species from Korean Peninsula and Park et al. (2011) expanded this database with specimens from Canadian National Collection of Insects. Besides identification these DNA sequences were used to find phylogenetic relationship amongst the species. There are some notable previous reports incorporating molecular data into phylogeny studies of Pentatomomorpha (Hua et al., 2008, Li et al., 2005).
Our search showed that efforts have been initiated to barcode true bugs from India (Rebijith et al., 2012) but were restricted to a few species from a single family Miridae. Therefore the present work was undertaken to identify more species using COI barcodes covering wider taxa range. For this, bug species were collected from the Pune and other parts of Western Ghats, which is a well-known biodiversity hotspot with thousands of plant and animal species; many of them being still unexplored. In spite of the fact that many plant bugs are economically very important and also considerable previous work (Distant, 1904) has been done on traditional taxonomy of this group in India, hardly any molecular data is available for these species. Therefore, the present study aims to check the utility of molecular techniques in identification of true bugs and in this context feasibility of DNA barcoding is explored.
Materials and methods
Analysis of seventy three taxa representing five superfamilies and ten families was carried out for barcoding studies of infraorder Pentatomomorpha. Voucher numbers of specimens used, their taxonomy, GenBank accession numbers and collection sites are shown in Table 1. All attempts were made to include more than one representative of all taxa, but sample availability was a major constraint. Sequences from congeneric specimens deposited by other workers were directly taken from GenBank. Two taxa (Rhynocoris ornatus and Rhynocoris marginatus) that belong to Cimicomorpha; a sister group of Pentatomomorpha, were used as outgroups. Accession numbers for these sequences are also included in Table 1.
Table 1.
Taxa used in the present study. Abbreviations: Amb, Amboli; Bgn, Bhugaon (area near Pune); Bhj, Bhaje (area near Pune); Khm, Kihim; Kyn, Koyna dam; Mgh, Meghalaya; Mul, Mulshi (area near Pune); Nsk, Nasik; Pun, Pune; Sol, Solapur; Tmh; Tamhini (area near Pune); and Vsp, Visapur (area near Pune).
| Classification | Taxa | Voucher no. | GenBank acc. no. | Locality |
|---|---|---|---|---|
| Pentatomoidea | ||||
| Pentatomidae | Carbula insocia (Walker) | B105 | GU247472 | Pun., India |
| Carbula insocia nymph | B103 | HQ333534 | Pun., India | |
| Carbula putonia (Jakovlev) | – | GQ292251 | – | |
| Carbula humerigeraa (Uhler) | – | GQ292241 | – | |
| Catacanthus incarnatus (Drury) | B18 | GU247474 | Pun., India | |
| Catacanthus incarnatus nymph | B263 | HQ236459 | Mul., India | |
| Halys dentatus (Fabricius) | B44 | GU247475 | Mul., India | |
| Halys sp. | B226 | HQ236461 | Sol., India | |
| Eocanthecona furcellata (Wolff) | B116 | GU247478 | Pun., India | |
| Eocanthecona furcellata nymph | B128 | GU247479 | Bhj, India | |
| Eocanthecona japanicolaa (Esaki and Ishihara) | – | GQ292274 | – | |
| Plautia crossota (Dallas) | B93 | GU247480 | Pun., India | |
| Plautia splendensa Distant | – | GQ292240 | – | |
| Plautia stali Scotta | – | JN087440 | – | |
| Bathycoelia indica Dallas | B265 | HQ236463 | Mul., India | |
| Halyomorpha picus (Fabricius) | B21 | GU247481 | Khm, India | |
| Halyomorpha picus nymph | B73 | GU247482 | Pun., India | |
| Halyomorpha halysa Stal | – | KF273401 | – | |
| Niphe subferruginea (Westwood) | B23 | GU247483 | Khm., India | |
| Nezara viridula nymph | B42 | GQ306225 | Bhj., India | |
| Nezara viridula (Linnaeus) | B225 | HQ236460 | Nsk., India | |
| Nezara viridulaa (Linnaeus) | – | JX548495 | Pun., India | |
| Placosternumalces (Stal) (Fabricius) | B261 | HQ236462 | – | |
| Placosternum esakiia (Miyamoto) | – | GQ292243 | – | |
| Dalpada sp. | B149 | GU247484 | Kyn, India | |
| Dinidoridae | Cyclopelta siccifolia (Westwood) | B53 | GU247485 | Pun., India |
| Coridius janus (Fabricius) | B110 | GU247486 | Pun., India | |
| Coridius brunneus (Thunberg) | B111 | GU247487 | Pun., India | |
| Coridius brunneus nymph | B113 | GU247488 | Pun., India | |
| Coridius sp. | B290 | HQ333535 | Mgh, India | |
| Coridius chinensisa (Dallas) | – | JQ387599 | – | |
| Coridius nigriventrisa (Westwood) | – | JQ387600 | – | |
| Megymenum distanti Kocorek & Ghate | B294 | HQ333537 | Pun., India | |
| Megymenum gracilicornea Dallas | – | GQ292282 | – | |
| Megymenum sp.a | – | AY252965 | – | |
| Scutelleridae | Solenostethium rubropunctatum (Guerin) | B187 | HQ236465 | Pun., India |
| Chrysocoris purpureus (Westwood) | B2 | GU247490 | Pun., India | |
| Choerocoris variegatusa Dallas | – | AY253006 | – | |
| Tessaratomidae | Tessaratoma sp. | B258 | HQ236466 | Pun., India |
| Tessaratoma papilosaa (Drury) | – | AY252948 | – | |
| Tessaratoma javanica (Thunberg) | – | KF534924 | – | |
| Coreoidea | ||||
| Alydidae | Leptocorisa acuta (Thunberg) | B17 | GU247495 | Bgn., India |
| Leptocorisa chinensisa Dallas | – | GQ292203 | – | |
| Nariscus fumosus (Distant) | B76 | GU247492 | Pun., India | |
| Nariscus fasciatus (Distant) | B30 | GU247491 | Pun., India | |
| Riptortus pedestris (Fabricius) | B224 | KJ685902 | Tmh., India | |
| Riptortus clavatus (Thunberg) | – | GQ292204 | – | |
| Riptortus clavatus (Thunberg) | – | KC510111 | – | |
| Heegeria tangirica (Saunders) | B83 | KJ685903 | Pun., India | |
| Rhopalidae | Leptocoris augur (Fabricius) | B190 | HQ236472 | Nsk., India |
| Leptocoris augur nymph | B191 | HQ236473 | Nsk., India | |
| Serinthea sp.a | – | AY253139 | – | |
| Coreidae | Plinachtus acicularis (Fabricius) | B13 | GU247496 | Pun., India |
| Plinachtus bicoloripesa Scott | – | GQ292218 | – | |
| Cletus rubidiventris (Westwood) | B26 | GU247497 | Khm., India | |
| Cletus sp.a | – | AY252993 | – | |
| Cletus schmidtia Kiritshenko | – | GQ292220 | – | |
| Petillopsis calcar nymph | B131 | GU247498 | Bhj., India | |
| Petillopsis calcar Dallas | B271 | HQ333539 | Tmh., India | |
| Physomerus parvulus Dallas | B244 | HQ236467 | Amb., India | |
| Physomerus grossipes (Fabricius) | B56 | GU247499 | Pun., India | |
| Acanthocoris scabrator (Fabricius) | B141 | GU247500 | Pun., India | |
| Acanthocoris sordidus (Thunberg) | – | GQ292219 | – | |
| Homoeocerus sp. | B63 | GU247508 | Mul., India | |
| Homoeocerus sp. | B152 | HQ236470 | Pun., India | |
| Prismatocerus prominulus (Dallas) | B202 | HQ236468 | Vsp., India | |
| Homoeocerus (Tliponius) unipunctatusa (Thunberg) | – | GQ292221 | – | |
| Anoplocnemis phasianus (Fabricius) | B208 | HQ236471 | Tmh., India | |
| Lygaeoidea | Lygaeus pandurus (Scopoli) | B61 | GU247502 | Pun., India |
| Lygaeidae | Lygaeus pandurus nymph | B20 | GU247503 | Pun., India |
| Lygaeus militaris (Fabricius) | B3 | GU247504 | Pun., India | |
| Lygaeus kalmiia Stal | – | GU013621 | – | |
| Lygaeus sjostedtia (Lindberg) | – | GQ292261 | – | |
| Metochus uniguttatus (Thunberg) | B54 | GU247506 | Pun., India | |
| Dieuches sp.a | – | AY253136 | – | |
| Dieuches leucoceras (Walker) | B65 | GU247505 | Pun., India | |
| Pyrrhocoroidea | ||||
| Pyrrhocoridae | Dysdercus evanescence Distant | B60 | GU247501 | Pun., India |
| Dysdercus koenigii (Fabricius) | B10 | GQ306227 | Pun., India | |
| Dysdercus sp.a | – | AY253049 | – | |
| Odontopus nigricornis Stal | B38 | GQ306226 | Pun., India | |
| Odontopus nigricornis nymph | B34 | GU247507 | Pun., India | |
| Aradoidea | Mezira granulataa (Say) | – | AY252962 | |
| Aradidae | ||||
| Cimicomorpha | Rhynocoris marginatus (Fabricius) | – | GQ229415 | – |
| Reduvidii | Rhynocoris ornatus (Uhler) | GQ292194 | – | |
Note: Classification follows Schuh and Slater [2].
Sequences obtained from GenBank.
Genomic DNA was extracted from ethanol-preserved specimens, usually from a leg. Samples were then dried, pinned, kept as voucher specimens and lodged in the Department of Zoology, Modern College, Pune, India. Photographs of 9 bug specimens, each representing one family, are shown in Fig. 1 and the remaining photographs are provided in supplementary information (Figures SM1 and SM2). DNA was extracted using DNeasy kit (Quiagen) as per the manufacturer's protocol. PCR was conducted in 25 μL volume containing 2 μL of DNA, 0.2 μL Taq Polymerase (Bangalore Genei), 1.25 μL 10 μM primers, 2.5 μL 2.0 mM dNTPs and 2.5 μL 10X buffer. The primer pair LCO1490 (5′-ggtcaacaaatcataaagatattgg-3′) and HCO2198 (5′-taaacttcagggtgaccaaaaaatca-3′) was used (Folmer et al., 1994) and amplification conditions were 1 cycle, 95 °C (5 min), 35 cycles, 94 °C (30 s), 47 °C (30 s), 72 °C (1:30 min), and 1 cycle 72 °C (10 min). PCR products were identified on 1% agarose gel electrophoresis with EtBr staining under UV light, purified by PEG-NaCl method (Sambrook et al., 1989) and sequenced in an automated sequencer (3730 DNA Analyser, ABI, Hitachi) using Big Dye Terminator cycle sequencing with same primers used for amplification. The sequences were checked for consistency, gaps, and non-sense codons and manually edited in CHROMAS Pro. Nucleotide database of GenBank was searched for COI sequences of true bug species from India. Obtained sequences were used for species identification using NCBI BLAST search. The sequences were aligned in Bioedit (Hall, 1999) and divergence amongst individuals, divergence at species, genus and family level were analysed by K2P model of base substitution in MEGA 5.1 (Tamura et al., 2007). Neighbour joining (NJ) analysis using K2P distance method was carried out in MEGA 5.1. The node supports were evaluated with 1000 bootstrap pseudo-replicates.
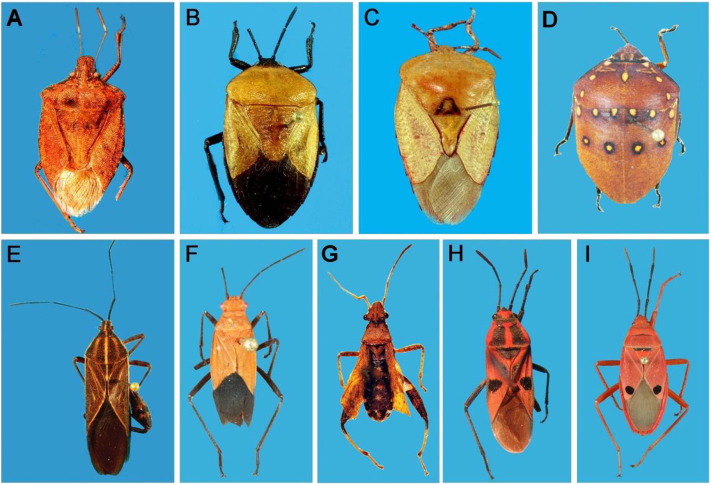
Fig. 1.
Photographs of Pentatomomorpha bugs representing nine families. A: Halyomorpha picus (Pentatomidae), B: Coridius brunneus (Dinidoridae), C: Tessaratoma sp. (Tessaratomidae), D: Solenostethium rubropunctatum (Scutelleridae), E: Physomerus parvulus (Coreidae), F: Leptocoris augur (Rhopalidae), G: Heegeria tangirica (Alydidae), H: Lygaeus militaris (Lygaeidae), and I: Odontopus nigricornis (Pyrrhocoridae).
Results
The 80 COI sequences representing 43 species and 35 genera were obtained. No stop codon or frame shifts were detected indicating that sequences were not pseudogenes (NUMTs). Sequences of more than 500 bp were obtained from all specimens. The final aligned data, after incorporating GenBank sequences, had 114 COI sequences of 542 bp length representing 73 species. Except for Nezara viridula and Riptortus pedestris, all the sequences from our studies were new addition to barcode database of this group as we could not find conspecific sequences when BLAST and nucleotide search was made (Supplementary information Table SM1).
A hierarchical increase in K2P mean divergence across different taxonomic levels was observed (Table 2). Intraspecific divergence averaged 0.4% (SE 0.2%) with a range of 0–2.1%, whilst interspecific distance showed an average of 11.7% (SE 1.3%) divergence (range 0–19.8%). Divergence amongst genera in family averaged 14.9% (SE 1.2%) with a range of 11.8–24.4%. The distance between different families was in the range of 18.0–30.2% with an average of 21% (SE 1.3%). Results from Jung et al. (2011) and Park et al. (2011) are also compared in Table 2. In the superfamilies, Pentatomoidea showed the highest mean divergence (19.6%) and Pyrrhocoroidea showed the lowest divergence (18.1%).
Table 2.
Sequence divergences (K2P) at various taxonomic levels.
| Present study |
Park et al. [20] |
Jung et al. [19] |
||||
|---|---|---|---|---|---|---|
| Range (%) | Mean dist (%) | Range (%) | Mean dist (%) | Range (%) | Mean dist (%) | |
| Within species | 0–2.1 | 0.4 | 0–7.72 | 0.74 | – | 0.8 |
| Amongst species | 0–19.8 | 11.7 | 0–24.80 | 10.67 | – | 12.6 |
| Amongst genera | 11.8–24.4 | 14.9 | 0–35.80 | 19.81 | – | 19.9 |
| Amongst families | 18.0–30.2 | 21.0 | 12.15–36.67 | 23.66 | – | – |
BLAST search carried out as an early test resulted in many of our sequences matching with the congeneric species from GenBank. This indicated that sequences are useful in diagnosing the genus if the database is robust. For those species where there was no sequence in database, the nearest match was always from the same family. Such sequences are of course new and are our contribution to the database. In some cases, such as some Coreidae (Physomerus, Homoeocerus) the BLAST search gave only a Rhopalidae match. In NJ tree (Fig. 2), closely related species and genera clustered cohesively and distinctly from the rest of the species. Except for Eocanthecona, nearest neighbour for all species was a congener in our data. Eocanthecona furcellata mtCOI sequence generated in our studies did not cluster with that of its congener Eocanthecona japanicola reported from Korea (GQ292274).
Fig. 2.

Phylogenetic tree of Pentatomomorpha bugs based on COI (K2P model) using Neighbour-Joining method. Numbers indicate the percentage of 1000 bootstrap replicates. Family and superfamily taxa according to Schuh–Slater are indicated on the right. Pen = Pentatomoidea, Pyr = Pyrrhocoroidea, Co = Coreoidea, and Ly = Lygaeoidea.
All members, except for four cases, of different species and genera showed distinct barcode in our studies. Two species of Leptocorisa; Leptocorisa acuta and Leptocorisa chinensis showed barcode sharing even though later species has been reported from Korea. Another instance of barcode sharing was observed in genus Nariscus where two of its species Nariscus fumosus and Nariscus fasciatus showed identical barcode. Two more cases showed little variation (< 2%) in barcode; COI sequence from an unidentified species of genus Tessaratoma in our study (not identified as javanica by us) showed 99% similarity with COI sequence from Tessaratoma javanica reported by other group (GenBank Accession number KF534924). Two species of genus Plautia; Plautia splendens and Plautia stali also showed low interspecies divergence (1.3%). On the other hand, two cases showed more than 2% intraspecific divergence. In the case of Nezara viridula; although no divergence was observed for haplotypes of our specimens, when COI sequence from GenBank (collected from USA; Accession number JX548495) was added for analysis, it showed 2.1% divergence indicating a genetically distinct population. Similar case was found in two Riptortus species; Riptortus pedestris and Riptortus clavatus also showed 2.1% divergence although they have been synonymised recently and treated as R. pedestris on the Coreoidea website (Livermore et al.).
A distinct barcode gap was observed for intra and interspecies divergence; at higher taxonomic levels, however, there was an overlap in sequence divergence. To resolve this problem, two more phylogenetic trees based on different methods (ML and MP) were constructed (Supporting information Figure SM3 and SM4). Although congeneric and conspecific taxa clustered cohesively in all trees, at family and superfamily level, however, there was no congruence in these trees. Only Pentatomidae, Lygaeidae and Pyrrhocoridae showed monophyly in NJ tree while other families showed problematic relationship.
Discussion
Plant bugs are known to damage a wide range of plants by sucking nutrients from them (Hori, 2000). Lygaeus pandurus, for example, is highly polyphagus and is reported from more than 10 plant families (Sweet, 2000) whilst Dysdercus koneigii is regarded as damaging pest of cotton (Schaefer and Ahmad, 2000). DNA barcoding may help in quick identification of such pests and therefore the present study focuses on this important insect group. It provides a greater representation of Pentatomomorpha bugs by adding more species from Western Ghats to the existing database.
Congeners clustered distinctly and COI sequences with same species name clustered cohesively in our present data. Only two cases of large intraspecific (> 2%) divergence were observed. Both N. viridula and R. pedestris are such a widespread species that it is quite likely to show considerable genetic variation in distant populations resulting in high intraspecies divergence. On the other hand, little variation in barcode for Tessaratoma javanica and an unidentified species of same genus in our study indicate the possibility of misidentification or poor genetic differentiation. Though our specimen is morphologically very similar to javanica, some of its taxonomic characters do not match with javanica and therefore we believe it to be different from T. javanica. As far as Nariscus (formerly Akbaratus) is concerned, Distant (1918) added two species fasciatus and fumosus to the genus Akbaratus which was later synonymized with Nariscus (Livermore et al., 2014); however, these two species described in Distant's Fauna were left out and are still not included in Nariscus. Identical barcode for these species indicate that they are not separate species and need to be synonymised. Little variation in two Plautia species; Plautia splendens and Plautia stali suggests that these species are very close, although these are considered as separate species (Pentatomoidea website of David Rider).
For species-level identification, the intraspecific divergence must be much less than the interspecies divergence. In this dataset, intraspecific divergences are usually less than 1.0% as observed for 97% of the taxa studied (only 2 notable exceptions), in contrast interspecific divergences always exceeded 3% (only 3% taxa showing less than 3% interspecies genetic divergence). Thus, distinct intra and interspecific divergence obtained in the present study indicates that barcode can be used for identification of true bugs at species level. However, at higher taxonomic level, COI-based trees are not satisfactory and may not resolve relationship clearly. Though the theme of the present paper is only to suggest the usefulness of COI sequences in identification, we have looked at the possible relationship here. Monophyly of family Pentatomidae was not supported in ML and MP trees; also some taxa of superfamily Pentatomoidea, for example, a dinidorid Cyclopelta always formed a separate cluster from its other family members in all trees indicating problematic relationship in Dinidoridae. Dinidoridae, Scutelleridae and Tessaratomidae are always close and Grazia et al. (2008) made similar observation based on morphology as well as molecular studies of this group. Superfamily Coreoidea and family Coreidae were polyphyletic as observed by other groups (Li et al., 2005, Park et al., 2011). Although, Pyrrhocoridae and Lygaeidae formed a coherent cluster, comments on superfamilies Pyrrhocoreoidea and Lygaeoidea cannot be made because no specimen beyond these two families could be incorporated in the present study.
Conclusions
Distinct barcode gap and NJ clustering analysis in the present data confirms that DNA barcoding is helpful in species level identification of true bugs. This technique may be used for quick identification of many more unexplored species from Western Ghats of India provided that mtCOI sequence database is rich and easily accessible. This can facilitate simple and rapid identification of serious pests and invasive species. The study contributes significantly to the barcode data of Pentatomomorpha bugs by adding sequences from indigenous true bug species of India and also calls for additional efforts to make more comprehensive reference library. It is unfortunate that hardly any sequence data is available in databases in spite of the fact that all these bugs are economically very important and need quick identification.
Acknowledgement
We thank the Department of Biotechnology, Government of India for providing financial support for this work in terms of postdoctoral fellowship to ST and fieldwork costs. We are thankful to the authorities of Modern College, Pune and the Director, NCCS, Pune for providing facilities. We are grateful to: Prof. C. Viraktamath (GKVK, Bangalore) Dr. W. R. Dolling (formerly of NHM, London), Prof. David Rider (North Dakota State University) Dr. H. Brailovsky (National Autonomous University of Mexico) and Dr. David Redei (Nankai University, China) for their help in identification of specimens by commenting on images or by providing useful literature and constant support. We appreciate help from our friends and students in specimen collection, especially Swapnil Gaikwad, Gauri Sathye, Shriraj Jakhalekar, Girish Pathak, Shaukat Inamdar and Prajakta Tembe. Thanks are also due to Shriraj and Dr. Sameer Padhye for their help in digitization of bug specimens and preparing illustration accompanying. We also thank two anonymous reviewers for their valuable comments to improve the quality of paper.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mgene.2014.09.006.
Contributor Information
Sanket Tembe, Email: sankettembe@gmail.com.
Yogesh Shouche, Email: yogesh@nccs.res.in.
H.V. Ghate, Email: hemantghate@gmail.com.
Appendix A. Supplementary data
Supplementary material.
References
- Damgaard J. MtDNA diversity and species phylogeny of western Palaearctic members of the Gerris lacustris group (Hemiptera:Heteroptera:Gerridae) with implications for DNA barcoding of water striders. Insect Syst. Evol. 2008;39:107–120. [Google Scholar]
- Distant W.L. Taylor and Francis; London Vol I and VII: 1904. The Fauna of British India, including Ceylon and Burma, Rhynchota. pp. 1904–1918. [Google Scholar]
- Dolling W.R. Natural History Museum Publication. Oxford University Press; Oxford: 1991. The Hemiptera. [Google Scholar]
- Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit 1 from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Grazia J., Schuh R.T., Wheeler W.C. Phylogenetic relationship of family groups in Pentatomoidea based on morphology and DNA sequences (Insecta: Heteroptera) Cladistics. 2008;24:932–976. doi: 10.1111/j.1096-0031.2008.00224.x. [DOI] [PubMed] [Google Scholar]
- Hall T.A. Bioedit: a user-friendly biological sequence alignment editor and analysis programme for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hori K. Possible causes of feeding disease symptoms resulting from the feeding of phytophagus heteroptera. In: Schaefer C.W., Panizzi A.R., editors. Heteroptera of Economic Importance. CRC Press; Boca Raton, Florida, USA: 2000. pp. 11–35. [Google Scholar]
- Hua J., Li M., Dong P., Cui Y., Xie Q., Bu W. Comparative and phylogenomic studies on the mitochondrial genomes of Pentatomomorpha (Insecta: Hemiptra: Heteroptera) BMC Genomics. 2008;9:610–624. doi: 10.1186/1471-2164-9-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Duwal R.K., Lee S. COI barcoding of true bugs (Insecta, Heteroptera) Mol. Ecol. Resour. 2011;11:266–270. doi: 10.1111/j.1755-0998.2010.02945.x. [DOI] [PubMed] [Google Scholar]
- Li H.M., Deng R.Q., Wang J.W., Chen Z.Y., Jia F.L., Wang X.Z. A preliminary phylogeny of the Pentatomomorpha (Hemiptera: Heteroptera) based on nuclear 18S rDNA and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2005;37:313–326. doi: 10.1016/j.ympev.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Livermore L.J.R., Lemaître V.A., Dolling W.R., Webb M.D. 2014. Coreoidea Species File Online. Version 5.0/5.0. (15 April, < http://Coreoidea.SpeciesFile.org>) [Google Scholar]
- Memon N., Meier R., Manan A., Su K. On the use of DNA sequences for determining the species limits of a polymorphic new species in the stink bug genus Halys (Heteroptera: Pentatomidae) from Pakistan. Syst. Entomol. 2006;31:703–710. [Google Scholar]
- Park D.S., Footit R., Maw E., Hebert P.D.N. Barcoding bugs: DNA-based identification of the true bugs (Insecta: Hemiptera: Heteroptera) PLoS ONE. 2011;6:e18749. doi: 10.1371/journal.pone.0018749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebijith K.B., Asokan R., Krishna N.K., Srikumar K.K., Ramamurthy V.V., Bhat P. DNA barcoding and development of species-specific markers for the identification of tea mosquito bugs (Miridae: Heteroptera) in India. Environ. Entomol. 2012;41:1239–1245. doi: 10.1603/EN12096. [DOI] [PubMed] [Google Scholar]
- Rider D. https://www.ndsu.edu/pubweb/~rider/Pentatomoidea/
- Sambrook J., Fritsch E.F., Maniatis T. 2nd edn. Cold Spring Harbor Laboratory; Cold Spring Harbor, NewYork: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Schaefer C.W., Ahmad I. Cotton strainers and their relatives (Pyrrhocoroidea: Pyrrhocoridae and Largidae) In: Schaefer C.W., Panizzi A.R., editors. Heteroptera of Economic Importance. CRC Press; Boca Raton, Florida, USA: 2000. pp. 271–307. [Google Scholar]
- Schuh R.T., Slater J.A. Classification and Natural History. Cornell University Press; Ithaca, NY: 1995. True Bugs of the World (Hemipera: Heteroptera) [Google Scholar]
- Sweet M.H. Seed and chinch bugs (Lygaeoidea) In: Schaefer C.W., Panizzi A.R., editors. Heteroptera of Economic Importance. CRC Press; Boca Raton, Florida, USA: 2000. pp. 143–264. [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA 5: molecular evolutionary genetics analysis (MEGA) software version 5. 0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Yao Y., Ren D., Rider D.A., Cai W. Phylogeny of the infraorder pentatomomorpha based on fossil and extant morphology, with description of a new fossil family from China. PLoS ONE. 2012;7(5):e37289. doi: 10.1371/journal.pone.0037289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.