Abstract
Breast cancer is the cancer that most commonly affects women worldwide. This type of cancer is genetically complex, but is strongly linked to steroid hormone signaling systems. Because microRNAs act as translational regulators of multiple genes, including the steroid nuclear receptors, single nucleotide polymorphisms (SNPs) in microRNA genes can have potentially wide-ranging influences on breast cancer development. Thus, this study was conducted to investigate the relationships between six SNPs (rs6977848, rs199981120, rs185641358, rs113054794, rs66461782, and rs12940701) located in four miRNA genes predicted to target the estrogen receptor (miR-148a, miR-221, miR-186, and miR-152) and breast cancer risk in Caucasian Australian women. By using high resolution melt analysis (HRM) and polymerase chain reaction- restriction fragment length polymorphism (PCR–RFLP), 487 samples including 225 controls and 262 cases were genotyped. Analysis of their genotype and allele frequencies indicated that the differences between case and control populations were not significant for rs6977848, rs66461782, and rs12940701 because their p-values are 0.81, 0.93, and 0.1, respectively, which are all above the threshold value (p = 0.05). Our data thus suggests that these SNPs do not affect breast cancer risk in the tested population. In addition, rs199981120, rs185641358, and rs113054794 could not be found in this population, suggesting that these SNPs do not occur in Caucasian Australians.
Keywords: Breast cancer, miR-148a, miR-221, miR-186, miR-152, SNP
Introduction
Breast cancer has become the most common cancer and the major cause of death among women worldwide. According to the WHO, about 1.38 million new cases of breast cancer, roughly 690,000 for developed and developing countries, were diagnosed in 2008. Specifically in Australia, breast cancer is the second most diagnosed cancer after prostate cancer for both sexes but it has the highest incidence among cancers in women only (Ferlay et al., 2010).
Breast cancer occurs when cells in a woman's breast tissue start to grow abnormally. Unlike many other diseases which have a single cause, such as infectious diseases, most cancers are usually consequences of the interaction between numerous factors comprising both genetic characteristics and lifestyle factors. Lately, researchers have begun to notice the role of micro RNA (miRNAs) and their polymorphisms in breast cancer. MicroRNAs (miRNAs) are small (usually from 18 to 25 nucleotide long), single stranded, and noncoding RNAs. These small miRNAs play crucial roles in a wide range of biological and pathological process such as cell cycle, differentiation, development, and metabolism as well as many diseases like cancer (Lu et al., 2005, Heneghan et al., 2009). Specifically, miRNAs can regulate gene expression at the posttranscriptional level by repressing translation of protein coding genes, or cleaving target mRNAs, depending on the degree of complementarity between the miRNA and its target mRNA (Jackson and Standart, 2007). It is estimated that miRNAs regulate more than 60% of the human genome and more than 1000 human miRNAs have been found (Friedman et al., 2009). It has been found that the abnormal expression of miRNAs may link to cancer risk. Oncogenic miRNAs, commonly termed oncomirs, are usually upregulated in cancer cells. Their targets are tumor suppressor genes and increases in their expression may lead to impeding the expression of those genes. Some examples are miR-21, miR-96, and miR-182 (Si et al., 2007, Guttilla and White, 2009). In contrast, tumor suppressor miRNAs (such as miR-205, miR-27b, miR-17-5p) often express at lower levels in cancer cells than in normal cells (Iorio et al., 2005, Tsuchiya et al., 2006, Hossain et al., 2006). Because these miRNAs act on genes that promote tumorigenesis (oncogenes), a decrease in their expression level will result in an increase in oncogene effects and development of cancer cells (Zhang et al., 2007). One of the causes of this dysregulation of miRNA expression is variations or single nucleotide polymorphisms (SNPs) in those genes (Slaby et al., 2012).
In this study, miR-148a, miR-221, miR-186, and miR-152 were chosen to examine the relationship between their polymorphisms and breast cancer risk. These miRNAs were chosen due to their function of targeting the estrogen receptor, which plays an important role in breast cancer development and progression. In addition, many studies have reported that while miR-148a and miR-152 were down regulated in breast cancer cells, miR-221 and miR-186 were up-regulated, indicating that they may be actively involved in breast cancer progression (Yu et al., 2011, Xu et al., 2013, Nassirpour et al., 2013, Zhou et al., 2008). Six single nucleotide polymorphisms (SNPs) – rs6977848, rs199981120, and rs185641358 in miR-148a; rs113054794 in miR-221; rs66461782 in miR-186 and rs12940701 in miR-152 – were tested in this research.
Materials and methods
Study population
This project was conducted using a Caucasian sporadic breast cancer population which was available at the Genomics Research Centre (GRC), Institute for Health and Biomedical Innovation, Queensland University of Technology, Australia. The population was composed of 262 cases from female Caucasian patients suffering from sporadic breast cancer. Sporadic nature of breast cancer was determined by eliminating any individual with a first degree relative with breast cancer, or any family history of other cancers. Controls (225 in total) were from healthy women and were matched to cases for sex, ethnicity and age within five years. No other breast cancer risk factors were controlled for. Controls were deemed ineligible if they had any family history of any cancer, including a first degree relative with breast cancer. All samples came from Caucasian women and the average age was 57 and 57.5 years for cases and controls, respectively.
SNP selection and genotyping
SNPs for the study were selected by identifying candidate miRNAs using the target prediction search at the microRNA.org database (http://www.microrna.org/microrna/searchGenes.do). Candidate miRNAs were selected based on strong mirSVR and PhastCons scores for binding to the estrogen receptor alpha, one of the main drivers of breast cell proliferation. Candidate miRNAs were then examined using the Gene database at NCBI (http://www.ncbi.nlm.nih.gov/gene/) to identify the presence of SNPs within them.
In order to detect candidate SNPs in this study, four sets of primers were designed using Primer3 (http://frodo.wi.mit.edu/) and checked for specificity through BLAST (http://blast.ncbi.nlm.nih.gov). SNPs in miR-148a, miR-221, and miR-186 were genotyped using high resolution melt (HRM) analysis on a RotorGene-Q (Qiagen, Australia). When initial experiments indicated that HRM could not provide reliable genotypes for the SNP in miR-152, restriction fragment length polymorphism (PCR–RFLP) was instead used to obtain genotypes. Primers and amplicon characteristics are summarized in Table 1.
Table 1.
Primers and their melting temperature.
| miR | SNP | Name | Sequence (5′ → 3′) | Tm | Product size (bp) |
|---|---|---|---|---|---|
| 148a | rs6977848 rs199981120 rs185641358 |
miR148a_F miR148a_R |
CTGGCGTCTGGAGCACTG AGGAAGACAGCACGTTTGGT |
58.6 56.9 |
136 |
| 221 | rs113054794 | miR221_F miR221_R |
CCCAGCAGACAATGTAGCTG ACTTGCAAGCTGAACATCCA | 55.9 55 |
93 |
| 186 | rs66461782 | miR186_F miR186_R |
TTGACATTCACATGCTTCAGG CCTTTTGGGCTTTCTGGTTT |
59.7 60.5 |
153 |
| 152 | rs12940701 |
miR152_F miR152_R |
TCTGTCATGCACTGACTGCTC GGGCATGCTTCTGGAGTCTA |
60.2 60.4 |
170 |
PCR amplification and HRM analyses were carried out in a 16.5 μL reaction containing the following: 2 μL of 20 ng/μL genomic DNA, 2 μL of 10 μM each forward and reverse primers, 2.4 μL of 5X GoTaq buffer, 0.8 μL of 5 μM dNTPs, 0.05 μL of 5 units/μL GoTaq (Promega, Australia), 0.9 μL of 5 μM Syto-9 dye, 2.1 μL of 25 mM MgCl2 and 4.25 μL dH2O (for miR-148a and miR-221); or 2.3 μL of 25 mM MgCl2 and 4.05 μL dH2O for miR-186. PCR conditions for amplifying miR-148a and miR-221 were as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, 61 °C for 15 s and 72 °C for 30 s. Melting analysis was carried out between 78 °C and 88 °C with 0.1 °C increments. The PCR protocol for miR-186 was similar but melting analysis was carried out from 68 °C to 87 °C, also with 0.1 °C increments. One negative control and positive controls of each genotype (confirmed by sequencing) were included in each run to ensure the accuracy of genotyping. Each sample was performed in duplicate.
The PCR–RFLP reaction was prepared slightly differently with a final volume of 25 μL: 1 μL of each forward and reverse primer at 10 μM, 5 μL GoTaq buffer (5X), 0.8 μL of dNTPs (5 μM), 0.1 μL of GoTaq Flexi DNA polymerase (Promega, Australia), 2 μL of DNA (20 ng/μL), the 1.8 μL of 25 mM MgCl2, and 13.3 μL dH2O. By using a Veriti Thermal Cycler (Applied Biosystems), the PCR reaction was run using the following protocol: 95 °C for 5 min, 40 cycles of 95 °C for 15 s, 65 °C for 15 s, 72 °C for 30 s, 72 °C for 5 min, and hold at 4 °C. Again, negative control reactions were also set up and performed alongside the experimental PCR. Products were checked by performing gel electrophoresis at 90 V for 30 min on a 2% agarose gel. Restriction reactions were carried out by adding 1 μL of buffer, 0.1 μL of DdeI (New England Biolabs, Australia), and 8.9 μL of PCR product in a PCR tube and incubating at 37 °C for 60 min and holding at 4 °C. The results were then obtained by running gel electrophoresis at 70 V for 60 min with a 4% agarose gel. A positive control for complete digestion (genotype TT) was included in each restriction reaction to verify cutting efficiency.
Analysis
Data from HRM was compared to positive control samples included in each run (where available). Samples where duplicates deviated from the positive control melt pattern by more than 0.125 °C were subject to repeat to confirm genotype, with repeatedly ambiguous samples removed from consideration in analysis. RFLP samples showing evidence of incomplete digestion were repeated, with samples showing repeated ambiguous digestion eliminated from the analysis. Data obtained from HRM and PCR–RFLP, analysis was tabulated into genotypes and allele frequencies in preparation for analysis. Analysis was carried out using a Chi-square test for genotype and allele frequencies using SPSS software version 20. Adherence to the Hardy–Weinberg equilibrium was also tested for each SNP and population using manual calculation.
Results
miR-221
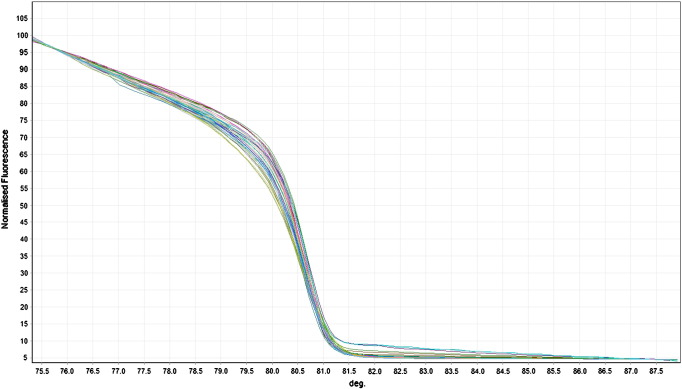
After obtaining melting data, 115 samples genotyped for miR-221 showed similar curves which were closely parallel to one another (Fig. 1). Thus, all samples appeared to have the same genotype which is homozygous CC, the genotype of the positive control sample. This also implies that SNP rs113054794 is either not present or highly rare in Caucasians.
Fig. 1.
Example of normalized melting curve for miR-221 SNP rs113054794. All curves have the same shape indicating that these samples have the same genotypes.
miR-148a
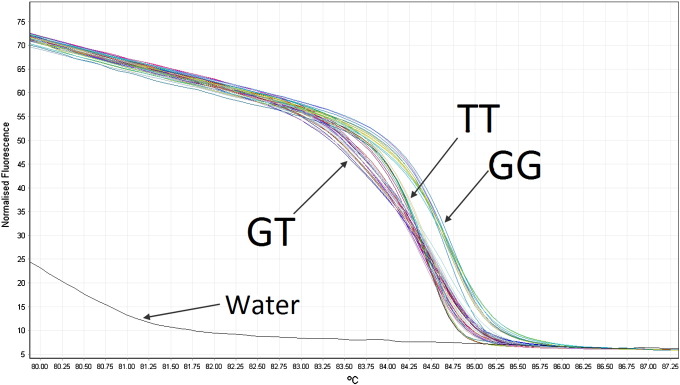
Despite obtaining optimal condition for the PCR reaction, only 427/487 samples were able to produce data for miR-148a, including 204 controls and 223 cases. Among the three targeted SNPs on this miRNA, only the presence of SNP rs6977848 could be detected. Based on our data, SNPs rs199981120 and rs185641358 occur in Caucasians at a very low frequency, or not at all, because all tested samples showed the AA genotype for both SNPs, containing only the ancestral A allele. This was confirmed along with variant genotypes in rs6977848 by sequencing 8 samples from the populations selected at random, in addition to the positive control samples.
A typical HRM result is shown in Fig. 2. Table 2 presents the genotyping results as well as statistical analysis for rs6977848 in control and case groups. Both populations proved to be in Hardy Weinberg equilibrium (p = 0.07 and 0.6 for cases and controls, respectively). Although the TT genotype was slightly more common in the breast cancer population than in the control population, Chi-square analysis of this data showed that there was no significant difference between these two groups both for genotypes (χ2 = 0.82, p = 0.67) and alleles (χ2 = 0.06, p = 0.81). Thus there is not a significant association between this SNP and breast cancer development.
Fig. 2.
Example of normalized melting curve for miR-148a SNP rs6977848. Three different genotypes (GG, GT, TT) were identified due to different curve shapes. Homozygous samples produce standard S-curves while heterozygotes show a flatter or multi-flexed curve.
Table 2.
Statistical analysis for SNP rs6977848- miR-148a.
| Genotypes | GG | GT | TT | G | T | HWE sig. |
|---|---|---|---|---|---|---|
| Cases 223 |
38 (17.0%) |
92 (41.3%) |
93 (41.7%) |
168 (37.7%) |
278 (62.3%) |
0.07 |
| Controls 204 |
32 (15.7%) |
93 (45.6%) |
79 (38.7%) |
157 (38.5%) |
251 (61.5%) |
0.60 |
| Chi-square | 0.82 | 0.06 | ||||
| p-Value | 0.67 (non-significant) | 0.81 (non-significant) | ||||
miR-186
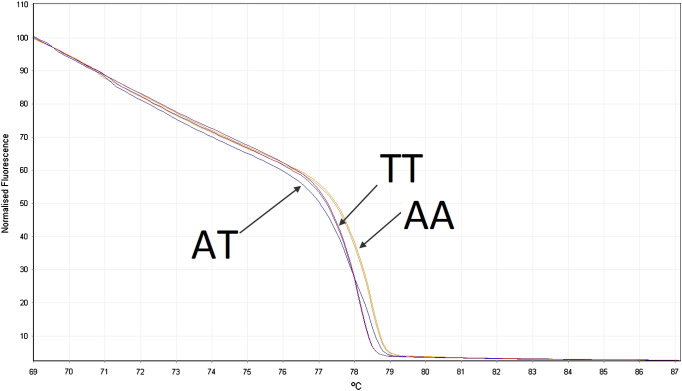
For miR-186, 393/487 samples were genotyped, including 187 controls and 206 cases. An example HRM result with three different genotypes can be seen in Fig. 3. The results obtained from HRM and Chi-square analysis are summarized in Table 3. HWE p values were 0.73 and 0.62, indicating that both control and case groups were in Hardy Weinberg equilibrium. Again, the TT genotype in cases seems to appear more frequently than in controls but with the significance higher than the α of 0.05, statistical analysis indicated that this SNP does not have a significant association with breast cancer risk.
Fig. 3.
Example of normalized melting curve for miR-186 SNP rs66461782. Three different genotypes (AA, AT, TT) were identified due to different curve shapes. Homozygous samples produce standard S-curves while heterozygotes show a flatter or multi-flexed curve.
Table 3.
Statistical analysis for SNP rs66461782- miR-186.
| Genotypes | AA | AT | TT | A | T | HWE sig. |
|---|---|---|---|---|---|---|
| Cases 206 |
129 (62.6%) |
67 (32.5%) |
10 (4.9%) |
325 (78.9%) |
87 (21.1%) |
0.73 |
| Controls 187 |
116 (62.0%) |
64 (34.2%) |
7 (3.7%) |
296 (79.1%) |
78 (20.9%) |
0.62 |
| Chi-square | 0.37 | 0.01 | ||||
| p-Value | 0.83 (non-significant) | 0.93 (non-significant) | ||||
miR-152
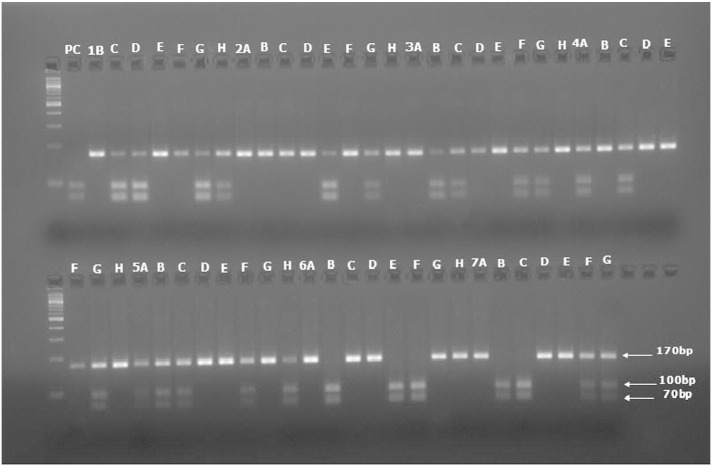
Fig. 4 illustrates RFLP results for miR-152. Table 4 shows the summarized results obtained after RFLP genotyping. Unlike the two other SNPs, the non-ancestral allele (T allele) seems to appear more often in controls than in cases. HWE p values were calculated for both cases and controls and both p values (HWE pcase = 0.95, HWE pcontrol = 0.15) were above 0.05 threshold value, indicating that both populations are in Hardy–Weinberg equilibrium. Despite the clear increase of T alleles in the controls, χ2 analysis of those frequencies showed an insignificant difference between cases and controls since both genotypes (χ2 = 3.79, p = 0.15) and alleles (χ2 = 2.73, p = 0.1) were both greater than the α. Thus, this polymorphism was also not associated with sporadic breast cancer.
Fig. 4.
Example of RFLP results for miR-152 SNP rs12940701. PC: positive control. The first lane is 100 bp ladder. PCR produces a single band of 170 bp, which remains undigested in CC homozygous individuals (e.g., 1B and 1E). Heterozygous samples show three bands at 170 bp, 100 bp, and 70 bp (e.g., 1C and 1H). Samples with only 100 bp and 70 bp bands were homozygous TT (e.g., 6E and 7B).
Table 4.
Statistical analysis for SNP rs12940701- miR-152.
| Genotypes | CC | CT | TT | C | T | HWE sig. |
|---|---|---|---|---|---|---|
| Cases 232 |
120 (51.7%) |
94 (40.5%) |
18 (7.8%) |
334 (72.0%) |
130 (28.0%) |
0.95 |
| Controls 205 |
87 (42.4%) |
100 (48.8%) |
18 (8.8%) |
274 (66.8%) |
136 (33.2%) |
0.15 |
| Chi-square | 3.79 | 2.73 | ||||
| p-Value | 0.15 (non-significant) | 0.10 (non-significant) | ||||
In addition, when comparing control genotype frequencies for those SNPs with those in a European population (HapMap and 1000 genome project) it was found that these populations corresponded closely or were not significantly different (this can be seen in Table 5). This would indicate that genotyping for these SNPs was accurate.
Table 5.
Genotype and allele frequencies for rs6977848, rs66461782, and rs12940701 in study vs. HapMap/1000 genomes data.
| p-Value | Genotyping result | HapMap/1000 genome | |||
|---|---|---|---|---|---|
| miR-148a | rs6977848 | G T |
0.56 (non-significant) | 38.5% 61.5% |
42.5% 57.5% |
| GG GT TT |
0.34 (non-significant) | 15.7% 45.6% 38.7% |
23.3% 38.3% 38.3% |
||
| miR-186 | rs66461782 | A T |
0.66 (non-significant) | 79.1% 20.9% |
76.5% 23.5% |
| AA AT TT |
0.56 (non-significant) | 62.0% 34.2% 3.7% |
60.0% 32.9% 7.1% |
||
| miR-152 | rs12940701 | C T |
0.34 (non-significant) | 66.8% 33.2% |
72.9% 27.1% |
| CC CT TT |
0.23 (non-significant) | 42.4% 48.8% 8.8% |
54.1% 37.6% 8.3% |
Data from http://www.ncbi.nlm.nih.gov and http://www.ensembl.org/index.html.
Discussion
In this study, we examined the genotype frequency of six SNPs in four miRNAs predicted to target the estrogen receptor alpha, specifically miR-148a, miR-221, miR-186, and miR-152, using HRM and RFLP methods. Of the SNPs genotyped, rs6977848, rs66461782, and rs12940701 were found to not show significant association with sporadic breast cancer risk. In addition, the SNPs rs199981120, rs185641358 and rs113054794 could not be detected in cases or controls, indicating that these SNPs are non-polymorphic or highly rare in Caucasian populations.
Previous work by our laboratory and others has shown that SNPs in micro-RNAs and their binding sites on genes have influence on breast cancer risk (Kontorovich et al., 2010, Pastrello et al., 2010, Smith et al., 2012; Wang et al., 2012; Zhang et al., 2011, Zheng et al., 2011). This includes both sporadic and familial forms, where polymorphisms in miRNAs have been found to alter the age of onset of tumors (Pastrello et al., 2010). On the other hand, other studies have failed to show association with miRNA SNPs and breast cancer risk, such as the large study by Cattuci and colleagues, who examined a large association population consisting of German and Italian women with breast cancer, finding no association between SNPs in miR-146a, miR196a2 and miR-499 and breast cancer risk (Catucci et al., 2010). It seems likely, therefore, that the impact of a given SNP will depend heavily on its functional effects on the miRNA itself and the degree to which the native miRNA affects the expression of cancer driving genes.
Several previous research projects have been carried out to investigate the relationships between the miRNAs targeted by this research and breast cancer. Using Taqman real-time polymerase chain reaction, the expression of these miRNAs was measured and compared between normal and breast cancer cell lines. By applying this method, Xu and colleagues found that miR-148a and miR-152 were dramatically down-regulated in breast cancer cells (p < 0.05). A consistent result was also achieved when they tested breast cancer tissue. In addition, lower levels of miR-148a and miR-152 were significantly correlated with breast cancer stage and breast cancer cases with lymph node metastasis (p < 0.05) (Xu et al., 2013). Meanwhile, miR-186 was found to be up-regulated in breast cancer cells as well as in cervical cancer cells (Zhou et al., 2008). Similarly, miR-221 was also up-regulated in breast cancer, especially in triple negative breast cancer cells (Nassirpour et al., 2013). This would imply that the miRNAs tested in this study have effective roles in breast cancer development, though functional research on their effects is needed to determine precisely what function they perform in breast cancer pathogenesis and whether their predicted targeting of the estrogen receptor is responsible.
This study is the first research that examines the effect of the targeted SNPs (rs6977848, rs199981120, rs185641358, rs113054794, rs66461782, and rs12940701) in these miRNAs in terms of breast cancer risk development. The results showed that the chosen single nucleotide polymorphisms in miR-148a (rs6977848), miR-186 (rs66461782) and miR-152 (rs12940701) were not associated with increased breast cancer risk. This implies that any aberrant expression of those miRNAs in breast cancer cells may be due to other SNPs at different loci or be caused by another effect, such as changes in DNA copy number in cancer cells or DNA methylation, which could also contribute to the different expression of the miRNAs observed in previous research. Similarly, it implies that these SNPs do not significantly affect the expression of the estrogen receptor in breast cancer, or that other factors allow the estrogen receptor to be expressed or function at unchanged levels, despite any difference in miRNA behavior these SNPs might cause.
It may be, however, that these SNPs do affect the behavior of these miRNAs in some way significant to the progression of breast cancer, but that they do not affect risk of developing the disease. Due to the lack of research on the functional effects of these SNPs, the likelihood of their having effects after breast cancer initiation cannot be properly assessed. Thus, such research is a logical next step, and may be useful in understanding the role that these miRNAs play in breast cancer progression, even if they have no effect on risk of breast cancer initiation. Our population is also one of limited size, and it is possible that there is an effect of these SNPs on breast cancer risk, but that it is relatively weak or interacts with other risk factors, and we have insufficient power to detect it. Despite our agreement with previously obtained genotype frequencies from HapMap and 1000 genomes data, further research using a larger population of cases and controls, preferably with further risk factor controls is advised.
In addition, our study is the first to examine the SNPs rs113054794, rs199981120 and rs185641358 and their effect on breast cancer risk. These SNPs have been annotated in the NCBI SNP database, but no frequency data is currently available for them. Our results indicate that these SNPs are extremely rare in Caucasian populations, occurring at rates much lower than 1%. While these SNPs are likely to be insignificant for breast cancer risk in Caucasians, they may be significant factors in other populations and research to determine their frequency and breast cancer risk profile is worth undertaking.
Acknowledgments
This work was supported by infrastructure purchased with Australian Government EIF Super Science Funds as part of the Therapeutic Innovation Australia — Queensland Node project.
Contributor Information
Lyn R. Griffiths, Email: Lyn.Griffiths@qut.edu.au.
Hue T. Nguyen, Email: nthue@hcmiu.edu.vn.
References
- Catucci I., Yang R. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum. Mutat. 2010;31(1):E1052–E1057. doi: 10.1002/humu.21141. [DOI] [PubMed] [Google Scholar]
- Ferlay J., Shin H.R. International Agency for Research on Cancer; Lyon, France: 2010. GLOBOCAN 2008 v2.0. Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10 [Internet] (Available from: http://globocan.iarc.fr) [Google Scholar]
- Friedman R.C., Farh K.K. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla I.K., White B.A. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J. Biol. Chem. 2009;284(35):23204–23216. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan H.M., Miller N. MicroRNAs as novel biomarkers for breast cancer. J. Oncol. 2009;2009:950201. doi: 10.1155/2010/950201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain A., Kuo M.T. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol. Cell. Biol. 2006;26(21):8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M.V., Ferracin M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Jackson R.J., Standart N. How do microRNAs regulate gene expression? Sci. STKE. 2007;2007(367):re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- Kontorovich T., Levy A. Single nucleotide polymorphisms in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in Jewish high-risk women. Int. J. Cancer. 2010;127(3):589–597. doi: 10.1002/ijc.25065. [DOI] [PubMed] [Google Scholar]
- Lu J., Getz G. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Nassirpour R., Mehta P.P. miR-221 promotes tumorigenesis in human triple negative breast cancer cells. PLoS One. 2013;8(4):e62170. doi: 10.1371/journal.pone.0062170. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pastrello C., Polesel J. Association between hsa-mir-146a genotype and tumor age-of-onset in BRCA1/BRCA2-negative familial breast and ovarian cancer patients. Carcinogenesis. 2010;31(12):2124–2126. doi: 10.1093/carcin/bgq184. [DOI] [PubMed] [Google Scholar]
- Si M.L., Zhu S. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Slaby O., Bienertova-Vasku J. Genetic polymorphisms and microRNAs: new direction in molecular epidemiology of solid cancer. J. Cell. Mol. Med. 2012;16(1):8–21. doi: 10.1111/j.1582-4934.2011.01359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.A., Jedlinski D.J. A genetic variant located in miR-423 is associated with reduced breast cancer risk. Cancer Genomics Proteomics. 2012;9(3):115–118. [PubMed] [Google Scholar]
- Tsuchiya Y., Nakajima M. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66(18):9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- Wang L., Liu W. A miRNA binding site single-nucleotide polymorphism in the 3'-UTR region of the IL23R gene is associated with breast cancer. PLoS One. 2012;7(12):e49823. doi: 10.1371/journal.pone.0049823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Jiang Y. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J. Mol. Cell Biol. 2013;5(1):3–13. doi: 10.1093/jmcb/mjs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Li Q. MiR-148a inhibits angiogenesis by targeting ERBB3. J. Biomed. Res. 2011;25(3):170–177. doi: 10.1016/S1674-8301(11)60022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Pan X. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Zhang L., Liu Y. Functional SNP in the microRNA-367 binding site in the 3′UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc. Natl. Acad. Sci. U. S. A. 2011;108(33):13653–13658. doi: 10.1073/pnas.1103360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Song F. Genetic variants at the miR-124 binding site on the cytoskeleton-organizing IQGAP1 gene confer differential predisposition to breast cancer. Int. J. Oncol. 2011;38(4):1153–1161. doi: 10.3892/ijo.2011.940. [DOI] [PubMed] [Google Scholar]
- Zhou L., Qi X. MicroRNAs miR-186 and miR-150 down-regulate expression of the pro-apoptotic purinergic P2X7 receptor by activation of instability sites at the 3′-untranslated region of the gene that decrease steady-state levels of the transcript. J. Biol. Chem. 2008;283(42):28274–28286. doi: 10.1074/jbc.M802663200. [DOI] [PMC free article] [PubMed] [Google Scholar]