Abstract
LIPE is an intracellular neutral lipase, which is capable of hydrolyzing a variety of esters and plays a key role in the mobilization of fatty acids from diacylglycerols. The objectives of this study were to characterize the genetic polymorphism of bovine LIPE gene and to evaluate the possible association between three SNPs in the coding regions of this gene with the fatty acid composition of meat in a cattle population. Forty-three unrelated animals from different cattle breeds were re-sequenced and 21 SNPs were detected over approximately 2600 bp, five of these SNPs were novel. Three SNPs were selected, on the basis of evolutionary conservation, to perform validation and association studies in a crossbred cattle population. Our results may suggest a possible association of SNP1 with contents of oleic acid and total monounsaturated fatty acids (p < 0.01), and SNP2 and SNP3 with Heneicosylic acid content (p < 0.01), may be helpful to improve the quality of meat and improve health.
Abbreviations: LIPE, hormone-sensitive lipase; SNP, single nucleotide polymorphism; n, number of samples; pb, base pairs; p, p-value; F1, first filial; F2, second filial; INTA, National Institute of Agricultural Technology; PCR, polymerase chain reaction; HWE, Hardy–Weinberg equilibrium; he, unbiased expected heterozygosity; ho, observed heterozygosity; MUFA, total monounsaturated fatty acids; C18:1c9, oleic acid; C21:0, heneicosylic acid; Nt, N-terminal; R, regulatory module; Ct, C-terminal; ALBP, adipocyte lipid binding protein; GNRHR, gonadotropin-releasing hormone receptor
Keywords: LIPE, Polymorphism, Bovine, Lipid content
Highlights
-
•
Twenty-one SNPs were detected by re-sequencing.
-
•
Five novel SNPs were found.
-
•
Three SNPs were significantly associated with the fatty acid composition of meat.
Introduction
The interest of consumers in food quality has increased in the recent years. This responds mainly to the large amount of available information about consumed foods and the effects foods have on health and the environment. Regarding meat, its lipid content and composition are interesting issues, since these factors affect its nutritional and organoleptic properties (Warriss, 2000).
The chemical composition of meat is complex and is determined by many extrinsic and intrinsic factors. It is known that the fatty acid composition of ruminant products is affected by microbial digestion, de novo synthesis, in situ desaturation and transfer of fatty acids among tissues (Martínez Marín et al., 2010). A percentage of the variability provided by these metabolic pathways may be attributed to the genetic variants of the pathway members. The easy detection and analysis of these variants are allowed currently by technological developments and advances in molecular genetics.
Many genes of the mentioned metabolic pathways have been identified. A better understanding of their genetic variants would allow us to adapt bovine meat to consumer preferences through selection programs (Baeza et al., 2013, Granneman et al., 2011). One of the genes involved in the pathways of transfer is the hormone-sensitive lipase (LIPE, previously called HSL) gene. LIPE is a key enzyme in the mobilization of fatty acids from acylglycerols (Lampidonis et al., 2011). This enzyme acts as an intracellular neutral lipase, which is capable of hydrolyzing a variety of esters (Holm, 2004). Until recently, LIPE was considered to be the main, if not exclusive, lipase mediating hormone-stimulated lipolysis (Yeaman, 2004). Adipocytes of LIPE null mice are largely incapable of releasing glycerol and have massive accumulation of cellular diacylglycerol, clearly demonstrating the importance of LIPE as a diglyceride lipase (Haemmerle et al., 2002). LIPE exhibits strong diglyceride hydrolase activity that is 10- and 5-fold greater than its activity against triglyceride and monoglyceride substrates, respectively (Belfrage et al., 1978). LIPE is highly expressed not only in adipose tissue, but also in the adrenal gland, ovary, testis, heart, skeletal tissue and muscle tissue, to a lesser extent (Qiao et al., 2007, Wang et al., 2004). It has also been observed that this enzyme is highly conserved among mammalian species such as pigs, humans, mice and rats (Yajima et al., 2007).
Since LIPE provides free fatty acids to the body directly, mutations in the gene may affect many traits that involve free fatty acids as metabolic precursors besides the fatty acid composition of meat (Hermo et al., 2008, Wang et al., 2014). Various single nucleotide polymorphisms (SNPs) have been reported in pigs, and other domestic species, but only a few of them have been further analyzed (Lei et al., 2005). Recently, two SNPs have been successfully associated with carcass and meat quality traits in Chinese Simmental-cross steers (Fang et al., 2013).
There is little information about genetic variants in other cattle breeds and the effects they may have on phenotypic qualities. Therefore, the objectives of this study were to characterize the genetic diversity of LIPE in cattle breeds of different origins, and evaluate the association between genotypes and meat lipid content and composition in crosses between Angus, Hereford and Limousin cattle breeds.
Materials and methods
Animal samples and DNA extraction
Two groups of samples were collected for the study: The first group comprised blood samples from 43 unrelated purebred animals [Angus (n = 5), Hereford (n = 5), Creole (n = 5), Wagyu (n = 5), Holstein (n = 5), Limousin (n = 4), Shorthorn (n = 5), Nelore (n = 4) and Brahman (n = 5)], which were used to identify polymorphisms in the bovine LIPE gene. The second sampling, used for SNP validation and statistical analyses, comprised meat samples from steers that have been involved in previous studies to evaluate crossbreeding systems under pasture grazing with strategic supplementation at the Experimental Station of the National Institute of Agricultural Technology (INTA, Balcarce, Argentina) (Villarreal et al., 2006). Animals (n = 260) from eight groups were sampled at slaughter; these steers were from 15 to 29 months old and were born between 2006 and 2010. The steers belonged to different genetic groups and included: purebred Angus – A – (n = 44) and Hereford – H – (n = 26) steers, F1 – A × H – reciprocal crossbreds and backcrosses (n = 110), F2 crossbreds – AH × AH and HA × HA – (n = 39), and a group of steers produced by mating Limousin – L – sires to both F1 crossbred cows (n = 41). The decision to sample this experimental population instead of other commercial cattle populations was based on the availability of reliable information in terms of phenotypic data, management and the genetic background of the animals.
DNA was isolated from blood lymphocytes using Wizard® Genomic DNA purification kit (Promega, Madison, WI, USA) following the instructions of the supplier, and from meat samples as previously described by Giovambattista et al. (2001).
Re-sequencing study
In order to amplify the coding regions of the bovine LIPE gene, nine primer pairs were designed (Table S2) based on the DNA sequence available in GenBank (Gene ID: 286879, AC_000175.1). PCRs were performed in a total volume of 25 μl, containing 20 mM of Tris–HCl (pH = 8.4), 50 mM of KCl, 2.5 mM of MgCl2, 100 mM of each dNTP, 0.5 U of Taq polymerase (Invitrogen, Carlsbad, CA), 0.4 mM of each primer, and 25–50 ng of DNA. The cycling conditions were: 3 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 40 s at 61 °C, and 40 s at 72 °C with a final elongation step of 10 min at 72 °C. Amplification products were purified with polyethylene glycol 8000 and sequenced in an automatic DNA sequencer MegaBACE 1000 (GE Healthcare), using DYEnamic ET Terminator Kit (GE Healthcare). Raw sequences were edited using Sequence Analyzer (GE Healthcare).
DNA sequences were aligned using CLUSTAL-X 2.1 software (Larkin et al., 2007). Variations were defined by direct comparison with the LIPE bovine sequence reported in GenBank.
SNP selection and genotyping
Three SNPs that were detected on Hereford, Angus and Limousin samples were selected for validation and association studies based on their location within the gene (conserved areas) and the nature of the mutation (residue character). Genotyping was performed using Sequenom platform (www.sequenom.com) by Neogen genotyping service (USA, www.neogen.com) in the second group of samples described in the Animal samples and DNA extraction section.
Statistical analysis
Linkage disequilibrium among the SNPs of LIPE was determined by PHASE (Li and Stephens, 2003) and visualized on HAPLOVIEW (Barrett et al., 2005). Allele frequencies and Hardy–Weinberg equilibrium (HWE) were estimated using GENEPOP 4 software (Rousset, 2008). The unbiased expected heterozygosity (he) was calculated using ARLEQUIN 3.1 (Schneider et al., 2000). For these analyzes, the steers were grouped depending on their breeds to estimate the allele frequencies: purebred Angus (A, n = 44), purebred Hereford (H, n = 26), steers that were 75% Angus (75A, n = 30), 75% Hereford (75H n = 24), 50% Hereford–50% Angus (50AH, n = 95) and Limousin crossbred steers (LX, n = 41). The genetic structure of the population is detailed in Table S1.
Meat quality measurement
To perform association studies, the fatty acid composition of meat from 179 animals of the A–H–L population (the first five slaughter groups) was determined. A block of meat corresponding to the Longissimus dorsi muscle (13th rib) was removed from the carcass 24 h after slaughter and kept at − 20 °C. Subcutaneous fat was removed from each sample. Total lipids of muscle samples were extracted according to Folch et al. (1957), purified by alkaline methylation and recovered on hexane. Fatty acid composition was determined using a gas chromatograph (Schimadzu GC14B). Fatty acid methyl ester (FAME) separation was performed on a 100 m × 0.25 mm capillary column (ResteK). The chromatograph was set at a temperature of 140 °C for 1 min, increased from 140 to 240 °C at 4 °C per min, and then held constant at 240 °C for 20 min. The injector and detector were kept at 260 °C. Data were recovered using GCSolultion Software and the amount of each fatty acid was quantified by the internal standard technique (Supelco 37 FAME MIX), expressed as percentage of total fatty acids. The following fatty acid measurements were taken: Myristoleic acid (C14:1); pentadecylic acid (C15:0); palmitic acid (C16:0); palmitoleic acid (C16:1); stearic acid (C18:0); oleic acid (C18:1c9); vaccenic acid (C18:1c11, C18:1t11); linoleic acid (C18:2c912); linolenelaidic acid (C18:2 t912); γ-linolenic acid (C18:3c6912); α-linolenic acid (C18:3c91215); eicosenoic acid (C20:1); eicosadienoic acid (C20:2); eicosatriynoic acid (C20:3c81114); arachidonic acid (C20:4); heneicosylic acid (C21:0); behenic acid (C22:0); docosenoic acid (C22:1); docosahexaenoic acid (C22:6); total saturated fatty acids (SFA); total monounsaturated fatty acids (MUFA); total polyunsaturated fatty acids (PUFA); total omega-6 fatty acids (ω− 6); and total omega-3 fatty acids (ω − 3). Total ether extract was expressed as the amount of fat in 100 g of fresh muscle excluding the external adipose tissue.
Association analysis between genotypes and meat composition traits
A mixed model analysis was conducted to evaluate the association between the fatty acid composition and the genotypes. This model considered the fixed effects of breed group; date of slaughter, which included the contemporary group (year of birth and time of slaughter within the year of birth) and molecular marker. The sire was nested within breed group and considered as a random effect. Longissimus muscle ether extract percentage was included as a covariable. All statistical analyses were performed using the MIXED procedure of SAS (SAS Inst. Inc., Cary, NC, 1998). When statistically significant (p < 0.05), least square means were reported and Bonferroni's mean separation test was used to determine differences between genotypes. The false discovery rate (FDR) was applied to control the proportion of incorrectly rejected null hypotheses due to multiple comparisons by using the Benjamini & Hochberg method (Benjamini and Hochberg, 1995) and considering 47 comparisons, since the third SNP was almost completely linked to SNP2. Additive effects were estimated by the difference between the two homozygous genotypes and the dominance effects were estimated by subtracting the average of solutions for homozygous genotypes from that for the heterozygous genotype. The contrasts and tests among or between means of genotypes were performed as described by Boldman et al. (1993).
Structural modeling
In order to observe the effects of the three selected SNPs on the protein structure, a 3D modeling study was performed using HHpred (Söding et al., 2005). Residues 316–756 of the amino acid sequence of LIPE were modeled according to similarities to known structures publicly available in the Protein Data Bank (PDB). This study was performed using the structure of a hyper-thermophilic carboxylesterase from the archaeon Archaeoglobus fulgidus (De Simone et al., 2001), a homologous enzyme, as the template: 1jji_A (E-value = 2.7E− 27).
Results
Re-sequencing analysis of the LIPE gene
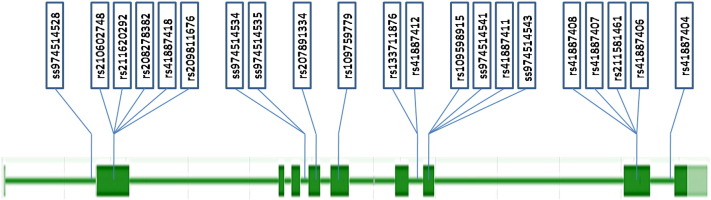
Twenty-one SNPs were identified by DNA sequencing and five of them were novel (Fig. 1 and Table 1). The first novel one (ss974514528) was 62 bp upstream of the first exon. The second (ss974514534) and third (ss974514535) were in the third intron, 61 bp away from each other. The last two (ss974514541 and ss974514543) were detected in exon 7 and were synonymous. The first was detected in the Holstein breed, unlike the other four, which were in Zebu breeds. Regarding the rest of the SNPs (16), five were observed in the first exon (rs210602748, rs211620292, rs208278382, rs41887418, rs209811676); one in the fourth (rs207891334); one in the fifth (rs109759779); two were in the sixth intron (rs133711876, rs41887412); two in the seventh exon (rs109598915, rs41887411); four in the eighth exon (rs41887408, rs41887407, rs211581461, rs41887406); and one in the eighth intron (rs41887404). Sixteen of the SNPs were transitions and five were transversions.
Fig. 1.
Scheme of the LIPE gene showing the polymorphisms detected.
Table 1.
Genetic variants detected in the LIPE gene by re-sequencing a panel of different cattle breeds. Chr Pos (UMD 3.1): chromosome position. AA: Angus, He: Hereford, Ho: Holstein, Sho: Shorthorn, Lim: Limousin, Wa: Wagyu, Ne: Nelore, Br: Brahman.
| Reference number | Chr Pos | Status | Region | Type | Change | Sequence | AA (monomorphic) | BB (monomorphic) | Polymorphic breeds |
|---|---|---|---|---|---|---|---|---|---|
| ss974514528 | 51217451 | Novel | Upstream | Non coding | CTGGGACCCC G/A GGGCCCAGTG | AA, Br, Cr, He, Lim, Ne, Sho, Wa | Ho | ||
| rs210602748 | 51217544 | Reported | Exon 1 | Missense | Arg × His | ATGGACCTGC G/A CACCATGACA | AA, Cr, He, Ho, Lim, Sho, Wa | Br, Ne | |
| rs211620292 | 51217620 | Reported | Exon 1 | Synonymous | CCGGGGAGAC G/A GCCCGGCGGC | He, Ne, Sho, Wa | AA, Br, Cr, Ho, Lim | ||
| rs208278382 | 51217635 | Reported | Exon 1 | Synonymous | GGCGGCTGAC G/A GGCGTCTTTG | AA, Cr, He, Ho, Lim, Sho, Wa | Ne | Br | |
| rs41887418 | 51217809 | Reported | Exon 1 | Synonymous | AATCGCGCTA T/C GTGGCCTCCA | Wa | Ne | AA, Br, Cr, He, Ho, Lim, Sho | |
| rs209811676 | 51217959 | Reported | Exon 1 | Synonymous | GGCTCTTCTT T/C GAGGGTGATG | AA, Cr, He, Ho, Lim, Sho, Wa | Br, Ne | ||
| ss974514534 | 51220859 | Novel | Intron 3 | Non coding | GGGCCCAGGG C/A GGGCACAGGA | AA, Br, Cr, He, Ho, Lim, Sho, Wa | Ne | ||
| ss974514535 | 51220921 | Novel | Intron 3 | Non coding | GGACCCCTGC C/A AGCAGTTCCT | AA, Cr, He, Ho, Lim, Sho, Wa | Br, Ne | ||
| rs207891334 | 51221010 | Reported | Exon 4 | Synonymous | GCCTGCCACC C/T GTCGCCTTTG | AA, Cr, He, Ho, Lim, Sho, Wa | Br, Ne | ||
| rs109759779 | 51221527 | Reported | Exon 5 | Synonymous | CCCTGGCCCC C/A GAGGCCCCCT | Br, Ne, Wa | AA, Cr, He, Ho, Lim, Sho | ||
| rs133711876 | 51222749 | Reported | Intron 6 | Non coding | TTGAGACTGG G/A CCCAGAAAGA | Br, Ne, Wa | AA, Cr, He, Ho, Sho | ||
| rs41887412 | 51222768 | Reported | Intron 6 | Non coding | GAGAGGAGAC T/C AACTCACCCA | Br, Ne, Wa | AA, Cr, He, Ho, Sho | ||
| rs109598915 | 51222827 | Reported | Exon 7 | Missense | Asp × Ala | GACCACCCCG A/C CTCAGACCAG | Br, Ne, Wa | AA, Cr, He, Ho, Lime, Sho | |
| ss974514541 | 51222849 | Novel | Exon 7 | Synonymous | AGGCGCTGGG T/C GTGATGGGGC | AA, Cr, He, Ho, Lim, Sho, Wa | Br, Ne | ||
| rs41887411 | 51222864 | Reported | Exon 7 | Synonymous | TGGGGCTCGT A/G CAGCGGGACA | AA, Lim | Br, Ne | Cr, He, Ho, Sho, Wa | |
| ss974514543 | 51222900 | Novel | Exon 7 | Synonymous | GAGACCTCCG G/C CTGGGCGCCT | AA, Cr, He, Ho, Lim, Sho, Wa | Br, Ne | ||
| rs41887408 | 51226081 | Reported | Exon 8 | Missense | Arg × Gln | GCCCTGACCC G/A GCCGGAGGGC | AA, Br, Cr, He, Ho, Lim, Ne, Sho, Wa | ||
| rs41887407 | 51226092 | Reported | Exon 8 | Missense | Pro × Ser | GCCGGAGGGC C/T CACTGGGAAC | Ne, Wa | AA, Br, Cr, He, Ho, Lim, Sho | |
| rs211581461 | 51226221 | Reported | Exon 8 | Missense | Ile × Val | ACCCTCAACC A/G TCAACTTCTT | He, Ho, Ne, Sho, Wa | AA, Br, Cr, Lim | |
| rs41887406 | 51226263 | Reported | Exon 8 | Missense | Glu × Lys | TGAAATGTCT G/A AGGCCCCAGA | Br, Ne, Wa | AA, Cr, He, Ho, Lim, Sho | |
| rs41887404 | 51226856 | Reported | Intron 8 | Non coding | CCCCGCCCCC [G/A] CAGGCCTGCG | Cr, Sho * |
One of the SNPs detected on the first exon was a missense mutation (Arg4His) and the other four were synonymous. The mutations in the fourth and fifth exons were also synonymous. One of the seventh exon SNPs was a missense (Asp497Ala), the others were synonymous. Lastly, the four SNPs found at the eighth exon were missense mutations (Arg560Gln, Pro564Ser, Ile607Val, Glu621Lys).
Mutation selection for genotyping
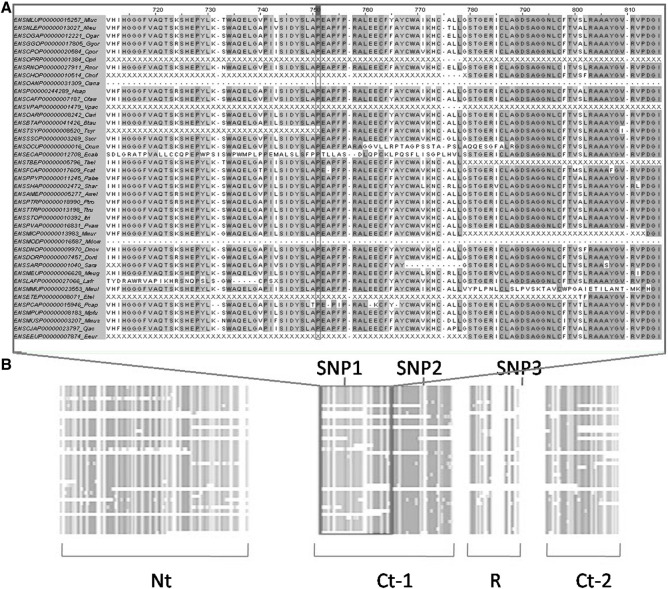
An analysis of residue conservation was performed to identify the most important areas or domains of the enzyme. The purpose of this analysis was to select three of the detected SNPs for further evaluation. This study was performed considering all mammalian sequences available, and the residues were colored according to the degree of conservation (Fig. 2). Basically four conserved regions were observed in the comparison: an N-terminal (Nt) region of about 280 residues; an intermediate region, of about 290 residues, composed by two segments; and a C-terminal region of about 100 residues. This is consistent with works of sequencing and structural modeling, which suggest a multi-domain enzyme structure (Osterlund et al., 1996, Osterlund et al., 1999, Smith et al., 1996). The Nt portion has no clear role to date. Its role has been suggested as a binding site for other proteins (ALBP) and lipids, and as a dimerization site (Shen et al., 2000, Shen et al., 2001, Smith et al., 2007). This portion is quite conserved in hormone-sensitive lipases of all mammals. The intermediate portion exhibits an α/β hydrolase fold, common among lipases (Contreras et al., 1996, Osterlund et al., 1996, Osterlund et al., 1997), and a regulatory module (R) that contains the phosphorylation sites involved in the activation of the enzyme (Anthonsen et al., 1998). The catalytic domain is composed of two interrupted fragments (Ct-1 and Ct-2) and contains the motif Gly-X-Ser-X-Gly, existent in most of the esterases and lipases, along with the other two members of the catalytic triad (Holm et al., 1994). The triad is composed of a serine, a carboxylic acid residue (Asp or Glu) and a histidine, in this order in the sequence. Besides the catalytic triad there is a putative histidine catalytic site (G-D-X-G family).
Fig. 2.
Analysis of residue conservation at the evolutionary level. Columns with residues exceeding 75% identification were colored light gray, and those who were above 80% with dark gray. (A) The area surrounding SNP1. A proline residue is observed at position 751 in every species that showed a homologous region. (B) Overall view of the enzyme. The framework indicates the area shown in panel A.
Considering the previously mentioned regions and their importance, three SNPs were selected for causing amino acid changes or falling into highly conserved areas. rs109759779 (SNP1) was synonymous, but it fell in a highly conserved amino acid position in mammals (above 80%). SNP1 involved a proline residue (386) located within the catalytic domain, 27 amino acids away from the catalytic serine. All the mammal species that had a region homologous to bovine LIPE showed a proline in this position, and because of that reason SNP1 became a candidate for our study. rs109598915 (SNP2) caused an amino acid change, aspartate to alanine, at the end of Ct-1. This SNP was in a region of about twelve residues that were not so conserved, but this region was in the midst of a widely conserved area and the nature of the amino acid change (negatively charged polar to non-polar neutral) was quite interesting. rs41887406 (SNP3) was located in a poorly conserved region between R and Ct-2, but caused a change of residue: a glutamate for a lysine (negatively charged to positively charged). In addition, this SNP has been recently associated with fatty-acid composition and various other traits in Chinese Simmental steers (Fang et al., 2013). Given these good features, SNP3 became our last candidate for further analysis. The intention was to select mutations that belonged to or approached the catalytic domain, the region with the most defined and clear role so far.
Statistical analysis
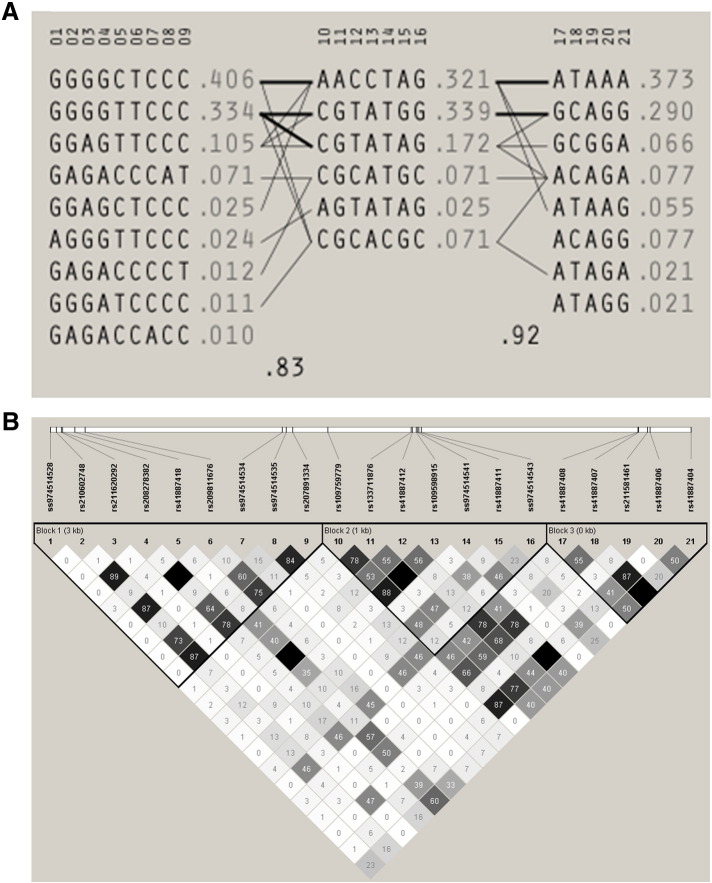
Three haplotype blocks were observed within the set of 44 sequenced samples (Fig. 3). These blocks were consistent with the relative positions of the SNPs within the gene; the first block extended from exon 1 to exon 4, the second extended from exon 5 to exon 7 and the third extended from exon 8 to exon 9. The majority of the SNPs in zebu were markedly linked, as expected.
Fig. 3.
Haplotypes (A) and linkage disequilibrium (B) within the set of re-sequencing samples. The r2 values are indicated inside the boxes. Blocks are indicated with thick lines.
Well-balanced allele frequencies were found in all the population groups of the three selected SNPs (minor allele frequency > 0.22). Their values ranged from 0.225 to 0.775 and are listed in Table 2. Estimates of observed (ho) and unbiased expected (he) heterozygosity for each SNP and group studied are given in Table 3. We observed high values of genetic diversity for every SNP within each group (he > 0.35) and within the whole population (he > 0.44). Expected heterozygosity values ranged from 0.35 for the Limousin crossbred group to 0.5046 for the 75% Angus group. On the other hand, the HWE test showed no significant deviations from the theoretical proportions (Table 3).
Table 2.
Allele frequencies of SNP1 (rs109759779), SNP2 (rs109598915) and SNP3 (rs41887406) in each group and the whole population (global). N: sample size.
| Locus |
Population |
||||||
|---|---|---|---|---|---|---|---|
| N = 44 |
N = 26 |
N = 30 |
N = 24 |
N = 95 |
N = 41 |
N = 260 |
|
| A | H | 75A | 75H | 50AH | LX | Global | |
| SNP1 | |||||||
| 1 | 66.25 | 71.74 | 57.69 | 76.09 | 63.89 | 77.5 | 67.77 |
| 2 | 33.75 | 28.26 | 42.31 | 23.91 | 36.11 | 22.5 | 32.23 |
| SNP2 | |||||||
| 1 | 39.77 | 42 | 48.28 | 35.42 | 45.21 | 28.05 | 40.66 |
| 2 | 60.23 | 58 | 51.72 | 64.58 | 54.79 | 71.95 | 59.34 |
| SNP3 | |||||||
| 1 | 60.23 | 55.77 | 51.67 | 64.58 | 53.72 | 71.95 | 58.69 |
| 3 | 39.77 | 44.23 | 48.33 | 35.42 | 46.28 | 28.05 | 41.31 |
Table 3.
Unbiased expected heterozygosity (he), observed heterozygosity (ho) and Hardy–Weinberg equilibrium p-value (HWE p-value) of SNP1 (rs109759779), SNP2 (rs109598915) and SNP3 (rs41887406) in each group and the whole population (global).
| Populations |
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP |
A |
H |
75A |
75H |
50AH |
LX |
Global |
||||||||||||||
| he | ho | HWE p value | he | ho | HWE p-value | he | ho | HWE p-value | he | ho | HWE p-value | he | ho | HWE p-value | he | ho | HWE p-value | he | ho | HWE p-value | |
| 1 | 0.45 | 0.53 | 0.48 | 0.41 | 0.48 | 0.63 | 0.50 | 0.54 | 0.71 | 0.37 | 0.39 | 1.00 | 0.46 | 0.50 | 0.50 | 0.35 | 0.40 | 0.65 | 0.44 | 0.48 | 0.15 |
| 2 | 0.48 | 0.52 | 0.76 | 0.50 | 0.52 | 1.00 | 0.51 | 0.55 | 0.72 | 0.47 | 0.46 | 1.00 | 0.50 | 0.56 | 0.22 | 0.41 | 0.46 | 0.46 | 0.48 | 0.53 | 0.20 |
| 3 | 0.48 | 0.52 | 0.76 | 0.50 | 0.50 | 1.00 | 0.51 | 0.57 | 0.72 | 0.47 | 0.46 | 1.00 | 0.50 | 0.56 | 0.22 | 0.41 | 0.46 | 0.46 | 0.49 | 0.53 | 0.21 |
A: purebred Angus; H: purebred Hereford; 75A: 75% Angus steers; 75H: 75% Hereford steers; 50AH: 50% Angus–50% Hereford steers; L: Limousin sire; LX: Limousin crossbred steers.
Association analysis
When individual tests were performed initially, SNP1 showed significant effects on MUFA and C18:1c9 (p = 0.0047, 0.0077) (Table 4). The MUFA value for the AC genotype was 2.19% higher than for the AA genotype, but samples with CC genotype showed no significant differences from the other groups, probably due to the low number of samples with this genotype and the high standard error. Regarding C18:1c9, the CA genotype showed a value 2.61% higher than the AA genotype. The SNP2 showed a significant effect on C21:0 (p = 0.0116), the AC genotype had a value 47.83% lower than the AA genotype. Samples with the CC genotype showed no significant differences from the other groups. A similar effect was observed on SNP3 (p = 0.0075) with AG and GG genotypes (51.09%), as both SNPs were almost completely linked. The three SNPs showed significant dominance effects (p < 0.05) in the tests of contrast. Additive effects were not significant (p > 0.05). No significant effects were observed on the rest of the measurements. At first sight, the SNPs seemed to be associated with the mentioned measurements, but when the Benjamini & Hochberg method for multiple comparisons was applied none of them remained significant.
Table 4.
Least Squares Means of fatty acid values and standard error (SE) for the genotypic classes based on the individual polymorphisms of the LIPE gene and test for dominance effect. n: number of samples, MUFA: monounsaturated fatty acids content, C18:1c9: oleic acid content, C21:0: Heneicosylic acid content. N: sample size.
| SNP/trait | Average of genotype1 | Dominance effect2 | Additive effect3 | ||
|---|---|---|---|---|---|
| SNP1 | AA (N = 71) | AC (N = 80) | CC (N = 17) | ||
| MUFA | 47.104 ± 0.278a | 48.137 ± 0.263b | 46.795 ± 0.539ab | 1.188 ± 0.373 (p = 0.002) | − 0.309 ± 0.604 (p = 0.610) |
| C18:1c9 | 39.483 ± 0.263a | 40.513 ± 0.249b | 39.594 ± 0.510ab | 0.975 ± 0.353 (p = 0.006) | 0.111 ± 0.571 (p = 0.846) |
| SNP2 | AA (N = 30) | AC (N = 92) | CC (N = 53) | ||
| C21:0 | 0.092 ± 0.014a | 0.136 ± 0.008b | 0.115 ± 0.010ab | 0.033 ± 0.011 (p = 0.004) | 0.024 ± 0.017 (p = 0.162) |
| SNP3 | GG (N = 30) | AG (N = 93) | AA (N = 52) | ||
| C21:0 | 0.092 ± 0.014a | 0.139 ± 0.008a | 0.115 ± 0.010ab | 0.035 ± 0.011 (p = 0.002) | − 0.022 ± 0.017 (p = 0.196) |
In the same row, least square means with different letters are significantly different according to Bonferroni's means separation test (p < 0.05).
Estimated by subtracting the average of solutions for homozygous genotypes from that for heterozygous genotype.
Estimated by the difference between the two homozygous genotypes.
Structural analysis
We attempted to generate structural models of the enzyme by using HHpred (Söding et al., 2005). These models were meant to evaluate the effect of the missense SNPs on the protein structure, but the limited availability of solved homologous structures did not allow a reliable modeling of the zones at which the SNPs were located. It was only possible to model the catalytic domain (residues 316–497, 620–756), which was shared by many esterases and lipases from other organisms. The 3D structure of this domain was constrainedly predicted using 1jji_A, the most highly homologous protein available on the data bank (Fig. 4). The model consisted of an α/β hydrolase fold: eight β sheets surrounded by α helices and a special closeness between the three catalytic residues (Ser424, Asp692 and His722), as we expected. The missense SNPs that we attempted to analyze were at the borders of the homologous regions, almost outside of the fold.
Fig. 4.
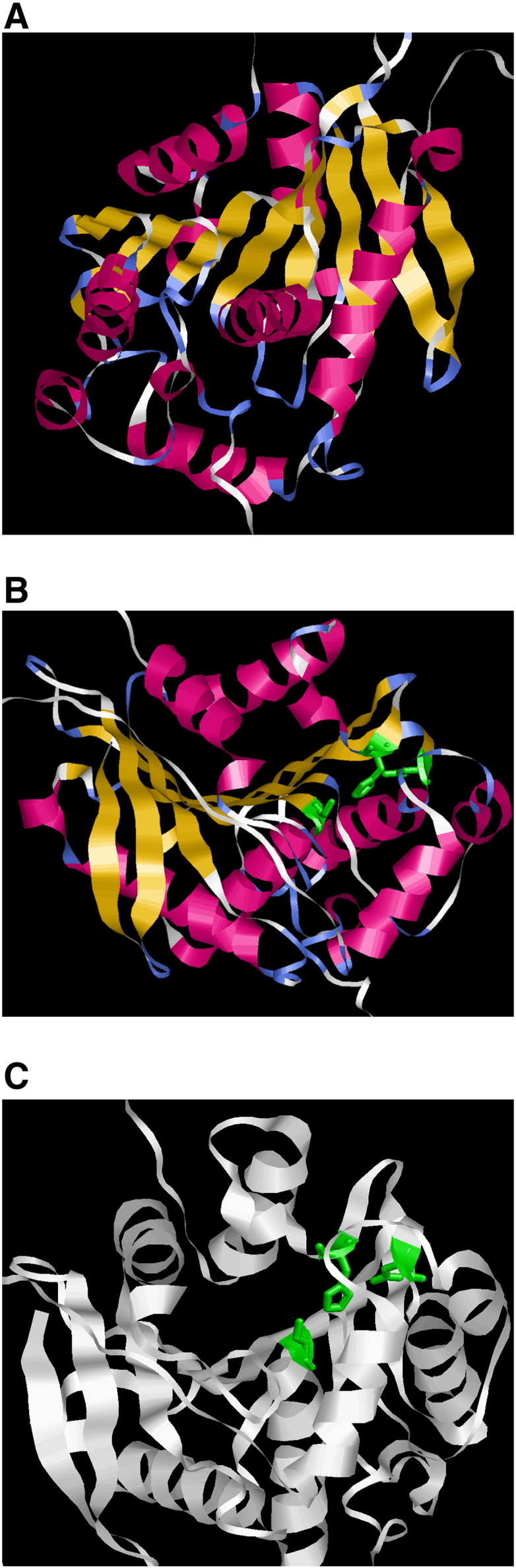
Molecular modeling of the putative catalytic domain of bovine LIPE. (A) The three-dimensional (3D) model contains the 316–497 and 620–756 amino acid residues, showing eight β sheets connected by α helices, which literally represent the core structural elements of the α/β hydrolase fold catalytic domain. (B) The catalytic triad residues (depicted in green) are illustrated in sticks. (C) Alternate view of the catalytic triad with the enzyme depicted in green.
Discussion
The LIPE gene was found to be quite variable compared to the average reported in cattle. Heaton et al. (2002) reported a density of about one SNP per 500 bp. A few years ago, The Bovine HapMap Consortium et al (2009) results showed one SNP every 714 bp for Angus or Holstein, and one SNP every 285 bp for Brahman. In this work, we detected 21 SNPs over 2600 bp, in other terms, one SNP every 123 bp. However, we should consider that two zebu breeds were included in the study, and nearly half of the SNPs (8) corresponded exclusively to these breeds, but the gene variability is still high. Lirón et al. (2011) reported one variant every 249 bp in the gonadotropin-releasing hormone receptor (GNRHR) gene by using a similar sample panel and the same re-sequencing strategy. This high variability could be related to the constitution of the enzyme, since the most critical areas for proper functioning seem to be quite punctual: residues H194 and E199 for ALBP interaction (Shen et al., 2001); S424, D692, H722 and possibly the aminoacids 191–199 for catalytic function (Holm et al., 1994, Shen et al., 2001); and the regulatory residues S552, S554 and S649 (Anthonsen et al., 1998). The missense SNPs were detected in exons 1 (1), 7 (1) and 8 (4). This is also consistent with the comparison made between species, which showed higher variability in the regulatory module of the enzyme.
To evaluate the effect of the detected genetic variants on the fatty acid composition of meat we performed an association study. For this, we selected three SNPs on the basis of type of mutation and site conservation. These SNPs showed high variability and were in equilibrium; hence they were suitable candidates for an association study. When individual tests were performed, we found apparent significant associations for SNP1 with total monounsaturated fatty acids and oleic acid, and for SNP2 and SNP3 with heneicosylic acid, although none of them remained significant once the FDR was implemented to control the false rejections of null-hypotheses due to multiple comparisons. These initial and suggestive associations showed dominance effects (p < 0.01) in the tests of contrast, so the homozygous and heterozygous genotypes of certain alleles seemed to behave differently than the other homozygous genotype. Dominance is expected and common among enzymes, since most of them exert their roles within metabolic routes that provide certain margins of action depending on the equilibrium of other parallel reactions and the availability of substrate, so slight increases in the activity caused by heterozygous genotypes may be sufficient to saturate reactions. In the case of LIPE, several scenarios can be imagined for the enzyme to affect the accumulation of oleic acid, monounsaturated fatty acids and heneicosylic acid. Previous works have reported that oleic acid is esterified mostly at sn-2 and sn-3 on the molecule of glycerol in cattle, while heneicosylic acid is esterified at positions sn-1 and sn-3. Since LIPE preferably hydrolyses the sn-3 position of diacylglycerides (Eichmann et al., 2012, Rodriguez et al., 2010), a single increase in activity may affect both fatty acids directly as a large portion of them is located at sn-3. Besides, LIPE can also hydrolyze triacylglycerides at sn-1 and sn-3 to a minor extent, so fatty acids at sn-2 may gradually accumulate. The rates and kinetics of triacylglyceride-hydrolysis by ATGL and re-esterification from diacylglycerides by DGAT1 and DGAT2 must also be considered. The real scenario may be much more complex and further studies are needed to fully elucidate the processes by which SNPs located in the enzymes of this route may affect the fatty acid regulation.
Other SNPs of LIPE were previously associated with lipid composition in cattle (Fang et al., 2013), mainly rs41887407. The C allele homozygotes of rs41887407 were associated with higher content of arachidic acid, dohono-c-Linolenic acid and eicosanoic acid in beef (p < 0.05). This SNP is very close to the SNP3 we analyzed (170 bp).
Since LIPE is directly involved in lipolysis, structural changes on the enzyme are expected to modify the fatty acid composition of tissues. These structural changes may reflect on the stereo/regioselectivity of the enzyme, activation or inhibition processes, and/or stability. SNP1 was synonymous, but changed the codon frequency from 20.4 (per thousand) for CCC to 14.6 for CCA, according to data available on http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=9913. It has been proposed that changes in codon frequencies affect the timing of post-translational folding of proteins (Kimchi-Sarfaty et al., 2007), especially in haplotypes, and this could contribute to the explanation of the effects caused by this SNP on the studied traits. SNP1 seems to be linked to rs133711876, rs109598915 and rs41887406. Structural changes in the proximity of the SNP may affect various features such as substrate selectivity or the binding to other proteins such as ALBP, which interacts with LIPE by the N-terminal domain and reduces the effect of inhibition by product (Shen et al., 2001). SNP2 and SNP3 were missense mutations, but we could not evaluate their effects by structural modeling. This happened because, unfortunately, no homologous structures were available to make reliable models of the particular regions at which the SNPs were found.
Conclusions
In conclusion, the LIPE gene is highly polymorphic, and our results may suggest a possible association of three SNPs with fatty acid composition in cattle. These SNPs should be evaluated in independent populations with in-vitro and in-vivo analyses to explain the mechanisms by which these polymorphisms may be involved in the mentioned trait.
There are increasingly larger amounts of information about the nutritional and therapeutic properties of the fatty acids. It is known that replacement of dietary saturated fatty acids for oleic acid and other unsaturated acids reduces the cardiovascular risk by reducing blood lipids, particularly LDL cholesterol (Lopez-Huertas, 2010). In recent years, researchers and producers have been trying to develop food products, such as milk, with lower saturated fatty acid content and higher proportions of beneficial acids such as oleic. It has also been demonstrated that oleic acid has anti-tumoral properties, since it plays a role in the activation of different intracellular pathways involved in carcinoma cell development (Carrillo et al., 2012). Our results may be helpful to improve the quality of meat and the health of consumers by genetic selection.
The following are the supplementary material related to this article.
Genetic structure of the population used to perform validation and association studies. N: number of samples; A: purebred Angus; H: purebred Hereford; 75A: 75% Angus steers; 75H: 75% Hereford steers; 50AH: 50% Angus–50% Hereford steers; L: Limousin sire; LX: Limousin crossbred steers.
The primer sequences and information of bovine LIPE gene.
Acknowledgments
Research for this paper was supported by grants from ANPCYT (Grant PICT2007-00482), CONICET (Grant PIP2010-2012 # 379), INTA (Grant Proyecto nacional PNPA 1126033), UNMDP (Grant AGR 456/14) and UNLP (Grant V168).
Contributor Information
Lilia Magdalena Melucci, Email: melucci.lilia@inta.gob.ar.
Guillermo Giovambattista, Email: ggiovam@fcv.unlp.edu.ar.
References
- Anthonsen M.W., Rönnstrand L., Wernstedt C., Degerman E., Holm C. Identification of novel phosphorylation sites in hormone-sensitive lipase that are phosphorylated in response to isoproterenol and govern activation properties in vitro. J. Biol. Chem. 1998;273:215–221. doi: 10.1074/jbc.273.1.215. [DOI] [PubMed] [Google Scholar]
- Baeza M.C., Corva P.M., Soria L.A., Pavan E., Rincon G., Medrano J.F. Genetic variants in a lipid regulatory pathway as potential tools for improving the nutritional quality of grass-fed beef. Anim. Genet. 2013;44(2):121–129. doi: 10.1111/j.1365-2052.2012.02386.x. [DOI] [PubMed] [Google Scholar]
- Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Belfrage P., Jergil B., Stralfors P., Tornqvist H. Identification and some characteristics of the enzyme protein of the hormone-sensitive lipase from rat adipose tissue. Adv. Exp. Med. Biol. 1978;101:113–126. doi: 10.1007/978-1-4615-9071-2_11. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Boldman K.G., Kriese L.A., Van Vleck L.D., Van Tassel L.A., Kachman S.D. USDA, ARS; Washington, DC: 1993. A Manual for Use of MTDFREML, a Set of Programs to Obtain Estimates of Variance and Covariances. [Google Scholar]
- Carrillo C., Cavia M.M., Alonso-Torre S.R. Antitumor effect of oleic acid; mechanisms of action: a review. Nutr. Hosp. 2012;27:1860–1865. doi: 10.3305/nh.2012.27.6.6010. [DOI] [PubMed] [Google Scholar]
- Contreras J.A., Karlsson M., Osterlund T., Laurell H., Svensson A., Holm C. Hormone-sensitive lipase is structurally related to acetylcholinesterase, bile salt-stimulated lipase, and several fungal lipases. J. Biol. Chem. 1996;271:31426–31430. doi: 10.1074/jbc.271.49.31426. [DOI] [PubMed] [Google Scholar]
- De Simone G., Menchise V., Manco G., Mandrich L., Sorrentino N., Lang D., Rossi M., Pedone C. The crystal structure of a hyper-thermophilic carboxylesterase from the archaeon Archaeoglobus fulgidus. J. Mol. Biol. 2001;314:507–518. doi: 10.1006/jmbi.2001.5152. [DOI] [PubMed] [Google Scholar]
- Eichmann T.O., Kumari M., Haas J.T., Farese R.V., Jr., Zimmermann R., Lass A., Zechner R. Stereo/regioselectivity of adipose triglyceride lipase, hormone-sensitive lipase, and diacylglycerol-O-acyltransferases. J. Biol. Chem. 2012;287:41446–41457. doi: 10.1074/jbc.M112.400416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X.B., Zhang L.P., Yu X.Z., Li J.Y., Lu C.Y., Zhao Z.H., Yang R.J. Association of LIPE gene E1-c.276C>T and E8-c.51C>T mutation with economical traits of Chinese Simmental cattle. Mol. Biol. Rep. 2013;41(1):105–112. doi: 10.1007/s11033-013-2842-6. [DOI] [PubMed] [Google Scholar]
- Folch J., Lees M., Sloane-Stanley H. A simple method for the isolation and purification of total lipids from animal tissue. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Giovambattista G., Ripoli M.V., Lirón J.P., Villegas Castagnasso E.E., Peral-García P., Lojo M.M. DNA typing in a cattle stealing case. J. Forensic Sci. 2001;46(6):1484–1486. [PubMed] [Google Scholar]
- Granneman J.G., Kimler V.A., Moore H.-P.H. Cell Biology Symposium: imaging the organization and trafficking of lipolytic effectors in adipocytes. J. Anim. Sci. 2011;89(3):701–710. doi: 10.2527/jas.2010-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle G., Zimmermann R., Hayn M., Theussl C., Waeg G., Wagner E., Sattler W., Magin T.M., Wagner E.F., Zechner R. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J. Biol. Chem. 2002;277:4806–4815. doi: 10.1074/jbc.M110355200. [DOI] [PubMed] [Google Scholar]
- Heaton M.P., Harhay G.P., Bennett G.L., Stone R.T., Grosse W.M., Casas E., Keele J.W., Smith T.P.L., Chitko-McKown C.G., Laegreid W.W. Selection and use of SNP markers for animal identification and paternity analysis in US Beef cattle. Mamm. Genome. 2002;13:272–281. doi: 10.1007/s00335-001-2146-3. [DOI] [PubMed] [Google Scholar]
- Hermo L., Chung S., Gregory M., Smith C.E., Wang S.P., El-Alfy M., Cyr D.G., Mitchell G.A., Trasler J. Alterations in the testis of hormone sensitive lipase-deficient mice is associated with decreased sperm counts, sperm motility, and fertility. Mol. Reprod. Dev. 2008;75(4):565–577. doi: 10.1002/mrd.20800. [DOI] [PubMed] [Google Scholar]
- Holm C. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Biochem. Soc. Trans. 2004;31(Pt 6):1120–1124. doi: 10.1042/bst0311120. [DOI] [PubMed] [Google Scholar]
- Holm C., Davis R.C., Osterlund T., Schotz M.C., Fredrikson G. Identification of the active site serine residue of hormone-sensitive lipase by site-specific mutagenesis. FEBS Lett. 1994;344:234–238. doi: 10.1016/0014-5793(94)00403-x. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Lampidonis A.D., Rogdakis E., Voutsinas G.E., Stravopodis D.J. The resurgence of Hormone-Sensitive Lipase (HSL) in mammalian lipolysis. Gene. 2011;477(1–2):1–11. doi: 10.1016/j.gene.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lei M.G., Wu Z.F., Deng C.Y., Dai L.H., Zhang Z.B., Xiong Y.Z. Sequence and polymorphism analysis of porcine hormone-sensitive lipase gene 5′-UTR and exon I. Yi Chuan Xue Bao. 2005;32(4):354–359. [PubMed] [Google Scholar]
- Li N., Stephens M. Modelling linkage disequilibrium, and identifying recombination hotspots using SNP data. Genetics. 2003;165:2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lirón J.P., Prando A., Ripoli M.V., Rogberg-Muñoz A., Posik D.M., Baldo A., Peral-García P., Giovambattista G. Characterization and validation of bovine Gonadotropin releasing hormone receptor (GNRHR) polymorphisms. Res. Vet. Sci. 2011;91:391–396. doi: 10.1016/j.rvsc.2010.09.024. [DOI] [PubMed] [Google Scholar]
- Lopez-Huertas E. Health effects of oleic acid and long chain omega-3 fatty acids (EPA and DHA) enriched milks. A review of intervention studies. Pharmacol. Res. 2010;61(3):200–207. doi: 10.1016/j.phrs.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Martínez Marín A.L., Pérez Hernández M., Pérez Alba L., Gómez Castro G., Carrión Pardo D. Lipid metabolism in ruminants. REDVET. 2010;11(08) [Google Scholar]
- Osterlund T., Daniellson B., Degerman E., Contreras J.A., Edgren G., Davis R.C., Schotz M.C., Holm C. Domain-structure analysis of recombinant rat hormone-sensitive lipase. Biochem. J. 1996;319:411–420. doi: 10.1042/bj3190411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund T., Contreras J.A., Holm C. Identification of essential aspartic acid and histidine residues of hormone-sensitive lipase: apparent residues of the catalytic triad. FEBS Lett. 1997;403:259–262. doi: 10.1016/s0014-5793(97)00063-x. [DOI] [PubMed] [Google Scholar]
- Osterlund T., Beussman D.J., Julenius K., Poon P.H., Linse S., Shabanowitz J., Hunt D.F., Schotz M.C., Derewenda Z.S., Holm C. Domain identification of hormone-sensitive lipase by circular dichroism and fluorescence spectroscopy, limited proteolysis, and mass spectrometry. J. Biol. Chem. 1999;274:15382–15388. doi: 10.1074/jbc.274.22.15382. [DOI] [PubMed] [Google Scholar]
- Qiao Y., Huang Z., Li Q., Liu Z., Hao C., Shi G., Dai R., Xie Z. Developmental changes of the FAS and LIPE mRNA expression and their effects on the content of intramuscular fat in Kazak and Xinjiang sheep. J. Genet. Genomics. 2007;34(10):909–917. doi: 10.1016/S1673-8527(07)60102-7. [DOI] [PubMed] [Google Scholar]
- Rodriguez J.A., Ben Ali Y., Abdelkafi S., Mendoza L.D., Leclaire J., Fotiadu F., Buono G., Carrière F., Abousalham A. In vitro stereoselective hydrolysis of diacylglycerols by hormone-sensitive lipase. Biochim. Biophys. Acta. 2010;1801:77–83. doi: 10.1016/j.bbalip.2009.09.020. [DOI] [PubMed] [Google Scholar]
- Rousset F. GENEPOP'007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Schneider S., Roessli D., Excoffier L. Genetics and Biometry Lab, Department of Anthropology, University of Geneva; 2000. Arlequin, a Software for Population Genetics Data Analysis. Ver 2.0. [Google Scholar]
- Shen W.J., Patel S., Hong R., Kraemer F.B. Hormone sensitive lipase functions as an oligomer. Biochemistry. 2000;39:2392–2398. doi: 10.1021/bi992283h. [DOI] [PubMed] [Google Scholar]
- Shen W.J., Liang Y., Hong R., Patel S., Natu V., Sridhar K., Jenkins A., Bernlohr D.A., Kraemer F.B. Characterization of the functional interaction of adipocyte lipid-binding protein with hormone-sensitive lipase. J. Biol. Chem. 2001;276(52):49443–49448. doi: 10.1074/jbc.M104095200. [DOI] [PubMed] [Google Scholar]
- Smith G.M., Garton A.J., Aitken A., Yeaman S.J. Evidence for a multi-domain structure for hormone-sensitive lipase. FEBS Lett. 1996;396:90–94. doi: 10.1016/0014-5793(96)01076-9. [DOI] [PubMed] [Google Scholar]
- Smith A.J., Thompson B.R., Sanders M.A., Bernlohr D.A. Interaction of the adipocyte fatty acid-binding protein with the hormone-sensitive lipase: regulation by fatty acids and phosphorylation. J. Biol. Chem. 2007;282:32424–32432. doi: 10.1074/jbc.M703730200. [DOI] [PubMed] [Google Scholar]
- Söding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Bovine HapMap Consortium et al Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science. 2009;324:528. doi: 10.1126/science.1167936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal E.L., Melucci L.M., Mezzadra C.A. 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, Brasil. 2006. Genetic Components for Slaughter and Meat Quality. Traits in the Angus–Hereford Crossing. [Google Scholar]
- Wang S.P., Chung S., Soni K., Bourdages H., Hermo L., Trasler J., Mitchell G.A. Expression of human hormone-sensitive lipase (LIPE) in postmeiotic germ cells confers normal fertility to LIPE-deficient mice. Endocrinology. 2004;145(12):5688–5693. doi: 10.1210/en.2004-0919. [DOI] [PubMed] [Google Scholar]
- Wang S.P., Wu J.W., Bourdages H., Lefebvre J.F., Casavant S., Leavitt B.R., Labuda D., Trasler J., Smith C.E., Hermo L., Mitchell G.A. The catalytic function of hormone-sensitive lipase is essential for fertility in male mice. Endocrinology. 2014 doi: 10.1210/en.2014-1031. [DOI] [PubMed] [Google Scholar]
- Warriss P.D. University of Bristol; Bristol. UK: 2000. Meat Science: An Introductory Text. School of Veterinary Science. (321 pp.) [Google Scholar]
- Yajima H., Kobayashi Y., Kanaya T., Horino Y. Identification of peroxisome-proliferator responsive element in the mouse HSL gene. Biochem. Biophys. Res. Commun. 2007;352(2):526–531. doi: 10.1016/j.bbrc.2006.11.054. [DOI] [PubMed] [Google Scholar]
- Yeaman S.J. Hormone-sensitive lipase—new roles for an old enzyme. Biochem. J. 2004;379:11–22. doi: 10.1042/BJ20031811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genetic structure of the population used to perform validation and association studies. N: number of samples; A: purebred Angus; H: purebred Hereford; 75A: 75% Angus steers; 75H: 75% Hereford steers; 50AH: 50% Angus–50% Hereford steers; L: Limousin sire; LX: Limousin crossbred steers.
The primer sequences and information of bovine LIPE gene.