Abstract
Introduction
Natural killer (NK) cells possess an antitumor activity against multiple myeloma cells proven by the susceptibility of plasmocytes to NK lysis. In the early stage of MM, the killing of MM cells is mediated by natural cytotoxicity receptors (NRC) and NKG2D-dependent pathway, while in the late stage, NK cells lose their killing potential against MM cells due to the high expression of HLA class I molecules on MM cells.
Aim
The aim of this paper is to study KIR expression of NK cells in MM patients and in healthy controls, to check for any association between KIR genotypes and MM.
Methods
KIR genotype was analyzed in 120 healthy Lebanese individuals and 34 MM patients using the KIR Genotyping SSP kit.
Results
KIR 2DS4*001/002 and KIR 2DS5 were found to be significantly more prevalent among MM patients as compared to controls. For MM patients, the AA, AB, and BB genotype frequencies were, respectively, 38.23%, 47.06% and 14.71% with an A:B ratio of 1.62:1. As for the healthy controls, the AA, AB, and BB genotype frequencies were, respectively, 39.17%, 50%, and 10.83% with an A:B ratio of 1.80:1.
Conclusion
The interesting observation of the significant presence of KIR2DS4 and KIR2DS5 genes more among multiple myeloma patients than controls is worth further clinical, translational as well as survival research studies in these cases.
Keywords: Myeloma, KIR, Genotype, Lebanon
Highlights
-
•
KIR genotyping has been implicated in a variety of clinical and immunological disorders.
-
•
This study is the first international research paper that describes the prevalence of KIR genes among Multiple Myeloma patients
-
•
Results were compared to a large healthy control population sample
Introduction
The role of natural killer cells (NK) in the control or progression of different diseases is becoming a source of interest for many researchers. NK cells, discovered 30 years ago, are large granulocytic lymphocytes yet they are neither T cells nor B cells, they belong to the innate immune system and they originate from CD34+ hematopoietic progenitor cells (Caligiuri, 2008). NK cells recognize target cells and react against them using inhibitory and activating killer immunoglobulin-like receptors (KIR).
The KIR gene is located on chromosome 19 and the locus includes up to 17 genes and pseudogenes (Boyton and Altmann, 2007). The nomenclature of a KIR gene indicates the number of immunoglobulin-like domains of the molecule (2D or 3D), the length of the cytoplasmic tail (L or S), and the number of the gene encoding a protein with this structure, while pseudogenes are designated by 2DP or 3DP (Gomez-Lozano et al., 2002). Based on the gene content, two groups of KIR haplotypes, A and B, have been identified in humans. Haplotype A encodes inhibitory receptors and consists of nine genes (3DL3, 2DL3, 2DP1, 2DL1, 3DP1, 2DL4, 3DL1, one activating (2DS4), 3DL2, and 2DL5) whereas haplotype B carries a variety of gene combinations and encodes more activating receptors as compared to haplotype A (3DL3, 2DS2, 2DL2, 2DL5B (inhibitory) 2DS3, 2DP1, 2DL1, 3DP1, 2DL4, 3DS1, 2DL5A (inhibitory), 2DS5, 2DS1, and 3DL2) (McQueen et al., 2007, Bashirova et al., 2011). Despite the high allelic polymorphism of KIR genes, almost all individuals possess the following genes referred to as framework loci: KIR3DL3, KIR3DP1, KIR2DL4, and KIR3DL2 with KIR2DL4 possibly encoding both inhibitory and activating receptors.
NK cells, through KIR and other receptors, play an important role in various diseases, including viral infection, autoimmune and inflammatory conditions, tumor immunity, and recurrent abortions (Boyton and Altmann, 2007). It has been shown that NK cells possess an antitumor activity against multiple myeloma cells proven by the susceptibility of plasmocytes to NK lysis (Frohn et al., 2002).
Multiple myeloma (MM) is a hematological malignancy characterized by the accumulation of plasma cells that secrete monoclonal immunoglobulins in the bone marrow (Fauriat et al., 2006). In the early stage of MM, the killing of a myeloma cell is mediated by natural cytotoxicity receptors (NRC) and NKG2D-dependent pathway, while in the late stage, NK cells lose their killing potential against MM cells due to the high expression of HLA class I molecules on the myeloma cells (Carbone et al., 2005).
In order to avoid the resistance of multiple myeloma cells to conventional chemotherapy and to achieve a better treatment outcome, thalidomide and bortezomib are being implicated in the treatment protocol (Hayashi et al., 2005). Studies have shown that tumor cell lysis is triggered by NK cells interleukin-2 (IL-2)-dependent activation as part of the molecular mechanism of thalidomide (Davies et al., 2001, Frohn et al., 2002, Hayashi et al., 2005).
A recent study showed that advanced clinical stage of multiple myeloma was associated with decreased NK cell activity and that those patients with higher NK cell activity (upon diagnosis) have better overall survival in comparison with those with low NK cell activity after receiving a special chemotherapy protocol (Jurisic et al., 2007).
This is the first study that investigates the association between KIR genotypes and multiple myeloma through KIR genotyping analysis among MM patients and healthy controls.
Materials and methods
Study population
This Institutional Review Board (IRB) approved study was performed at the American University of Beirut Medical Center (AUBMC), a major Lebanese tertiary-care center. KIR genotype was analyzed for 34 multiple myeloma cases using the KIR Genotyping SSP kit. The 120 healthy controls were deducted from the major study by Mahfouz et al. (2006) describing the KIR prevalence in the healthy Lebanese population mainly from donors of a bone marrow transplantation center representing all ethnicities and districts in the country.
DNA extraction and KIR genotyping
PEL-FREEZ kits (Pel-freez/Dynal, Norway) were used for DNA extraction from 2 to 3 ml of collected peripheral blood. The DNA material was properly labeled and stored at − 80 °C. Based on our IRB committee rules and study approval protocols, confidentiality was strictly observed for all analyzed samples.
Primer mixes were purchased from PEL-FREEZ/DYNAL (PEL-FREEZ/DYNAL company, Oslo, Norway) as part of the KIR Genotyping SSP kit which is a PCR-based method designed to detect the absence or presence of the following 16 gene loci of KIR (variants also tested): 2DL1, 2DL2, 2DL3, 2DL4, 2DL5, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DL1, 3DL2, 3DL3, 3DS1, 2DP1, and 3DP1. Two variants for KIR2DL5 were typed KIR2DL5A*001 and KIR2DL5B*002/003/004 and two variants for KIR2DS4 were tested and reported as KIR2DS4*001/002 and KIR2DS4*003-006. In addition, two variants for the pseudogene KIR3DP1 were tested: KIR3DP1*001/002/004 and KIR3DP1*003.
KIR genotyping was performed as recommended by the manufacturer and all steps were strictly followed. Briefly, 25 μl of DNA was added to 150 μl of PCR buffer and 2.4 μl of Taq DNA polymerase and dispensed as aliquots of 8 μl into a supplied 96-well plate for a total reaction volume of 23 μl in each well (reaction + paraffin oil). The thermocycling steps include an initial heating step at 95 °C for 1 min, followed by 30 cycles of 94 °C for 20 s, 63 °C for 20 s, and 72 °C for 90 s. A final holding step was performed at 4 °C. Electrophoresis of the 2% agarose gel was performed in ethidium bromide and visualization was performed under UV light transillumination.
Statistical analysis
We utilized direct counting for the observed genotype frequencies of KIR. SPSS 15.0 was used to conduct statistical analysis. Genotypic analysis was expressed as number and frequency. Chi-square was used to test for association between group (case vs control) and genetic profile. P-value less than 0.05 was considered statistically significant.
Results
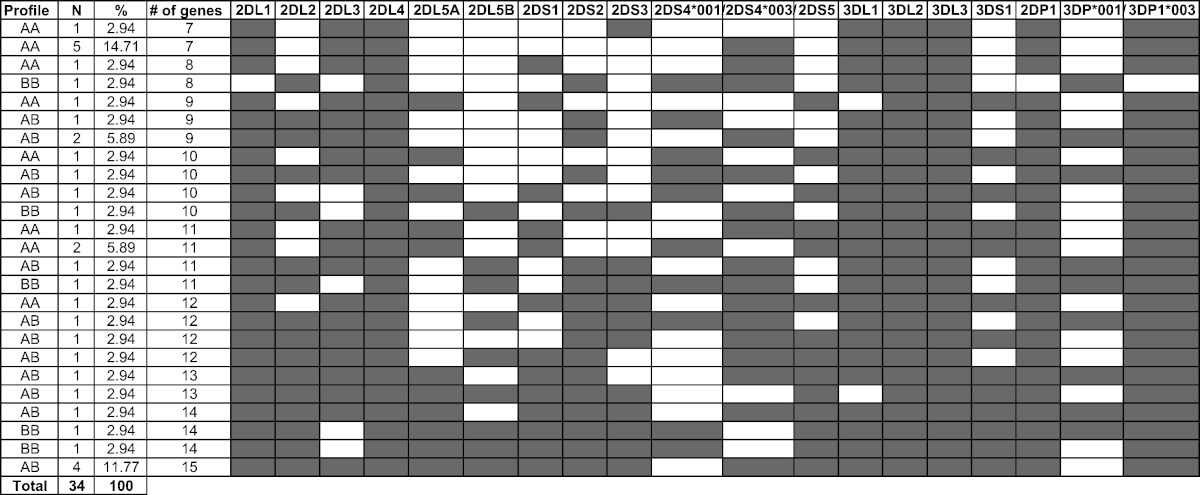
KIR genotypic profile distribution among the 34 Lebanese patients with MM is shown in Table 1. The content of KIR genes ranged from 7 to 15 and as per Table 4, the AA, AB, and BB genotypes frequencies were, respectively, 38.23%, 47.06% and 14.71% with an A:B ratio of 1.62:1.
Table 1.
KIR genotypic profile distribution among the 34 Lebanese patients with multiple myeloma.
N refers to the number of individuals with the corresponding genotypic profile.
Table 4.
Frequency distribution of AA, AB, and BB genotypes among MM patients and controls.
| Genotype | Controls |
MM patients |
||
|---|---|---|---|---|
| N | % | N | % | |
| AA | 47 | 39.17 | 13 | 38.23 |
| AB | 60 | 50 | 16 | 47.06 |
| BB | 13 | 10.83 | 5 | 14.71 |
| Total | 120 | 100 | 34 | 100 |
| Haplotype | Controls | MM patients |
|---|---|---|
| A | 154 | 42 |
| B | 86 | 26 |
| A:B ratio | 1.80:1 | 1.62:1 |
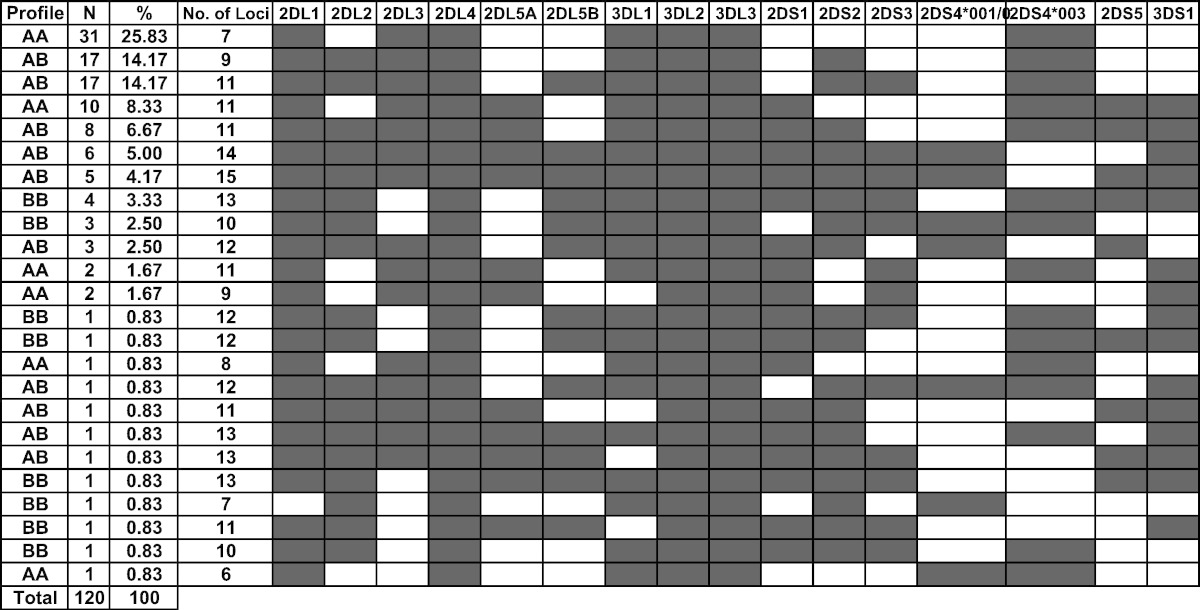
Table 2 shows the distribution of different KIR genotypes among the 120 healthy Lebanese controls. The content of KIR genes ranged from 6 to 15 and, as per Table 4, the AA, AB, and BB genotypes frequencies were, respectively, 39.17%, 50%, and 10.83% with an A:B ratio of 1.80:1.
Table 2.
KIR genotypic profile among the 120 healthy Lebanese controls.
N refers to the number of individuals with the corresponding genotypic profile.
Table 3 shows the distribution of different KIR genes among the 120 controls and the 34 MM patients. KIR3DL2, KIR3DL3, and KIR3DP1*003 were present in all individuals. KIR2DS4*001/002 and KIR2DS5 were found to be significantly (with a p-value of 0.04 and 0.007, respectively) more prevalent among MM patients as compared to controls.
Table 3.
Distribution of KIR genes among MM patients and healthy controls. A p-value < 0.05 was considered significant.
| Healthy |
MM |
p-value | |
|---|---|---|---|
| N = 120 | N = 34 | ||
| 2DL1 | 119 (99.2%) | 33 (97.1%) | 0.39 |
| 2DL2 | 73 (60.8%) | 20 (58.8%) | 0.49 |
| 2DL3 | 106 (88.3%) | 28 (82.4%) | 0.39 |
| 2DL4 | 120 (100%) | 34 (100%) | 1 |
| 2DL5A | 38 (31.7%) | 15 (44.1%) | 0.22 |
| 2DL5B | 45 (37.5%) | 12 (35.3%) | 0.49 |
| 2DS1 | 49 (40.8%) | 17 (50.0%) | 0.22 |
| 2DS2 | 73 (60.8%) | 21 (61.8%) | 0.54 |
| 2DS3 | 45 (37.5%) | 15 (44.1%) | 0.31 |
| 2DS4*001/002 | 20 (16.7%) | 11 (32.4%) | 0.04 |
| 2DS4*003/007 | 99 (82.5%) | 24 (70.6%) | 0.1 |
| 2DS5 | 34 (28.3%) | 18 (52.9%) | 0.007 |
| 3DL1 | 115 (95.8%) | 32 (94.1%) | 0.65 |
| 3DL2 | 120 (100%) | 34 (100%) | – |
| 3DL3 | 120 (100%) | 34 (100%) | – |
| 3DS1 | 45 (37.5%) | 17 (51.5%) | 0.11 |
| 2DP1 | 116 (96.7%) | 33 (97.1%) | 1 |
| 3DP*001/002/004 | 33 (27.5%) | 10 (29.4%) | 0.49 |
| 3DP1*003 | 120 (100%) | 34 (100%) | – |
Discussion
The international literature is adding up more information related to the relationship between KIR genes and different diseases and clinical entities where several studies have reported either a resistance or susceptibility to these entities based on the KIR genotypic profiles.
Our study data are consistent with the presence of the two major KIR haplotypes, group A and group B with predominance of the group A haplotype among the multiple myeloma patients similar to its prevalence among the healthy individuals. In addition, the frequency distribution of AA, AB, and BB genotypes was 38.23%, 47.06%, and 14.71% respectively reflecting the low occurrence of BB homozygous state among the myeloma patients which is also the case in the healthy individuals. All our genotypes included between 7 and 15 genes with an average number of 11 KIR loci per patient (excluding the pseudogenes KIR2DP1 and KIR3DP1) which is comparable to the 11 KIR loci in the general healthy population (Mahfouz et al., 2006).
The distribution of the KIR genotypes (AA, AB, and BB) does not seem to be different among patients with multiple myeloma as compared to the healthy controls which is also applicable to the haplotype distribution. And since our current study does not include clinical data related to the Myeloma patients involved as it is a genetic prevalence study, no major phenotypic–genotypic correlations can be deduced at this level. However, it reports an interesting observation of a significant increase of KIR2DS4 and KIR2DS5 genes among multiple myeloma patients as compared to healthy individuals. The role of these two activating KIR genes has not been elucidated or described in the pathogenesis of multiple myeloma which is worth further clinical and translational research in order to better understand the significance of this expression more specifically. As a typical example, KIR2DS4 and KIR2DS5 can be assessed in correlation with outcome of bone marrow transplantation in these patients or maybe a direct association with the disease development and prognostication itself.
It has been previously well reported that the balance between the inhibitory and activating signals regulates the function of natural killer cells and the current study is showing an increase of two major activating KIR genes with potential contributory roles.
Our study is the first to report on the KIR genes prevalence among multiple myeloma patients and compared it to a large healthy control population and will definitely serve as a template for other researchers to build on their clinical, translational and survival studies.
References
- Bashirova A.A., Thomas R., Carrington M. HLA/KIR restraint of HIV: surviving the fittest. Annu. Rev. Immunol. 2011;29:295–317. doi: 10.1146/annurev-immunol-031210-101332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyton R.J., Altmann D.M. Natural killer cells, killer immunoglobulin-like receptors and human leucocyte antigen class I in disease. J. Transit. Immunol. 2007;149(1):1–8. doi: 10.1111/j.1365-2249.2007.03424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caligiuri M.A. Human natural killer cells. Blood. 2008;112(3):461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone E., Neri P., Mesuraca M., Fulciniti M.T., Otsuki T., Pende D. HLA class I, NKG2D, and natural cytotoxicity receptors regulate multiple myeloma cell recognition by natural killer cells. Blood. 2005;105:251–258. doi: 10.1182/blood-2004-04-1422. [DOI] [PubMed] [Google Scholar]
- Davies F.E., Raje N., Hideshima T. Thalidomide and immunomodulatory derivatives augment natural killer cell cytotoxicity in multiple myeloma. Blood. 2001;98:210–216. doi: 10.1182/blood.v98.1.210. [DOI] [PubMed] [Google Scholar]
- Fauriat C., Mallet F., Olive D., Costello R.T. Impaired activating receptor expression pattern in natural killer cells from patients with multiple myeloma. Leukemia. 2006;20:732–733. doi: 10.1038/sj.leu.2404096. [DOI] [PubMed] [Google Scholar]
- Frohn C., Hoppner M., Schlenke P., Kirchner H., Koritke P., Luhm J. Anti-myeloma activity of natural killer lymphocytes. Br. J. Haematol. 2002;119:660–664. doi: 10.1046/j.1365-2141.2002.03879.x. [DOI] [PubMed] [Google Scholar]
- Gomez-Lozano N., Gardiner C.M., Parham P., Vilches C. Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics. 2002;54:314–319. doi: 10.1007/s00251-002-0476-2. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Hideshima T., Akiyama M. Molecular mechanisms whereby immunomodulatory drugs activate natural killer cells: clinical application. Br. J. Haematol. 2005;128:192–203. doi: 10.1111/j.1365-2141.2004.05286.x. [DOI] [PubMed] [Google Scholar]
- Jurisic V., Srdic T., Konjevic G., Markovic O., Colovic M. Clinical stage-depending decrease of NK cell activity in multiple myeloma patients. Med. Oncol. 2007;24(3):312–317. doi: 10.1007/s12032-007-0007-y. [DOI] [PubMed] [Google Scholar]
- Mahfouz R., Rayes R., Mahfoud Z., Bazarbachi A., Zaatari G. Distribution of killer cell immunoglobulin-like receptors genotypes in the Lebanese population. Tissue Antigens. 2006 Jul;68(1):66–71. doi: 10.1111/j.1399-0039.2006.00605.x. [DOI] [PubMed] [Google Scholar]
- McQueen K.L., Dorighi K.M., Guethlein L.A., Wong R., Sanjanwala B., Parham P. Donor–recipient combinations of group a and B KIR haplotypes and HLA class I ligand affect the outcome of HLA-matched, sibling donor hematopoietic cell transplantation. Hum. Immunol. 2007;68(5):309–323. doi: 10.1016/j.humimm.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]