Abstract
Summary: We have developed an integrated molecular network learning method, within a well-grounded mathematical framework, to construct differential dependency networks with significant rewiring. This knowledge-fused differential dependency networks (KDDN) method, implemented as a Java Cytoscape app, can be used to optimally integrate prior biological knowledge with measured data to simultaneously construct both common and differential networks, to quantitatively assign model parameters and significant rewiring p-values and to provide user-friendly graphical results. The KDDN algorithm is computationally efficient and provides users with parallel computing capability using ubiquitous multi-core machines. We demonstrate the performance of KDDN on various simulations and real gene expression datasets, and further compare the results with those obtained by the most relevant peer methods. The acquired biologically plausible results provide new insights into network rewiring as a mechanistic principle and illustrate KDDN’s ability to detect them efficiently and correctly. Although the principal application here involves microarray gene expressions, our methodology can be readily applied to other types of quantitative molecular profiling data.
Availability: Source code and compiled package are freely available for download at http://apps.cytoscape.org/apps/kddn
Contact: yuewang@vt.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Modeling biological networks is an important tool in systems biology to study the orchestrated activities of gene products in cells (Mitra et al., 2013). Significant rewiring of these networks provides a unique perspective on phenotypic transitions that can occur in biological systems. Thus, instead of asking ‘which genes are differentially expressed’, a more interesting question is ‘which genes are differentially connected?’ (Hudson et al., 2012; Mitra et al., 2013). To systematically characterize selectively activated or deactivated regulatory components and mechanisms, the modeling tools must effectively distinguish significant rewiring from random background fluctuations. Although specific biological networks cannot be constructed by the existing knowledge alone, novel incorporation of prior knowledge into data-driven approaches can improve the robustness and biological relevance of network inference.
Differential dependency network (Zhang et al., 2009; Zhang and Wang, 2010) and its knowledge-fused extension (Tian et al., 2011, 2014) KDDN have been developed to infer biological networks with significant rewiring by integrating experimental data and biological knowledge. The unique and attractive features of the KDDN software tool are as follows: (i) it is easy to use with a user-friendly graphic user interface (GUI) and cutting-edge performance; (ii) both conserved and differential biological networks can be inferred via efficient closed-form numerical solutions; (iii) model parameters are determined statistically aligned with the expected performance; (iv) prior knowledge (condition specific or non-specific) is incorporated for inferring dependency networks under different conditions; and (v) statistical significance on the differential connections and the type I error rate are rigorously assessed.
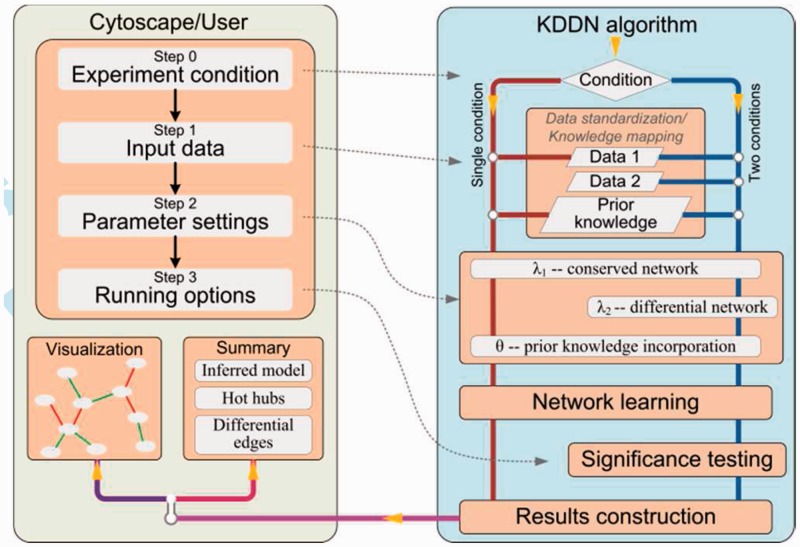
The KDDN Cytoscape 3.x app adapts and extends recent KDDN algorithms in the literature (Tian, et al., 2011, 2014; Zhang et al., 2009; Zhang and Wang, 2010) (Fig. 1). Via a user-friendly GUI, users can easily install KDDN software and perform network analysis by just a few clicks. KDDN software has been tested thoroughly on both synthetic and real gene expression data, and has been successfully applied to a wide range of research projects, including yeast oxidative stress response, breast cancer recurrence, muscular dystrophy and estrogen exposures. Helpful tips for users to use the software in an efficient manner yet with fully informed limitations are provided in Supplementary Information.
Fig. 1.
Architecture and flowchart of KDDN software
2 DESCRIPTION
2.1 Methods and software
KDDN algorithm jointly learns the conserved biological network and statistically significant rewiring across different conditions. Condition-specific data and prior knowledge are quantitatively fused via an extended Lasso model with -1 regularized convex optimization formulation (Tian et al., 2014). Based on the unique nature of the problem, we derive an efficient closed-form solution for the embedded subproblem solved by the block-wise coordinate descent algorithm. We conduct the computational complexity analysis on the KDDN algorithm (Supplementary Information). As existing knowledge is often non-specific or imperfect, KDDN uses a ‘minimax’ strategy to maximize the benefit of prior knowledge while confining its negative impact under the worst-case scenario. Furthermore, KDDN matches the values of model parameters to the expected false-positive rates on network edges at a specified significance level, and assesses edge-specific P-values on each of the differential connections (Tian et al., 2014).
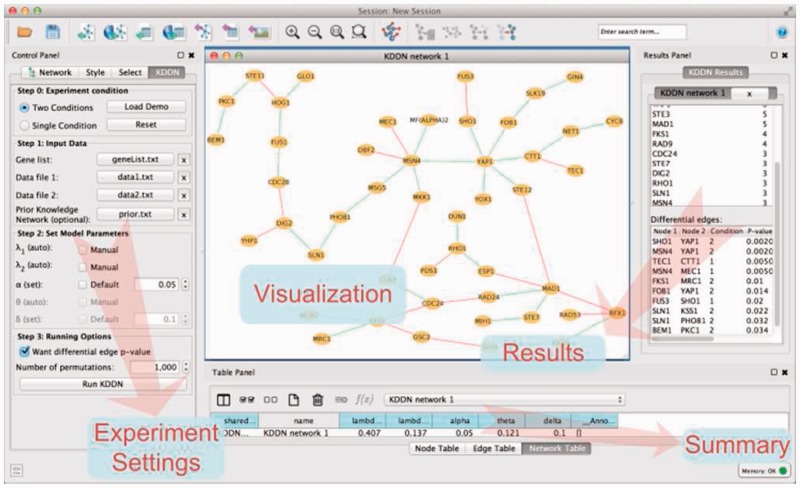
KDDN is implemented as an open-source cross-platform Cytoscape 3.x app with parallel computing capability in Java. The control panel will guide users to navigate through the experiment settings that allow flexible configuration of the analytic tasks. KDDN software takes the quantitative expression values of relevant genes/proteins as input, and incorporates the prior knowledge applicable to either or both conditions (e.g. KGML files downloaded from KEGG pathway Web site can be directly imported as prior knowledge). In addition to inferring common and/or significant rewiring with edge-specific P-values, conventional differential analysis is simultaneously performed allowing users to compare expression fold changes and network rewiring side by side. The constructed networks are visualized seamlessly in Cytoscape, and detailed numerical results together with model parameters are presented in dedicated panels (Fig. 2). All experimental settings and results can be conveniently exported for further analysis.
Fig. 2.
KDDN software interface allowing for user-computer interaction, network/differential analysis, and result presentation/export
2.2 Case study
We applied KDDN to analyze the network rewiring in budding yeast Saccharomyces cerevisiae in response to oxidative stress, focused on cell cycle-related genes. Integrating the prior biological knowledge in the KEGG yeast pathway and gene expression data (GSE7645), the significant differential networks constructed by KDDN are given in Figure 2, where red edges indicate the connections existing under control and green edges indicate the connections created under stress, exclusively.
Oxidative stress is a harmful condition in cell, due to the failure of the antioxidant defense system to effectively remove reactive oxygen molecules and other oxidants (Lee et al., 2011). The result shows that Yap1, Rho1 and Msn4 are among differential hubs and at the center of the network response to oxidative stress; they are activated under oxidative stress and many connections surrounding them are created (green edges). Yap1 is a major transcription factor that responds to oxidative stress; Msn4 is considered as a key responder to environmental stresses, including oxidative stress; Rho1 is known to resist oxidative damage and facilitate cell survival; Ctt1 acts as an antioxidant, and it coordinates with Yap1 to protect cells from oxidative stress. The stress-induced interaction between Hog1 and Fus1, detected by KDDN, is also observed in an earlier study. The biologically plausible results suggest not only the network rewiring as a mechanistic principle in determining phenotypes but also KDDN’s ability to detect them efficiently and correctly.
Detailed descriptions of method, software and more case studies are included in the Supplementary Information.
3 DISCUSSION
The KDDN Cytoscape app presents an integrated software tool to construct biological networks and detect significant rewiring across different conditions. Supported by a well-grounded mathematical framework, KDDN integrates the abundant biological knowledge and condition-specific experimental data to depict the overall dependency networks and their dynamics. Tested on both simulations and real gene expression data, KDDN outperforms peer methods (Supplementary Information) and demonstrates its effectiveness in revealing significant topological changes. We expect KDDN to be a useful tool for performing differential network analysis in many biological contexts (Mitra et al., 2013).
Funding: National Institutes of Health, under Grants [CA160036, CA164384, NS29525, CA149147, HL111362].
Conflict of interest: none declared.
Supplementary Material
REFERENCES
- Hudson NJ, et al. Beyond differential expression: the quest for causal mutations and effector molecules. BMC Genomics. 2012;13:356. doi: 10.1186/1471-2164-13-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ME, et al. The rho1 GTPase acts together with a vacuolar glutathione S-conjugate transporter to protect yeast cells from oxidative stress. Genetics. 2011;188:859–870. doi: 10.1534/genetics.111.130724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K, et al. Integrative approaches for finding modular structure in biological networks. Nat. Rev. Genet. 2013;14:719–732. doi: 10.1038/nrg3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, et al. Knowledge-fused differential dependency network models for detecting significant rewiring in biological networks. BMC Systems Biol. 2014;8:87. doi: 10.1186/s12918-014-0087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, et al. 2011. Knowledge-guided differential dependency network learning for detecting structural changes in biological networks. Proceedings of the 2nd ACM Conference on Bioinformatics, Computational Biology and Biomedicine, ACM BCB 2011, pp. 254–263. [Google Scholar]
- Zhang B, et al. Differential dependency network analysis to identify condition-specific topological changes in biological networks. Bioinformatics. 2009;25:526–532. doi: 10.1093/bioinformatics/btn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang Y. 2010. Learning structural changes of Gaussian graphical models in controlled experiments. Proceedings of the 26th Conference on Uncertainty in Artificial Intelligence, UAI 2010, pp. 701–708. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.