Abstract
Motivation: The development of cost-effective next-generation sequencing methods has spurred the development of high-throughput bioinformatics tools for detection of sequence variation. With many disparate variant-calling algorithms available, investigators must ask, ‘Which method is best for my data?’ Machine learning research has shown that so-called ensemble methods that combine the output of multiple models can dramatically improve classifier performance. Here we describe a novel variant-calling approach based on an ensemble of variant-calling algorithms, which we term the Consensus Genotyper for Exome Sequencing (CGES). CGES uses a two-stage voting scheme among four algorithm implementations. While our ensemble method can accept variants generated by any variant-calling algorithm, we used GATK2.8, SAMtools, FreeBayes and Atlas-SNP2 in building CGES because of their performance, widespread adoption and diverse but complementary algorithms.
Results: We apply CGES to 132 samples sequenced at the Hudson Alpha Institute for Biotechnology (HAIB, Huntsville, AL) using the Nimblegen Exome Capture and Illumina sequencing technology. Our sample set consisted of 40 complete trios, two families of four, one parent–child duo and two unrelated individuals. CGES yielded the fewest total variant calls (), the highest Ts/Tv ratio (3.02), the lowest Mendelian error rate across all genotypes (0.028%), the highest rediscovery rate from the Exome Variant Server (EVS; 89.3%) and 1000 Genomes (1KG; 84.1%) and the highest positive predictive value (PPV; 96.1%) for a random sample of previously validated de novo variants. We describe these and other quality control (QC) metrics from consensus data and explain how the CGES pipeline can be used to generate call sets of varying quality stringency, including consensus calls present across all four algorithms, calls that are consistent across any three out of four algorithms, calls that are consistent across any two out of four algorithms or a more liberal set of all calls made by any algorithm.
Availability and implementation: To enable accessible, efficient and reproducible analysis, we implement CGES both as a stand-alone command line tool available for download in GitHub and as a set of Galaxy tools and workflows configured to execute on parallel computers.
Contact: trubetskoy@uchicago.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Whole-exome sequencing (WES) has quickly become an affordable approach to identifying rare variants contributing to disease. Over the past 5 years the number of published papers that were indexed in PubMed with the key words ‘exome sequencing’ has increased 200-fold, representing a clear trend in human genetics. The utility of WES for revealing biological mechanisms depends on the genetic architecture of the phenotype in question, the quality of the sequencing technology and, to a significant extent, the analytic methods used to identify and genotype variations in sequence. In recent years, several methods have been developed to analyze raw WES data, including Atlas-SNP2 (Challis et al., 2012), GATK (DePristo et al., 2011; McKenna, 2010a), SeqEM (Martin et al., 2010), FreeBayes (Garrison and Marth, 2012), SAMtools (Li et al., 2009a), Dindel Albers et al. (2011), SOAPsnp (Li et al., 2009b) and Varscan2 (Koboldt et al., 2012), among others.
These methods represent substantial effort and expertise in the analysis of next generation sequencing (NGS) data, including both whole-exome and whole-genome sequencing. Here we present a natural extension of these individual algorithms that integrates their relative strengths into a consensus-calling approach, which we call CGES. This algorithm, developed as a collaborative effort between the Department of Medicine and the Computation Institute at the University of Chicago, takes advantage of the diverse variant-calling strategies of four existing algorithms (GATKv2.8, Atlas-SNP2, SAMtools and FreeBayes) in an open-source, freely available and user-friendly analysis platform.
While all variant-calling programs seek to optimize performance relative to some core properties of sequencing data (such as read depth and allele count), they often differ along other dimensions. We chose to base our consensus-based pipeline on GATKv2.8, SAMtools, Altas-SNP2 and FreeBayes, as these four algorithms use complementary approaches. For example, all algorithms include genotype and indel likelihood models, but the models themselves differ with respect to information used and weights given to such information [see O’Rawe et al. (2013) and Yu and Sun (2013) for complete comparison]. Furthermore, FreeBayes allows explicit parameterization if there are known copy number variants in a sample, while GATKv2.8 provides sophisticated filtering options that can be trained on a given dataset. Together, these programs constitute a suite of algorithms that attempt to integrate as much information as possible, including prior variant observations, linkage disequilibrium structure and structural variation, to reduce both type I and type II errors.
In brief, CGES first runs each algorithm separately and then combines the resulting four collections of genotype calls to create three possible output sets, typically of increasing size but lower average quality: consensus calls (i.e. calls made by all four algorithms), partial consensus calls (e.g. those made by three or more algorithms) and the union of all calls (calls made by one or more algorithms). CGES also harmonizes quality scores from each algorithm to provide QC reports and publication quality plots (CGES-QC tool).
CGES and CGES-QC together form a multi-step pipeline and must perform multiple program invocations, as shown in Figure 1. We use the Galaxy platform (Goecks et al., 2010) to combine implementations for each branch of the pipeline and for CGES itself. Our use of Galaxy has the benefit of democratizing NGS data analysis, as Galaxy reduces the computational expertise needed to run an NGS pipeline and can run on public clouds such as those operated by Amazon Web Services, Google and Microsoft. The links to the Galaxy workflows can be found in Supplementary Table S4. We have applied the CGES consensus-calling approach to real-world exome data collected on subjects with autism and their family members. We use the results of this study to demonstrate the power of the CGES approach and provide project-level, variant-level, sample-level and family-based quality metrics across all algorithms. Additionally, we provide known rare variant rediscovery rates, and an estimate of the PPV of each algorithm based on previously identified and lab-validated de novo variation.
Fig. 1.
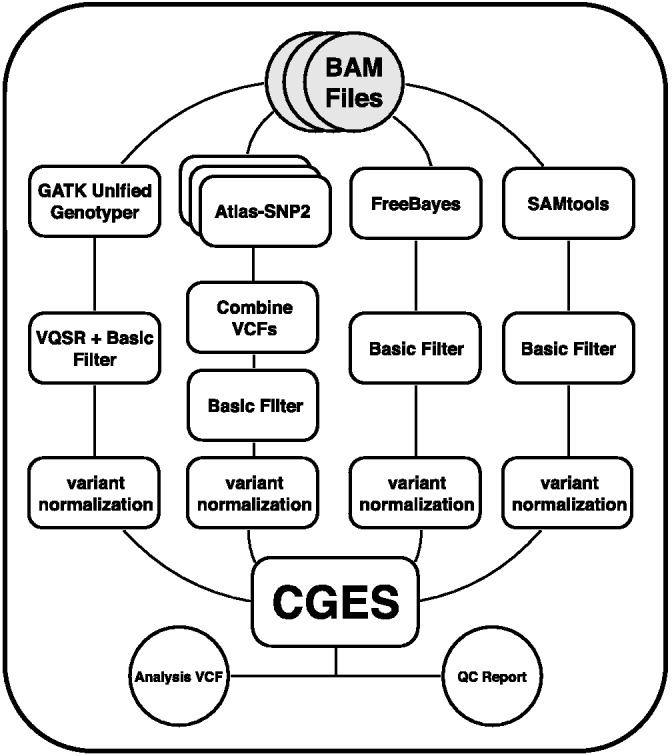
High-level schematic of the CGES pipeline running from top to bottom. Each of the four branches applies a separate caller to the same input alignment (BAM) files. The basic filter consists of a BED target file defined by the capture, and a minimum QUAL of 10. Variant normalization entails standardizing the representation of more complex alleles such as indels. The final product is a multi-sample variant call format (VCF) file and associated quality metrics
2 METHODS
2.1 Samples
To test the robustness of the CGES consensus-calling algorithm in the context of real-world data, we used binary alignment (BAM) files from 132 individuals representing 40 complete trios, two families of 4 and 2 additional unrelated individuals recruited from the Autism Center of Excellence study at the University of Illinois at Chicago, Vanderbilt University or Tufts-New England Medical Center. Probands were assessed with the Autism Diagnostic Interview (ADI-R), the Autism Diagnostic Observation Schedule by Western Psychological Services (ADOS-WPS) and clinical evaluation. We included families in this study if the probands met diagnostic criteria for autism or autism spectrum disorder on both the ADI-R and ADOS-WPS (Berument et al., 1999; Le Couteur et al., 1989).
2.2 NGS
Sequencing for the majority of samples was performed at the HAIB (Hunstville, AL) as a part of the NIH Autism Sequencing Consortium (dbGAP Accession Number: phs000298.v1.p1; Liu et al., 2013; Neale, 2012) and for the remainder of samples at HAIB as a part of the University of Illinois and Vanderbilt Autism Center for Excellence study. Methods used for WES and alignment are described in depth in Supplementary Materials and previous publications (Neale et al., 2012). In brief, samples were sequenced at Hudson Alpha Biotechnology Institute using a paired-end approach with NimbleGen exome capture followed by Illumina HiSeq 2000 sequencing.
2.3 Determination of parameter values for consensus-calling algorithms
Figure 1 illustrates the variant-calling schema used by CGES. The four variant-calling algorithms used in this analysis are implemented in previously published programs GATK v2.8 (DePristo et al., 2011; McKenna, 2010a), SAMtools (Li et al., 2009a), Atlas-SNP2 (Challis et al., 2012) and FreeBayes (Garrison and Marth, 2012). As these methods are described in depth in their primary publications, here we describe only their parameterization and implementation within the Galaxy framework.
As shown in Figure 1, we implement variant calling for each of the four programs within an independent subpipeline or branch. Within each branch, we select parameter values for exome sequence data as follows. For GATK, we followed best practices published by the Broad Institute (http://www.broadinstitute.org/gatk/guide/best-practices). We determined optimal parameters empirically for all callers (Supplementary Methods) using QC data from our project to iteratively develop a set of project-specific best practices. As the variant calls from each branch serve as the substrate for CGES, it is important that parameters for each branch are optimized for best performance. Thus, we strongly recommend that investigators review the parameters for each caller, and ensure that they are appropriately determined for a given dataset.
2.4 CGES calling pipeline
The CGES pipeline (Fig. 1) takes as input BAM files generated and provided by a sequencing center. The BAM files do not require any preprocessing before entering the CGES workflows, but are subject to the preprocessing steps required by each branch algorithm. Each branch and the CGES workflow include SNP and indel calling. All 132 samples were run together through GATKv2.8, FreeBayes and SAMtools.
The first step in running the CGES pipeline is to run the GATKv2.8 Unified Genotyper across all chromosomes, with separate multi-threaded processes for each chromosome. The resulting VCF files are then combined and variant recalibration is applied. We train the GATK gaussian mixture model on 1000 Genomes, HapMap and dbSNP135 data hosted by the Broad Institute. After performing variant quality score recalibration (VQSR), variants are then filtered down to a VQSR-based quality tranche (99.9%). A simple filter is then applied to remove genotype calls that do not satisfy an on-target status criterion or that have a minimum QUAL score of <10.
The FreeBayes and SAMtools workflows are similar to the GATK workflow in that we first parallelize variant detection by chromosome. During this step, we implement the chosen detection and genotyping parameters described in Supplementary Materials. This results in one multi-sample VCF file per chromosome. We then combine VCFs, and finally filter based on a minimum QUAL of 10 and on-target status.
Unlike the other callers, Atlas-SNP2 calls variation per sample instead of across multiple samples simultaneously. This required a sample-based parallelization scheme in which we spawn a separate Atlas-SNP2 process for each sample. The Galaxy Atlas-SNP2 tool was therefore developed with a Swift (Wilde et al., 2011) backend enabling parallelization across samples. We then follow the Atlas-SNP2 protocol for creating an initial multi-sample VCF file from individual sample VCF files (http://sourceforge.net/projects/atlas2/). Finally, we applied an on-target filter and compiled resulting variants into a multi-sample Atlas-SNP2 VCF file to produce the final multi-sample VCF file.
Finally, each multi-sample VCF file is normalized by Variant tool (Vt) (http://genome.sph.umich.edu/wiki/Vt). Variant normalization by Vt ensures that such complex variation, including indels and single nucleotide variants (SNVs) within indels, is represented with the most parsimonious set of variations with respect to the reference sequence. Applying Vt to all branches allows results from each branch to be standardized, so that complex variants may be compared between datasets.
2.5 CGES
We use a two-stage voting scheme to generate consensus genotypes. First, we identify the variant positions (irrespective of genotype) that agree among a specified number of callers. At this step, a user can specify a level of concordance among callers (e.g. three of four, or consensus). CGES considers variants to be uniquely identified by any difference from the reference sequence at a given chromosomal position. The algorithm then proceeds to consider genotypes within these consensus sites. Genotypes that do not agree are set as missing and flagged as discordant for downstream quality analysis. Stringency thresholds for genotype concordance can be set independently for each stage, conditional on the fact that genotype concordance cannot be stricter than site concordance. As each caller uses slightly different priors and weights for SNV and indel likelihood determination, and as the underlying truth is unknown, each caller is given an equal vote at both stages of voting.
2.6 CGES-QC
We have also developed CGES-QC, a tool for the calculation, comparison and visualization of sample-based, variant-based and project-based QC metrics across all branches of the consensus genotyper. CGES-QC incorporates QC calculations from PLINK, VCFtools and scripts developed in-house to perform analyses and output publication quality plots. Unless otherwise noted to refer to indels, QC metrics are reported with respect to single nucleotide variants.
2.7 Project-based QC
Project-based QC results include the total number of variants called, Transition-Transversion ratio (Ts/Tv), EVS variant rediscovery rate, 1KG variant rediscovery rate, Genome in a Bottle (GiaB) rediscovery rate and the genotype Mendelian error rate (gMER). The total number of variants called is limited to the user-specified settings and refers to the total number of variants present in the output VCF. The Ts/Tv ratio is a routinely reported QC measure for sequence data and refers to the ratio of transitions (G↔A or C↔T) to transversions (G↔C or A↔T). Based on previously reported analyses, the Ts/Tv ratio is expected to be 2.1 for whole-genome sequencing and 2.6–3.3 for exome sequence data (DePristo et al., 2011). Low Ts/Tv ratios represent technical artifacts, and a randomly generated set of variants yields a Ts/Tv ratio of 0.5 (Zook et al., 2014). The EVS, GiaB and 1KG rediscovery rates represent the total number of variants in a VCF that have been previously identified in those sequencing projects. Finally, for each variant site called, there may be anywhere from one individual with a sequence variation to N individuals with variant genotypes (where N = sample size) present in the VCF file. Therefore, we calculate a ‘genotype Mendelian error rate’, which is the total number of MEs in a VCF file divided by the total number of genotypes with the potential for Mendelian inconsistency (i.e. offspring genotypes with parental genotypes known) in a VCF file. This measure describes the proportion of all offspring genotypes that are inconsistent with parental genotypes present in the VCF.
2.8 Sample-based QC
Sample-based QC results include the F-statistic per sample, trio Mendelian error rate (tMER) and genotype concordance/discordance per sample. The F-statistic is calculated using the classic Wright formula one minus the ratio of observed heterozygote genotypes to expected heterozygote genotypes according to Hardy–Weinberg equilibrium (Danecek et al., 2011). This statistic provides a red flag for both sample contamination (extreme heterozygosity) and consanguinity (extreme homozygosity). We calculate Mendelian errors per trio, which we defined as the number of MEs in an offspring (given by the trio) divided by the total number of genotypes in the offspring (Purcell et al., 2007). This metric is useful for determining whether there are any trios that require further attention that may be due to sample mismatch or large copy number variants. In the case of a contaminated sample, the F-statistic and the tMER can be used jointly to determine whether the contamination came from a relative or an unrelated sample. Finally, CGES genotype discordance rates are calculated per sample defined as the proportion of all genotypes in a given sample that are flagged as discordant across any of the four calling algorithms.
2.9 Variant-based QC
Variant-based QC includes calculation of the variant site Mendelian error rate (vMER), variant site missingness distributions and minor allele frequency distributions. The vMER is defined as the number of variant sites where it is possible to have a Mendelian error (i.e. the total number of variant sites in probands for which both parents are also genotyped). This metric provides a bird’s eye view of the general sensitivity and specificity of each calling algorithm. A more inclusive approach to genotype calling will allow a higher number of sites to contribute at least one ME, while a stricter approach will result in fewer sites with at least one ME. Moreover, the vMER may increase with the inclusion of genomic regions that are difficult to sequence. We believe this is an important quality metric for custom capture exome sequencing and will be important for whole-genome sequencing. The vMER as implemented in the CGES-QC tool can also be calculated within a subset of samples or by genomic region to prioritize regions for further analysis.
At each variant site there are a small proportion of genotype calls that cannot be made (i.e. flagged as ‘missing’) because a given algorithm does not receive enough information to accurately determine the genotype. In the CGES algorithm, the resulting conflicts between branches are also flagged and described as ‘missing’. Conflicts between branches consist of any conflicting genotype calls, including scenarios in which one caller contributes a missing call. We defined the missingness rate per variant site as the number of missing genotypes at a site divided by the total number of genotypes at that site. We then calculated this missingness rate across all variant sites for all branches and provide the distributions. Finally, we provide the minor allele frequency distributions across all variant sites for each algorithm.
2.10 PPV estimated from de novo variant calls
A set of predicted de novo variants was predicted based on the Broad Institute de novo filtering practices (Supplementary Methods) provided by the Hudson Alpha Genomic Services Laboratory. A total of 54 variants from 31 samples were predicted de novo and validated as true positives on resequencing with Sanger sequencing methods. Twenty variants (37.0%) were predicted de novo but did not validate with Sanger sequencing and were, therefore, classified as false positives. This set of laboratory-validated true-positive and false-positive de novo variants then served as a benchmark to determine the PPV of CGES and each branch algorithm. The position and validation status of each variant is included in Supplementary Table S3. PPV was calculated as the ratio of true positives detected to the sum of true positives and false positives detected.
2.11 Indel QC
In addition to SNV consensus calls, users can generate identical consensus indel calls by requiring that all four algorithms agree. An investigator may alternatively require three out of four or two out of four algorithms agree. We identified 2122 identical consensus calls across all four algorithms, 4093 across three out of four algorithms, 5514 across two out of four algorithms and a union call set of 13 410 indels. As a measure of indel quality, we have calculated the EVS (82.3%) and GiaB (14.1%) rediscovery rates for each branch and consensus. It is important to note that the rediscovery rates can be used as a general measure of dataset quality, and may help to detect a branch that has been poorly parameterized. However, the rediscovery rate will also be influenced by the number of unique variants detected in a sample, which may increase as a function of sequence depth, caller sensitivity and population.
2.12 Availability of the CGES pipeline to investigators
There are three primary ingredients needed for the successful use of the CGES pipeline: (i) the user-supplied files necessary for analysis (i.e. raw BAMs, target BED file and reference files), (ii) the branch and CGES software and (iii) a computational infrastructure capable of handling the demands of the software. The input and reference files are user supplied and are routinely made available by NGS centers. There are multiple accessibility points for the CGES and branch software. First, we have made the enhanced version of these tools (e.g. CGES, FreeBayes, GATKv2.8, SAMtools, Atlas-SNP2) available in the public Galaxy toolshed (Blankenberg et al., 2010; Giardine, 7; Goecks, 2010a), so that community members can download the tools into their own respective Galaxy instances. We have also made the workflows (i.e. descriptions of the pipelines including various arguments used in the execution of the tools) available online (Supplementary Table S4). Second, the University of Chicago Computation Institute maintains and updates the pipelines under the Globus Genomics service offering (Madduri et al., 2013). Finally, the code for the CGES tool has been provided as a stand-alone command line tool (Supplementary Table S4). The computational infrastructure for the analysis performed in this article was developed by the Globus Genomics initiative (http://www.globus.org/genomics/), led out of the Computation Institute (a joint institute between the University of Chicago and Argonne National Laboratory). The analysis of the autism trios described here was conducted on the Amazon Web Services public cloud. Investigators wishing to run the CGES software will require a local server capable of parallelization and analysis, or access to cloud computing space such as that offered by Amazon, Digital Ocean or Azure.
3 RESULTS
3.1 CGES-QC results
QC analysis and descriptive statistics of the consensus dataset showed that the highest quality call set was obtained by using overlap of all callers together. Using the parameters for single callers described in the Supplementary Materials, we found that, as expected, the strict consensus of all calls made by CGES yielded the fewest total variant calls (; Table 1, Fig. 2). CGES calls resulted in the highest Ts/Tv ratio (3.02) (Table 1, Fig. 3). CGES calls resulted in the lowest gMER (0.028%) (Table 1, Fig. 4) across all genotypes and the lowest vMER (0.92%) (Supplementary Table S2, Fig. 4). CGES results contained a total count of 18 466 404 genotypes representing 139 897 variant sites across 132 individuals, with 667 020 discordant genotypes resulting in CGES, a discordance rate of 3.61%.
Table 1.
Set-based QC results
| Call set | Number of variants | gMER (%) | Ts/Tv | EVS rediscovery (%) | 1KG rediscovery (%) | GiaB (%) | PPV (%) |
|---|---|---|---|---|---|---|---|
| CGES | 139 897 | 0.0282 | 3.02 | 89.3 | 84.1 | 16.9 | 96.1 |
| Atlas-SNP2 | 214 149 | 0.245 | 2.12 | 72.7 | 69.0 | 11.8 | 92.3 |
| FreeBayes | 149 230 | 0.474 | 2.67 | 80.4 | 76.8 | 12.4 | 94.7 |
| GATKv2.8 | 149 230 | 0.271 | 2.95 | 88.0 | 84.1 | 16.1 | 93.1 |
| SAMtools | 193 945 | 0.802 | 2.57 | 80.6 | 77.2 | 13.1 | 89.2 |
Fig. 2.
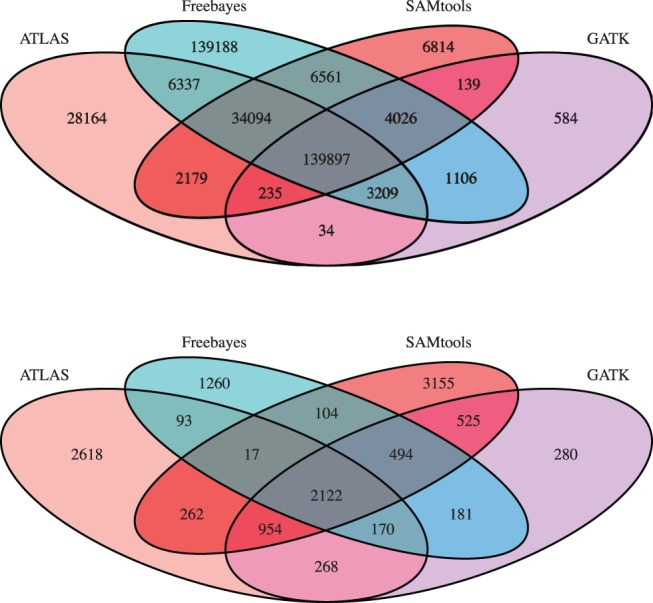
Venn diagram of variant sites and their overlap between constituent call sets. We report both SNVs (top) and indels (bottom). CGES variants can be produced from the intersection of any two- or three-constituent sets, or from the union of all calls
Fig. 3.
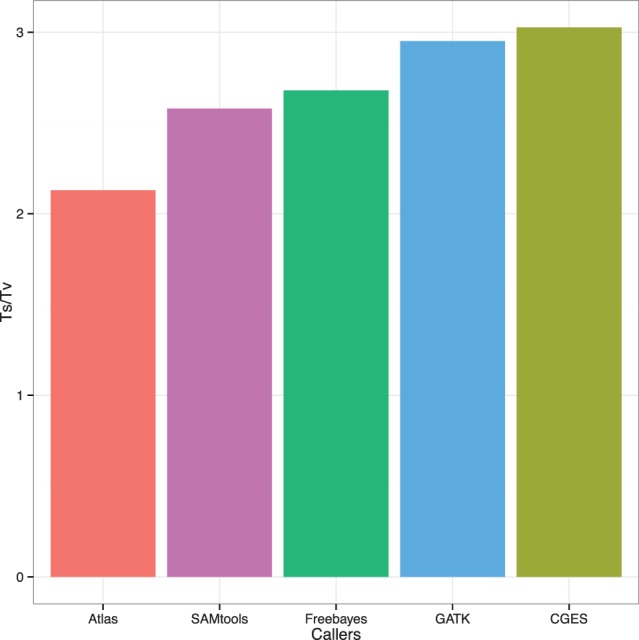
Transition/transversion mutation ratio for different call sets. This ratio has been observed to lie between 2.6 and 3.3 for coding regions in the human genome (DePristo et al., 2011). The Ts/Tv ratio was 3.02 for the CGES calling algorithm compared with 2.95 for GATK, 2.67 for FreeBayes, 2.57 for SAMtools and 2.12 for Atlas-SNP2
Fig. 4.
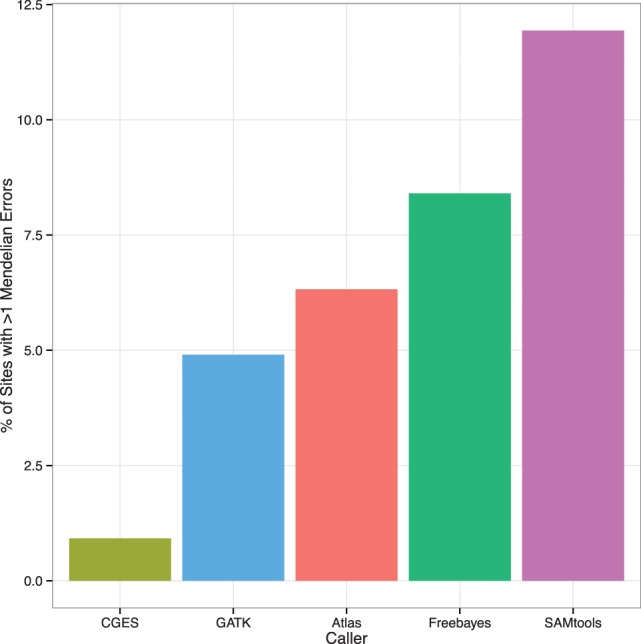
The variant site Mendelian error rate (vMER). The vMER is calculated as the total number of MEs in a VCF file divided by the total number of genotypes with the potential for Mendelian inconsistency (i.e. offspring genotypes with parental genotypes known) in a VCF file. This measure describes the proportion of all offspring genotypes that are inconsistent with parental genotypes present in the VCF
We also evaluated the distribution of calls across the minor allele frequency (MAF) spectrum. We show spectra as empirical cumulative distributions to facilitate comparisons between CGES and the constituent branches (Supplementary Fig. S3). We found that 89.3% of CGES consensus results were previously identified in the EVS project, and 84.1% of CGES consensus results were previously indentified in the 1KG project and 16.9% of results were identified in the GiaB project (Table 1, Supplementary Fig. S6). CGES calls exhibited the highest proportion of variant rediscovery out of the total number of calls compared with any single caller dataset. Finally, we examined Ts/Tv ratios as a function of MAF (Supplementary Fig. S5), finding that within the CGES full consensus calls, the Ts/Tv ratio was consistently >3.0 regardless of MAF. CGES generated a set of identical consensus indels by requiring that the indel match exactly by start position and alternative allele present. The identical consensus set included 2122 indels called by all four programs (Fig. 2).
In addition, we observed a small number of variant positions (N = 369) for which the calls were either homozygous reference or discordant, meaning that at these positions the callers never agreed on a variant genotype in any sample. These highly unreliable variant positions have been included in the Supplementary Materials as a potential black list of exome sequencing variants (Supplementary Table S2).
Lastly, we generated a set of variants for more liberal levels of agreement among algorithms: variants observed in three of four callers, two of four callers and a union of all observed variation. The observed Ts/Tv ratios ranged from 3.02 for the consensus set to 2.00 for the union set (Fig. 5, Supplementary Table S5). The observed gMER ranged from 0.0282% for the consensus set to 0.466% for the union set.
Fig. 5.
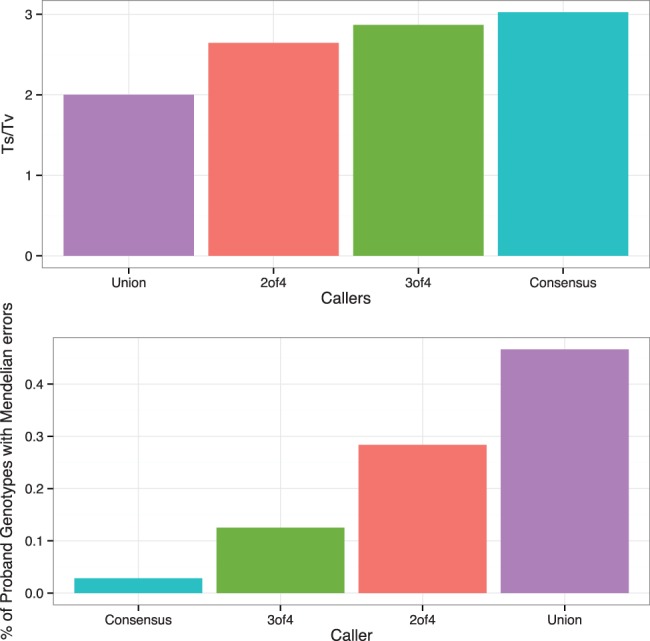
We generate sets of genotypes with different levels of agreement among callers: agreement among all (consensus), agreement among three of four, agreement among two of four and a union of all called variation. Relative to other levels of agreement, consensus has the highest Ts/Tv (top) and the lowest gMER (bottom)
During the course of our analyses, we identified a sample with extreme deviation on the F-statistic () suggesting that this sample showed extreme heterozygosity. On review of the branch call sets, we found that the same sample deviated significantly from the rest of the samples according to every algorithm and showed evidence of extreme heterozygosity (Supplementary Fig. S4). A review of laboratory records showed that this sample had been previously noted as possibly contaminated, and contamination was subsequently confirmed using microsatellite markers. The contaminated sample was removed for the remainder of the analyses provided here, but is retained in Supplementary Figure S4 to illustrate the usefulness of the F-statistic as a QC measure.
3.2 Comparison of CGES predicted and laboratory validated de novo calls
A Sanger sequencing validated set of de novo true positives and false positives was used to test the PPV of the CGES algorithm and its constituent branches. CGES demonstrated the highest PPV (96.1%), which was an improvement over constituent call sets (Table 1).
4 DISCUSSION
We have presented a novel variant-calling approach based on an ensemble of variant-calling algorithms, which we call CGES. CGES uses a two-stage voting scheme among four algorithm implementations to identify variant sites and determine genotypes. In addition to presenting the consensus approach, we have described its application to real-world exome data collected on a sample of autism trios and singletons. We provide project-based, sample-based and variant-based quality metrics across all algorithms, as well as an estimate of the PPV of each algorithm and CGES. Finally, we provide a Galaxy-based implementation of CGES and its constituent parts. Taken together, the results show that Galaxy-CGES provides a robust, flexible and user-friendly approach to exome sequence variant calling. Additionally, these results provide a strong rationale for further development of ensemble methodology in the analysis of NGS data. CGES is not limited to exome data and could in principle be used for whole-genome sequencing, although this application has not yet been tested.
The full CGES consensus-calling algorithm produced the highest quality output but the smallest number of genotypes: 139 897 SNVs in total in our example data. Leveraging the strengths of all callers produced a dataset with the highest Ts/Tv ratio (3.02), the lowest vMER (0.92%), the lowest gMER (0.028%), the highest EVS rediscovery percentage (89.3%), the highest 1KG rediscovery percentage (84.1%), the highest GiaB rediscovery rate (16.9%) and the highest de novo PPV (96.1%).
Consensus approaches for NGS variant detection can be particularly useful when the downstream analysis (i.e. rare variant transmission distortion test (TDT), pathway analysis or de novo filtering) is reliant on a low false-positive rate. However, there may be scenarios in which the preferred strategy is to maximize the rate of true positives even at the expense of a higher false-positive rate: for example, when performing segregation analysis in large extended families. In that case, it may be more fruitful to use the union of all calls from all branches. Additionally, when identifying de novo variants one may wish to use the consensus of all calls in probands and the union of all calls in parents as an added stringency filter to reduce false positives. It is important to stress that there is no ‘one size fits all’ approach to sequencing analysis. The shift in the balance between type I and type II errors is inherent in the stringency with which a consensus dataset is created. Therefore, investigators are given the option of requiring any level of caller overlap (i.e. union of all calls, two-caller consensus, three-caller consensus or full consensus) simultaneously providing the lowest possible false-positive and lowest possible false-negative datasets. The best approach for variant calling depends entirely on the type of data and the downstream analytic plans. As new methods are continually being developed, it is our hope that this report, in conjunction with other consensus efforts (Zook et al., 2013), will help set the tone for an open discussion on the importance of unifying different approaches.
It is important to note the limitations of the analyses presented here. One important limitation is that the quality metrics from each branch are not directly comparable, as their optimization strategies differed. We optimized calling for each branch of CGES to reflect reasonable real-world parameter decisions and not for the sake of a comparison among methods, which has been recently published (O’Rawe et al., 2013). For example, FreeBayes allowed us to set many parameters (Supplementary Methods) based on the raw data descriptive statistics and our previous sequencing experience. Atlas-SNP2, on the other hand, offered relatively fewer parameter options (Supplementary Methods). As best practices have been published for GATKv2.8, we used these guidelines verbatim. Ultimately, the performance of each branch can differ dramatically based on parameters set by the user. Of course, the better the branch calls, the higher quality the final consensus calls will be. Additionally, it may be possible to use concordance between callers as a guiding metric when iterating to the optimal parameters for each branch.
In addition to providing the description of the pipeline and the resultant data, we have provided multiple accessibility modalities. The code for the CGES and CGES-QC algorithms is open source and available through GitHub (Supplementary Table S4). For investigators who do not wish to invoke the command line, we have provided CGES and its constituent branches in a user-friendly Galaxy environment. Finally, for researchers without institutional computational infrastructure, or simply for those who wish to outsource the computing but retain control over the scientific aspects of analysis, the pipeline is available and will be sustained through Globus Genomics (http://www.globus.org/genomics/).
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to acknowledge the individuals who participated in research at the Autism Center for Excellence (University of Illinois, Chicago). The authors also wish to thank Steve Guter (UIC) for assistance with data management, Bo Lui for early work on Galaxy wrappers, Kelley Moore (UIC) and Kathleen Hennessy (UIC) for expert technical assistance, Braden Boone (HAIB), Jack Wimbish (HAIB) and Shawn Levey (HAI) for sequencing and expert advice. The authors are grateful to Erik Garrison for intellectual contributions on review of this manuscript and for technical advice pertaining to FreeBayes. L.K.D. also wishes to posthumously thank George Stephen Karatheodoris for intellectual contributions to this work. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health. The authors are grateful to Amazon Web Services, Inc., for an award of Amazon Web Services time that facilitated early experiments.
Funding: This work was supported in part by P50 HD055751 (EHC), U19 GM61393 (NJC), P60 DK20595 (NJC), P50 MH094267 (NJC), R01 MH089482 (JSS), R24HL085343 (IF), by a Lever Award from the Chicago Biomedical Consortium and by the US Department of Energy under contract DE-AC02-06CH11357. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number KL2TR000431 (L.K.D.).
Conflict of interest: none declared.
REFERENCES
- Albers CA, et al. Dindel: accurate indel calls from short-read data. Genome Res. 2011;21:961–973. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berument SK, et al. Autism screening questionnaire: diagnostic validity. Br. J. Psychiatry. 1999;175:444–451. doi: 10.1192/bjp.175.5.444. [DOI] [PubMed] [Google Scholar]
- Blankenberg D, et al. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 2010 doi: 10.1002/0471142727.mb1910s89. Chapter 19, Unit 19.10.1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis D, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907. 2012:1–9. [Google Scholar]
- Giardine B, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, et al. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Couteur A, et al. Autism diagnostic interview: a standardized investigator-based instrument. J. Autism Dev. Disord. 1989;19:363–387. doi: 10.1007/BF02212936. [DOI] [PubMed] [Google Scholar]
- Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009a;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009b;19:1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, et al. Analysis of rare, exonic variation amongst subjects with autism spectrum disorders and population controls. PLoS Genet. 2013;9:e1003443. doi: 10.1371/journal.pgen.1003443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madduri RK, et al. 2013 Experiences in building a next-generation sequencing analysis service using galaxy, globus online and Amazon web service. In Proceedings of the Conference on Extreme Science and Engineering Discovery Environment: Gateway to Discovery. p. 34. ACM. [Google Scholar]
- Martin ER, et al. SeqEM: an adaptive genotype-calling approach for next-generation sequencing studies. Bioinformatics. 2010;26:2803–2810. doi: 10.1093/bioinformatics/btq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rawe J, et al. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med. 2013;5:28. doi: 10.1186/gm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde M, et al. 2011 Swift: A language for distributed parallel scripting. Parallel Computing, 39, 633–652. [Google Scholar]
- Yu X, Sun S. Comparing a few SNP calling algorithms using low-coverage sequencing data. BMC Bioinformatics. 2013;14:274. doi: 10.1186/1471-2105-14-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook JM, et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nature biotechnology. 2014;32:246–251. doi: 10.1038/nbt.2835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


