Abstract
HIV-1, the agent of the AIDS pandemic, is an RNA virus that reverse transcribes its RNA genome (gRNA) into DNA, shortly after its entry into cells. Within cells, retroviral assembly requires thousands of structural Gag proteins and two copies of gRNA as well as cellular factors, which converge to the plasma membrane in a finely regulated timeline. In this process, the nucleocapsid domain of Gag (GagNC) ensures gRNA selection and packaging into virions. Subsequent budding and virus release require the recruitment of the cellular ESCRT machinery. Interestingly, mutating GagNC results into the release of DNA-containing viruses, by promo-ting reverse transcription (RTion) prior to virus release, through an unknown mechanism. Therefore, we explored the biogenesis of these DNA-containing particles, combining live-cell total internal-reflection fluorescent microscopy, electron microscopy, trans-complementation assays and biochemical characterization of viral particles. Our results reveal that DNA virus production is the consequence of budding defects associated with Gag aggregation at the plasma membrane and deficiency in the recruitment of Tsg101, a key ESCRT-I component. Indeed, targeting Tsg101 to virus assembly sites restores budding, restricts RTion and favors RNA packaging into viruses. Altogether, our results highlight the role of GagNC in the spatiotemporal control of RTion, via an ESCRT-I-dependent mechanism.
INTRODUCTION
In lentiviruses, such as HIV-1, the nucleocapsid protein (NC) encoded by Gag is formed of small basic domains flanking two highly conserved CCHC zinc fingers (ZFs) (Supplementary Figure S1). The multifunctional NC acts throughout the virus replication cycle. During the early stage, fidelity and completion of viral DNA synthesis rely upon multiple interactions between NC, reverse transcriptase (RT) and the viral nucleic acids involved in recombination, consequently fueling the genetic diversity of virus progeny. There is an extensive body of work showing the importance of the NC ZFs in chaperoning the RTion in vitro (reviewed in (1,2)). In vivo the conserved ZFs contribute to RT activity, proviral DNA stability and integration into the host genome (reviewed in (3,4)). However, the precise role of the NC ZFs during RTion has been difficult to evaluate in newly infected cells.
At the late stage of replication, the primary well-established role of NC is to specifically recognize and bind to the genomic RNA (gRNA), which is thought to start the Gag assembly process in infected cells. During these steps, NC is part of the Gag polyprotein (GagNC), which will later be cleaved into freestanding proteins by the viral protease during viral particle maturation. Indeed, HIV buds as immature, non-infectious virions and proteolysis of its main structural component, Gag, and its enzymatic component, GagPol, is required for morphological maturation and infectivity (5). Proteolytic processing starts during assembly and is completed after budding (6,7). Protease activation needs to be coordinated with assembly and budding to prevent a destructive premature digestion of Gag in producer cells. Mistiming assembly can make Gag vulnerable to protease processing before budding can occur, as reported for a Gag mutant with NC deleted (8). Indeed, NC ZFs are important determinants in trafficking, assembly and budding (9,10). Biogenesis of viral particles requires thousands of Gag and hundreds of GagPol precursor proteins (11,12), 8–10 Env trimers (13,14) and two copies of gRNA, as well as some key cellular co-factors that must converge at the assembly sites at the plasma membrane (PM) with finely regulated timing. Virus genesis starts with a short step (<10 min) of assembly of the Gag shell recruiting cytosolic and recently membrane-attached Gag molecules. The events of budding and release from individual assembly sites constitute the major part of the duration (∼25 min) of virion formation (15,16). GagNC appears to be central to the genesis of all retroviruses. HIV-1 Gag contains three other domains in addition to NC: (i) the N-terminal matrix (MA) domain, targeting Gag to the PM, incorporating Env into virions and harboring RNA-binding ability; (ii) the capsid (CA) domain, driving Gag–Gag interactions and (iii) the C-terminal p6 region, recruiting host ESCRT (endosomal sorting complex required for transport) machinery necessary for particle budding and release of particles (17–19). p6 harbors motifs known as late domains. Its primary late domain motif, PTAP, binds the ESCRT-I component Tsg101 (20,21), which in turn recruits the ESCRT-III machinery enabling final pinching off of particles and subsequent recycling of the ESCRT components (reviewed in (18)). Ultimately, Tsg101 is specifically incorporated into HIV-1 particles (22). Additionally, Gag-Tsg101 interaction enhances the recruitment of the host protein tetherin, which could restrict HIV-1 release when viral accessory protein Vpu is absent (23–25). Two accessory late domains were found in Gag p6 (26,27) and in NC ZFs (28,29) both interacting with the ESCRT-associated factor Alix. However, the role of Alix in HIV-1 biogenesis is still unclear since its depletion did not significantly impair HIV egress in transformed epithelial cell lines (30,31).
Recently, several groups have reported that RTion could also occur at the late steps of virus replication (called late RTion), especially when NC is mutated. Surprisingly, mutating GagNC ZF motifs induces viral DNA packaging and the production of DNA-containing viruses, a consequence of RTion activation in producer cells prior to virus release (Supplementary Figure S1) (3,32,33). Deletion of the distal ZF motif (ΔZF2 mutant) had a drastic effect, resulting in the formation of viral particles containing a 100-fold increase in viral DNA content (33). Thus, NC appears to play a key role in the spatiotemporal control of RTion throughout the virus life cycle, by delaying RT activity during late steps within producer cells, by avoiding HIV-1-DNA particle production and by chaperoning the RTion process during early steps in target cells (reviewed in (34)). Interestingly, a late-occurring RTion step is also a property of the hepadnaviruses and foamy retroviruses. In contrast to HIV-1, foamy viruses undergo to a large extent RTion late in their life cycle within the virus producer cell, whereas an early occurring step of RTion is only observable upon virus entry in target cells at low multiplicity of infection (35) (Supplementary Figure S1). When, where and how DNA synthesis takes place in producer cells are therefore key issues that we addressed in this study. We used the HIV-1 ΔZF2 mutant that exhibits strong enhancement of late RTion and leads to the release of DNA-HIV particles (33). Real-time visualization of ΔZF2 DNA-virus formation in live cell was performed using total internal reflection fluorescent microscopy (TIRFM)—a technique well-suited to study events at the PM at the scale of individual virion (15,16,36–38). Combined with electron microscopy (EM) that provides higher resolution images, our experiments revealed ΔZF2 particle aggregation at the PM forming large patches, which correspond to budding defects and abnormal morphology of released particles. We also used in situ Duolink assays, siRNA interference and trans-complementation assays to demonstrate that biogenesis defects are due to abnormal uptake of Tsg101 during ΔZF2 virion formation. Indeed, targeting Tsg101 to the PM restored proper budding and decreased the level of viral DNA in mutant particles and cells. In contrast, Tsg101 downregulation increased viral DNA synthesis in wild-type (WT) HIV producer cells. In addition, pulldown assays revealed an in vitro interaction between NC domain and Tsg101. Taken together, this work provides the first evidence of a connection between late RT activity and HIV biogenesis.
MATERIALS AND METHODS
Additional details of the experimental procedures are provided in the supplemental Text S7.
TIRFM imaging
HeLa cells, cultured in Dulbecco's modified Eagle's medium without phenol red and supplemented with 10% FBS, were transfected 14 h before imaging using FuGene® (accor-ding to the manufacturer's protocol) and two HIV-1 pNL4-3Δenv clones, with untagged and YFP tagged Gag at molar ratio 1:1. For anti-RT treatment, 50 μM zidovudine (AZT) was added 3 h before transfection and maintained until analysis. For analysis of virus-like particle (VLP) formation, cells were transfected for 6 h with an untagged and a YFP-tagged synGag plasmid lacking the pol gene (1:1). For trans-complementation assays, 0.5 μg of a third plasmid, pENXΔp6 (WT or ΔZF2), fused or not fused to Tsg101 (or 0.5 μg of an empty vector as control), was co-transfected. Analysis by TIRF microscopy is described in Text S7.
Analysis of Tsg101-Gag interaction in cells
To study Tsg101-Gag recruitment by colocalization, HeLa cells were transfected with pCR3.1cherry-Tsg101 (0.2 μg) and pNL4.3MA-YFP WT, ΔZF2 or L− with its respective untagged counterpart pNL4.3Δenv at a molar ratio of 0.2:1:1. Cells were fixed 20 h post-transfection in 4% paraformaldehyde/phosphate buffered saline (PBS) for 20 min at room temperature, washed three times in PBS and imaged with a confocal Leica SP5-SMD microscope equipped with a 63x/1.4 oil immersion objective and Argon-488-nm/DSSP 561-nm lasers. For comparative purposes, exposure times were set equal for all the conditions tested. In addition, Gag–Tsg101 interactions were visualized in situ by using the Duolink technique that reveals whether two proteins might interact in the cell (39). HeLa cells were transfected 20 h with cherry-Tsg101 and untagged pNL4-3Δenv WT, ΔZF2 or L- (at the molar ratio 0.2:1). Total Gag and Tsg101 expression in cells was monitored by western blotting (Text S7). Duolink signals were visualized at the PM using TIRFM and fluorescent spots quantified using Image J software. Duolink assays are described in more detail in Text S7.
Electron microscopy
293T cells were co-transfected with HIV-1 molecular clones (pNL4-3) and pENX vectors for trans-complementation assays and collected 24 h post-transfection for EM. Cells were fixed with 2.5% glutaraldehyde in 0.1-M phosphate buffer for 1 h at 4°C and were post-fixed with 1% osmium tetroxide (Electron Microscopy Sciences Inc.) for 1 h at 4°C, and 0.5% tanic acid (Merck) for 30 min at 4°C. Dehydration was obtained with a graded series of ethanol solutions (from 70 to 100%) for 30 min, before embedding in resin at 60°C for 24 h (Embed-812, Electron Microscopy Sciences Inc.). Ultrathin sections were cut with a Reichert Ultracut microtome (Leica) and stained with 2% uranyl acetate for 20 min at room temperature and lead citrate for 4 min at room temperature. Samples were observed with a Hitachi H7100 transmission electron microscope
Analysis of viral production
293T cells were co-transfected with 2 μg of pNL4-3 plasmid and 0.5 μg of pENX plasmids by the calcium phosphate precipitation method in a 6-well plate. After 48 h post-transfection, the culture media was ultracentrifuged to concentrate virions in order to monitor the virus production by using the p24 ELISA Innotest HIV kit (Innogenetics) according to the manufacturer's protocol. For analysis of Tsg101 recruitment into virions, cells were transfected with 8 μg of complete molecular clones (pNL4-3 WT or ΔZF2) in a T75 flask. Virions were purified, quantitated by ELISA and protein content analyzed by western blotting (Text S7).
Transfection, siRNA and quantitative RT-PCR and PCR
293T cells were cotransfected with pNL4-3Δenv (WT or ΔZF2) and pNL4-3MA-YFP-Δenv (WT or ΔZF2) plasmids (4 μg of each plasmid) and complemented with pENX plasmid (2 μg) by the calcium phosphate precipitation method in a T75 flask. The supernatant was harvested 20 h after transfection and virions were purified by ultracentrifugation through a 20% sucrose cushion to extract viral RNA and DNA. DNA was extracted from cells with DNAzol (MRC). Viral RNAs were reverse transcribed using an oligo(dT) primer with the Expand RT (Roche). A control experiment was systematically performed without RT to control the absence of DNA contamination. Full-length RNA (FL) and multispliced cDNA forms (MS) corresponding to the RTion of the spliced viral RNAs were amplified with the SYBR Green kit (Roche) using the RotorGene (Labgene) system. A standard curve was generated from 103 to 106 copies of pNL4-3 plasmid. The amount of viral MS-DNA was normalized to the GAPDH DNA level. In RNA interference experiments, 293T cells were cotransfected with Tsg101 siRNA (Tsg101-6Flexitube siRNA, Qiagen) or negative control siRNA duplex (Eurogentec) and the pNL4-3Δenv plasmid (2 μg) by lipofectamine in a 6-well plate. Cell lysates were harvested after 48 h. Gag and endogenous Tsg101 levels were analyzed by western blotting (Text S7) and DNA was extracted from cells and analyzed by qPCR as described above.
GST pulldown assay
Interactions of glutathione S-transferase (GST) fusion proteins with Tsg101 were examined in GST pulldown assays by following the protocol described in (40). Briefly, the pGEX vector empty or carrying the coding sequences of HIV-1 p15NCp1p6, p9NCp1, p9NCΔZF2p1, p7NC or p6 was transformed and expressed in BL21pLysS E. coli (Stratagene) and the GST fusion proteins immobilized on glutathione-Sepharose beads (GE Healthcare). The beads were then incubated for 4 h at 4°C with lysates of 293T cells transiently expressing non-coding gRNA (pNL4-3GagKO: see Text S7), followed by an extensive washing. Bound proteins were eluted by boiling and analyzed by SDS-PAGE. Tsg101 was detected by western blotting with an anti-Tsg101 antibody (Santa Cruz) and GST fusion proteins were visualized with Coomassie blue staining.
RESULTS
Deletion of NC ZF2 induces the formation of large Gag assemblies at the PM of living cells
Deletion of either one or two highly conserved ZF motifs of HIV-1 GagNC induced activation of a late-occurring RTion step and release of abnormal viruses with high viral DNA content (Figure S1) (reviewed in (34)). The most dramatic effect was obtained by deleting the distal ZF2 motif producing 100% particles containing viral DNA species. Therefore, in this study, we focus on this HIV-1 mutant (ΔZF2), in order to investigate a potential link between late RTion activity and virus assembly defects. Indeed, a delayed virus assembly could give enough time to complete DNA synthesis prior to virus egress. We then studied the dynamics of the biogenesis of these DNA-containing particles using live-cell imaging TIRFM to monitor the formation of individual virions at the PM, from the assembly of the Gag shell to budding and virus release.
We first investigated the effect of ZF2 deletion on the initial steps in this process, from the recruitment of Gag at the PM to the nucleation of the assembly sites. To generate fluorescently labeled HIV-1 particles, an HIV molecular clone with a deficient env gene (pNL4-3Δenv) was co-expressed with a fluorescent version (pNL4-3MA-YFP-Δenv) at a molecular ratio of 1:1 (Figure 1A and B) (41). The NC ZF2 sequence was deleted in these two constructs. To determine the kinetics of ΔZF2 HIV assembly, we monitored in real time the fluorescence intensity of puncta generated by Gag accumulation at the PM. Fluorescence increased steadily as assembly proceeded until it reached a plateau indicating the end of the assembly process (15,16). Our results showed that both ΔZF2 and WT viruses required an average of ∼8 min to complete assembly of the Gag shell, with total assembly times of 8.3 ± 0.4 min and 8.1 ± 0.3 min, respectively. This indicates that the ZF2 deletion did not change the rate of Gag accumulation at the PM (Figure S2) and did not alter the early assembly step. We also investigated the dynamics of Gag at the PM by tracking hundreds of ΔZF2 puncta at the PM. Our results revealed that the ZF2 deletion reduced the mobility of Gag clusters, suggesting defects in events later than the assembly step (Figure S3). Indeed, as previously reported for the well-characterized Gag p6 mutant (called L−; Figure 1A) that blocks the pinching off of particles from the PM (42), Gag assembly could actually be achieved with normal kinetics but associated with defective virus budding and release (15). We therefore examined whether the ZF2 deletion could affect these two later steps. We observed the formation of large fluorescent patches at the PM 14 h post-transfection with the ΔZF2 mutant, while a punctuated pattern was observed with WT particles. Sizes of TIRFM signals were not increased when cells were transfected with higher amounts of WT HIV (4 or 6 μg), excluding that Gag overexpression could contribute to larger patches (data not shown). The area of individual fluorescent patches was measured in live cells producing WT, L− or ΔZF2 viruses (Figure 1C). The scatter plots clearly showed that the distribution of fluorescent areas for L- and ΔZF2 was significantly different from that observed with the WT (P < 0.001, as determined by the Mann–Whitney U-test) (Figure 1D). The mean ± SEM values of the fluorescent areas were 2 ± 0.1 and 2.45 ± 0.1 μm2 for L− and ΔZF2, respectively, both values higher than that measured for WT (1.44 ± 0.07 μm2) (Figure 1D). However, the shapes of the ΔZF2 patches slightly differed from those of L− which are more filamentous as observed in the pictures. Analysis of the movies indicates that fluorescent ΔZF2 areas grew by clustering of small puncta at the PM (Figure S4, upper panel). In addition, we observed the coalescence of individual patches forming larger domains (Figure S4, lower panel). These enlarged patches containing ΔZF2 Gag could correspond to membrane platforms due to budding defects and/or to viruses retained at the membrane.
Figure 1.
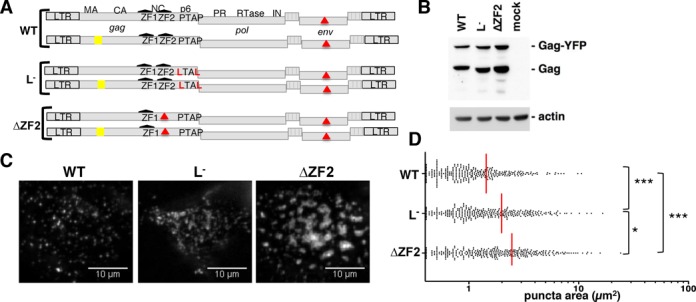
Study of ΔZF2 HIV-1 biogenesis in living cells by TIRFM. (A) Series of pairs of HIV-1 vectors (untagged/tagged-YFP) expressed in HeLa cells to visualize biogenesis of WT and mutant HIV-1 at the cell surface. YFP was incorporated into the stalk region of MA as previously described (41) (Text S7). The two ZFs of the NC are depicted in black and mutations are in red. (B) Anti-CA western blotting analysis of total Gag and Gag-YFP in cells used in TIRFM experiments (Text S7). (C) Representative TIRFM images showing Gag expression at the surface of live cells captured 14 h after transfection. For each virus, at least 50 cells with >50 events/cell were analyzed from three independent transfections. (D) Scatter plot of the area of 300 individual fluorescent puncta randomly selected. Red line indicates the mean value of the area. *P < 0.1; ***P < 0.001, as determined by the Mann–Whitney U-test.
Altogether, these results indicated a role for GagNC ZF2 in Gag behavior at the PM. GagNC via its ZF2 motif could endorse late-domain function, since similar defects were observed between ΔZF2 and p6-defective (L-) mutants in the later steps of virus genesis.
Large patches of ΔZF2 Gag at the PM are not caused by late RTion activation
We next attempted to determine the sequential order of the GagNC functions in virus assembly and late RTion. We examined in particular whether the defect in virion biogenesis of ΔZF2 HIV is a consequence or a cause of the late RTion activation. If the late RTion takes place before virion biogenesis, it is possible that the packaging of viral DNA instead of gRNA could disturb the overall assembly process. Conversely, defective budding characterized by slow kinetics of particle release might allow time for RTion to take place prior to release. To study the link between these two hypotheses, we analyzed by TIRFM the biogenesis of ΔZF2 HIV in producer cells treated with AZT, a well-characterized inhibitor of RT. Although AZT inhibits late RTion activity in ΔZF2 producer cells (33), the pattern of ΔZF2 viruses at the PM remained unchanged (mean value = 2.5 ± 0.1 μm2) (Figure 2). Moreover, TIRFM analyses were also performed with Gag alone (without enzymatic Pol proteins) in the ΔZF2 or WT version to produce VLPs. Similar to entire viruses, cherry-tagged and untagged Gag plasmids were co-transfected (ratio 1:1) and under these conditions, Gag proteins assembled with areas of 2.45 ± 0.1 and 1.5 ± 0.08 μm2 for ΔZF2 and WT VLP, respectively (Figure 2). These results revealed that patches formed with ΔZF2 were insensitive to the RT inhibitor and to the presence of Pol proteins. Therefore, patch formation is not the consequence of the late RTion enhancement but rather the cause of late RTion activation. Indeed, these assembly defects could impact on proteolytic maturation of Gag and GagPol precursors and consequently generate premature RT activity. The first evidence of a mistimed processing has already been reported, with the accumulation of Gag intermediates (p49, p41 or p25) in cells producing ΔZF2 Gag (9). Similarly, anti-RT western blotting was performed to analyze GagPol processing (Figure S5). While the anti-RT pattern was unchanged in WT and ΔZF2 virus particles, with similar p66:p51 ratios, ΔZF2-producing cells harbored an accumulation of GagPol (p113p6/PR/RT/IN) intermediates as well as a slight increase of mature RT proteins (p66/p51) (Figure S5). Whether these defects, in particular the presence of small amounts of mature RT in the cell, are sufficient to achieve late RTion remains to be demonstrated.
Figure 2.
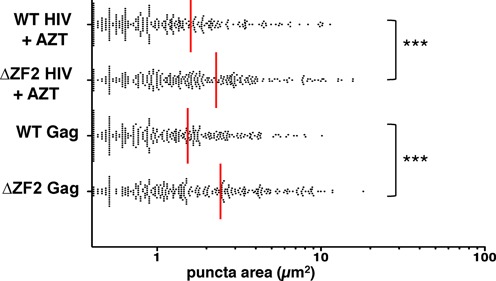
Budding defects of ΔZF2 HIV are not the consequence of late RTion activation. Budding events were studied by TIRFM and analyzed as performed in Figure 1, in the absence of late RT activity. Biogenesis of WT and ΔZF2 viruses in cells was indeed measured in the presence of AZT. For VLP formed by Gag alone (without Pol proteins), cells were transfected 6 h with Gag plasmids. ***P < 0.001, as determined by the Mann–Whitney U-test.
ZF2 deletion impairs the recruitment of Tsg101 at the PM and its incorporation into virions
The late-domain of p6 is required to recruit the cellular ESCRT machinery that is crucial for virus release (15). Gagp6 contains a PTAP motif that recruits Tsg101, a key subunit of the ESCRT-I complex during virus budding (42). The LTAL substitution of the L− variant prevents Tsg101 interaction, blocking the recruitment of the ESCRT-I components at the PM as well as the assembly of ESCRT-III complexes (20,21). To determine whether the GagNC also contributes to Tsg101 recruitment at the PM, we examined the colocalization of Tsg101 and Gag at the sites of HIV assembly. HeLa cells were co-transfected with untagged and YFP-tagged HIV with a mCherry-Tsg101 construct. HIV Gag and Tsg101 were imaged by confocal microscopy (Figure 3A). As expected, WT HIV efficiently recruited Tsg101 at the PM as shown by the colocalization of the two signals, while L− did not (43,44). Interestingly, ΔZF2 HIV weakly colocalized with Tsg101 at the PM, while the capacity of ΔZF2Gag to reach the PM was not impaired (Figure 3A). These data indicated that ZF2 deletion could impair HIV-1 late assembly events through defects in Tsg101 recruitment. To provide further evidences of GagNC-Tsg101 interplay in cells, the Duolink technique, which is based on a proximity ligation assay, was carried out (39). A positive fluorescent signal in Duolink experiments demonstrated the spatial proximity between two proteins (<40 nm). Duolink was performed on HeLa cells co-transfected with Tsg101 and HIV (WT, ΔZF2 or L−) and examined with TIRFM (Figure 3B). The level of expression of Gag and Tsg101 was monitored by western blotting (Figure 3C). Clear positive signals were observed at the PM when Tsg101 was cotransfected with WT HIV, revealing many Gag-Tsg101 interactions in cells, corroborating previous reports (20,45). Interestingly, the quantitative analysis revealed a significant reduced number of positive signals with ΔZF2 and L− (Figure 3B), highlighting the deficiency for ΔZF2 mutant in interacting with Tsg101. As a consequence of this defect, a lower amount of endogenous Tsg101 was found associated with ΔZF2 viruses released from cells transfected with pNL4-3ΔZF2 as compared to WT particles (Figure 3D). All together, these results revealed the role of the NC's ZF in the recruitment of Tsg101 at the virus assembly sites and ultimately inside viral particles. Until now, this role was only ascribed to the PTAP motif of Gag p6 domain.
Figure 3.
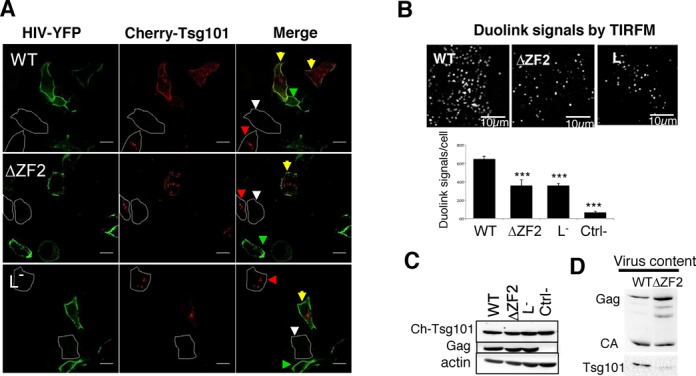
Recruitment of Tsg101 to HIV assembly sites and its incorporation into virions. (A) Representative confocal images of HeLa cells expressing mCherry-Tsg101 and untagged/tagged-YFP HIV (ratio 0.2:1:1). Yellow arrows show cells co-transfected with HIV and Tsg101, while green and red arrowheads show cells transfected only with HIV or Tsg101, respectively. White arrowheads indicate non-transfected cells. (B) Insitu Duolink experiments. Duolink immunolabeling generates fluorescent signals when Gag and Tsg101 proteins are in close proximity (≤40 nm). HeLa cells were cotransfected with cherry-Tsg101 and pNL4-3Δenv (WT, ΔZF2 or L−) for 20 h. A similar experiment was conducted without the anti-Tsg101 primary antibody as a negative control. Cells were imaged by TIRFM and the number of fluorescent Duolink spots per cell was measured using ImageJ. The histograms represent the average ± SD of the mean values of the Duolink signal per cell calculated from two independent experiments (n ≥ 27 cells) (more details in Text S7). ***P < 0.0001 using the Student's t-test. (C) Expression of Tsg101 and Gag proteins in cells used in Duolink assays was monitored by western blotting. (D) Incorporation of Tsg101 into virions. 293T cells were transfected with pNL4-3 (WT or ΔZF2) during 48 h. The production of virions was quantitated using p24 Elisa and normalized amounts of virions were loaded onto SDS-PAGE. Tsg101 and Gag proteins were detected by western blotting (Text S7).
Tsg101 restores an accurate pattern of ΔZF2 particles at the cell surface and increases virus production
To directly address the question of whether the GagNC ZF2 performs the PTAP-like function, we took advantage of trans-complementation assays initially developed by Bieniasz's team to show the role of the PTAP motif in Tsg101 recruitment (44). These assays are based on a proviral DNA mutant lacking p6 and pol genes (ENXΔp6) and generating a chimeric GagΔp6 protein (Gagc) that retains multimerization and membrane anchorage abilities but lacks p6 function. When fused to Tsg101 and co-expressed with the L− variant, the chimeric Gagc-Tsg101 was able to rescue the release of the L− virus (44). Here, we used a similar approach to investigate whether Tsg101 can restore accurate sizes of the GagΔZF2 assemblies at the PM using TIRFM (Figure 4A). First, the ENXΔp6 plasmid was deleted from its ZF2 sequence to produce a chimeric ΔZF2Gagc protein prior to its fusion to Tsg101 (ΔZF2Gagc-Tsg101). The two constructs, YFP-tagged (pNL4-3-MA-YFPΔZF2) and untagged (pNL4-3ΔZF2) used to observe ΔZF2 HIV-1 genesis (Figure 1A), were co-transfected into HeLa cells with either ΔZF2Gagc as a control or ΔZF2 Gagc-Tsg101 (Figure 4A). ΔZF2 virus biogenesis was then analyzed by TIRFM (Figure 4B) as described above. As expected, ΔZF2Gagc had no effect on ΔZF2 assembly as indicated by the presence of large patches at the PM (mean size = 2.7 ± 0.2 μm2). Similarly, co-expression of Gagc had no effect on WT assembly (wtHIV + wtGagc). Interestingly, the mean value of the surface of fluorescent areas when ΔZF2 virus assembled in the presence of the ΔZF2Gagc-Tsg101 was similar to the WT one (1.4 ± 0.06 μm2). Then, to further analyze the rescue of ΔZF2 genesis by Tsg101, we quantified the virus release during the trans-complementation assays. To do so, viruses were concentrated by ultracentrifugation of the culture supernatants and serially diluted for quantitation by an HIV-1 capsid (p24) ELISA assay (Figure 4C). As expected, deletion of ZF2 led to a 5-fold decrease in viral production compared to the WT. Interestingly, co-expression of chimeric ΔZF2Gagc-Tsg101 also increased up to 3-fold the production of ΔZF2 viruses, but without reaching the WT HIV levels. Of note virus production could not be entirely restored, likely because the presence of large Gag-Tsg101 proteins in capsids could generate unstable virions in the medium. This is supported by the decrease in WT HIV production induced by the co-expression of wtGagc-Tsg101 (Figure 4C), whereas no effect was observed on the shapes of fluorescent areas (Figure 4B).
Figure 4.
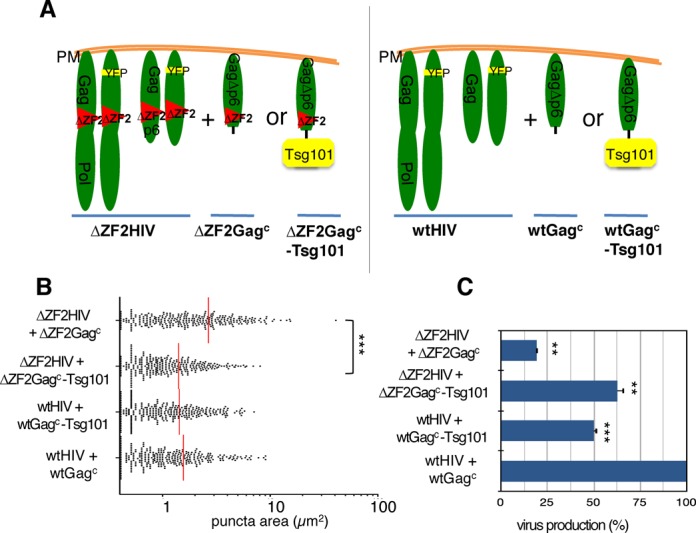
Late assembly of ΔZF2 HIV-1 and viral production were trans-complemented by targeting Tsg101 to the PM (A) trans-complementation strategy. A chimeric ΔZF2Gagc protein was used to target Tsg101 to the PM (ΔZF2Gagc-Tsg101) and to restore ΔZF2 virus biogenesis (left panel). Similar experiments were conducted with WT proteins (right panel). A protocol similar to the one used in Figure 1 was used to transfect HeLa cells with the two pairs of proviral plasmids and trans-complementation was performed with a third pENX vector series encoding chimeric Gagc proteins deleted of p6 (Text S7). (B) Cells were observed by TIRFM 14 h post-transfection and the area of a thousand individual virus puncta was determined. Data are presented as a scatter plot of the areas of individual puncta. ***P < 0.001 using the Mann–Whitney U-test. (C) Virus production in trans-complementation experiments. Virus-containing supernatants of transfected cells were collected and the amounts of viruses were determined by p24 ELISA. Values were normalized to the level of WT particles measured in the context of trans-complementation (wtHIV + wtGagc). **P < 0.01 using the Student's t-test.
In conclusion, these results provided evidence for the requirement of Tsg101 at assembly sites in order to rescue ΔZF2 virus biogenesis. These data strongly support a role for GagNC in the recruitment of Tsg101 at assembly sites for a proper budding, since ESCRT-I components are required for budding events.
EM reveals defects in late budding events of ΔZF2 HIV-1
To visualize the budding and release mechanisms of ΔZF2 DNA-HIV at the PM at a higher resolution, producer cells were imaged by EM. Many budding events were observed with WT HIV even if many viruses displayed immature capsid morphologies. Only a few mature capsid structures with typical conical cores were observed, probably due to the short time after transfection (24 h) (Figure 5A). Cells producing ΔZF2 HIV showed aberrant budding structures, with sticky and large particles bound to each other at the PM, and the immature particles harbored aberrant and heterogeneous capsid structures (Figure 5B). No mature particles were observed with ΔZF2 HIV. Interestingly, co-expression of ΔZF2Gagc-Tsg101 restored proper budding structures and many individual immature particles were observed in the extracellular space (Figure 5C). In conclusion, the large fluorescent areas imaged by TIRFM most likely corresponded to defective budding events and to bursts of large ΔZF2 viruses remaining bound to the PM. In addition, the trans-complementation by Tsg101 confirmed the late-domain function endorsed by the ZF2 motif in GagNC.
Figure 5.
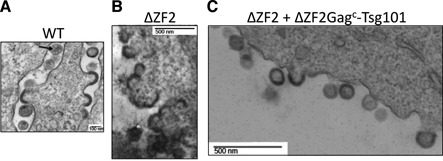
Analysis of the late stages of HIV genesis during trans-complementation experiments. 293T cells expressing pNL4-3 WT (A), pNL4-3 ΔZF2 (B) and ΔZF2 + ΔZF2Gagc-Tsg101 (C) were imaged by electron microscopy (EM). (A) shows WT HIV budding events of immature and mature (arrow) particles. ΔZF2 HIV clearly exhibits budding defects with ‘sticky’ particles with an abnormal morphology (B). The co-expression of ΔZF2 with Tsg101 restores accurate budding with the release of many immature particles (C).
HIV-1 NC controls late RTion activity via its late-domain function to produce RNA viruses
In view of the positive trans-complementing effects of Tsg101 in ΔZF2 virus biogenesis and release, we could not exclude that Tsg101 significantly influenced the late RTion and then the virion content of the ΔZF2 HIV-1. Because ΔZF2 HIV-1 packages high levels of viral DNA instead of gRNA, the nucleic acid content of viral particles produced during the trans-complementation assay was monitored by real-time RT-PCR and PCR to determine viral RNA and DNA contents, as previously described (33) (Figure 6A). Viral spliced cDNAs were found to represent an ideal marker for HIV-1 DNA quantitation due to the lack of plasmid DNA contamination (33) and because viral spliced RNAs are reverse transcribed with similar efficiencies as gRNA (46). A 2-log increase in the amount of the multi-spliced cDNA forms (MS) was observed for the ΔZF2 particles (33). Similarly, in the context of trans-complementation, high levels of intravirion DNA combined with a decrease of gRNA content were found for the ΔZF2 HIV produced in the presence of chimeric ΔZF2Gagc (Figure 6A). When ΔZF2 virions were co-assembled with ΔZF2Gagc -Tsg101, the DNA content of the trans-complemented virions decreased while the gRNA level increased. Deleting the ZF2 sequence could also generate a cis-effect at the RNA level by inactivating the RNA packaging enhancer signal (named GRPE), which overlaps with the NC ZF2 sequence (47), although this role remains controversial (48). Thus, in this case the ΔZF2 gRNA molecules could not be packaged as efficiently as the WT gRNA and this could explain why a complete rescue of ΔZF2 was not achieved by Tsg101. Furthermore, we also examined whether the reduction in the amount of DNA in ΔZF2 particles induced by Tsg101 could result from a decreased activity of the late RTion in producer cells. After cell lysis, DNA samples were normalized to the cellular GAPDH gene copy number determined by specific qPCR and viral DNA levels were determined as previously described (33) (Figure 6B). When expressed in trans, the chimeric ΔZF2Gagc-Tsg101 strongly reduced the DNA content in ΔZF2 producing cells, revealing that proper virus formation prevents late RTion activation and DNA packaging. This supports the notion that late RTion activation is a consequence of defects in late-budding steps.
Figure 6.
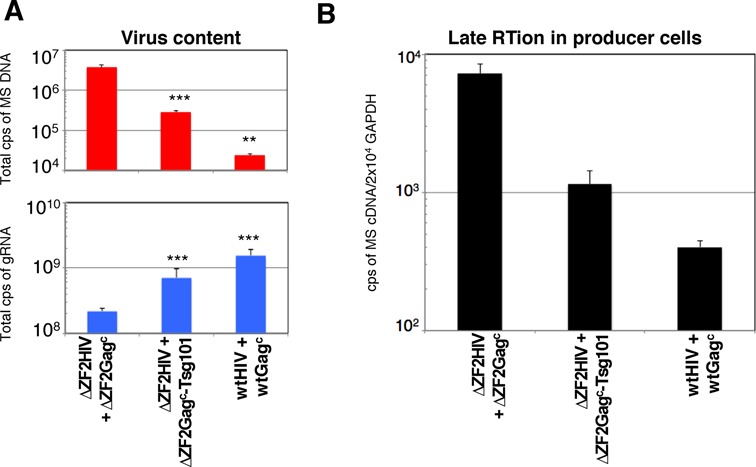
Tsg101 strengthens the control of late RTion in the producer cells. (A) Effect of Tsg101 on nucleic acid content of virions. Virus-containing supernatant produced in the trans-complementation experiments were collected and the viral nucleic acid content was monitored by RT-qPCR and qPCR. Results are presented as the number of DNA (top) or RNA (bottom) copies measured in the same sample of total virus produced. The virus populations from at least three independent transfections were analyzed and values plotted using a at log10 scale and displayed as mean ± SEM. ***P < 0.001. (B) Quantitation of the late RTion activity in producer cells. Amount of viral MS cDNA produced by late RTion in producer cells was determined by qPCR as in (A). The number of DNA copies was normalized to cellular GAPDH gene and displayed as mean ± SEM.
In order to further investigate the link between virion biogenesis and late RTion with infectious WT HIV-1, we used siRNA interference to analyze late RTion activation of WT HIV when Tsg101 expression was downregulated. Activity of the siRNA was first controlled by following endogenous Tsg101 levels in cells transfected for 48 h with Tsg101 siRNA versus negative-control siRNA. Transfection of 50-nM Tsg101 siRNA reduced by ∼60% the expression level of endogenous Tsg101 (Figure 7A). Then, cells were cotransfected with siRNA and pNL4-3Δenv for 48 h and late RTion activities determined as before. Interestingly, the late RT activity of WT HIV was specifically increased by siRNA-mediated knockdown of Tsg101 expression (Figure 7B), showing that Tsg101 prevents the HIV from reverse transcribing its genome into DNA prior to virus release.
Figure 7.
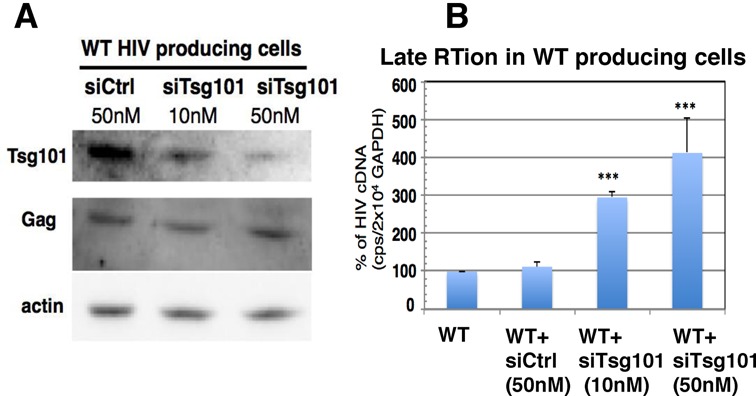
Tsg101 depletion activates late RTion in WT HIV producing cells. (A) Downregulation of Tsg101 expression by siRNA. 293T cells were cotransfected for 48 h with pNL4-3Δenv and 10 or 50 nM siRNA targeting or not Tsg101. Cell lysates were analyzed by anti-Tsg101 and anti-p24 western blots. (B) siRNA-mediated Tsg101 downregulation activated late RTion in cells producing WT HIV. Cells were similarly transfected and late RTion activity was determined as described in Figure 6 and compared to late RTion activity measured in WT HIV producer cells without siRNA. ***P < 0.001.
Furthermore, in order to investigate whether blocking virus biogenesis at a different stage could also activate late RTion, we analyzed the effects of two critical mutations (residues WM184/185 substituted by AA) in the Gag capsid domain (CA): these residues are essential for infectivity, viral assembly and maturation (for review (49)). More precisely, these two mutations weaken Gag–Gag interactions (50), slow the assembly process (51) and form immature capsids with defects within the spherical shell (52). As expected, the producer cells accumulated some Gag intermediates (p41MACAp2 and p25CAp2), revealing a mist-iming of Gag processing, and no viruses were released in the medium (Figure S6A). Duolink assays combined with TIRFM showed that the Gag mutant (pBruWM184/185) reached the PM and could efficiently recruit Tsg101 as the WT HIV did (Figure S6-B). Interestingly, these mutations did not activate the late RTion in producer cells (Figure S6-C), indicating that defects in assembly and maturation did not necessarily lead to the activation of late RTion. Taken together, these results support the idea that TSG101 is recruited to the assembly sites and can regulate RT activity in cell.
NC contributes to Gag–Tsg101 interaction in vitro
The interaction between the Gag p6 region and Tsg101 and its role in virus budding and release have been clearly established (reviewed in (18)). However, the defect in Tsg101 packaging into ΔZF2 particles (Figure 3D) suggested that Tsg101 could also bind other Gag regions. We thus performed an in vitro pulldown assay to examine whether the NC domain of Gag could also interact with Tsg101. The NC domain (the principal nucleic acid-binding domain) requires RNA to interact with Alix (an ESCRT-associated protein) during virus budding since the absence of RNA abrogated binding of GST-p15NCp1p6 to Alix (40). Thus, we tested NC-Tsg101 interaction in the presence of RNA. In addition, during Gag processing, the NC sequence is found within p15NCp1p6, p9NCp1 and mature p7NC proteins. All these proteins, as well as p6, were then expressed in bacteria as GST-fusion proteins, bound to glutathione-sepharose beads and incubated with extracts from 293T cells expressing non-coding HIV gRNA. Tsg101, naturally expressed in 293T, was detected associated with all the NC forms (GST- p7NC, p9NCp1, p15NCp1p6) but not with the GST alone (Figure 8A). In addition, a similar assay performed with an NCp1 fusion protein deleted from its ZF2 motif (GST- p9NCΔZF2p1) showed the loss of the Tsg101 interaction (Figure 8B), suggesting that NC provides a Tsg101 binding site. In the case of the p15NCp1p6 protein, two shorter bands were reproducibly observed after Coomassie staining, likely corresponding to proteolysis of its C-terminus. Compared with GST-p6, the p15NCp1p6 protein appeared to bind Tsg101 with higher efficiency, indicating that the Gag–Tsg101 interaction is optimal in the presence of both NC and p6 domains (Figure 8C).
Figure 8.
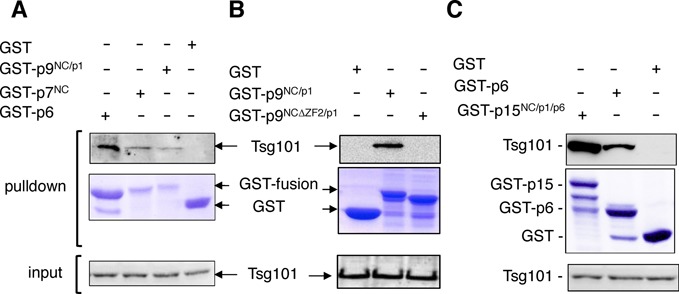
NC domain binds Tsg101 in vitro. GST, GST-p6, GST-p7NC, GST-p9NCp1, GST- p9NCΔZF2p1 or GST-p15NCp1p6 fusion protein was expressed in E. coli, captured on glutathione-conjugated beads, and subsequently incubated with lysates from 293T cells naturally expressing Tsg101 but transfected to produce non-coding HIV gRNA. Captured proteins and cell lysates were analyzed by SDS-PAGE and western blotting using an anti-Tsg101 antibody. GST fusion proteins in pulldown samples were visualized by Coomassie blue staining. (A) Tsg101 interaction with p7 or p9 fusion proteins. (B) Effect of the ZF2 deletion in the p9-Tsg101 interaction. (C) Comparative analysis of p15 and p6 ability to bind Tsg101.
DISCUSSION
Our results reveal a tight connection between virus assembly and late RTion, two events that require the highly conserved ZF2 motif of NC. In agreement with the fact that ZF2 is less critical than ZF1 for the nucleic acid chaperone activity of NC and for gRNA packaging (53,54), deleting ZF2 does not abolish or change RTion efficiency and fidelity, but rather modifies RTion localization and timing during the virus life cycle (33).
In infected cells, early after infection, RTion occurs in the microenvironment of disassembled capsids in the cytoplasm (high dNTP concentration, core condensation) and is intimately linked to uncoating, the process through which the viral core is reorganized all along its trafficking toward the nucleus (2). Recent data on HIV-1 suggest that uncoating and RTion may influence each other and that proper uncoating is required for efficient RTion (55). It is also thought that RTion depends on correct capsid assembly and maturation that occur before infection (56). The precise timing of late RTion in ΔZF2-producing cells is not known yet. Late RTion has been reported to be completed in producer cells before virion release, as addition of an RT inhibitor blocks RTion in producer cells but not in purified virions (3,33). The present study strongly indicates that late RTion starts shortly before or concomitantly to virus formation. The efficient and complete RTion performed by ΔZF2 HIV during its genesis supports the notion that, like the disassembly step, the virus assembly provides a microenvironment conducive to RTion at a particular stage. In contrast to the early RTion process, a correct formation precludes late RTion since the rescue of the ΔZF2 GagNC late-domain function was accompanied by the recovery of late RTion control.
Late- and early-occurring RTion differ in many aspects and, importantly, the players present in the RTion complexes (RTC) are in different maturation states. In contrast to standard RTC in early stages, late RTion can rely on at least three molecular players, Gag, GagPol and gRNA that are not mature or are incompletely matured. During assembly, the gRNA has not yet been fully matured. Its maturation consists of a thermodynamically stable RNA–RNA association likely promoted by initial annealing with the tRNA primer (reviewed in (57)). Proteolytic processing of the Gag and GagPol precursors starts during assembly and is completed after budding (6,7). Thus, the spatiotemporal coordination between protease activation, assembly and budding is critical. For instance, slowed assembly caused by an almost complete deletion of NC (ΔNC or NX) induces Gag cleavage by protease before effective particle formation (8). Interestingly, this phenotype was not obtained with more subtle NC mutations. Substitutions in the ZFs (H33C or H44C) did not accelerate proteolytic processing of Gag in the cell. In addition, blocking processing in these NC mutants (in combination with the PR mutation) did not entirely eliminate the production of DNA-containing particles (3). The authors concluded that extensive late RTion exhibited by these ZF mutants was not due to a faster Gag and/or GagPol processing (3). In the case of the ΔZF2 mutant, the role of the accumulation of Gag and GagPol intermediates in late RTion, the consequence of a mistimed processing, remains to be demonstrated and will require a better understanding of ΔZF2 maturation kinetics. It is conceivable that the small increase of mature RT in the producer cell is sufficient to achieve late RTion. It is unlikely but not totally excluded that GagPol precursor might confer late RTion even if the mechanism is not clearly established yet (3,58–60). Finally, late RTion in the presence of Gag precursor also appears unlikely, since Gag precursor, unlike mature NC, could inhibit DNA elongation by the RT through a ‘roadblock’ mechanism (61). However, slowed assembly and premature processing, as exhibited by the well-characterized CA mutant (WM184/185) (49), are not sufficient to activate late RTion. Despite severe defects in virus production, WM184/185 Gag is still able to reach the PM, to recruit the ESCRT-I subunit Tsg101 and to prevent late RTion, suggesting a functional link between the control of late RTion and the Tsg101 protein recruitment.
TIRF imaging and EM analysis of ΔZF2 virus biogenesis within producer cells revealed no alteration of early Gag nucleation process, but severe budding defects of DNA-HIV particles. Therefore ΔZF2 Gag forms large patches at the PM with reduced mobility. Once again, the phenotype appeared quite different with complete deletion of the NC domain which inhibited Gag multimerization and impeded Gag coalescence at the PM (62). Since budding defects of ΔZF2 HIV still persisted with Gag alone or when RT was inactive, we can assert that budding defects are not a consequence of late RTion but rather its cause. Clearly, the budding defect is detrimental to the late RTion control, as confirmed by our Tsg101-siRNA interference assays. Furthermore, we provided original evidence using in vitro and in vivo approaches that Tsg101 is required for proper budding and release of ΔZF2 particles and induces packaging of RNA instead of DNA. The increase in the size of Gag patches due to the ZF2 deletion and counterbalanced by Tsg101 trans-complementation suggests a requirement for Tsg101 to limit the growth of Gag/GagPol assembly and to switch to budding. Because ΔZF2 budding defects were associated with a low expression ofTsg101 at the PM and within virions, we focused on the role of NC in the Gag–Tsg101 interaction. GST-pulldown assays showed for the first time that the NC domain could interact in vitro with Tsg101 in the presence of gRNA. As reported for NC–Alix interactions (40), the negatively charged RNA could play a critical role by bridging interactions between NC and Tsg101. Gag–Tsg101 interactions were enhanced in the presence of both NC and p6 domains, indicating that NC contributes together with p6 to Tsg101 recruitment at the HIV assembly sites. These results correlate with a previous study (20) showing that Tsg101 immunoprecipitated Gag precursor as well as the NCp1p6 and NC forms in extracts of cells transfected with Gag. The NC–Tsg101 interactions reinforce the specific role of Tsg101 in late RTion regulation, different from other ESCRT members acting downstream of Tsg101. This novel interaction between NC and Tsg101 is in addition to the one described between NC and Alix (10,28). Thus, NC could supplement the Tsg101-binding PTAP and Alix-binding LYPXnL domains of p6. It will be interesting to determine the importance of Tsg101 dual interactions between NC and p6 in HIV-1 replication.
Altogether, our data support a model bringing together the two functions of GagNC in virus budding through Tsg101 recruitment and in the control of late RTion. The requirement for Tsg101 recruitment at the assembly sites inhibiting late RTion could arise from at least two non-exclusive mechanisms: (i) Tsg101 interacting with Gag could prevent the formation of the RTC on the gRNA template by steric hindrance. As a result, the RT could not reach the 5′ UTR of the gRNA including the primer-binding site to initiate the reaction (63). This is supported by Tsg101 recruitment together with Gag to a position closer to the center of Gag assemblages than Alix and the ESCRT-III proteins (64); (ii) efficient recruitment of Tsg101 at the PM through interplay between NC and p6 confers budding with proper kinetics of particle release. The aggregation and reduced motility of Gag at the PM observed for ΔZF2 particles could provide a time sufficient for RTion before the virus release. This model also relies on kinetics of PR-mediated cleavage during budding that needs to be further understood.
The precise location where HIV late RTion takes place in producer cells constitutes a central issue that also requires further investigation. Even though the cytosolic late RTion would benefit from a microenvironment with a large pool of available dNTPs, our results support its localization at the PM during the last step of particle formation where gRNA, Gag, GagPol, Tsg101 and other viral and host cell cofactors of virus biogenesis converge (65–68).
The new implication of Tsg101 in this regulation mechanism may explain why GagNC of gamma-retroviruses, such as the prototypic murine leukemia virus (MLV), does not play a similar role in late RTion control since MLV does not require Tsg101 for budding. In contrast to HIV, deletion of the unique ZF of MLV GagNC did not cause late RTion activation in producer cells (69).
The occurrence of late RTion in HIV-producing cells might explain the presence of viral DNA in HIV-1 particles isolated from the peripheral blood and semen of HIV-1-infected patients (70–72). A better understanding of this new role of the lentiviral NC and Tsg101 in the spatio-temporal control of RT activity is important to several aspects of HIV-1 replication such as assembly/disassembly, recombination and pathogenesis as well as to the development of new antiviral drugs against NC (73,74). In keeping with the importance of this process, a recent anti-HIV strategy targeting the NC ZFs has yielded promising results in a macaque model (75).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank B. Yu, N. Jouvenet and V. Baldin for technical assistance and N. Jouvenet, P. Bieniasz and J.R. Lingappa for providing plasmids. We thank D. Muriaux and O. Coux for reagents. Antiserum against HIV-1 p17, p24 (H183) and RT were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. We are grateful to the Montpellier RIO Imaging staff and to S.M. Simon for the TIRF microscopy and the Montpellier Genomic collection platform for the Gateway technology. Images of electron microscopy have been realized using the facilities of CRIC, Montpellier. We thank J.M. Jacqué and K. Wright for the critical reading of the manuscript.
Footnotes
The authors wish it to be known that, in their opinion, the second and third authors should be regarded as Joint Second Authors.
FUNDING
French National Agency for Research on AIDS and Viral Hepatitis (ANRS); ANRS [to P.J.R., P.R., M.F.]. Funding for open access charge: French National Agency for Research on AIDS and Viral hepatitis (ANRS).
Conflict of interest statement. None declared.
REFERENCES
- 1.Levin J.G., Mitra M., Mascarenhas A., Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7:754–774. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyonnais S., Gorelick R.J., Heniche-Boukhalfa F., Bouaziz S., Parissi V., Mouscadet J.F., Restle T., Gatell J.M., Le Cam E., Mirambeau G. A protein ballet around the viral genome orchestrated by HIV-1 reverse transcriptase leads to an architectural switch: from nucleocapsid-condensed RNA to Vpr-bridged DNA. Virus Res. 2013;171:287–303. doi: 10.1016/j.virusres.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas J.A., Bosche W.J., Shatzer T.L., Johnson D.G., Gorelick R.J. Mutations in human immunodeficiency virus type 1 nucleocapsid protein zinc fingers cause premature reverse transcription. J. Virol. 2008;82:9318–9328. doi: 10.1128/JVI.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darlix J.L., Godet J., Ivanyi-Nagy R., Fosse P., Mauffret O., Mely Y. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J. Mol. Biol. 2011;410:565–581. doi: 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- 5.de Marco A., Muller B., Glass B., Riches J.D., Krausslich H.G., Briggs J.A. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog. 2010;6:e1001215. doi: 10.1371/journal.ppat.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan A.H., Swanstrom R. Human immunodeficiency virus type 1 Gag proteins are processed in two cellular compartments. Proc. Natl Acad. Sci. U.S.A. 1991;88:4528–4532. doi: 10.1073/pnas.88.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karacostas V., Wolffe E.J., Nagashima K., Gonda M.A., Moss B. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology. 1993;193:661–671. doi: 10.1006/viro.1993.1174. [DOI] [PubMed] [Google Scholar]
- 8.Ott D.E., Coren L.V., Shatzer T. The nucleocapsid region of human immunodeficiency virus type 1 Gag assists in the coordination of assembly and Gag processing: role for RNA-Gag binding in the early stages of assembly. J. Virol. 2009;83:7718–7727. doi: 10.1128/JVI.00099-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grigorov B., Decimo D., Smagulova F., Pechoux C., Mougel M., Muriaux D., Darlix J.L. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology. 2007;4:54. doi: 10.1186/1742-4690-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dussupt V., Javid M.P., Abou-Jaoude G., Jadwin J.A., de La Cruz J., Nagashima K., Bouamr F. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009;5:e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briggs J.A., Simon M.N., Gross I., Krausslich H.G., Fuller S.D., Vogt V.M., Johnson M.C. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 12.Jacks T., Power M.D., Masiarz F.R., Luciw P.A., Barr P.J., Varmus H.E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 13.Bachrach E., Dreja H., Lin Y.L., Mettling C., Pinet V., Corbeau P., Piechaczyk M. Effects of virion surface gp120 density on infection by HIV-1 and viral production by infected cells. Virology. 2005;332:418–429. doi: 10.1016/j.virol.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 14.Zhu P., Chertova E., Bess J., Jr, Lifson J.D., Arthur L.O., Liu J., Taylor K.A., Roux K.H. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl Acad. Sci. U.S.A. 2003;100:15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanchenko S., Godinez W.J., Lampe M., Krausslich H.G., Eils R., Rohr K., Brauchle C., Muller B., Lamb D.C. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009;5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jouvenet N., Bieniasz P.D., Simon S.M. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goff S.P. In: Fields Virology. 5th edn. Knipe D. M., Howley P. M., editors. Vol. 2. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. pp. 1999–2070. [Google Scholar]
- 18.Votteler J., Sundquist W.I. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013;14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bieniasz P.D. The cell biology of HIV-1 virion genesis. Cell Host Microbe. 2009;5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.VerPlank L., Bouamr F., LaGrassa T.J., Agresta B., Kikonyogo A., Leis J., Carter C.A. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc. Natl Acad. Sci. U.S.A. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrus J.E., von Schwedler U.K., Pornillos O.W., Morham S.G., Zavitz K.H., Wang H.E., Wettstein D.A., Stray K.M., Cote M., Rich R.L., et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 22.Stuchell M.D., Garrus J.E., Muller B., Stray K.M., Ghaffarian S., McKinnon R., Krausslich H.G., Morham S.G., Sundquist W.I. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem. 2004;279:36059–36071. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
- 23.Neil S.J., Zang T., Bieniasz P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 24.Grover J.R., Llewellyn G.N., Soheilian F., Nagashima K., Veatch S.L., Ono A. Roles played by capsid-dependent induction of membrane curvature and Gag-ESCRT interactions in tetherin recruitment to HIV-1 assembly sites. J. Virol. 2013;87:4650–4664. doi: 10.1128/JVI.03526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janvier K., Pelchen-Matthews A., Renaud J.B., Caillet M., Marsh M., Berlioz-Torrent C. The ESCRT-0 component HRS is required for HIV-1 Vpu-mediated BST-2/tetherin down-regulation. PLoS Pathog. 2011;7:e1001265. doi: 10.1371/journal.ppat.1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strack B., Calistri A., Craig S., Popova E., Gottlinger H.G. AIP1/ALIX Is a Binding Partner for HIV-1 p6 and EIAV p9 Functioning in Virus Budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 27.von Schwedler U.K., Stuchell M., Muller B., Ward D.M., Chung H.Y., Morita E., Wang H.E., Davis T., He G.P., Cimbora D.M., et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 28.Popov S., Popova E., Inoue M., Gottlinger H.G. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J. Virol. 2008;82:1389–1398. doi: 10.1128/JVI.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bello N.F., Dussupt V., Sette P., Rudd V., Nagashima K., Bibollet-Ruche F., Chen C., Montelaro R.C., Hahn B.H., Bouamr F. Budding of retroviruses utilizing divergent L domains requires nucleocapsid. J. Virol. 2012;86:4182–4193. doi: 10.1128/JVI.07105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujii K., Munshi U.M., Ablan S.D., Demirov D.G., Soheilian F., Nagashima K., Stephen A.G., Fisher R.J., Freed E.O. Functional role of Alix in HIV-1 replication. Virology. 2009;391:284–292. doi: 10.1016/j.virol.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Serrano J., Yarovoy A., Perez-Caballero D., Bieniasz P.D. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl Acad. Sci. U.S.A. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Didierlaurent L., Houzet L., Morichaud Z., Darlix J.L., Mougel M. The conserved N-terminal basic residues and zinc-finger motifs of HIV-1 nucleocapsid restrict the viral cDNA synthesis during virus formation and maturation. Nucleic Acids Res. 2008;36:4745–4753. doi: 10.1093/nar/gkn474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houzet L., Morichaud Z., Didierlaurent L., Muriaux D., Darlix J.L., Mougel M. Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic Acids Res. 2008;36:2311–2319. doi: 10.1093/nar/gkn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mougel M., Houzet L., Darlix J.L. When is it time for reverse transcription to start and go. Retrovirology. 2009;6:24. doi: 10.1186/1742-4690-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zamborlini A., Renault N., Saib A., Delelis O. Early reverse transcription is essential for productive foamy virus infection. PLoS One. 2010;5:e11023. doi: 10.1371/journal.pone.0011023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baumgartel V., Ivanchenko S., Dupont A., Sergeev M., Wiseman P.W., Krausslich H.G., Brauchle C., Muller B., Lamb D.C. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat. Cell Biol. 2011;13:469–474. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- 37.Krementsov D.N., Rassam P., Margeat E., Roy N.H., Schneider-Schaulies J., Milhiet P.E., Thali M. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic. 2010;11:1401–1414. doi: 10.1111/j.1600-0854.2010.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin J., Sherer N.M., Heidecker G., Derse D., Mothes W. Assembly of the murine leukemia virus is directed towards sites of cell-cell contact. PLoS Biol. 2009;7:e1000163. doi: 10.1371/journal.pbio.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gullberg M., Göransson C., Fredriksson S. Duolink-‘In-cell Co-IP’ for visualization of protein interactions in situ. Nat. Methods. 2011;8 [Google Scholar]
- 40.Sette P., Dussupt V., Bouamr F. Identification of the HIV-1 NC binding interface in Alix Bro1 reveals a role for RNA. J. Virol. 2012;86:11608–11615. doi: 10.1128/JVI.01260-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jouvenet N., Neil S.J., Bess C., Johnson M.C., Virgen C.A., Simon S.M., Bieniasz P.D. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang M., Orenstein J.M., Martin M.A., Freed E.O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goff A., Ehrlich L.S., Cohen S.N., Carter C.A. Tsg101 control of human immunodeficiency virus type 1 Gag trafficking and release. J. Virol. 2003;77:9173–9182. doi: 10.1128/JVI.77.17.9173-9182.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin-Serrano J., Zang T., Bieniasz P.D. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 45.Van Engelenburg S.B., Shtengel G., Sengupta P., Waki K., Jarnik M., Ablan S.D., Freed E.O., Hess H.F., Lippincott-Schwartz J. Distribution of ESCRT machinery at HIV assembly sites reveals virus scaffolding of ESCRT subunits. Science. 2014;343:653–656. doi: 10.1126/science.1247786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houzet L., Morichaud Z., Mougel M. Fully-spliced HIV-1 RNAs are reverse transcribed with similar efficiencies as the genomic RNA in virions and cells, but more efficiently in AZT-treated cells. Retrovirology. 2007;4:30. doi: 10.1186/1742-4690-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chamanian M., Purzycka K.J., Wille P.T., Ha J.S., McDonald D., Gao Y., Le Grice S.F., Arts E.J. A cis-acting element in retroviral genomic RNA links Gag-Pol ribosomal frameshifting to selective viral RNA encapsidation. Cell Host Microbe. 2013;13:181–192. doi: 10.1016/j.chom.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikolaitchik O.A., Hu W.S. Deciphering the role of the Gag-Pol ribosomal frameshift signal in HIV-1 RNA genome packaging. J. Virol. 2014;88:4040–4046. doi: 10.1128/JVI.03745-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bell N.M., Lever A.M. HIV Gag polyprotein: processing and early viral particle assembly. Trends Microbiol. 2013;21:136–144. doi: 10.1016/j.tim.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 50.von Schwedler U.K., Stray K.M., Garrus J.E., Sundquist W.I. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 2003;77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lanman J., Sexton J., Sakalian M., Prevelige P.E., Jr Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J. Virol. 2002;76:6900–6908. doi: 10.1128/JVI.76.14.6900-6908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Briggs J.A., Wilk T., Welker R., Krausslich H.G., Fuller S.D. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gorelick R.J., Chabot D.J., Rein A., Henderson L.E., Arthur L.O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zargarian L., Tisne C., Barraud P., Xu X., Morellet N., Rene B., Mely Y., Fosse P., Mauffret O. Dynamics of linker residues modulate the nucleic acid binding properties of the HIV-1 nucleocapsid protein zinc fingers. PLoS One. 2014;9:e102150. doi: 10.1371/journal.pone.0102150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hulme A.E., Perez O., Hope T.J. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc. Natl Acad. Sci. U.S.A. 2011;108:9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reicin A.S., Ohagen A., Yin L., Hoglund S., Goff S.P. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J. Virol. 1996;70:8645–8652. doi: 10.1128/jvi.70.12.8645-8652.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paillart J.C., Shehu-Xhilaga M., Marquet R., Mak J. Dimerization of retroviral RNA genomes: an inseparable pair. Nat. Rev. Microbiol. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 58.Cen S., Huang Y., Khorchid A., Darlix J.L., Wainberg M.A., Kleiman L. The role of Pr55(gag) in the annealing of tRNA3Lys to human immunodeficiency virus type 1 genomic RNA. J. Virol. 1999;73:4485–4488. doi: 10.1128/jvi.73.5.4485-4488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaplan A.H., Manchester M., Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J. Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peng C., Chang N.T., Chang T.W. Identification and characterization of human immunodeficiency virus type 1 gag-pol fusion protein in transfected mammalian cells. J. Virol. 1991;65:2751–2756. doi: 10.1128/jvi.65.5.2751-2756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu T., Datta S.A., Mitra M., Gorelick R.J., Rein A., Levin J.G. Fundamental differences between the nucleic acid chaperone activities of HIV-1 nucleocapsid protein and Gag or Gag-derived proteins: biological implications. Virology. 2010;405:556–567. doi: 10.1016/j.virol.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen A.K., Sengupta P., Waki K., Engelenburg S.B., Ochiya T., Ablan S.D., Freed E.O., Lippincott-Schwartz J. MicroRNA binding to the HIV-1 Gag protein inhibits Gag assembly and virus production. Proc. Natl Acad. Sci. U.S.A. 2014;111:E2676–E2683. doi: 10.1073/pnas.1408037111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sleiman D., Goldschmidt V., Barraud P., Marquet R., Paillart J.C., Tisne C. Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions. Virus Res. 2012;169:324–339. doi: 10.1016/j.virusres.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 64.Bleck M., Itano M.S., Johnson D.S., Thomas V.K., North A.J., Bieniasz P.D., Simon S.M. Temporal and spatial organization of ESCRT protein recruitment during HIV-1 budding. Proc. Natl Acad. Sci. U.S.A. 2014 doi: 10.1073/pnas.1321655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henriet S., Sinck L., Bec G., Gorelick R.J., Marquet R., Paillart J.C. Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription. Nucleic Acids Res. 2007;35:5141–5153. doi: 10.1093/nar/gkm542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malim M.H., Emerman M. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 67.Martin-Serrano J., Neil S.J. Host factors involved in retroviral budding and release. Nat. Rev. Microbiol. 2011;9:519–531. doi: 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- 68.Weissenhorn W., Gottlinger H. Essential ingredients for HIV-1 budding. Cell Host Microbe. 2011;9:172–174. doi: 10.1016/j.chom.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 69.Chamontin C., Yu B., Racine P.J., Darlix J.L., Mougel M. MoMuLV and HIV-1 nucleocapsid proteins have a common role in genomic RNA packaging but different in late reverse transcription. PLoS One. 2012;7:e51534. doi: 10.1371/journal.pone.0051534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang H., Bagasra O., Niikura M., Poiesz B.J., Pomerantz R.J. Intravirion reverse transcripts in the peripheral blood plasma on human immunodeficiency virus type 1-infected individuals. J. Virol. 1994;68:7591–7597. doi: 10.1128/jvi.68.11.7591-7597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang H., Dornadula G., Pomerantz R.J. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenviroments: an important stage for viral infection of nondividing cells. J. Virol. 1996;70:2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang H., Zhang Y., Spicer T.P., Abbott L.Z., Abbott M., Poiesz B.J. Reverse transcription takes place within extracellular HIV-1 virions: potential biological significance. AIDS Res. Hum. Retroviruses. 1993;9:1287–1296. doi: 10.1089/aid.1993.9.1287. [DOI] [PubMed] [Google Scholar]
- 73.Breuer S., Chang M.W., Yuan J., Torbett B.E. Identification of HIV-1 inhibitors targeting the nucleocapsid protein. J. Med. Chem. 2012;55:4968–4977. doi: 10.1021/jm201442t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waheed A.A., Freed E.O. HIV type 1 Gag as a target for antiviral therapy. AIDS Res. Hum. Retroviruses. 2011;28:54–75. doi: 10.1089/aid.2011.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wallace G.S., Cheng-Mayer C., Schito M.L., Fletcher P., Jenkins L.M., Hayashi R., Neurath A.R., Appella E., Shattock R.J. Human immunodeficiency virus type 1 nucleocapsid inhibitors impede trans infection in cellular and explant models and protect nonhuman primates from infection. J. Virol. 2009;83:9175–9182. doi: 10.1128/JVI.00820-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


