Abstract
High-throughput sequencing reveals an abundance of microRNA-sized fragments derived from larger non-coding RNAs. Roles for these small RNAs in gene silencing are suggested by their co-precipitation with Argonaute, the microRNA effector protein, though the extent to which they suppress gene expression endogenously remains unclear. To address this, we used luciferase reporters to determine the endogenous functionality of small RNAs from a diverse range of sources. We demonstrate small RNAs derived from snoRNAs have the capacity to act in a microRNA-like manner, though we note the vast majority of these are bound to Argonaute at levels below that required for detectable silencing activity. We show Argonaute exhibits a high degree of selectivity for the small RNAs with which it interacts and note that measuring Argonaute-associated levels is a better indicator of function than measuring total expression. Although binding to Argonaute at sufficient levels is necessary for demonstrating microRNA functionality in our reporter assay, this alone is not enough as some small RNAs derived from other non-coding RNAs (tRNAs, rRNAs, Y-RNAs) are associated with Argonaute at very high levels yet do not serve microRNA-like roles.
INTRODUCTION
MicroRNAs (miRNAs) are small non-coding RNA (ncRNA) molecules that function as the specificity component of the RNA-induced Silencing Complex (RISC), which bind in a sequence-specific manner to mRNAs, resulting in the repression of target transcripts through translational inhibition and RNA destabilization. The canonical biogenesis pathway is well established with mature miRNAs processed from polyadenylated precursor RNAs, involving cleavage by the endonuclease Drosha to produce a pre-miRNA hairpin in the nucleus, which is then cleaved in the cytoplasm by Dicer, to produce the mature miRNA (reviewed in (1)). The discovery of novel miRNAs has been accelerated by advances in high-throughput sequencing, with over 2000 human miRNAs now catalogued in miRbase, the repository of annotated miRNA sequences (mirbase.org). Analysis of high-throughput sequencing data has also revealed an abundance of many other miRNA-sized fragments derived from various ncRNAs including from transfer RNA (tRNA) (2–23), ribosomal RNA (rRNA), (12,21–23), small nucleolar RNA (snoRNA), (12,20–35), small nuclear RNA (snRNA) (12,21,23), vault RNA (vtRNA) (21,36–38) and yRNA (39–42). Such miRNA-sized fragments have been observed throughout all kingdoms of life (33,34,43,44), but it remains unclear which fragments represent genuine functional miRNAs and which are merely degradation intermediates. Evidence is accumulating to suggest at least some of these non-canonical miRNA-sized RNAs do have a function. Firstly, some are produced by precise cleavages that are conserved across evolution (34,45). Secondly, production of some small RNAs is induced by specific factors, or occurs in a tissue-restricted manner (45,46). Thirdly, small RNA expression is often decoupled from that of the parent ncRNA (13,34,45). Fourthly, a number of these small RNAs are co-precipitated with Argonaute (AGO), the miRNA-binding component of RISC (21,46,47). Most importantly, functions have been reported for a number of small RNA fragments, including signalling roles in hypoxia and starvation-induced stress, as sequence-independent inhibitors of translation and in AGO-dependent, miRNA-like gene repression (6,8,9,17,19,24,25). The diversity of origins of these small RNAs suggests the possibility of a functional ‘miRNAome’ that is far broader than that currently recognized, though the actual contribution of most small RNAs to gene silencing remains uncertain, with often conflicting reports in the literature and a reliance in many of these studies upon artificial overexpression to demonstrate function.
From analysing high-throughput sequencing from MDA-MB-231 breast cancer cells, we also have observed a plethora of small RNAs (∼18–27nt in length) derived from a range of larger ncRNAs, many of which are bound to AGO. The abundance of these individual fragments varies over a wide range, prompting us to question which of these miRNA-sized molecules participate in miRNA-like gene regulation at their endogenous levels. We find that gene silencing is limited to a subset of small RNAs. Some snoRNA-derived fragments act in a miRNA-like manner, but the vast majority are not bound to AGO at high enough levels to produce measurable miRNA activity. In contrast, some small RNAs derived from tRNAs, Y-RNAs and snRNAs are associated with AGO at quite high abundance but do not mediate target repression, suggesting these AGO-associated RNAs may have other functions. Using high-throughput sequencing to compare the profile of small RNAs bound to AGO to that of the total population of small RNAs in the cell, we find AGO displays a high degree of selectivity in binding. We find that quantitating small RNAs bound to AGO provides a better indication of miRNA-like function than quantitating total small RNA levels. This is exemplified by the analysis of miR-374a, where the 5p arm is primarily bound by AGO and is highly functional whereas the more abundant 3p arm is mostly not bound to AGO and is less active as a miRNA.
MATERIALS AND METHODS
AGO:miRNA immunoprecipitation
AGO:miRNA immunoprecipitation was performed as previously described (48,49) based upon the HITS-CLIP (high-throughput sequencing, cross-linked immunoprecipitation) technique originally described (50). MDA-MB-231 cells were utilized, grown in DMEM + 10% FCS (Gibco). The pan anti-AGO 2A8 antibody (Millipore) was used for immunoprecipitation.
Reporter construct design and cloning
PsiCHECK2 dual luciferase reporters (Promega) were constructed to contain a single copy of a perfectly complementary target site of the small RNA sequences derived from miRBase (www.mirbase.org) and from our high-throughput sequencing. Oligonucleotides (Geneworks, Sigma) were designed with XhoI and NotI overhangs, annealed and ligated into psiCHECK2 using XhoI and NotI restriction sites.
Luciferase reporter assay
psiCheck vectors were transfected into MDA-MB-231 cells in 24-well plates using lipofectamine 2000 (Invitrogen). Reporter titrations were performed with 5 × 5-fold serial dilutions (starting with a final concentration of 100 nM). Luciferase reporter assays were performed using a dual luciferase kit (Promega) and read with Promega GloMax multi detection system. Relative luciferase activity was calculated as the ratio of Renilla to firefly luciferase activity. ‘Control’ lanes indicate the activity of the empty psiCHECK2 vector. All reporter assays were performed as independent biological triplicates, with error bars representing standard error of mean. miRNA mimics and inhibitors were provided by Ambion as double-stranded and single-stranded RNAs, respectively.
Bioinformatic analyses
Barcoded libraries were sequenced using Illumina GAII at Geneworks and HiSeq 2000 at the ACRF Cancer Genomics Facility as described (49). Adapters were removed using cutadapt (http://code.google.com/p/cutadapt) with 3′ quality cut-off of 28, maximum error rate of 0.2, minimum read length of 18 nt and a minimum adapter overhang of 2 nt. Low complexity reads were removed using prinseq-lite v0.19.5 (51) ‘dust’ method with a threshold of 40. To correct for post-transcriptional modifications of tRNAs, where present, the sequence 5′-CCA-3′ was trimmed from the 3′ end of reads where the trimmed read mapped to a tRNA. For multimapping, reads were mapped to the UCSC iGenomes Homo sapiens genome (build hg19) using bowtie2 v2.2.1 (52) with sensitive parameters for end-to-end alignment, allowing 0 mismatches in the seed region and reporting up to 100 alignments per read (flags: –sensitive –end-to-end -N 0 -k 100). Sequence data from other laboratories (21,46) were also processed in the same way. Data described as ‘non-multimapped’ or mapped by BWA (v0.6.1-r104) (53) was performed using default parameters. Analyses used iGenomes Ensembl gene annotations (build GRCh37.68), tRNA gene annotation from UCSC table browser and yRNAs were defined as the genes RNY1, RNY3, RNY4 and RNY5. Analyses were performed both for all transcripts and for only the most abundant transcript per gene and alignments were analysed in a strand-specific manner. Analyses were performed using the HTSeq package (http://dx.doi.org/10.1101/002824) in python: source code is available in the repository at https://bitbucket.org/sacgf/2014_thomson_nar. Read counts were taken from single HITS-CLIP (49) and RNA-Seq libraries (deposited as a GEO dataset). Additional AGO immunoprecipitation libraries (prepared in the absence of RNAse treatment) in Supplementary Figure S3 are previously published (21,46).
Primers annealed for reporter cloning.
| miR-140-5p | TCGAGATTTAAATCTACCATAGGGTAAAACCACTGGC |
| GGCCGCCAGTGGTTTTACCCTATGGTAGATTTAAATC | |
| miR-340-3p | TCGAGATTTAAATGCTATAAAGTAACTGAGACGGAGC |
| GGCCGCTCCGTCTCACTTACTTTATAGCATTTAAATC | |
| miR-200c-3p | TCGAGATTTAAATTCCATCATTACCCGGCAGTATTAGC |
| GGCCGCTAATACTGCCGGGTAATGATGGAATTTAAATC | |
| miR-141-3p | TCGAGATTTAAATCCATCTTTACCAGACAGTGTTAGC |
| GGCCGCTAACACTGTCTGGTAAAGATGGATTTAAATC | |
| miR-126-3p | TCGAGATTTAAATCGCATTATTACTCACGGTACGAGC |
| GGCCGCTCGTACCGTGAGTAATAATGCGATTTAAATC | |
| miR-34b-3p | TCGAGATTTAAATATGGCAGTGGAGTTAGTGATTGGC |
| GGCCGCCAATCACTAACTCCACTGCCATATTTAAATC | |
| miR-130b-5p | TCGAGATTTAAATGTAGTGCAACAGGGAAAGAGTGC |
| GGCCGCACTCTTTCCCTGTTGCACTACATTTAAATC | |
| miR-200b-3p | TCGAGATTTAAATTCATCATTACCAGGCAGTATTAGC |
| GGCCGCTAATACTGCCTGGTAATGATGAATTTAAATC | |
| miR-374a-3p | TCGAGATTTAAATAATTACAATACAATCTGATAAGGC |
| GGCCGCCTTATCAGATTGTATTGTAATTATTTAAATC | |
| miR-374a-5p | TCGAGATTTAAATCACTTATCAGGTTGTATTATAAGC |
| GGCCGCTTATAATACAACCTGATAAGAGATTTAAATC | |
| miR-15b-5p | TCGAGATTTAAATTGTAAACCATGATGTGCTGCTAGC |
| GGCCGCTAGCAGCACATCATGGTTTACAATTTAAATC | |
| miR-25-5p | TCGAGATTTAAATTCAGACCGAGACAAGTGCAATGGC |
| GGCCGCCATTGCACTTGTCTCGGTCTGAATTTAAATC | |
| miR-16-5p | TCGAGATTTAAATCGCCAATATTTACGTGCTGCTAGC |
| GGCCGCTAGCAGCACGTAAATATTGGCGATTTAAATC | |
| miR-222-3p | TCGAGATTTAAATAGAGACCCAGTAGCCAGATGTAGCTGC |
| GGCCGCAGCTACATCTGGCTACTGGGTCTCTATTTAAATC | |
| miR-30b-5p | TCGAGATTTAAATAGCTGAGTGTAGGATGTTTACAGC |
| GGCCGCTGTAAACATCCTACACTCAGCTATTTAAATC | |
| miR-125b-5p | TCGAGATTTAAATTCCCTGAGACCCTAACTTGTGA |
| GGCCGCAGGGACTCTGGGATTGAACACTATTTAAATC | |
| miR-21-5p | TCGAGATTTAAATTCAACATCAGTCTGATAAGCTAGC |
| GGCCGCTAGCTTATCAGACTGATGTTGAATTTAAATC | |
| miR-27a-3p | TCGAGATTTAAATGCGGAACTTAGCCACTGTGAAGC |
| GGCCGCTTCACAGTGGCTAAGTTCCGCATTTAAATC | |
| SNORD99 | TCGAGATTTAAATTCTCAGTCCCATATCCGCATTTCTGC |
| GGCCGCAGAAATGCGGATATGGGACTGAGAATTTAAATC | |
| SNORD52 | TCGAGATTTAAATAGTCAGAACTTAGTTTTGACATGC |
| GGCCGCATGTCAAAACTAAGTTCTGACTATTTAAATC | |
| SNORD31 | TCGAGATTTAAATGGCTCAGAAAATACCTTTCAGTCACACATTGC |
| GGCCGCAATGTGTGACTGAAAGGTATTTTCTGAGCCATTTAAATC | |
| SCARNA7 | TCGAGATTTAAATAACCTAGTCTAGTGTCCTGGTAAAAGC |
| GGCCGCTTTTACCAGGACACTAGACTAGGTTATTTAAATC | |
| SNORD97 | TCGAGATTTAAATTGCCCTCATATCTCATAATCTTCGCTGC |
| GGCCGCAGCGAAGATTATGAGATATGAGGGCAATTTAAATC | |
| SNORD98 | TCGAGATTTAAATGAGTTCAGTTCATTGTGTTCCACACTGC |
| GGCCGCAGTGTGGAACACAATGAACTGAACTCATTTAAATC | |
| SNORD14 | TCGAGATTTAAATGCAACCAATCATCATAGTGAGC |
| GGCCGCTCACTATGATGATTGGTTGCATTTAAATC | |
| SNORD25 | TCGAGATTTAAATTTCCTATGATGAGGACCTTTT |
| GGCCGCAAGGATACTACTCCTGGAAAACATTTAAATC | |
| SNORD56 | TCGAGATTTAAATGACGAAAAATATTGCCATCATTGC |
| GGCCGCAATGATGGCAATATTTTTCGTCATTTAAATC | |
| SNORA33 | TCGAGATTTAAATCACAAACACAATCAGGTAAGCAGATTCGC |
| GGCCGCGAATCTGCTTACCTGATTGTGTTTGTGATTTAAATC | |
| Y3-stem | TCGAGATTTAAATAAACACCACTGCACTCGGACCAGCCGC |
| GGCCGCGGCTGGTCCGAGTGCAGTGGTGTTTATTTAAATC | |
| Y3-loop | TCGAGATTTAAATGGAGAAGGAACAAAGAAATCTGTAACTGC |
| GGCCGCAGTTACAGATTTCTTTGTTCCTTCTCCATTTAAATC | |
| U3 | TCGAGATTTAAATCTACACGTTCAGAGAAACTTCTCTAGTGC |
| GGCCGCACTAGAGAAGTTTCTCTGAACGTGTAGATTTAAATC | |
| 5s_rRNA | TCGAGATTTAAATGACGAGATCGGGCGCGTTCAGGGTGGTATGC |
| GGCCGCATACCACCCTGAACGCGCCCGATCTCGTCATTTAAATC | |
| vtRNA | TCGAGATTTAAATAAAGAACTGTCGAAGTAACCGCTGAGCTAAAGCCAGCCCGC |
| GGCCGCGGGCTGGCTTTAGCTCAGCGGTTACTTCGACAGTTCTTTATTTAAATC | |
| tRNA Cys(GCA)′ | TCGAGATTTAAATAGCTCAGTGGTAGAGCATTTGACTGGC |
| GGCCGCCAGTCAAATGCTCTACCACTGAGCTATTTAAATC | |
| tRNA His(GTG) | TCGAGATTTAAATTTAGTACTCTGCGTTGTGGCCGC |
| GGCCGCGGCCACAACGCAGAGTACTAAATTTAAATC | |
| tRNA Gln(CTG) | TCGAGATTTAAATAATGGTTAGCACTCTGGACTCTGGC |
| GGCCGCCAGAGTCCAGAGTGCTAACCATTATTTAAATC | |
| tRNA Gly(TCC) | TCGAGATTTAAATAGTGGTGAGCATAGCTGCCTTCCGC |
| GGCCGCGGAAGGCAGCTATGCTCACCACTATTTAAATC | |
| SNORD25 mimic | UUCCUAUGAUGAGGACCUUUU |
| SNORD56 inhibitor | GACGAAAAATATTGCCATCATT |
| SNORA33 inhibitor | CACAAACACAATCAGGTAAGCAGATTC |
| vtRNA1_1 inhibitor | GAACTGTCGAAGTAACCGCTGAGCTAAA |
RESULTS
Small RNAs of diverse origin bind AGO
We chose to examine the repertoire of miRNA-like small RNAs in the breast cancer cell line, MDA-MB-231, comparing the relative abundances of small RNAs in total cellular RNA to their relative abundances in the AGO-associated small RNA population. A major difficulty that is often overlooked in the analysis of ncRNA fragments comes from their duplication in the genome, with many ncRNAs present at dozens of loci. The default settings of mapping software (such as Bowtie and BWA), which are most commonly used, distribute these multi-mapping reads proportionately across the loci, leading to an under-representation of expression levels for any given multi-mapping sequence. To avoid this error, we ensured that identical reads which map to multiple locations in the genome were appropriately counted as a single RNA (see the Materials and Methods section for details). This revealed higher read counts (particularly for transcripts mapping to tRNAs, rRNAs and snRNAs) associating with AGO than would otherwise be appreciated using conventional mapping parameters (Figure 1a and Supplementary Figure S1). We found that in addition to established miRNAs, other classes of ncRNA are also processed into small fragments (<30 nucleotides), many of which are associated with AGO (Figure 1b and c). We note the choice of sequence aligner (Bowtie or BWA) had little impact on the small RNAs identified (Supplementary Figure S2).
Figure 1.
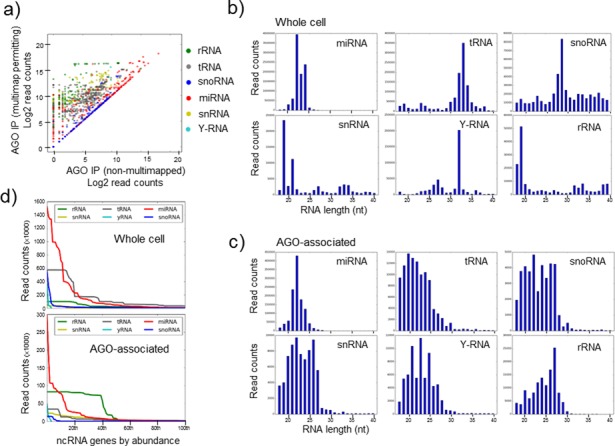
Argonaute binds small RNA from diverse sources. (a) Comparison of the transcript levels of small RNA genes using standard (default) mapping parameters, where multi-mapping reads are proportionately distributed across loci (horizontal axis), to custom parameters that permit multi-mapping (vertical axis) and reads are tallied independently for all individual sequences. For each gene, only the most abundant transcript is shown as a representative (for comparison, all transcripts are represented in Supplementary Figure S1). This highlights genes and small RNA classes whose expression may be under-estimated in standard analyses. (b,c) The relative abundance of transcripts of various lengths derived from diverse sources in both (b) the whole cell and (c) the AGO immunoprecipitate from MDA-MB-231 cells. The vertical axis represents read counts and the horizontal axis the length of the small RNA transcript. (d) For each class of small RNA, the 100 most abundant ncRNA genes for each class are ordered on the horizontal axis according to their abundance, which is plotted as read count on the vertical axis. This reveals both the predominant association of a subset of miRNAs with AGO and the association of small RNAs derived from other sources which individually may be in excess of many lowly and moderately expressed miRNAs.
AGO-bound small RNAs were found using the HITS-CLIP methodology in which protein:RNA complexes are crosslinked with UV irradiation and longer RISC-associated mRNAs are partially digested with RNAses prior to purification. We also re-analysed additional data sets from other cell lines in which AGO:RNA complexes were purified in the absence of cross-linking or enzymatic digestion (Supplementary Figure S3). Regardless of the methodology used, small RNAs derived from processing of various larger ncRNAs were consistently found associated with AGO. MiRNAs represented the most abundant class (Figure 1d), with a small subset of miRNAs representing the majority of AGO-bound transcripts. For example, the 15 most abundant miRNAs represent 77% of all miRNAs and ∼45% of the total AGO-associated small RNAs that were mapped to established classes of ncRNA. However, tRNA-derived RNAs were also quite abundant (representing ∼7% of AGO-associated RNAs), as were various examples of snoRNA, snRNA, rRNA and Y-RNA derived fragments that in some cases were individually more abundant than many well-characterized (but low to moderately expressed) miRNAs.
Assessing small RNAs from diverse sources for miRNA-like activity
Given the abundance and variety of origins of AGO-bound small RNAs, we sought to examine the extent to which the various classes of ncRNA may act as miRNAs at their endogenous levels. We employed luciferase reporter assays to establish the level of expression required for known miRNAs to be detectably functional. We constructed luciferase reporter genes with single perfectly complementary 3′UTR target sites for various known miRNAs and measured the luciferase activity produced after their transient transfection into MDA-MB-231 cells. We found that strongly expressed miRNAs (with >2000 reads in this experiment) strongly repressed reporter activity, whereas miRNAs with <1000 reads did not show observable reporter repression (Figure 2a), indicating there is a threshold level below which a miRNA is ineffective. Thus, of the 270 AGO-associated miRNAs that we detected by sequencing in MDA-MB-231 cells, only approximately the top 20% were expressed at sufficient level to detectably suppress the activity of a perfectly complementary reporter. The inability of modestly expressed miRNAs to modulate gene expression was similarly noted in a high-throughput study that reported ∼60% of detectable miRNAs within cells displayed no discernable suppressive activity (54), which in turn supported earlier work in which lowly expressed miRNAs (<100 copies/cell) were found to have little regulatory capacity (55).
Figure 2.
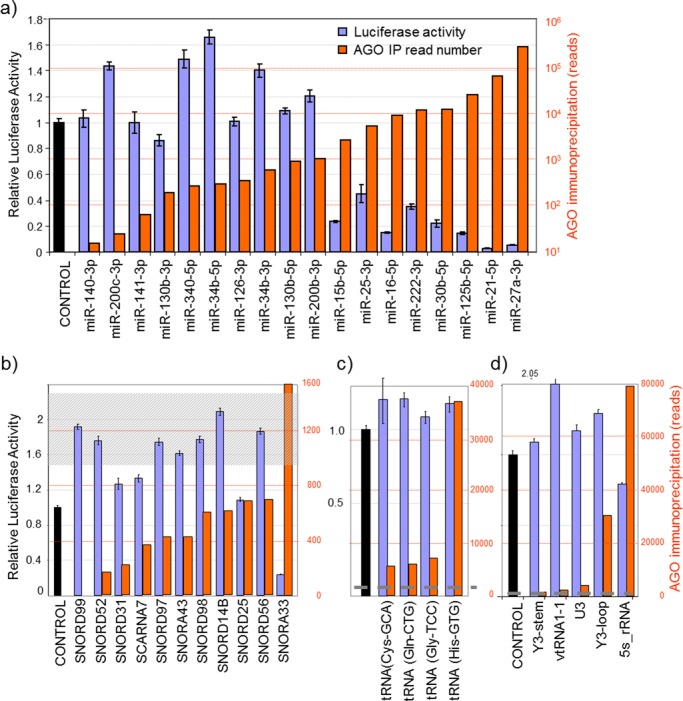
Comparing the abundance of AGO-bound small RNAs with their capacity to suppress reporters of fully complementary sequence. MDA-MB-231 cells were transfected with luciferase reporters containing sites fully complementary to small RNAs of (a) miRNA, (b) snoRNA, (c) tRNA or (d) diverse origin. Relative luciferase activity was determined 48 h post-transfection (blue bars). Orange bars represent the read numbers of these small RNAs associated with AGO from high-throughput sequencing. Luciferase assays were performed as biological triplicates with error bars depicting standard error of mean. The shaded region in (b)/dotted line in (c, d) indicates the number of sequencing reads required for established miRNAs to mediate detectable reporter suppression. Most snoRNA-derived fragments are present at low levels, whilst the small RNAs chosen (in c, d) are all above the expression level required for detectable function.
MiRNA-sized molecules derived from both snoRNAs (termed snoRNA-derived RNAs or sdRNAs) and tRNAs (tRNA-derived RNAs or tdRNAs) are readily detected in small RNA-sequencing and AGO-immunoprecipitation libraries. Although most individual sdRNAs and tdRNAs were present in the MDA-MB-231 cells at levels below the ∼1000 read threshold required for established miRNAs to suppress their reporter, certain sdRNAs and tdRNAs were expressed above this level, which prompted us to test their capacity to suppress reporter genes in a miRNA-like manner. To assess the repressive effects of miRNA-sized RNAs derived from snoRNAs, we again constructed luciferase reporters with single perfectly complementary sites in the 3′UTR and tested their activity in MDA-MB-231 cells. Consistent with the activity threshold observed for miRNAs, we found none of the sdRNAs with expression below 1000 reads had any repressive effect on their corresponding reporter, whereas the reporter for the only sdRNA that was expressed above this threshold (from SNORA33) was repressed, indicating this endogenous sdRNA functions as a miRNA (Figure 2b). A SNORD25-derived fragment that is expressed at insufficient levels for endogenous silencing was also capable of strongly suppressing reporter activity when cells were transiently transfected with a mimic of the corresponding sequence (Supplementary Figure S4). Thus, although their low abundance argues against widespread miRNA-like effects for most sdRNAs, they are nevertheless capable of functioning in this manner if present at sufficiently high level, in agreement with a dual miRNA/snoRNA classification sometimes bestowed.
Similarly, although most tdRNAs were present at low level, a subset was detected well above the 2000 read level required to detect activity of recognized miRNAs (Figure 2c). We constructed reporters for four such tdRNAs and tested their activity in MDA-MB-231 cells. None of the reporters for these tdRNAs was repressed, including the reporter for tRNA-His(GTG), a tdRNA expressed at levels equivalent to highly abundant (and highly repressive) miRNAs (Figure 2c). Thus, we find tdRNAs do not typically act as miRNAs and their association with AGO must be of a different nature from that of miRNAs and sdRNAs.
We also constructed reporter genes for several other miRNA-sized RNAs derived from Y-RNA (RNY3), vtRNA (vtRNA1–1), snRNA (U3) and 5S rRNA. Fragments derived from these ncRNAs have been previously noted in small RNA sequencing data sets, leading to their dual annotation. For example, miR-886 and vtRNA2–1 map to the same locus, miR-1975 and miR-1979 map within Y-RNAs and miRs -691, -712, -714 and -715 map within rRNAs. These full-length ncRNAs serve various functions. The U3 snRNA participates in pre-rRNA processing (56), Y-RNAs have roles in DNA replication and RNA quality control (57) and vtRNAs are components of vault ribonucleoprotein particles, which are cytoplasmic complexes of ill-defined function that are associated with resistance to chemotherapy and poor cancer prognosis (58). The capacity of some of these fragments to silence genes has been previously tested, but with varied outcomes. For example, Y-RNA fragments have been reported to not possess miRNA-like activity (39), vtRNA-derived fragments were reported to act as miRNAs (36,38) and U3-derived small RNAs displayed a variable capacity to silence reporter constructs between cell lines (24). We found at their endogenous levels of expression, only the small RNA derived from 5S rRNA displayed repressive activity, and this was very modest given the high level at which it is bound to AGO (Figure 2d). No repression was detectable for small RNAs derived from Y-RNAs, vtRNAs or U3 snRNA (Figure 2d). Taken together, this demonstrates that small RNAs derived from sources other than miRNAs and snoRNAs do not generally serve miRNA-like functions, despite some fragments being bound to AGO at levels in excess of that required to detect the suppressive activity of established miRNAs.
Testing the sensitivity of luciferase reporter assays
The correlation we observed between miRNA levels and the suppression of reporter activity demonstrates the utility of this reporter assay, but it is possible that the sensitivity of the assay may be dampened by the high levels of reporter transcripts expressed from the transient transfection, with the reporter mRNAs effectively acting as miRNA sponges (54,59,60). To test this possibility we repeated the reporter assays using serially diluted reporter plasmids for the abundant miRNA miR-21 and for miR15b-5p, which is expressed at a level only slightly above the threshold required for the suppression of reporter activity. We found that the ratio of firefly to renilla luciferase for miR-21 was already repressed to maximal levels and was unaffected by dilution, whilst repression of the miR-15–5p reporter, which remained detectable, was modestly increased as the plasmid was diluted (Figure 3), indicating that reduction of plasmid levels can enhance sensitivity of the assay for less abundant miRNAs. Similarly, when we tested the effect of titration of the reporter plasmid for the most abundant sdRNA (from SNORA33), we found that the reporter was strongly repressed at all levels of plasmid (Figure 3). We found that dilution of the reporter plasmid for the sdRNA derived from SNORD56, which is present at levels just below that required to detect repression by miRNAs (Figure 3), revealed repression at the lowest plasmid concentration (Figure 3). This underscores both an improved capacity of the assay to detect miRNA-like function, and further indicates the miRNA-like roles that sdRNAs can play. Importantly however, even with this increased sensitivity, no miRNA-like effect was observed for any tdRNA, Y-RNA, vtRNA or snRNA tested. In further support of their miRNA-like activities, co-transfection of sequestering ‘anti-miRs’ partially de-repressed a SNORA33 reporter (and that of miR-21 and miR-125b) (Supplementary Figure S5). Taken together, these data further suggest that sdRNAs are capable of functioning as miRNAs though most individual sdRNAs are expressed at very low levels and hence are unlikely to play prominent roles in endogenous gene repression, whilst the abundant small RNAs derived from tRNA, snRNA, vault and Y-RNAs do not serve miRNA-like roles at all.
Figure 3.
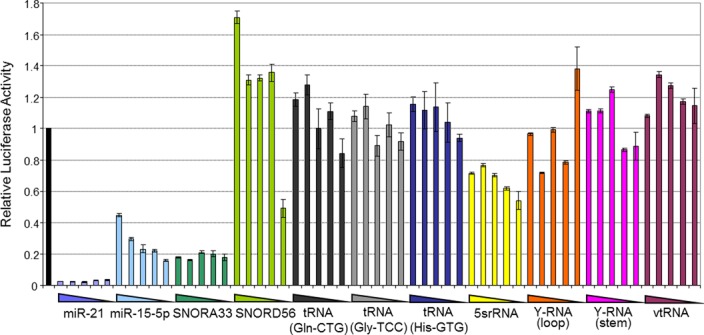
Higher sensitivity reporter assays do not demonstrate miRNA-like roles for other AGO-bound small RNAs. MDA-MB-231 cells were transfected with 5 × 5-fold serial dilutions of luciferase reporter plasmids containing sites fully complementary to a number of small RNAs assayed in Figure 2. Luciferase activity was determined in triplicate, 48 h post-transfection. Error bars depict standard error of mean. Greater suppression of luciferase reporters with reduced levels of transfected plasmid (for miR-15b-5p and SNORD56) indicates an increased capacity for this assay to detect miRNA-like function, though reporters for most other (non-miRNA/snoRNA-derived) RNAs are still not repressed, re-affirming their inability to perform gene suppressive roles.
AGO-binding is a better indicator of functionality than total RNA abundance
The capacity of small RNAs to act as miRNAs is dependent upon their association with AGO. However, determination of small RNA abundance is mostly done independently of AGO binding, and the entry of new miRNAs into miRbase does not require AGO association to be demonstrated. Whilst the levels of AGO-associated miRNAs are generally correlated to the overall cellular levels, for some miRNAs the correlation is poor (Figure 5 and Supplementary Figure S2). Curiously, for certain pre-miRNAs, when the total and AGO-associated abundances of the 5p and 3p miRNAs are compared, it is the less abundant arm that is most associated with AGO. This is of interest because it is the AGO-associated arm that determines the miRNA target profile. This applies for example to miR-374a (Figure 4), where although the 3p arm is the more abundant in the cell, it is the 5p arm that is the more AGO-associated. We examined the relative functionality of the miR-374a-5p and -3p arms using luciferase reporter assays and confirmed that it is the -5p arm (which is most associated with AGO) that is functionally significant in MDA-MB-231 cells (Figure 4b). Repressive activity of the -3p arm was detectable, but only upon transfection of significantly reduced levels of reporter. AGO binding may therefore be an important regulatory step and there is a need to consider AGO association when assessing the functional ‘miRNAome’. In agreement with a recent report (46), our data indicate measuring the levels of AGO-bound miRNAs provides a better indication of function than measuring total miRNA level within cells (compare Figure 2 and Supplementary Figure S6).
Figure 5.
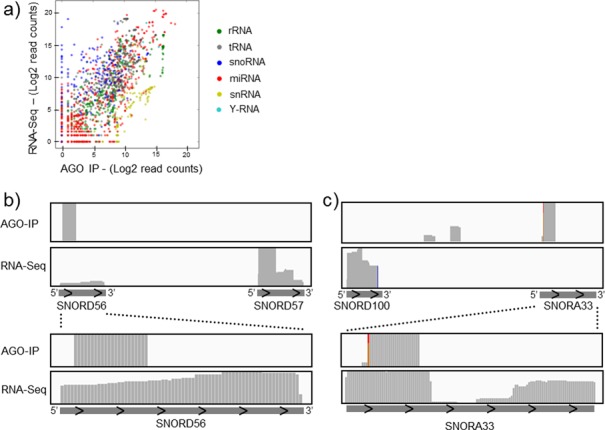
Argonaute selectively binds small RNAs. (a) The abundance of small RNA transcripts identified within the cell (vertical axis) or the AGO immunoprecipitate (horizontal axis) is compared, with colour indicating RNA class. A representative transcript is shown for each gene. (b, c) Read abundance (vertical axis) for the AGO immunoprecipitation and whole cell RNA sequencing is shown for various snoRNA and rRNA-derived reads. The position along the transcript is represented on the horizontal axis plotted 5′–3′. SNORD56 (b) and SNORA33 (c) derived reads are enlarged in the lower panels.
Figure 4.
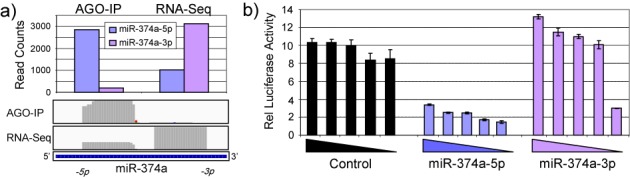
Argonaute binding is a better indicator of miRNA function than total expression. (a) Read numbers associated with the 5p and 3p arms of miR-374a are indicated in the AGO immunoprecipitation and whole cell RNA sequencing by histogram (above) and plotting of read abundance across the miR-374a locus (below). (b) MDA-MB-231 cells were transfected with 5 × 5-fold serial dilutions of luciferase reporter plasmids containing sites fully complementary to the 5p or 3p arm of miR-374a. Luciferase activity was determined in triplicate, 48 h post-transfection. Error bars depict standard error of mean. Results demonstrate the most functionally active arm of miR-374a represents the arm most associated with AGO rather than the arm predominantly expressed within cells.
Whilst many tdRNAs and sdRNAs were less associated with AGO than may be predicted from their total expression, snRNA-derived fragments were surprisingly highly associated with AGO (Figure 5a). Though snRNA-derived fragments do not appear to serve miRNA-like roles, this observation is consistent with a role for AGO in splicing and with the identification of AGO in snRNA-containing spliceosomal complexes (61). Just as for miRNAs, we observed distinct preferences for the association of various other small RNAs with AGO. For example, specific small RNA fragments derived from SNORA33 and SNORD56 (which we demonstrate suppress reporter genes) are effectively bound by AGO whilst far more abundant small RNAs derived from neighbouring snoRNAs are not (Figure 5b and c). In accordance with previously reported AGO-binding preference (62), the 5′ position of miRNAs was enriched for U and A nucleotides. Similar A/U enrichment was noted for other classes of small AGO-bound RNAs as well (Supplementary Figure S7).
Taken as a whole, our data indicate that it is a subset of miRNA-sized RNAs that are actually bound to AGO and that measuring the abundance of AGO-bound small RNAs provides a better indication of their functional level as miRNAs than simply measuring their total abundance within cells. Along with a small but growing body of literature, our results indicate that functionally significant target suppression is limited to a subset of well-expressed small RNAs and that the vast majority of sdRNAs (and miRNAs) are expressed at levels insufficient to mediate such function.
DISCUSSION
Though the presence of small RNA fragments derived from larger ncRNAs is widely observed, there is conflicting evidence regarding their capacity to fulfil miRNA-like roles within cells at their endogenous levels of expression. We characterized the abundance of small AGO-bound RNAs within MDA-MB-231 cells and found that sequences correspond to not only miRNAs but also to tRNAs, snoRNAs, Y-RNAs and rRNAs amongst other sources. We sought to investigate how both the abundance and origins of these small RNAs reflect their capacity to silence target genes.
Full length snoRNAs direct covalent rRNA modification, with H/ACA box snoRNAs directing pseudouridylation and C/D box snoRNAs directing 2′-O-methylation (63). A third class, small-cajal body RNAs (scaRNA), direct both the pseudouridylation and methylation of spliceosomal snRNA (64). In 2008, unbiased high-throughput sequencing of the human transcriptome revealed the accumulation of miRNA-sized sdRNAs (23). Subsequently, sdRNAs have now been reported across metazoa, plants, yeast and protozoa (31,34), with conserved cleavage sites implying specific roles (45). SdRNAs and miRNAs share a common evolutionary history, with similarities in regard to their size, secondary structure of the pre-cursor RNA, genomic localization, AGO association and the involvement of Drosha and/or Dicer in their processing (33). Not surprisingly therefore, sdRNAs have been implicated in miRNA-like gene regulation, even though they generally associate with AGO at low levels compared to more abundant miRNAs (26). Possibly for this reason, endogenous gene silencing has rarely been demonstrated through the use of sdRNA-inhibitors and even when inhibitors have been used, only single targets have been described (25,28,35). A number of sdRNAs were reported to exhibit a variable capacity to act as miRNAs between cells, though it is unclear if this reflects intrinsic differences in the function of these molecules, or merely reflects differential expression levels (24). To address the gene-suppressive roles of sdRNAs and other small RNAs, we constructed a series of complementary luciferase reporters and assayed the capacity of the endogenous small RNAs to silence luciferase expression. We found that sdRNAs were generally capable of acting in a miRNA-like manner, but most sdRNAs are expressed at levels below that required for detectable suppressive activity. We speculate that in other cell lines, or under conditions in which the expression of specific sdRNAs increases, these too may play more prominent gene suppressive roles.
As with snoRNAs, high-throughput sequencing has revealed a plethora of tdRNAs, which have been variously classified depending upon their location relative to the parent tRNA. Mature tRNAs are produced through 5′ and 3′ cleavage (by RNAseP and tRNAseZ), followed by 3′CCA addition to create an amino acid binding site. tdRNA sequences may then be derived from the resulting 5′-leader and 3′-trailer sequences from the pre-tRNA, or from further cleavage of the D- and TψC-loops to generate 5′ and 3′ fragments from mature tRNAs. The most prominent of these fragments have been termed tRNA-regulatory fragments (tRFs): tRF-1 (3′-trailer), tRF-3 (3′-mature) and tRF-5 (5′-mature) (reviewed in (44)). So-called ‘tRNA-halves’ may also originate from cleavage within anticodon loops, generating fragments ∼30–50 nt in size which have been reported in organisms ranging from prokaryotes to plants and humans (5,9,11,18,19). tRNA-halves increase in response to stress, and along with other fragments (tRF-5), function in activating cytoprotective stress responses through the global inhibition of translation (8). The capacity of tdRNAs to act as miRNAs is controversial. Studies have failed to demonstrate miRNA-like roles for examples of tRF-1 (6,10) or tRF-5 fragments (17) despite their capacity to associate with AGO. Silencing has been reported for several tRF-3s, though the repressive capacity of single chosen examples was either modest (6), or only demonstrated with overexpression (15). For any of the tdRNAs we selected, no miRNA-like activity was detectable, in keeping with reports of their weak co-immunoprecipitation with AGO (compared to miRNAs) (2) and their failure to co-precipitate MOV10, an RNA helicase that is required for RISC function (6).
The system we employ entails artificial fully complementary reporters, with the capacity to detect miRNA-like function affected by the amount of the transiently transfected reporter vector (Figure 3). As such, the failure of a reporter to be silenced by endogenous levels of the corresponding small RNA does not necessarily mean the small RNA in question has no miRNA-like role and it is possible that lowly expressed miRNAs are still functional, but are sequestered by endogenous targets and not available for the silencing of transfected reporters. However, there is a strong relationship between the level of AGO-bound miRNA and reporter activity (Figure 2) which strongly suggests small RNAs produced from rRNAs, tRNAs and Y-RNAs do not have miRNA-like roles because they display no suppressive activity despite being at levels well in excess of many miRNAs that do silence reporter genes.
Stem loop structures are characteristic of primary miRNAs, which are successively processed into small miRNAs that are loaded into RISC. Stem-loops are also characteristic of the termini of snoRNAs, so it is not surprising that the majority of snoRNA-mapping reads are derived from these ends (Supplementary Figure S8). With regard to tRNAs, we find that small RNAs derived from the 5′ ends of mature tRNAs (tRF-5) are the most abundant class within MDA-MB-231 cells. We find that AGO associates both with these fragments and with tdRNAs derived from internal locations (Supplementary Figure S8). Both classes of tdRNA are represented in our reporter assays in which neither class were shown to display miRNA-like properties. We also note that small RNAs derived from miRNA hairpins or from snoRNAs have typically well-defined boundaries (particularly at the 5′ end), whilst the ends of small RNAs derived from tRNAs, snRNAs, Y-RNAs and the like are ill-defined, with start and end positions often distributed across half a dozen nucleotides (Supplementary Figure S8).
It is not known why these smRNAs do not possess miRNA-like roles, though we note miRNAs function in the context of RISC, and therefore must be bound and presented by AGO proteins in such a manner as to interact with target transcripts. This is achieved through the anchoring of miRNAs by the AGO Mid and PAZ domains, aligning the miRNA along a narrow cleft from which the seed nucleotides are exposed and available for target interaction (65). The nature of the physical interaction between AGO and these other small RNAs is not defined and may reflect a mode of AGO association separate from the miRNA-binding cleft.
If not functioning as miRNAs, it begs the question what these AGO-associated fragments are doing. Whilst miRNA-independent roles have been postulated for some of these fragments (8,17,19), our findings may also point to miRNA-independent roles for the AGO proteins themselves (66). Recently, AGO has been implicated in heterochromatinization (67), alternate splicing (68), double-stranded break repair (69) and, in prokaryotes, genomic defence by DNA-guided DNA interference (70). Given the nature of known AGO-interacting proteins, the slicer activity of AGO2 and the plethora of interacting small RNAs presented here, it is plausible that AGO proteins may be playing more extensive RNA-processing roles than currently recognized. Supporting this speculation, AGO2-mediated cleavage of the passenger arm is required for miR-451 production (71–73) and in Arabidopsis thaliana, AGO4 is present within cajal bodies, sites at which snRNAs are modified by pseudouridylation and 2′-O-Methylation (74,75). This is of particular interest as we note despite U3 (an snRNA) not acting in a miRNA-like manner, snRNA-derived fragments are strongly enriched in the AGO immunoprecipitate. A similar situation may occur for Y-RNAs, and it is noteworthy that of the handful of known Y-RNA binding proteins, several have been co-precipitated in human cells by anti-AGO antibodies, including hnRNP-K, IGFBP2, nucleolin and La (76).
Our data highlight the selective nature of small RNA:AGO association and demonstrate a strong relationship between the abundance of an AGO-bound miRNA and its endogenous functionality. For this reason sdRNAs are not likely to play extensive miRNA-like roles, though for more abundant small RNAs derived from other classes of ncRNA, their failure to suppress gene expression is due to other factors. The selective nature of AGO binding underscores the importance of profiling the expression of AGO-bound small RNAs, whilst being mindful that the mere presence of a small RNA associated with AGO is not necessarily a good indication of a miRNA-like function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council of Australia [1026191 to G.J.G.]; Florey Foundation; National Breast Cancer Foundation [PF-09-03 to C.P.B.]; National Health and Medical Research Council of Australia [1008327]; Association for International Cancer Research [22-1170]; National Collaborative Research Program, EMPathy Breast Cancer Network, National Breast Cancer Foundation (Australia) [CG-10-04].
Conflict of interest statement. None declared.
REFERENCES
- 1.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole C., Sobala A., Lu C., Thatcher S.R., Bowman A., Brown J.W., Green P.J., Barton G.J., Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emara M.M., Ivanov P., Hickman T., Dawra N., Tisdale S., Kedersha N., Hu G.F., Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Silva M.R., Frugier M., Tosar J.P., Correa-Dominguez A., Ronalte-Alves L., Parodi-Talice A., Rovira C., Robello C., Goldenberg S., Cayota A. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol. Biochem. Parasitol. 2010;171:64–73. doi: 10.1016/j.molbiopara.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Haiser H.J., Karginov F.V., Hannon G.J., Elliot M.A. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008;36:732–741. doi: 10.1093/nar/gkm1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haussecker D., Huang Y., Lau A., Parameswaran P., Fire A.Z., Kay M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh L.C., Lin S.I., Kuo H.F., Chiou T.J. Abundance of tRNA-derived small RNAs in phosphate-starved Arabidopsis roots. Plant Signal. Behav. 2010;5:537–539. doi: 10.4161/psb.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jochl C., Rederstorff M., Hertel J., Stadler P.F., Hofacker I.L., Schrettl M., Haas H., Huttenhofer A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008;36:2677–2689. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y.S., Shibata Y., Malhotra A., Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y., Luo J., Zhou H., Liao J.Y., Ma L.M., Chen Y.Q., Qu L.H. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z., Ender C., Meister G., Moore P.S., Chang Y., John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 2012;40:6787–6799. doi: 10.1093/nar/gks307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao J.Y., Ma L.M., Guo Y.H., Zhang Y.C., Zhou H., Shao P., Chen Y.Q., Qu L.H. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3’ trailers. PLoS One. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loss-Morais G., Waterhouse P.M., Margis R. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biol. Direct. 2013;8:6. doi: 10.1186/1745-6150-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maute R.L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K., Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl Acad. Sci. U.S.A. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reese T.A., Xia J., Johnson L.S., Zhou X., Zhang W., Virgin H.W. Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. J. Virol. 2010;84:10344–10353. doi: 10.1128/JVI.00707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobala A., Hutvagner G. Small RNAs derived from the 5’ end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10:553–563. doi: 10.4161/rna.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson D.M., Lu C., Green P.J., Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamasaki S., Ivanov P., Hu G.F., Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langenberger D., Cakir M.V., Hoffmann S., Stadler P.F. Dicer-processed small RNAs: rules and exceptions. J. Exp. Zool. B Mol. Dev. Evol. 2013;320:35–46. doi: 10.1002/jez.b.22481. [DOI] [PubMed] [Google Scholar]
- 21.Burroughs A.M., Ando Y., de Hoon M.J., Tomaru Y., Suzuki H., Hayashizaki Y., Daub C.O. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158–177. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zywicki M., Bakowska-Zywicka K., Polacek N. Revealing stable processing products from ribosome-associated small RNAs by deep-sequencing data analysis. Nucleic Acids Res. 2012;40:4013–4024. doi: 10.1093/nar/gks020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawaji H., Nakamura M., Takahashi Y., Sandelin A., Katayama S., Fukuda S., Daub C.O., Kai C., Kawai J., Yasuda J., et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brameier M., Herwig A., Reinhardt R., Walter L., Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ender C., Krek A., Friedlander M.R., Beitzinger M., Weinmann L., Chen W., Pfeffer S., Rajewsky N., Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 26.Kishore S., Gruber A.R., Jedlinski D.J., Syed A.P., Jorjani H., Zavolan M. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome Biol. 2013;14:R45. doi: 10.1186/gb-2013-14-5-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kishore S., Khanna A., Zhang Z., Hui J., Balwierz P.J., Stefan M., Beach C., Nicholls R.D., Zavolan M., Stamm S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum. Mol. Genet. 2010;19:1153–1164. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W., Saraiya A.A., Wang C.C. The profile of snoRNA-derived microRNAs that regulate expression of variant surface proteins in Giardia lamblia. Cell. Microbiol. 2012;14:1455–1473. doi: 10.1111/j.1462-5822.2012.01811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ono M., Scott M.S., Yamada K., Avolio F., Barton G.J., Lamond A.I. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011;39:3879–3891. doi: 10.1093/nar/gkq1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saraiya A.A., Li W., Wang C.C. Transition of a microRNA from repressing to activating translation depending on the extent of base pairing with the target. PLoS One. 2013;8:e55672. doi: 10.1371/journal.pone.0055672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saraiya A.A., Wang C.C. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott M.S., Avolio F., Ono M., Lamond A.I., Barton G.J. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott M.S., Ono M. From snoRNA to miRNA: dual function regulatory non-coding RNAs. Biochimie. 2011;93:1987–1992. doi: 10.1016/j.biochi.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taft R.J., Glazov E.A., Lassmann T., Hayashizaki Y., Carninci P., Mattick J.S. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan Y.Z., Zhou A., Hu Z., Yu A.M. Small nucleolar RNA-derived microRNA hsa-miR-1291 modulates cellular drug disposition through direct targeting of ABC transporter ABCC1. Drug Metab. Dispos. 2013;41:1744–1751. doi: 10.1124/dmd.113.052092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hussain S., Sajini A.A., Blanco S., Dietmann S., Lombard P., Sugimoto Y., Paramor M., Gleeson J.G., Odom D.T., Ule J., et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255–261. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee K., Kunkeaw N., Jeon S.H., Lee I., Johnson B.H., Kang G.Y., Bang J.Y., Park H.S., Leelayuwat C., Lee Y.S. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA. 2011;17:1076–1089. doi: 10.1261/rna.2701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Persson H., Kvist A., Vallon-Christersson J., Medstrand P., Borg A., Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat. Cell Biol. 2009;11:1268–1271. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 39.Meiri E., Levy A., Benjamin H., Ben-David M., Cohen L., Dov A., Dromi N., Elyakim E., Yerushalmi N., Zion O., et al. Discovery of microRNAs and other small RNAs in solid tumors. Nucleic Acids Res. 2010;38:6234–6246. doi: 10.1093/nar/gkq376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicolas F.E., Hall A.E., Csorba T., Turnbull C., Dalmay T. Biogenesis of Y RNA-derived small RNAs is independent of the microRNA pathway. FEBS Lett. 2012;586:1226–1230. doi: 10.1016/j.febslet.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 41.Rutjes S.A., van der Heijden A., Utz P.J., van Venrooij W.J., Pruijn G.J. Rapid nucleolytic degradation of the small cytoplasmic Y RNAs during apoptosis. J. Biol. Chem. 1999;274:24799–24807. doi: 10.1074/jbc.274.35.24799. [DOI] [PubMed] [Google Scholar]
- 42.Smalheiser N.R., Lugli G., Thimmapuram J., Cook E.H., Larson J. Endogenous siRNAs and noncoding RNA-derived small RNAs are expressed in adult mouse hippocampus and are up-regulated in olfactory discrimination training. RNA. 2011;17:166–181. doi: 10.1261/rna.2123811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pederson T. Regulatory RNAs derived from transfer RNA. RNA. 2010;16:1865–1869. doi: 10.1261/rna.2266510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuck A.C., Tollervey D. RNA in pieces. Trends Genet. 2011;27:422–432. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Scott M.S., Ono M., Yamada K., Endo A., Barton G.J., Lamond A.I. Human box C/D snoRNA processing conservation across multiple cell types. Nucleic Acids Res. 2012;40:3676–3688. doi: 10.1093/nar/gkr1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flores O., Kennedy E.M., Skalsky R.L., Cullen B.R. Differential RISC association of endogenous human microRNAs predicts their inhibitory potential. Nucleic Acids Res. 2014;42:4629–4639. doi: 10.1093/nar/gkt1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamakawa N., Okuyama K., Ogata J., Kanai A., Helwak A., Takamatsu M., Imadome K., Takakura K., Chanda B., Kurosaki N., et al. Novel functional small RNAs are selectively loaded onto mammalian Ago1. Nucleic Acids Res. 2014;42:5289–5301. doi: 10.1093/nar/gku137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomson D.W., Bracken C.P., Szubert J.M., Goodall G.J. On measuring miRNAs after transient transfection of mimics or antisense inhibitors. PLoS One. 2013;8:e55214. doi: 10.1371/journal.pone.0055214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bracken C.L.X., Wright J.A., Pillman K.A., Lawrence D., Salmanidis M., Anderson M.A., Dredge K., Gregory P.A., Tsykin A., Neilsen C., et al. Genome-wide identification of miR-200 targets reveals a regulatory network controlling cell invasion. EMBO J. 2014;33:2040–2056. doi: 10.15252/embj.201488641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chi S.W., Zang J.B., Mele A., Darnell R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mullokandov G., Baccarini A., Ruzo A., Jayaprakash A.D., Tung N., Israelow B., Evans M.J., Sachidanandam R., Brown B.D. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown B.D., Gentner B., Cantore A., Colleoni S., Amendola M., Zingale A., Baccarini A., Lazzari G., Galli C., Naldini L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 56.Culver G.M. Sno-capped: 5’ ends of preribosomal RNAs are decorated with a U3 SnoRNP. Chem. Biol. 2002;9:777–779. doi: 10.1016/s1074-5521(02)00171-0. [DOI] [PubMed] [Google Scholar]
- 57.Verhagen A.P., Pruijn G.J. Are the Ro RNP-associated Y RNAs concealing microRNAs? Y RNA-derived miRNAs may be involved in autoimmunity. Bioessays. 2011;33:674–682. doi: 10.1002/bies.201100048. [DOI] [PubMed] [Google Scholar]
- 58.Stadler P.F., Chen J.J., Hackermuller J., Hoffmann S., Horn F., Khaitovich P., Kretzschmar A.K., Mosig A., Prohaska S.J., Qi X., et al. Evolution of vault RNAs. Mol. Biol. Evol. 2009;26:1975–1991. doi: 10.1093/molbev/msp112. [DOI] [PubMed] [Google Scholar]
- 59.Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haraguchi T., Ozaki Y., Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J., Hu J., Corey D.R. Expanding the action of duplex RNAs into the nucleus: redirecting alternative splicing. Nucleic Acids Res. 2012;40:1240–1250. doi: 10.1093/nar/gkr780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu H.Y., Yan Z., Xu Y., Hu H., Menzel C., Zhou Y.H., Chen W., Khiatovich P. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics. 2009;10:413. doi: 10.1186/1471-2164-10-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henras A.K., Dez C., Henry Y. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 2004;14:335–343. doi: 10.1016/j.sbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 65.Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 67.Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S.I., Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Allo M., Buggiano V., Fededa J.P., Petrillo E., Schor I., de la Mata M., Agirre E., Plass M., Eyras E., Elela S.A., et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 69.Wei W., Ba Z., Gao M., Wu Y., Ma Y., Amiard S., White C.I., Rendtlew Danielsen J.M., Yang Y.G., Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 70.Swarts D.C., Jore M.M., Westra E.R., Zhu Y., Janssen J.H., Snijders A.P., Wang Y., Patel D.J., Berenguer J., Brouns S.J., et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheloufi S., Dos Santos C.O., Chong M.M., Hannon G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D., et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang J.S., Maurin T., Robine N., Rasmussen K.D., Jeffrey K.L., Chandwani R., Papapetrou E.P., Sadelain M., O'Carroll D., Lai E.C. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl Acad. Sci. U.S.A. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li C.F., Pontes O., El-Shami M., Henderson I.R., Bernatavichute Y.V., Chan S.W., Lagrange T., Pikaard C.S., Jacobsen S.E. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 75.Pontes O., Li C.F., Costa Nunes P., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 76.Landthaler M., Gaidatzis D., Rothballer A., Chen P.Y., Soll S.J., Dinic L., Ojo T., Hafner M., Zavolan M., Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


