Abstract
Phosphatidylinositol phosphate (PIP) second messengers relay extracellular growth cues through the phosphorylation status of the inositol sugar, a signal transduction system that is deregulated in cancer. In stark contrast to PIP inositol head group phosphorylation, changes in phosphatidylinositol (PI) lipid acyl chains in cancer have remained ill-defined. Here, we apply a mass spectrometry-based method capable of unbiased high-throughput identification and quantification of cellular PI acyl chain composition. Using this approach we find that PI lipid chains represent a cell-specific fingerprint and are unperturbed by serum-mediated signaling in contrast to the inositol head group. We find that mutation of Trp53 results in PIs containing reduced-length fatty acid moieties. Our results suggest that the anchoring tails of lipid second messengers form an additional layer of PIP signaling in cancer that operates independently of PTEN/PI3-Kinase activity, but is instead linked somehow to p53.
Introduction
The phosphatidylinositol phosphates (PIPs) emerged as central to cellular signaling subsequent to the revelation that the pattern of their phosphorylation status formed a code for intracellular signal transduction (Whitman et al., 1988). They serve pivotal roles in normal cellular processes and a broad spectrum of pathologies (Aoyagi and Matsui, 2011; Courtney et al., 2010; Liu and Bankaitis, 2010). The un-phosphorylated phosphatidylinositol (PI) forms the scaffold upon which multiple kinase and phosphatase-dependent events occur which generate the PIP second messengers of growth control. They signal through multiple axes, including the PI3K/AKT pathway, which is aberrant in a majority of cancers. PIs are glycerophospholipids, composed of a glycerol central moiety with two fatty acid esters, a phosphate ester in the third position and an inositol ring (‘head group’) bound to the phosphate group (Figure. S1A). Variable and combined phosphorylation of PIs on the 3′, 4′, 5′ positions of inositol generates seven distinct variants. Extracellular ligand stimulated PI-3 kinase activation generates PI(3,4,5)P3 which promotes cell survival and growth (Cantley, 2002). The tumor suppressor PTEN, frequently inactive in cancer (Hollander et al., 2011), antagonizes PI-3 kinase function by its conversion of PI(3,4,5)P3 to PI(4,5)P2. In addition to malignant scenarios, PIPs mediate growth cues in developmental and other biological processes (Clague et al., 2009; Comer and Parent, 2007; Moss, 2012). Thus, the ‘head group’ status of these lipid second messengers has taken center stage in investigations of PIP biology while the lipid tails have been assumed to play a limited role in signaling. Here, we report the application of mass spectrometry (MS)-based high-throughput method for global analysis of PIs. With this application, we dissected the dynamic nature of the PI signaling scaffold upon which second messengers are formed, demonstrating that in response to mutation of p53, reduced-length fatty acid moieties are present in cellular PIs.
Results
Global assessment of cellular PI content
To analyze global PI content, we adapted electrospray ionization, multiple reaction monitoring (ESI-MRM) methodologies for the assessment of the total compliment of PI variants present in cells and tissue. Identification of the most abundant PI species present in each cell line was possible via interrogation of ions with 700–1200 m/z with signals corresponding to anticipated PI masses (Figure S1B, C) clearly identifiable. For prominent PI species, both molecules containing minor differences in isotopic composition and molecules varying by differences in saturation status could be discerned (Figure S2F). Identification of the mono (phosphatidylinositol phosphate- PIP) and bis (phosphatidylinositol bisphosphate- PIP2) forms of the most abundant PI species was achieved. We termed these phosphatidylinositol families “triplets” (Figure S1C-left panel). Each additional phosphate moiety contributes an increased 80 m/z, such that the PI for any particular species may be given as ‘N’ m/z, the PIP as N+80 m/z and the PIP2 N+160 m/z. Of note, in our analysis phosphatidylinositol triphosphate, PIP3, was not apparent at levels above those of background. However, subsequent fragmentation of ions with a predicted m/z (i.e. N+240 m/z) corresponding to anticipated PIP3 masses generated PIP3-specific ions (Milne et al., 2005; Wenk et al., 2003), demonstrating the presence of these molecules, although at abundances with signal-to-noise ratios significantly below 3:1 (data not shown). In order to positively identify lipid species, fragmentation of parent ions corresponding to anticipated PI m/z was performed (Figure S1C-right panel). These detailed fragmentation spectra allowed unambiguous identification of parent ions, with identification of both the fatty acid and head-group components of lipids, consistent with previous descriptions of PI fragmentation spectra (Hsu and Turk, 2000; Ivanova et al., 2009). Fragmentation patterns were compared to known phospholipid standards to confirm their phosphoinositide-derived nature (data not shown).
To enable high-throughput identification and quantification of individual PIs, we developed a method employing automated sample injection, fragmentation and ion measurement. MRM, using anticipated PI fragment ions as parent masses and calculated daughter ions, corresponding to inositol head-group and acyl chain configurations observed in preliminary PI fragmentation experiments, permitted broad assessment and quantification of PI species (Figure S1E). The abundances of twenty anticipated PI variants were measured in tandem to generate robust quantification data for each individual molecule. Initial experiments interrogated ions in the 300–1300 m/z range. In no experiments did we observe PI species <800 m/z and >920 m/z, as determined by ion fragmentation of parent masses. Therefore, in subsequent analyses we restricted our analysis to PI molecules in this mass range (Figure S1B). It should be noted, however, that although in none of the initial tests of cultured cell lines were PIs containing acyl chains of >20 carbons observed, some tissue types, such as rat liver (Patton et al., 1982), have been shown to contain low amounts of PIs with fatty acid components of more than 20 carbons. Resultantly, for detailed assessment of previously unexamined tissues using this method pilot scans interrogating masses greater than 900 m/z could be performed.
Total ion chromatograms (Figure S1D) highlight that the extraction protocol employed does not generate PI-pure lipid mixtures. However, assessment of PI-specific daughter ions, and not putative parent ion masses, enables specific measurement of PI abundance, providing the molecular specificity of this method. All potential acyl chain saturation states (including hypothetical variants) for each predicted PI are assessed with no prior understanding of molecular configuration required. For example, PI 889 m/z corresponds to the molecular configuration 38:2 (a total acyl chain composition harboring 38 carbon atoms and two double bonds). From the empirical formula, it is not possible to determine acyl chain length/saturation status. To overcome this, all potential fatty acid configurations that the molecule may comprise were measured simultaneously: 18:0, 18:1, 18:2, 20:0, 20:1, 20:2. Of note, in our analysis we failed to observe acyl chains with odd numbered carbon atoms. These configurations are presumably not utilized or generated by cells, or if so, in abundances below the level of detection. In summary, this adapted method enabled unbiased, PI-specific quantification in an automated, high throughput manner and can be performed in the absence of any prior molecular knowledge of the target lipid(s) of interest. Fragmentation of lipid standards comprising 16:0/18:1 and 18:0/20:4 fatty acids revealed comparable fragmentation and ion generation, suggesting that PI fragmentation across molecular species did not vary as a function of fatty acid length (Figure S1F), thereby allowing direct comparison of quantified ions between PI species.
Electrospray ionization techniques have been employed previously with great success in identification of PI, PIP, PIP2 and PIP3 (Milne et al., 2005; Wenk et al., 2003). However, previous reports have not attempted to quantify these molecules. A more advanced lipid extraction/analysis procedure with incorporation of phosphate methylation steps and high performance liquid chromatography has also been developed for the assessment of phosphatidylinositol phosphates, including PIP3 (Clark et al., 2011). However, to date, the ability to globally quantify phosphatidylinositol phosphates has remained elusive. In our hands, the quantification of PIP and PIP2 content was achieved, albeit with large observed deviations between samples (data not shown). At present, it is unclear whether these deviations are due to biological phenomena or lower abundances of the molecules being assessed. Furthermore, analysis of lipid standards demonstrated that ionization of equivalent molar amounts of PI, PIP, PIP2 and PIP3 did not generate equivalent ion abundances (data not shown), further complicating the direct comparison of phosphatidylinositides of disparate phosphorylation status.
PI lipid tail fingerprints
Assessment of PIs was performed across multiple cells line with high reproducibility achieved across biological replicates. Analysis of cells of both human and mouse origin revealed that disparate cancer cell lines possessed unique PI characteristics- a PI fingerprint (Figure 1). In addition, it was apparent that the cell lines tested did not harbor individual PIs, defined by acyl chain composition, in isolation, but certain species and their closely related sibling molecules differentiated by saturation state of the fatty acid moieties. Quantification of the four most abundant PI species (Figure S2A) revealed that strikingly ~40–50% of the total PI composition of cells is composed of just two PI masses. Approximately 70% of the total PI content in cells was composed of the four most abundant PI masses in any single cell line. Human and murine cells demonstrated no remarkable differences in this analysis.
Figure 1. PI characteristics are unique to disparate cell lines and are dictated by genetics and not growth factors.
(A) Representation of quantification of phosphoinositide abundances in the common laboratory cell lines NIH3T3 and HeLa. Graphs are derived from two independent lipid preparations and measurements. Individual sample measurements were derived from three measurements of the same sample. The sum carbon content of both lipid fatty acids is displayed as ‘number of carbon atoms’ [left vertical axis]. Relative ion counts are normalized to the ion of observed highest abundance [right vertical axis]. (B) Assessment of PI the characteristics of the HCT116 cells in both normal (10% serum) and serum starved (0.1%) conditions. (C) Assessment of two genetically engineered cell lines derived from mouse pancreas tumors. Cancers were initiated with identical genetic lesions (Kras and p53 mutations) from independent mice with identical backgrounds. (D) Assessment of two independent and distinct human pancreatic adenocarcinoma cell lines. For all graphs, mean measurements of two biological replicates are displayed (error bars: ± S.D.).
Assessment of the saturation status of PI lipid chains demonstrated that in the cell very few of these lipids are polyunsaturated. These data also revealed that saturation of cellular PIs in murine and human cells do not differ significantly. Of note, however, NIH3T3 and Capan-2 (a pancreatic cancer-derived line) cells harbored fewer monounsaturated PIs and higher levels of 3 or 4 double bond-containing PIs than the other lines assessed (Figure S2B).
Analysis of the carbon content, which reflects fatty acid chain length, revealed that the majority of PIs possessed 34, 36 or 38 carbon atoms in their fatty acid components (Figure S2C). NIH3T3 and Capan-2 lines contained a higher proportion of 38-carbon PIs comparative to the other cells tested.
PI acyl chain content is not affected by extracellular ligands but is dependent on serum-derived essential fatty acids
In order to assess whether the PI fingerprint of each cultured cell line was altered in response to extracellular ligands, cultured cells were serum starved for 24 hours before PI lipid analysis. Unlike in the case of PI head-group phosphorylation, removal of serum-provided extracellular ligands had no effect on PI lipid chain composition (Figure 1B). Comparison of the abundances of the twenty quantified PI masses in normal culture and 24-hour serum-starved conditions identified no change in PI abundances at a statistically significant level (Figure S2D, HCT116 cells shown). Testing of PI characteristics in response to 24-hour serum withdrawal in multiple additional cell lines confirmed the lack of response to extracellular ligands (data not shown).
We then sought to investigate PI composition when cells are grown in the absence of fatty acids- another component provided by FBS in tissue culture contexts. To this end, 3 cultured cell lines (HeLa, NIH3T3 and PC3) were grown in medium supplemented with delipidated FBS. Subsequent to 3-days growth in delipidated medium all cell types exhibited a reduced proliferation compared to control experiments supplemented with normal FBS (Figure S3A). In the case of NIH3T3 cells, lack of lipid supplementation led to an overall reduction in cell number with most of the plated cells perishing. PI characteristics were then investigated. For all lines, in both lipidated and de-lipidated scenarios, the 38:3 PI species was in the vast majority composed of PIs containing 18:0 and 20:3 fatty acids, with a limited amount of 18:1/20:2 variants detectable (data not shown). For the 38:4 species, all cells harboured mostly 18:0/20:4 PIs, but with some 18:1/20:3 species present in both lapidated and de-lipidated contexts (data not shown).
Some fatty acid molecules incorporated into PIs, such as linoleic acid (18:2), are not produced by the cell and in tissue culture scenarios must be provided in serum. Linoleic acid, for instance is utilized as a precursor in the production of arachidonic acid (20:4) (Steinberg et al., 1956). Although some variations in total cellular PI content (Figure S3B) are observed after removing lipids from culture medium, the greatest effect appeared to be on cell viability/proliferation. Our initial studies suggest that cells lack a large degree of plasticity regarding their fatty acid composition in response to environmental changes and where cells are unable to acquire the optimal fatty acid configurations for lipid production their viability is compromised. In the absence of linoleic acid in culture medium, we assume that levels of this acid, and arachadonic acid, were remnants from the original cell material plated at the start of the experiment.
Testing the conservation of PI fingerprints
In order to determine some of the underlying parameters responsible for a cell’s PI fingerprint, we generated cultured cell lines with known genetic lesions and well characterized origins. Genetically engineered mice were generated harboring pancreas-specific Kras and Trp53 mutations. These genetic lesions were sufficient to generate pancreatic neoplasia in these animals and from these malignancies culture lines were generated from two animals with identical strain background and identical tumor-initiating/driving genetic lesions (Figure 1C).
Using these two lines, we were able to address the question: do tumor cell lines, from the same genetically modified cancerous organ, but different individuals, retain the same PI fingerprint? Quantification of the PI abundances in the two lines and statistical testing revealed that the PI lipid fingerprints were not statistically significantly different (Figure 1C and Figure S2E). Furthermore, comparison of the saturation status and acyl chain carbon content of the PI content of the cell lines confirmed an unaltered PI content in these disparate tumor-derived cell culture lines. These data suggested that cancer from different individuals but triggered by the same mutations, share common PI acyl chain characteristics thereby indicating the potential use of PI fingerprints in diagnostic contexts.
Our analysis also includes the study of the HCT116 and KO22 culture lines (Figure 1B and S2G). HCT116 cells were originally derived from a colorectal cancer metastasis. KO22 cells were engineered from HCT116 cells by knock out of the PTEN tumor suppressor gene (Lee et al., 2004). These two cell lines also showed the same PI profile. This result suggested that PTEN-status, responsible for regulation of PIP headgroup phosphorylation, had no effect on PI lipid tail features. In addition, the finding suggested that two closely related but genetically distinct cancer cell lines, which were cultured independently for a decade since divergence, retained similar lipid profiles. On the whole, these data support the notion that the PI content of cells is fixed and not significantly altered by extensive passaging in tissue culture.
Next we tested two human pancreatic cancer-derived lines, Capan-2 and MiaPaCa-2, both classified as pancreatic carcinomas (Kyriazis et al., 1986; Yunis et al., 1977), which showed distinct PI fingerprints (Figure 1D). This observation suggests that organ of origin does not dictate cellular PI characteristics for cancer cells. It is plausible in the instance of these two pancreatic carcinomas that independent genetic events, which contributed to their malignant development, precipitated the divergence of disparate PI profiles. Conversely, the genetically engineered cell lines, obtained from mouse pancreas adenocarcinomas (Figure 1C), of identical genetic lineage, presented indistinguishable PI fingerprints.
Non-malignant cells harbor primarily 38-carbon PIs
To disentangle factors that may cause cells to harbor PIs specific acyl chain composition, we used primary mouse embryonic fibroblasts (MEFs). Strikingly, assessment of wildtype primary MEFs revealed that they harbored a PI profile dominated by 38 carbon acyl chain content (Figure 2), highly similar to that seen in NIH3T3 cells, and in stark contrast to all cancer cell lines tested (Fig 1A-right panel, 1B, 1C, 1D, S2G). Most primary tissues from mouse displayed the same type of dominant 38 carbon lipid chain content in their PIs (Figure 2), suggesting that it represents a normal state with the exception of brain and testicle (Figure S2F) which also possessed a single, unique, additional PI compound (835 and 809 m/z respectively).
Figure 2. Non-malignant cells and tissue harbour almost exclusively 38-carbon lipids.
Assessment of PI characteristics from primary wildtype MEFs and mouse tissue obtained a resection. For MEFs, mean measurements of two biological replicates are displayed (error bars: ± S.D.). For tissues, mean measurements of technical triplicates are displayed (error bars: ± S.E.M.). Animals from which tissue was taken were healthy, 7-week old wildtype males. Blood was entire fluid containing both serum and cellular components.
p53 mutation status dictates changes in PI fingerprints
We then went on to analyze mouse-derived pancreatic cancer cell lines, as shown in Figure 1C, but in a comparative context of other genetic lesions and normal, non-cancerous pancreas tissue (Figure 3). Normal mouse pancreas demonstrated a PI fingerprint similar to wildtype MEFs and NIH3T3 cells: almost exclusively 38 carbon acyl chain content. In contrast, the pancreatic cancer cells, generated from pancreas engineered to achieve pancreas specific expression of KrasG12D and Trp53 R172H mutation from the endogenous alleles demonstrated a significant shift to lower PI carbon content. Analysis of pancreas-derived tumor lines generated harboring solely the oncogenic KrasG12D mutation, in the absence of Trp53 alteration, retained a PI profile indistinguishable from normal pancreas tissue.
Figure 3. Initiating genetic events contribute to changes in PI characteristics in cancer progression.
(A) Comparison of PI characteristics from normal mouse pancreas (mean measurements of technical triplicates are displayed, error bars: ± S.E.M.) and two pancreas-cancer-derived tumor lines of each of two distinct genotypes, initiated with the genetic lesions shown (mean measurements of two biological replicates are displayed, error bars: ± S.D.). (B) Assessment of PI the characteristics of p53-null MEFs. Mean measurements of two biological replicates are displayed (error bars: ± S.D.).
To confirm that shifts in PI lipid composition are not simply the result of increased proliferation, we analyzed the PI fingerprint of the primary p53Δ/Δ MEF cell line (Figure 3B, generated using the Cre-loxP/viral system (Chen et al., 2005)). As a result of this p53 loss-dependent immortalization, these cells have significantly enhanced proliferation compared to wildtype MEFs (Chen et al., 2005). Irrespective of their increased growth rates however, p53-null MEFs did not share the PI fingerprint of Trp53 +/R172H, Kras+/G12D cells, these data suggested that changes in the population of cellular PI acyl chain moieties correlates with p53 mutation, but not loss.
To further explore the relationship between p53 mutation and PI characteristics, we sought to correlate p53 mutation status with cellular PI content. In all cell lines harboring TP53 mutation we identified a corresponding high level of 36-carbon PIs. Two cell lines absent TP53 mutation did not exhibit this shift in PI composition: the prostate cancer metastasis-derived line PC3, and the pancreatic cancer-derived line Capan-2. Sequencing of PC3 DNA demonstrated heterozygous loss of one allele of TP53 and a frameshift mutation in the remaining allele, rendering this line TP53-null (Figure 4B). Sequencing of the mutation cluster region of TP53 (exons 5-8) of Capan-2 DNA revealed no TP53 mutations (data not shown), in agreement with previous reports describing the TP53-wildtype nature of this line (Caldas et al., 1994; Loukopoulos et al., 2004). The lipid fingerprint of this line lacked high levels of 34 and 36-carbon PIs (Figure 1D, left panel). Resultantly, genetic analysis of cultured cancer cells pointed to a link between TP53 mutation status and increased 34 and 36-cabon PI content.
Figure 4. Tp53 mutation is associated with modulation of PI content.
(A) PCR amplification of p53 exons 5-8 in selected cancer cell lines. Sequencing primers produced a single amplicon used for subsequent sequencing. (B) Sequence traces of selected cancer cell lines showing identified mutations and corresponding PI characteristics. All identified mutations were identified in the DNA-binding domain of p53.
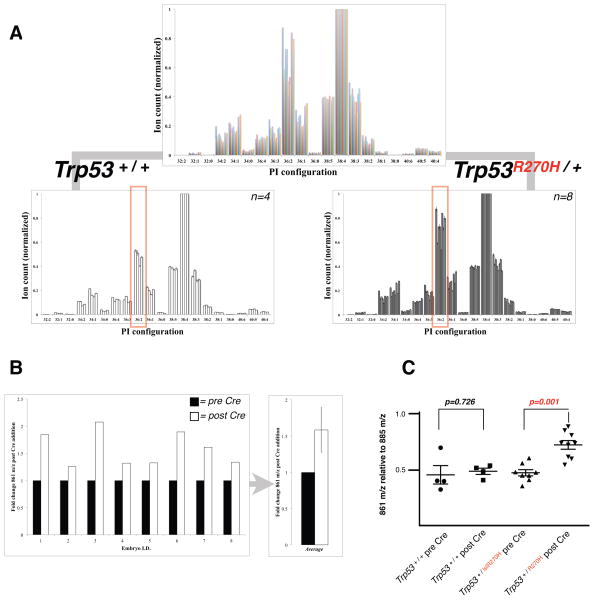
In order to assess if p53 mutation alone could cause the transition of cellular PIs from 38 to 36/34 carbon acyl chain length, we employed a lox-stop-lox Trp53R270H MEF system. Cre recombinase causes recombination at the Trp53 locus to express the mutant Trp53 gene from the endogenous Trp53 promoter (Tuveson and Hingorani, 2005). We harvested a panel of littermate primary MEFs of the two genotypes Trp53+/+, which served as wildtype control MEFs before and after Cre-mediated recombination, and Trp53lslR273H/+ A total of 12 primary cells of these two genotypes were cultured and independently infected or not at passage 2. The PI content of the cells was then determined six days subsequent to infection and analyzed in blinded experiments on 24 samples (12 littermate MEF samples with or without viral addition). When lipid data from Cre-recombined cells were plotted stratified by Trp53 status, variability in 36-carbon acyl chain content (relative to 38 carbon content) became apparent (Figure 5A). Un-blinding revealed that the eight Trp53 mutant MEF lines had higher 36 carbon content lipids than their Cre infected Trp53 wildtype littermate controls. When assessing the recombination effect on a line by line level (Figure 5B), we found that mutant Trp53 expression induced an approximate 50% increase in 36 carbon content PI lipids on average, an increase observed in every before/after Cre paired comparison. Importantly, no significant effect on the relative abundance of the dominant 36 carbon PIs was observed after viral infection of the wildtype cells. Furthermore, the uninfected Trp53 +/lslR273H MEFs, which only express one copy of wildtype Trp53, also showed no significant change in 36-carbon PIs compared to either treated or untreated wildtype controls (Figure 5C). Of note, fragmentation of the 36-carbon PIs that were enriched subsequent to expression of mutant Trp53 showed that >80% of this lipid were 18:1/18:1 with <15% of 18:0/18:2 composition (data not shown). This observation is suggestive of an increase in production of, or incorporation of, oleic acid (18:1) into PI-destined molecules as being prominent in the shift we observe.
Figure 5. Trp53 mutation modulates PI fingerprints.
(A) Infection of Trp53 wildtype and Trp53wildtype/lslR270H MEFs with adenovirus encoding Cre recombinase and lipid analysis was performed blinded. In total 12 cell lines were generated to form genetic biological replicates. Correlation of Trp53 genotype with lipid status demonstrates that levels of 36-carbon PIs (861 m/z) increase subsequent to Trp53 R270H expression. (B) Analysis of 8 biological replicates of Cre-treated Trp53wildtype/lslR270H MEFs demonstrates that all lines exhibit an increase of PI 861 m/z. 861 m/z abundance in Cre-treated cells is displayed as fold-change relative to the same line prior to Cre addition. (C) Quantification of PI 861 m/z relative to the most abundant PI species (885 m/z) pre and post Cre addition (T-test, 2 tailed, whiskers: ± S.D.).
Shifts to shorter acyl chain content subsequent to p53 mutation do not occur in all phospholipids
To further shed light on the mechanism of the shift to 36-carbon PIs seen in malignancies and subsequent to p53 mutation, we developed a method for the analysis of phosphatidylcholine (PC) (Figure S4). Analysis of a panel of cultured cell lines, including those that harbored almost exclusively 38-carbon PI content (NIH3T3, PC3, Capan-2) demonstrated no remarkable difference between the lines tested in terms of PC spectra. Unlike PIs, PC content in all the cells tested was primarily composed of 34 and 36-carbon lipids (Figure S5). No apparent change across lines was observed, irrespective of TP53 status. These data suggest that the observed alterations in acyl chain content subsequent to p53 mutation is not characteristic of all phospholipids.
PC and PI share a common precursor: phosphatidic acid (PA). Synthesis of PI from PA occurs via two enzymatic steps involving first cytidine diphosphate diacylglycerol synthase and then cytidine diphosphate diacylglycerol inositol phosphatidyltransferase (‘phosphatidylinositol synthase’). Generation of PC from PA involves two sequential enzymatic steps requiring phosphatidic acid phosphatase then cholinephosphotransferase (Holthuis and Menon, 2014). It is unclear as to how exactly the latter synthesis steps of PC and PI might contribute to different acyl chain moieties being present in these different lipid types subsequent to p53 mutation. It might be plausible, however, that incorporation of different fatty acids into PA leads to accumulation of distinct pools of this precursor lipid, localized in different membrane regions in the cell. If enzymes required for the synthesis of PI and PC have differential access to different PA pools, this may in part explain differences in the fatty acid content seen in these lipid types in the context of p53 mutation. Alternatively, PI remodelling does occur such that the composition of mature PIs can be altered after their formation (Darnell et al., 1991). Investigation of the influence of Tp53 mutation on enzymes responsible for such remodelling may further define the step at which acyl chain composition changes in PI molecules occurs.
Discussion
The PIP second messengers, consist of lipid tails linked to the inositol head group, upon which information is presented. In the case of phosphoinositide biology, research into the functional roles of the lipid tails has lagged behind the study of how information is encoded. Many biological information systems, most famously nucleic acids, consist of structural backbones that carry chemically coded additional information. As was the case in understanding and correlating DNA backbone methylation to disease progression (Feinberg and Vogelstein, 1983), quantitative, reproducible technology is a critical prerequisite for the deciphering of biological roles that can be attributed to the PIP scaffolds. It is intriguing that the PIP acyl chains have the potential for far greater molecular flexibility than the inositol sugar head group: only 7 discrete phosphorylation states are known for PIPs. In contrast, the variety of lipids that can be added to two positions on the glycerol linker, considering saturation status, double bond position, as well as carbon content, is in far excess of the combinatorial possibilities that are mediated by the currently known PIP modifying kinases and phosphatases.
Using mass spectrometry, we have defined the spectrum of phosphoinositide second messenger lipid tails in a rapid, reproducible and high-throughput manner. Our results show that (1) cultured cancer cell lines present their own unique PI characteristic ‘fingerprints’. (2) The PI lipid tails of cells are limited to very few species by mass, which have a total acyl chain carbon content of 34, 36 or 38 atoms. (3) Non-cancerous tissue exhibits largely 38 carbon species. (4) In contrast, malignant cells contain the shorter acyl chain lipids totaling 34 and 36 carbons. (5) The shift to lower carbon content PI lipid tails can be caused by p53 mutation.
We observed the shift to low carbon content when comparing the two pancreatic cancer cell lines MiaPaCa-2 and Capan-2. It has been reported that the Capan-2 cells, which show dominant 38 carbon content PIs, retain wildtype p53 status (Caldas et al., 1994; Loukopoulos et al., 2004), a finding that we confirmed in our analysis. In contrast, the MiaPaCa-2 cells with high 34 and 36 carbon PIP lipid content harbor a point mutation in the Tp53 gene codon 248 and loss of the remaining wildtype allele. Note that both cell lines harbor mutant KRAS (Aoki et al., 2000; Berrozpe et al., 1994; Kita et al., 1999; Loukopoulos et al., 2004; Moore et al., 2001). Furthermore, sequencing of a panel of cancer cell lines has provided further evidence that Tp53 mutation is correlated to modulatory effects on cellular PI composition. Employing a lox-stop-lox mutantTrp53 MEF system allowed the unambiguous identification of mutant Trp53 as a modulator of cellular PI content. Our studies in no way indicate that p53 is the sole effector molecule responsible for alterations in PI characteristics, but they are indicative of this gatekeeper gene regulating lipid-specific pathways. HCT116 cells are Tp53 wildtype (Lee et al., 2004) and yet possess abundant 36-carbon PIs. Similarly, HeLa cells also possess wildtype p53 genes and a significant amount of 36-carbon PIs. However, this cervical cancer cell line is positive for human papillomavirus 18, which is known to induce outcomes that are similar to p53 mutation itself (Kessis et al., 1993). Resultantly, it is likely that multiple aberrations, which may be acquired during malignancy, can give rise to alterations in PI cellular content of which p53 mutation is one. Although p53 activity has hitherto not been linked to PIP biology and the associated signaling, mutant Tp53 protein has been described to increase the expression of genes encoding components of the mevalonate pathway, thereby altering cellular cholesterol levels and promoting breast cancer (Santos and Schulze, 2012). Whether the effects of mutant p53 on PIs observed in our studies is due to aberrant target gene promoter binding remains to be shown. Of note, all mutation types observed in the DNA-binding domain of p53 seem associated with the lipid-changing activity observed in our study.
The PI analysis methodology we present can be achieved within minutes, e.g. during an ongoing operation (Balog et al., 2013), which is in stark contrast to the time frame for gene sequencing. Thus it could assist intra-operative pathology calls with molecular information on tissue to guide surgeons on the needed extent of resection around a lesion.
It still remains to be determined if and how the different lipid chain lengths could be causally involved in malignancy. Lipid ‘packing defects’ contribute to the identity of lipid membranes (Bigay and Antonny, 2012). Thus the change in PIP lipid tails could cause differential cellular localization of PIP signaling. Another intriguing hypothesis could involve the phosphatidylinositol transfer proteins (reviewed in (Cockcroft, 2012)), which are among the proteins that can selectively interact with PIPs based on their lipid composition. Phosphatidylinositol transfer protein α (PITP) is an intracellular lipid transporter that transfers both PIP and phosphatidylcholine and has been shown to preferentially use PIs with shorter acyl chains (Hunt et al., 2004).
The alterations in PI acyl chain configuration measured in our studies raises the intriguing question of how these changes are, if at all, represented in the primary signaling phosphoinositides; primarily PIP3. Stimulation of cells by different agonists results in different molecular configurations of PIP3 being generated, which is highly suggestive of phosphoinositides of specific acyl configuration being converted to the triple phosphate form by specific head group-catalyzing enzymes (Wakelam, 2014). It has also been hypothesized that alternate acyl chain compositions on phosphoinositides confer different orientation of the lipid head group relative to the membrane (Wakelam, 2014). Although singular lipid binding domains may not realize altered binding tendencies in different cases of head group orientation, the interaction of domains such as the phsophoinositide 3 phosphate-binding FYVE motif, which in the case of Hrs, for example, functions in tandem, may experience differing binding characteristics in such scenarios. Further understanding of the specific functional relevance of PI acyl chain configuration and determination of the prevalence of these molecules being converted into signaling-active PIP3 via subsequent head group modifications would provide a fascinating insight into the role of PI tail chemistry in signaling and disease.
Taken together, our findings identify a rapid and reliable high throughput method to test the value of PI lipid tails as biomarkers for p53 status in tissue. Seeing these effects in pancreatic malignancies, where PI 3-Kinase/AKT signaling through PIPs is thought to play a minor role next to K-RAS and p53 alterations, points to a second, lipid-driven layer of PIP signaling in cancer. Our observations point to mutation of Tp53 as an important factor in remodeling of cellular PIs. Investigation of the exact mechanism by which this genetic change gives rise to this phenomenon would provide fascinating insight into the effects of tumor suppressor mutations in lipid composition. If an exact enzyme can be identified which is responsible for mediating such changes, inhibition of this protein and observation of the cellular response in terms of malignant transformation may provide a hitherto unconsidered approach for treating Tp53-mutant malignancies. Furthermore, elucidation of the exact influence that regulation of PI backbones has on PI3K/AKT-mediated signaling will create a new understanding of how p53 mutation modulates this cancer-related signaling axis.
Experimental procedures
Generation of murine pancreatic tumor-derived cell lines
Cell lines were derived from animals transgenic for Pdx1-Cre, carrying the Kras lsl-G12D/+ alleles. Double mutant mice also harbored the Trp53 lsl-R172H/+ alleles (Tuveson and Hingorani, 2005).
Lipid analysis
Lipid extraction from tissue culture material has been described previously (Milne et al., 2005). Extracted lipids were analyzed by a Vantage triple-stage quadrupole mass spectrometer (Thermo Scientific, NC, USA) in negative ion mode with the Heated Electrospray Ionization (HESI-II) ion source.
PCR amplification and sequencing of p53 exons 5-8
PCR reactions, primer sequences and amplification conditions and Sanger dideoxysequencing methodology have been described previously (Park et al., 2010).
Extensive descriptions of methodology are included in the supplemental information.
Supplementary Material
Acknowledgments
We thank C. Ruse for help with lipid quantification procedures and the members of the Trotman lab for valuable discussion and help with experimental protocols and reagents. L.C.T. is a Research Scholar of the American Cancer Society and is supported by the Pershing Square Sohn Foundation and the Department of Defense (W81XWH-13-PCRP-IDA). This work was graciously supported by grants to L.C.T from the ACS (RSG-14-069-01-TBE), the NIH (CA137050), the Robertson Research Fund of Cold Spring Harbor Laboratory and by support to D.J.P., D.A. and L.C.T through the NIH Cancer Center Support Grant 5P30CA045508. D.A.T. is an investigator of the Lustgarten Foundation for Pancreatic Cancer Research, and is supported by the Cold Spring Harbor Laboratory Association, the David Rubinstein Center for Pancreatic Cancer Research at MSKCC, and the NIH 1R01CA190092-01; 5P30CA45508-26; 5P50CA101955-07. I.I.C. is supported by the Damon Runyon Cancer Research Foundation (DRG-2165-13), and D.D.E. by a T32 grant from the NIH (5T32CA148056).
Footnotes
Author Contributions
AN, DJP, DT and LCT designed the study. AN, GB, DE, IICC, TH, KW and SB performed experiments. AN and LCT wrote the manuscript.
The authors declare that they have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoki Y, Hosaka S, Tachibana N, Karasawa Y, Kawa S, Kiyosawa K. Reassessment of K-ras mutations at codon 12 by direct PCR and sequencing from tissue microdissection in human pancreatic adenocarcinomas. Pancreas. 2000;21:152–157. doi: 10.1097/00006676-200008000-00008. [DOI] [PubMed] [Google Scholar]
- Aoyagi T, Matsui T. Phosphoinositide-3 kinase signaling in cardiac hypertrophy and heart failure. Current pharmaceutical design. 2011;17:1818–1824. doi: 10.2174/138161211796390976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balog J, Sasi-Szabó L, Kinross J, Lewis M, Muirhead L, Veselkov K, Mirnezami R, Dezsö B, Damjanovich L, Darzi A, et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Science translational medicine. 2013;5 doi: 10.1126/scitranslmed.3005623. [DOI] [PubMed] [Google Scholar]
- Berrozpe G, Schaeffer J, Peinado M, Real F, Perucho M. Comparative analysis of mutations in the p53 and K-ras genes in pancreatic cancer. International journal of cancer Journal international du cancer. 1994;58:185–191. doi: 10.1002/ijc.2910580207. [DOI] [PubMed] [Google Scholar]
- Bigay J, Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Developmental cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Caldas C, Hahn S, da Costa L, Redston M, Schutte M, Seymour A, Weinstein C, Hruban R, Yeo C, Kern S. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nature genetics. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- Cantley L. The phosphoinositide 3-kinase pathway. Science (New York, NY) 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Chen Z, Trotman L, Shaffer D, Lin HK, Dotan Z, Niki M, Koutcher J, Scher H, Ludwig T, Gerald W, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague M, Urbé S, de Lartigue J. Phosphoinositides and the endocytic pathway. Experimental cell research. 2009;315:1627–1631. doi: 10.1016/j.yexcr.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Clark J, Anderson K, Juvin V, Smith T, Karpe F, Wakelam M, Stephens L, Hawkins P. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nature methods. 2011;8:267–272. doi: 10.1038/nmeth.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft S. The diverse functions of phosphatidylinositol transfer proteins. Current topics in microbiology and immunology. 2012;362:185–208. doi: 10.1007/978-94-007-5025-8_9. [DOI] [PubMed] [Google Scholar]
- Comer F, Parent C. Phosphoinositides specify polarity during epithelial organ development. Cell. 2007;128:239–240. doi: 10.1016/j.cell.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Courtney K, Corcoran R, Engelman J. The PI3K pathway as drug target in human cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:1075–1083. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Osterman DG, Saltiel AR. Fatty acid remodelling of phosphatidylinositol under conditions of de novo synthesis in rat liver microsomes. Biochim Biophys Acta. 1991;1084:279–291. doi: 10.1016/0005-2760(91)90070-x. [DOI] [PubMed] [Google Scholar]
- Feinberg A, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Hollander M, Blumenthal G, Dennis P. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nature reviews Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JC, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510:48–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- Hsu F, Turk J. Characterization of phosphatidylinositol, phosphatidylinositol-4-phosphate, and phosphatidylinositol-4,5-bisphosphate by electrospray ionization tandem mass spectrometry: a mechanistic study. Journal of the American Society for Mass Spectrometry. 2000;11:986–999. doi: 10.1016/S1044-0305(00)00172-0. [DOI] [PubMed] [Google Scholar]
- Hunt A, Skippen A, Koster G, Postle A, Cockcroft S. Acyl chain-based molecular selectivity for HL60 cellular phosphatidylinositol and of phosphatidylcholine by phosphatidylinositol transfer protein alpha. Biochimica et biophysica acta. 2004;1686:50–60. doi: 10.1016/j.bbalip.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Ivanova P, Milne S, Myers D, Brown H. Lipidomics: a mass spectrometry based systems level analysis of cellular lipids. Current opinion in chemical biology. 2009;13:526–531. doi: 10.1016/j.cbpa.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessis TD, Slebos RJ, Nelson WG, Kastan MB, Plunkett BS, Han SM, Lorincz AT, Hedrick L, Cho KR. Human papillomavirus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:3988–3992. doi: 10.1073/pnas.90.9.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita K, Saito S, Morioka C, Watanabe A. Growth inhibition of human pancreatic cancer cell lines by anti-sense oligonucleotides specific to mutated K-ras genes. International journal of cancer Journal international du cancer. 1999;80:553–558. doi: 10.1002/(sici)1097-0215(19990209)80:4<553::aid-ijc12>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Kyriazis A, Kyriazis A, Sternberg C, Sloane N, Loveless J. Morphological, biological, biochemical, and karyotypic characteristics of human pancreatic ductal adenocarcinoma Capan-2 in tissue culture and the nude mouse. Cancer research. 1986;46:5810–5815. [PubMed] [Google Scholar]
- Lee C, Kim JS, Waldman T. PTEN gene targeting reveals a radiation-induced size checkpoint in human cancer cells. Cancer research. 2004;64:6906–6914. doi: 10.1158/0008-5472.CAN-04-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bankaitis V. Phosphoinositide phosphatases in cell biology and disease. Progress in lipid research. 2010;49:201–217. doi: 10.1016/j.plipres.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukopoulos P, Kanetaka K, Takamura M, Shibata T, Sakamoto M, Hirohashi S. Orthotopic transplantation models of pancreatic adenocarcinoma derived from cell lines and primary tumors and displaying varying metastatic activity. Pancreas. 2004;29:193–203. doi: 10.1097/00006676-200410000-00004. [DOI] [PubMed] [Google Scholar]
- Milne S, Ivanova P, DeCamp D, Hsueh R, Brown H. A targeted mass spectrometric analysis of phosphatidylinositol phosphate species. Journal of lipid research. 2005;46:1796–1802. doi: 10.1194/jlr.D500010-JLR200. [DOI] [PubMed] [Google Scholar]
- Moore P, Sipos B, Orlandini S, Sorio C, Real F, Lemoine N, Gress T, Bassi C, Klöppel G, Kalthoff H, et al. Genetic profile of 22 pancreatic carcinoma cell lines. Analysis of K-ras, p53, p16 and DPC4/Smad4. Virchows Archiv: an international journal of pathology. 2001;439:798–802. doi: 10.1007/s004280100474. [DOI] [PubMed] [Google Scholar]
- Moss S. How actin gets the PIP. Science signaling. 2012;5 doi: 10.1126/scisignal.2002839. [DOI] [PubMed] [Google Scholar]
- Park JY, Mitrou PN, Keen J, Dahm CC, Gay LJ, Luben RN, McTaggart A, Khaw KT, Ball RY, Arends MJ, et al. Lifestyle factors and p53 mutation patterns in colorectal cancer patients in the EPIC-Norfolk study. Mutagenesis. 2010;25:351–358. doi: 10.1093/mutage/geq012. [DOI] [PubMed] [Google Scholar]
- Patton GM, Fasulo JM, Robins SJ. Separation of phospholipids and individual molecular species of phospholipids by high-performance liquid chromatography. J Lipid Res. 1982;23:190–196. [PubMed] [Google Scholar]
- Santos C, Schulze A. Lipid metabolism in cancer. The FEBS journal. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- Steinberg G, Slaton WH, Jr, Howton DR, Mead JF. Metabolism of essential fatty acids. IV. Incorporation of linoleate into arachidonic acid. J Biol Chem. 1956;220:257–264. [PubMed] [Google Scholar]
- Tuveson D, Hingorani S. Ductal pancreatic cancer in humans and mice. Cold Spring Harbor symposia on quantitative biology. 2005;70:65–72. doi: 10.1101/sqb.2005.70.040. [DOI] [PubMed] [Google Scholar]
- Wakelam MJ. The uses and limitations of the analysis of cellular phosphoinositides by lipidomic and imaging methodologies. Biochim Biophys Acta. 2014;1841:1102–1107. doi: 10.1016/j.bbalip.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Wenk M, Lucast L, Di Paolo G, Romanelli A, Suchy S, Nussbaum R, Cline G, Shulman G, McMurray W, De Camilli P. Phosphoinositide profiling in complex lipid mixtures using electrospray ionization mass spectrometry. Nature biotechnology. 2003;21:813–817. doi: 10.1038/nbt837. [DOI] [PubMed] [Google Scholar]
- Whitman M, Downes C, Keeler M, Keller T, Cantley L. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- Yunis A, Arimura G, Russin D. Human pancreatic carcinoma (MIA PaCa-2) in continuous culture: sensitivity to asparaginase. International journal of cancer Journal international du cancer. 1977;19:128–135. doi: 10.1002/ijc.2910190118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.