Abstract
NAC proteins are plant-specific transcription factors (TFs). Although they play a pivotal role in regulating distinct biological processes, TFs in maize are yet to be investigated comprehensively. Within the maize genome, we identified 152 putative NAC domain-encoding genes (ZmNACs), including eight membrane-bound members, by systematic sequence analysis and physically mapped them onto ten chromosomes of maize. In silico analysis of the ZmNACs and comparison with similar genes in other plants such as Arabidopsis, rice, and soybean, revealed a similar NAC sequence architecture. Phylogenetically, the ZmNACs were arranged into six distinct subgroups (I–VI) possessing conserved motifs. Phylogenetic analysis using stress-related NAC TFs from Arabidopsis, rice, and soybean as seeding sequences identified 24 of the 152 ZmNACs (all from Group II) as putative stress-responsive genes, including one dehydration-responsive ZmSNAC1 gene reported earlier. One drought-tolerant genotype (HKI577) and one susceptible genotype (PC13T-3) were used for studying the expression pattern of the NAC genes during drought stress. qRT-PCR based expression profiles of 11 genes predicted to be related to stress confirmed strong differential gene expression during drought stress. Phylogenetic analyses revealed that ZmNAC18, ZmNAC51, ZmNAC145, and ZmNAC72, which were up-regulated in the tolerant genotype and down-regulated in the susceptible genotype, belonged to the same group to which also belong other drought-responsive genes, namely SNAC1, OsNAC6, ANAC019, and ANAC055, which act as a transcriptional activator and are strongly induced under stress from various abiotic sources. Differentially expressed ZmNAC genes, alone or in combination with each other or with other type(s) of TFs, may control the general cellular machinery and regulate stress-responsive downstream genes. Alternatively, they may serve as a platform to regulate a broad set of genes, which are subsequently fine-tuned by specific regulators. This genome-wide identification and expression profiling opens new avenues for systematic functional analysis of new members of the NAC gene family, which may be exploited in developing lines that are better adapted to drought.
Keywords: NAC family, Phylogenetic relationship, Gene expression, Drought stress, Maize
Introduction
Drought, cold, and salinity are major forms of stress from abiotic sources that adversely affect plant growth and productivity (Nakashima et al., 2012), of which drought is considered as the most devastating. Water is one of the significant limiting factors and affects maize at all stages of its growth. Maize is especially sensitive to drought at the reproductive stage, particularly between tassel emergence and early grain-filling (Grant et al., 1989). Drought stress during this period reduces kernel size and thus lowers grain yield significantly (Bolanos and Edmeades, 1993a, Bolanos and Edmeades, 1993b). Plants adapt to drought stress at physiological, biochemical, and molecular levels by activating a number of defense mechanisms that increase the plant's tolerance to water deficit.
Transcription factors (TFs) are key proteins that regulate gene expression at the transcription level by interacting with promoter elements of stress genes resulting in over-expression of many functional genes. Many TFs have been characterized as drought-responsive TFs and include WRKY (Rushton et al., 2012), zinc finger (Huang et al., 2009), AP2/ERF2 (Sakuma et al., 2002), MYB (Abe et al., 1997), ZmDREB2A (Qin et al., 2007), and NAC (Tran et al., 2004).
NAM, ATAF, and CUC (NAC) proteins belong to a plant-specific TF superfamily, possess a highly conserved N-terminal DNA binding domain (NAC) and a variable C-terminal transcription regulation region (TRR), and can activate or suppress the transcription of multiple target genes. The C-terminal regions of some NAC TFs also contain transmembrane motifs (TMs), which anchor to the plasma membrane (Tran et al., 2004, Nakashima et al., 2007). Several NAC genes have been identified in different species: 117 non-redundant putative NAC genes in Arabidopsis (Nuruzzaman et al., 2010), 151 putative NAC or NAC like genes in rice (Nuruzzaman et al., 2010), and 152 non-redundant genes in soybean (Le et al., 2011). In Arabidopsis ANAC019, ANAC055, and ANAC072 were induced by drought, salinity and low temperature (Tran et al., 2004), and OsNAC10 and OsNAC6 have been reported as responsive to drought stress in rice (Ohnishi et al., 2005, Jeong et al., 2010).
Research in Arabidopsis indicates that NAC TFs recognize the CACG core motif that acts as a drought-responsive NAC recognition sequence (NACRS). Wu et al. (2009) reported that Arabidopsis plants over-expressing the abiotic stress-responsive gene ATAF1, which is homologous to RD26, showed greater drought tolerance. These reports indicate that NAC TFs are important in stress tolerance and that their over-expression has a potential biotechnological application in improving stress tolerance in plants.
However, only a few NAC members have been characterized in maize. ZmSNAC1 has shown drought tolerance in transgenic Arabidopsis (Zimmama and Veer, 2005, Lu et al., 2012). In maize, 56 transcription families were predicted, of which the plant-specific NAC transcription family consists of more than 190 putative members. The importance of NAC TF family proteins in drought tolerance prompted us to carry out a genome-wide systematic analysis of the ZmNAC (Zea mays NAC) TF family for (a) identifying complete NAC domain containing gene sequences in silico and identifying gene orthologs and drought responsive genes by phylogenetic analysis of the NAC genes and (b) exploring the expression pattern of NAC genes using a set of contrasting maize genotypes under drought stress.
Materials and methods
Retrieval and curating of data
The NAC domain containing protein sequences of Zea mays L. was collected from the Plant Transcription Factor Database ver. 2.0 as well as by exploring the B73 maize genome (release 5b.60) for the hidden Markov model (HMM) profile of the NAC domain (PF02365) downloaded from the Pfam database using HAMMER (ver. 3.0). All redundant sequences were removed and the collected data were further curated by examining the presence of the conserved NAC domain in protein sequences with the help of Pfam (http://pfam.sanger.ac.uk/), SMART (http://smart.embl-heidelberg.de/), and InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/) web server (Quevillon et al., 2005, Letunic et al., 2012, Finn et al., 2013). The protein sequences were also investigated for their physical position in the maize genome. Proteins belonging to the same location were aligned, and protein sequences were rejected if they were completely aligned to any other sequence and shorter in length than that sequence. Finally, 152 non-redundant and complete NAC-domain-containing protein sequences were selected for further analysis. The membrane-bound ZmNAC members were predicted using TMHHM server ver.2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
Phylogenetic analysis and identification of conserved motifs
The finalized 152 NAC-domain-containing maize protein sequences and 11 published NAC proteins from Arabidopsis, rice, and soybean were aligned with a gap open penalty of 10 and a gap extension penalty of 0.2 using ClustalW. The result of the sequence alignment was used for constructing an unrooted phylogenetic tree by the neighbor-joining method. The confidence level of monophyletic groups was estimated using bootstrap analysis of 1000 replicates. Multiple sequence alignment (MSA) and phylogenetic analysis were performed using MEGA 5.10.
A MEME utility (http://meme.nbcr.net/meme4_1/cgi-bin/meme.cgi) was used for displaying the motifs of ZmNAC proteins from maize, Arabidopsis, rice, and soybean (Bailey et al., 2009). The parameters were set as follows. Occurrences of a single motif: zero or one per sequence, motifs width range: 10–200 amino acids, and maximum number of motifs to find: 100. For other parameters, the default values were retained. InterPro (http://www.ebi.ac.uk/interpro/) and the Pfam database were adopted to annotate the MEME motifs (http://meme.sdsc.edu).
Plant materials, treatments, and collection of tissues
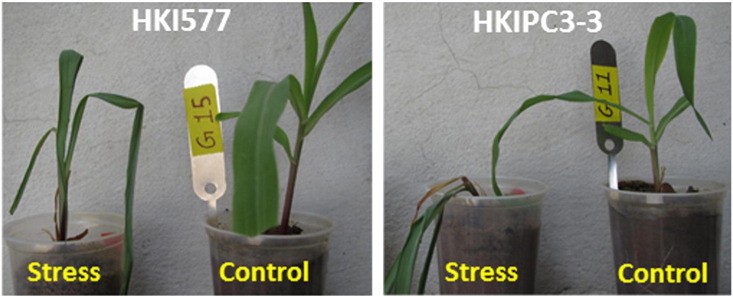
Two maize inbred lines, HKI577 (drought tolerant) and HKIPC3-3 (drought susceptible), were used. Plants were grown under natural conditions in plastic cups filled with a mixture of sand and clay. Before subjecting the plants to drought stress, they were watered daily to field capacity. Drought stress was imposed on 21-day-old plants by withholding water for 7 days (severe stress). Control plants were irrigated daily to field capacity. Samples for RNA extraction were collected from both the groups twice: on the day the treatment began and 7 days later, when the treatment was terminated (Fig. 1).
Fig. 1.
Response of HKI577 (drought tolerant) and HKIPC3-T3 (drought susceptible) genotypes to well-watered control and severe drought stages.
RNA isolation, DNase I treatment, and cDNA synthesis
Total RNA was isolated from the harvested samples using Qiagen RNeasy columns (Qiagen, Hilden, Germany) followed by DNase I treatment to remove contamination with any genomic DNA. Concentrations were determined with a Thermo Scientific NanoDrop 1000 spectrophotometer (Thermo Scientific, Delaware, USA). The cDNA was synthesized using a ProtoScript first strand cDNA synthesis kit (BioLabs, Massachusetts, USA) from 1 μg of total RNA according to the manufacturer's protocol. The reverse transcription reaction was carried out at 44 °C for 60 min followed by 92 °C for 10 min.
Quantitative real-time PCR and statistical analyses
Gene-specific primers were designed using IDT PrimerQuest (http://www.idtdna.com/scitools/applications/primerquest/default.aspx) and an 18s RNA primer was used as the internal control (Nakashima et al., 2007). The primer combinations used here for quantitative real-time PCR (qRT-PCR) analysis specifically amplified only one desired band. Dissociation curve testing carried out for each primer pair showed only one melting temperature for all samples. The qRT-PCR reactions were carried out at 95 °C for 5 min followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C for 1 min. Expression data for the NAC TFs were normalized by subtracting the mean reference gene CT value from individual CT values of the corresponding target genes (ΔCT). The fold-change value was calculated using the expression 2− ΔΔCT where ΔΔCT is the difference between the ΔCT condition of interest and ΔCT control. The primer sets used for studying the NAC TFs' expression profile are given in Supplementary Table S1.
Results and discussion
Identification and chromosomal distribution of ZmNAC members
The ZmNAC members in maize were predicted by three independent tools using genome-wide screening of the results of the search for ZmSNAC1 protein sequence. Each tool provided different numbers of putative ZmNAC TFs within its public database. The highest number of ZmNAC TFs (219) was predicted by NCBI BLASTp, followed by Plant TFDB (190) and phytozome (180) — as an initial step, we collected the sequences for all. The collected protein sequences were curated to remove redundancy, and only those protein sequences that had the complete NAC domain were selected. All of the alternative splice variants were carefully checked, and splice variants encoding the longest reading frames were selected. This left us with 152 members for the subsequent sequence alignment and phylogenetic analysis. Among the ZmNAC members, 8 were found to be membrane-bound members (Table 1).
Table 1.
Details of membrane-bound ZmNAC proteins.
| S No | ZmNAC | Tm helix (trans membrane region) |
|---|---|---|
| 1 | ZmNAC45 | 488–510 |
| 2 | ZmNAC62 | 636–658 |
| 3 | ZmNAC69 | 633–655 |
| 4 | ZmNAC108 | 677–699 |
| 5 | ZmNAC118 | 487–509 |
| 6 | ZmNAC136 | 581–603 |
| 7 | ZmNAC149 | 624–666 |
| 8 | ZmNAC119 | 487–509 |
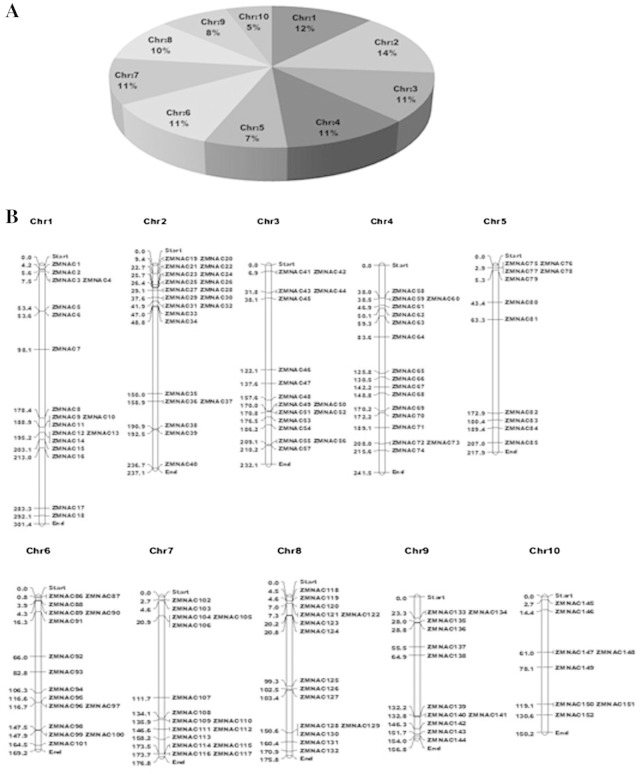
In silico mapping of ZmNACs on the maize genome indicated uneven distribution of the genes on all 10 chromosomes (Fig. 2). Of these, chromosome 2 had the highest number of ZmNACs (22) and chromosome 10, the lowest (8). The pattern of their distribution on individual chromosomes also revealed some physical regions with high accumulation of ZmNAC gene clusters. For example, ZmNAC genes located on chromosomes 2 and 7 appear to be congregating at the lower and upper ends of the arms respectively (Fig. 2). A uniform nomenclature for all the ZmNAC genes identified in this work has been adapted based on the order of the chromosomes to facilitate communication. Detailed information on, and structures of, representative ZmNACs can be found in Table S2.
Fig. 2.
Distribution of 152 ZmNAC genes on maize chromosomes. (A) Percentage of ZmNAC genes on each maize chromosome showing their distribution abundance. (B) Physical locations of each ZmNAC gene on the ten maize chromosomes (positions in Mb).
Phylogenetic analysis of the ZmNAC TF family
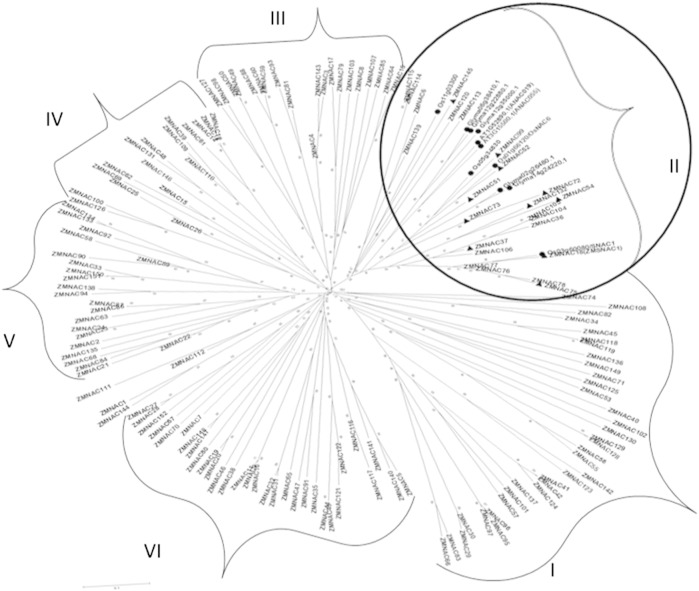
To examine the structure and phylogenetic relationships of ZmNAC TFs identified in our study, a combined phylogenetic tree was constructed with the aligned ZmNACs and 11 published sequences of NAC domains from Arabidopsis, rice, and soybean (Fig. 3). All well-known NAC domain proteins bind specifically to the CATGTG motif of the promoter region (Tran et al., 2004) or act as a functional motif or activation domain (Oh et al., 2005). The conserved NAC domain consisting of about 150 residues located in the N-terminal region and NAC domains is more closely associated with stress responses than C-terminal region. Examination of the phylogenetic tree suggested that the ZmNAC TFs can be classified into six major groups: Group I and Group VI, 32 TFs each; Group III, 20 TFs; Group IV, 17 TFs; and Group V, 21 TFs. This leaves Group II, which was noteworthy in that alone contained all the published stress-related NAC TFs. This group had 35 TFs, of which 24 were ZmNACs and 11 were published stress-related TFs of NAC family. These published TFs included OsNAC6, a stress responsive NAC from rice that has been reported as responsive to drought stress (Ohnishi et al., 2005, Jeong et al., 2010), soybean GmNAC085 that has been highly induced by dehydration (Le et al., 2011), and maize that ZmSNAC1 has shown to develop drought tolerance in transgenic Arabidopsis (Lu et al., 2012); we considered Group II of the phylogenetic tree as drought-responsive group. Under drought stress, SNAC1 (Hu et al., 2006) and OsNAC6 (Nakashima et al., 2007) from rice are highly expressed in guard cells of transgenic rice: the result is many more closed stomata, which, in turn, reduces the amount of water lost through transpiration without altering the rate of photosynthesis (Hu et al., 2006). Two more, ANAC019 and ANAC055, were from Arabidopsis (Tran et al., 2004). Induced under dehydration stress, these bind to a region containing the MYC-like sequence of the ERD1 promoter. As with Arabidopsis, rice, and soybean, our data identified a diversified ZmNAC family in maize (Fang et al., 2008, Nuruzzaman et al., 2010). The main purpose of phylogenetic analysis of ZmNAC TFs was to predict abiotic stress-responsive ZmNAC TFs that could be subsequently prioritized further in plant functional studies. Earlier reports have shown that phylogenetic analysis provides strong evidence for predicting stress-related functions of several gene families, including TF families. Phylogenetic analysis of the rice OsNAC families with their orthologs from other plant species, whose stress responsive expression patterns and functions are known, resulted in a nearly perfect match between sequence conservation and functions or expression patterns (Fang et al., 2008, Zhang et al., 2008). Among the ANACs and OsNACs, many proteins involved in regulating physiological and biochemical responses are associated with resistance to stress from various abiotic sources, including drought (Tran et al., 2010). For instance, Arabidopsis ANAC019 and ANAC055 and rice SNAC1/OsNAC002 and OsNAC6/SNAC2//OsNAC048 act as positive regulators in plant response to drought stress (Tran et al., 2007). The statistical significance of the phylogenetic analysis was confirmed through bootstrap analysis of 1000 replicates. A good number of internal branches had high bootstrap values, pointing to the derivation from a common ancestor of statistically reliable pairs of possible homologous proteins sharing similar functions. On the basis of sequence alignments and phylogenetic analyses, we identified 11 putative stress-related ZmNAC TFs that were in the same group that also contained known stress-related NAC TFs from Arabidopsis, rice, and soybean.
Fig. 3.
Phylogenetic relationships among maize NAC proteins. The sequences were aligned by ClustalW at MEGA5.10 and the unrooted phylogenetic tree was deduced by the neighbor-joining method. The proteins were classified into six distinct groups (I–VI). Triangles represent selected putative drought stress genes and circles represent published drought stress genes.
Identification of motifs and their distribution
The MEME server was used for exploring motif distribution in 152 NAC proteins in maize and 11 published NAC proteins in Arabidopsis, rice, and soybean. Thirty-two putative motifs were present in the NAC family of maize (Table S3) of which 13 motifs had known functions: motifs 1–5, 7–9, and 12 encoded the NAC/NAM domain; motif 13, the DUF1296 domain; motif 18, the DUF3739 domain; motif 21, the GHPB domain; and motif 28, the zinc-finger domain. Of the 152 ZmNAC TFs, all except ZmNAC40 and ZmNAC116 had at least four NAC domain-encoding motifs, and 114 shared a highly conserved typical NAC domain containing five consensus subdomains (motifs 2, 4, 1, 5 and 3) in the same order. Based on annotated motifs, the 152 ZmNAC proteins were divided into 9 groups (Fig. S1). Group I had 104 ZmNAC and all the 11 published NAC proteins. Among the total 125 proteins in this group, 83 ZmNAC and 5 published NAC proteins started with NAC subdomain motif 2, and 21 ZmNAC and 6 published NAC proteins started with different un-annotated motifs (9, 14, 25). Such motif sequence conservation and variation among the proteins indicate functional equivalence and diversification respectively (Puranik et al., 2012). In Group II, the motifs additionally contained the DUF3739 domain in their TRR. ZmNAC108 transmembrane transcription factor (TMTF) had three functional motifs, namely NAC, motif 18, and motif 21. As reported earlier, members of a particular subfamily show a tendency to have a constant motif composition (with slight variations) (Hu et al., 2010), and some motifs are deleted or duplicated within particular clades. Groups III, VII, and VIII had different types of functional motifs: motif 21, motif 13, and motif 28 respectively. In Group IV, motif 1 was replaced by motif 9; in Group V, motifs 1 and 3 were replaced by motifs 9 and 12 respectively; and in Group VI, motifs 2 and 4 were replaced by motifs 8 and 7 respectively. Group IX contained 12 ZmNAC TFs having randomly distributed motifs. ZmNAC40 had the least number of motifs (2) and ZmNAC54, the highest (12). Motif deletion or duplication within a conserved domain of a protein may be crucial to disposing of undesirable regions and developing only those that are essential for developing a particular phenotype. Although the functions of most of the motifs remain to be determined, most of them do not have homologous sequences in the InterPro database (http://www.ebi.ac.uk/interpro/, release 43.1). The motif composition of these unknown NAC sequences may provide clues for further functional analysis of these TFs.
Expression patterns of predicted ZmNAC genes during dehydration stress
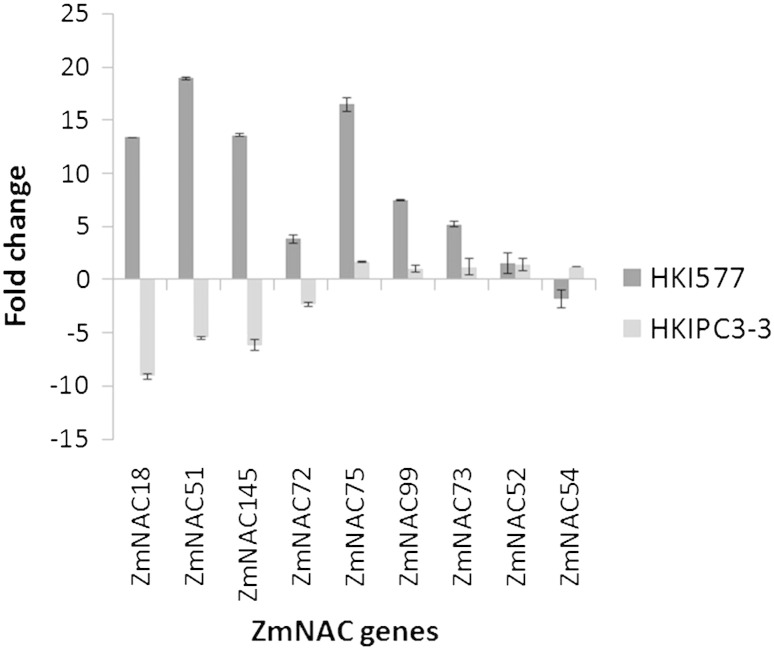
Gene expression patterns can provide vital clues for determining gene function. To identify candidate dehydration-responsive ZmNAC genes for engineering drought-tolerant maize genotypes, we carried out qRT-PCR analysis of 11 predicted stress-related ZmNAC genes with gene-specific primers in 21-day-old maize plants subjected to dehydration stress. We included a previously reported stress-responsive gene ZmNAC18 (ZmSNAC1) in the qRT-PCR analysis to serve as a positive reference for the verification of our results analysis. All the genes differed in their expression in drought-susceptible and drought-tolerant genotypes. ZmNAC18, ZmNAC51, and ZmNAC145 were up-regulated in the tolerant genotype and down-regulated in the susceptible genotype (Fig. 4). Attempts at over-expressing TFs that show higher expression under drought stress in tolerant genotypes than that in the susceptible genotype (Hayano-Kanashiro et al., 2009) have led to greater drought tolerance in several crops. In general, the genes involved in protecting plants from drought stress through stress perception, signal transduction, transcriptional regulatory networks in cellular responses, or tolerance to dehydration were found to be up-regulated in drought-tolerant genotypes (Guo et al., 2009). It is noteworthy that phylogenetic analysis showed that these three genes (ZmNAC18, ZmNAC51, and ZmNAC145) were closely linked to SNAC1 and OsNAC6 in rice, which have been reported as drought responsive and which act as transcriptional activators and regulate the genes that encode proteins (such as protein kinases and phosphatases) involved in regulating stress responses and also regulate the production of osmolytes (such as sorbitol transporter, exoglucanase, galactosidase, and glycosyltransferase). In drought-tolerant maize, genes encoding hormones, aquaporins, heat-shock proteins, and detoxification enzymes were induced to a greater extent than those in drought-susceptible maize (Hayano-Kanashiro et al., 2009). For example, gene ZmNAC51 was up-regulated 18-fold in the tolerant genotype. This gene, which has a sequence similar to that of SNAC1 and controls the expression of ERD1, has two novel cis-acting elements, namely a MYC-like sequence (CATGTG) and anrps1-site-1-like sequence (CACTAAATTGT-CAC), which are involved in dehydration stress. The drought-responsive gene ZmNAC18 (ZmSNAC1) was up-regulated 13-fold in the tolerant genotype and down-regulated 5-fold in the susceptible genotype. ZmNAC51 was in the same clade that contained the soybean drought-responsive genes GmNAC011 and GmNAC109. ZmNAC75 ranked second in terms of its expression in the drought-tolerant genotype (Fig. 4). ZmNAC drought stress group was analyzed in silico for any miRNA degradation site. Out of 24 ZmNACs, only two were found to have miRNA degradation site. ZmNAC54 was found to be cleaved by zma-miR164 family and zma-miR408b-5p cleaved ZMNAC72. ZmNAC54 was down-regulated in the tolerant genotype and up-regulated in the susceptible genotype. Fang et al. (2014) reported that over expression of Oryza miR164-targeted NAC genes (OMTN) caused increased drought sensitivity in rice plant. Under drought stress, reactive oxygen species (ROS) are generated in large amounts; these are nullified by the enzymes that prevent cellular damage and protect the plant from stress. Genes concerned with primary metabolic processes such as photosynthesis are down-regulated under drought stress (Wang et al., 2003). Differentially expressed ZmNAC genes, acting singly or together with each other or with other TFs, may control the general cellular machinery and regulate stress-responsive downstream genes and serve as a platform to regulate a broad set of genes, which are subsequently fine-tuned by specific regulators.
Fig. 4.
Comparison of expression pattern of ZmNAC gene, predicted to be related to stress, in 21-day-old plants of drought-tolerant (black) and drought-susceptible (gray) genotypes under dehydration stress. A previously reported stress-responsive gene, ZmNAC18 (ZmSNAC1), was taken as a positive reference for qRT-PCR analysis.
Conclusion
A comprehensive genome-wide analysis identified 152 NAC TFs including 8 membrane-bound members in maize. Based on the comparison of phylogenetic relationships, all the 152 ZmNAC TFs, along with 11 well-known stress-responsive NAC TFs in Arabidopsis, rice, and soybean, fell into six distinct groups. Group II was found to be the stress group and contained all the known stress-responsive NAC TFs along with 24 ZmNAC TFs that may act as candidate TFs. Motif analysis revealed functional equivalence among members of a group and diversification between groups. Furthermore, expression profiles by qRT-PCR showed that the candidate ZmNACs differed in their expression in tolerant and susceptible genotypes under drought stress. Overall, this genome-wide identification and expression profiling open new avenues for a systematic functional analysis of four novel members of NAC gene family as candidates for stress-responsive genes: ZmNAC51, ZmNAC145, ZmNAC72, and the previously reported ZMNAC18 (ZmSNAC1). The expression of stress-responsive genes is regulated largely by TFs, which, in turn, are subjected to intricate regulation at the chromatin level, RNA level, and protein level. This understanding will prove useful in improving drought tolerance since these differentially expressed genes are probably involved in drought tolerance in maize.
Abbreviations
- ZmNAC
NAC transcription factor of maize.
- ANAC
NAC transcription factor of Arabidopsis.
- OsNAC
NAC transcription factor of rice.
- GmNAC
NAC transcription factor of soybean.
- TF
transcription factor.
- qRT-PCR
quantitative real time PCR.
Acknowledgments
The authors sincerely acknowledge the ICAR network project on Transgenics in Crops (Maize Functional Genomics Component, Grant Number 21-22) for funding the study.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mgene.2014.05.001.
Appendix A. Supplementary data
Supplementary materials.
References
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos J., Edmeades G.O. Eight cycles of selection for drought tolerance in lowland tropical maize. 1. Responses in grain yield, biomass, and radiation utilization. Field Crop Res. 1993;31:233–252. [Google Scholar]
- Bolanos J., Edmeades G.O. Eight cycles of selection for drought tolerance in tropical maize. II. Responses in reproductive behavior. Field Crop Res. 1993;31:253–268. [Google Scholar]
- Fang Y., You J., Xie K., Xie W., Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics. 2008;280:547–563. doi: 10.1007/s00438-008-0386-6. [DOI] [PubMed] [Google Scholar]
- Fang Y., Xie K., Xiong L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J. Exp. Bot. 2014 doi: 10.1093/jxb/eru072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., Sonnhammer E.L., Tate J., Punta M. Pfam: the protein families database. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R.F., Jackson B.S., Kiniry J.R., Arkin G.F. Water deficit timing effects on yield components in maize. Agron. J. 1989;81:61–65. [Google Scholar]
- Guo P., Baum M., Grando S., Ceccarelli S., Bai G., Li R., von Korff M., Varshney R.K., Graner A., Valkoun V. Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J. Exp. Bot. 2009;60:3531–3544. doi: 10.1093/jxb/erp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayano-Kanashiro C., Calderon-Vazquez C., Ibarra-Laclette E., Herrera-Estrella L., Simpson J. Analysis of gene expression and physiological responses in three Mexican maize landraces under drought stress and recovery irrigation. PLoS One. 2009;4:e7531. doi: 10.1371/journal.pone.0007531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Dai M., Yao J., Xiao B., Li X., Zhang Q., Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R., Qi G., Kong Y., Kong D., Gao Q., Zhou G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010;10:145. doi: 10.1186/1471-2229-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X.Y., Chao D.Y., Gao J.P., Zhu M.Z., Shi M., Lin H.X. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009;23:1805–1817. doi: 10.1101/gad.1812409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J.S., Kim Y.S., Baek K.H., Jung H., Ha S.H., Do Choi Y., Kim M., Reuzeau C., Kim J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010;153:185–197. doi: 10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le D.T., Nishiyama R., Watanabe Y., Mochida K., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011;18:263–276. doi: 10.1093/dnares/dsr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Ying S., Zhang D.F., Shi Y.S., Song Y.C., Wang T.Y., Li Y. A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep. 2012;31:1701–1711. doi: 10.1007/s00299-012-1284-2. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Tran L.S., Van Nguyen D., Ito Y., Hayashi N., Shinozaki K., Yamaguchi-Shinozaki K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51:617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Takasaki H., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta. 2012;1819:97–103. doi: 10.1016/j.bbagrm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M., Manimekalai R., Sharoni A.M., Satoh K., Kondoh H., Ooka H., Kikuchi S. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Oh S.J., Song S.I., Kim Y.S., Jang H.J., Kim S.Y., Kim M., Kim Y.K., Nahm B.H., Kim J.K. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 2005;138:341–351. doi: 10.1104/pp.104.059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T., Sugahara S., Yamada T., Kikuchi K., Yoshiba Y., Hirano H.Y., Tsutsumi N. OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Gene Genet. Syst. 2005;80:135–139. doi: 10.1266/ggs.80.135. [DOI] [PubMed] [Google Scholar]
- Puranik S., Sahu P.P., Srivastava P.S., Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17:1360–1385. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Qin F., Kakimoto M., Sakuma Y., Maruyama K., Osakabe Y., Tran L.S., Shinozaki K., Yamaguchi-Shinozaki K. Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 2007;50:54–69. doi: 10.1111/j.1365-313X.2007.03034.x. [DOI] [PubMed] [Google Scholar]
- Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:116–120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton D.L., Tripathi P., Rabara R.C., Lin J., Ringler P., Boken A.K., Langum T.J., Smidt L., Boomsma D.D., Emme N.J., Chen X., Finer J.J., Shen Q.J., Rushton P.J. WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 2012;10:2–11. doi: 10.1111/j.1467-7652.2011.00634.x. [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Liu Q., Dubouzet J.G., Abe H., Shinozaki K., Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.S., Nakashima K., Sakuma Y., Osakabe Y., Qin F., Simpson S.D., Maruyama K., Fujita Y., Shinozaki K., Yamaguchi-Shinozaki K. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 2007;49:46–63. doi: 10.1111/j.1365-313X.2006.02932.x. [DOI] [PubMed] [Google Scholar]
- Tran L.S., Nishiyama R., Yamaguchi-Shinozaki K., Shinozaki K. Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops. 2010;1:32–39. doi: 10.4161/gmcr.1.1.10569. [DOI] [PubMed] [Google Scholar]
- Wang W., Vinocur B., Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- Wu Y., Deng Z., Lai J., Zhang Y., Yang C., Yin B., Zhao Q., Zhang L., Li Y., Yang C., Xie Q. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009;19:1279–1290. doi: 10.1038/cr.2009.108. [DOI] [PubMed] [Google Scholar]
- Zhang G., Chen M., Chen X., Xu Z., Guan S., Li L.C., Li A., Guo J., Mao L., Ma Y. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.) J. Exp. Bot. 2008;59:4095–4107. doi: 10.1093/jxb/ern248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmama R., Veer W. Pattern formation in the monocot embryo as revealed by NAM and CUC3 orthologues from Zea mays L. Plant Mol. Biol. 2005;58:669–685. doi: 10.1007/s11103-005-7702-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials.