Abstract
Horizontal dissemination of the genes encoding extended spectrum beta-lactamases (ESBLs) via conjugative plasmids is facilitating the increasingly widespread resistance of pathogens to beta-lactam antibiotics. However, there is relatively little known about the regulatory factors and mechanisms that govern the spread of these plasmids. Here, we carried out a high-throughput, transposon insertion site sequencing analysis (TnSeq) to identify genes that enable the maintenance and transmission of pESBL, an R64 (IncI1)-related resistance plasmid that was isolated from Escherichia coli O104:H4 linked to a recent large outbreak of gastroenteritis. With a few exceptions, the majority of the genes identified as required for maintenance and transmission of pESBL matched those of their previously defined R64 counterparts. However, our analyses of the high-density transposon insertion library in pESBL also revealed two very short and linked regions that constitute a previously unrecognized regulatory system controlling spread of IncI1 plasmids. In addition, we investigated the function of the pESBL-encoded M.EcoGIX methyltransferase, which is also encoded by many other IncI1 and IncF plasmids. This enzyme proved to protect pESBL from restriction in new hosts, suggesting it aids in expanding the plasmid's host range. Collectively, our work illustrates the power of the TnSeq approach to enable rapid and comprehensive analyses of plasmid genes and sequences that facilitate the dissemination of determinants of antibiotic resistance.
INTRODUCTION
The emergence of new bacterial pathogens and the spread of determinants of antibiotic resistance and virulence is often mediated by mobile genetic elements such as bacteriophages and plasmids (1,2). Escherichia coli O104:H4, which caused a large outbreak of bloody diarrhea with a high prevalence of associated hemolytic–uremic syndrome (HUS) in Germany in late Spring 2011 (3,4), provides a recent dramatic example of how horizontal transmission of mobile elements can underlie the emergence of a new pathogen. The O104:H4 outbreak strain was classified as an enteroaggregative E. coli (EAEC) because of its pattern of adherence to cultured cells and the presence of a plasmid (pAA) that encoded the fimbriae that mediate this type of adherence (5). Whole-genome sequencing revealed that the strain contains an unusual amalgam of mobile elements that account for its pathogenicity and its extensive antibiotic resistance profile against a variety of beta-lactams (5–9). In contrast to typical EAEC strains, the outbreak strain contains a prophage encoding Shiga toxin, a potent inhibitor of protein synthesis that is known to cause HUS (10). Furthermore, this strain contains an 88.5-kb plasmid—pESBL-EA11 (henceforth referred to as pESBL)—that encodes two extended spectrum beta-lactamases (ESBL), bla-CTX-M-15 and bla-TEM, which hydrolyze penicillins and extended spectrum cephalosporins (1,8,9,11). Phylogenetic and comparative genomic analyses suggested that pESBL was acquired relatively recently by a Shiga toxin-producing, pAA-bearing precursor of the outbreak strain (8,12,13), as it is not present in other highly related Shiga toxin-producing EAEC O104:H4 non-outbreak strains (12,14).
Successful horizontal dissemination of plasmids such as pESBL in bacterial populations typically depends on factors associated with four principal processes: replication, maintenance, transfer and establishment. Replication and maintenance factors bestow upon plasmids the capacity to replicate and segregate, enabling stable propagation of the plasmid DNA to daughter cells. Transfer factors enable the translocation of plasmid DNA between bacterial cells by conjugation, a complex and highly regulated process that depends on direct cell-to-cell contact requiring specialized ‘Type IV secretion systems’ (15,16). Establishment factors counteract host factors that antagonize acquisition of foreign DNA, e.g. host restriction systems.
The DNA sequence of pESBL is similar to that of pEC_Bactec, an IncI1 incompatibility group plasmid that was isolated from a horse with arthritis and also bears bla-CTX-M-15 and bla-TEM-1 (8,9,13,17). Additional IncI1 plasmids of animal and human origin, isolated from diverse locations, have also been reported to carry ESBLs (18,19). The genome sequence of pESBL also has a similar organization to those of R64 and ColIb, well-studied IncI1 plasmids originally isolated from Salmonella enterica typhimurium and Shigella sonnei, respectively, although the latter plasmids lack genes encoding ESBLs. While the ESBL plasmid is more than 30 kb smaller than R64, it has readily identifiable homologs of known R64 genes that mediate the cardinal processes of plasmid replication, maintenance and transfer. For example, pESBL's RepZ, which functions as a replication initiator and controls the copy number of the plasmid (20,21), is nearly identical to its homolog in R64. Furthermore, the four principal R64 transfer region clusters, which mediate conjugative pilus biogenesis, conjugative DNA processing, general conjugative functions and transfer regulation, are also found in pESBL (see below).
pESBL also appears to encode factors that may promote its establishment in new hosts. For example, both R64 and pESBL encode ardA, which blocks the activity of the restriction endonuclease EcoK1 (22). Both plasmids also encode M.EcoGIX (referred to as YfbB in R64 (20)), an unusual methyltransferase (MTase) that was recently shown to methylate only one of the double-strand DNAs, but whose biological significance remains unknown (23). However, since DNA methylation is a common mechanism by which plasmids can escape digestion by host-encoded restriction endonucleases, the presence of these MTases may expand pESBL's host range.
While there have been several studies of the transfer and replication machinery of R64 and ColIb-P9 (20,24,25), knowledge of the regulatory processes that control the spread of this large group of plasmids remains incomplete and to date there have been no reports of the molecular factors that mediate and control spread of IncI1 plasmids that carry ESBLs, such as pESBL. Furthermore, pESBL contains ∼12 kb of sequence encompassing 13 open reading frames (ORFs) that are not present in the prototypical R64 plasmid, most of which have no discernable homologs. Thus, our goal in this work was to assess the importance of these uncharacterized sequences, as well as the remainder of pESBL's coding and non-coding regions, for the plasmid's replication and transmission.
We utilized transposon (Tn) insertion site sequencing (aka TnSeq) (26,27) to carry out a high-throughput genetic analysis of the factors required for pESBL replication and transmission. The ESBL plasmid was extremely stable in a host E. coli strain, and relatively few genes proved essential for its promulgation in the host cell. However, we found that 32 of 97 genes contribute to its capacity for transmission to new hosts. Most, but not all, of the genes that are required for pESBL transfer are important for R64 transfer as well. In addition to identifying pESBL loci that are required for its transfer, we found two very short regions in the plasmid that appear to regulate its transfer. Transposon-mediated disruption of these regions, whose precise roles remain to be elucidated, augments the levels of mRNA encoding pESBL transfer genes, suggesting that these loci encode a heretofore unrecognized regulatory system controlling spread of IncI1 plasmids. Lastly, we investigated the function of the pESBL-encoded M.EcoGIX MTase, which is also encoded by many IncI1 and IncF plasmids. This enzyme proved to protect pESBL from restriction in new hosts, suggesting it aids in the expansion of the host range of this plasmid.
MATERIALS AND METHODS
Strains, plasmids and media
A Δstx::gent mutant of the E. coli O104:H4 outbreak strain C227-11 (6,23) was used as the source of pESBL for this study. E. coli K-12 strains used here have different antibiotic resistance profiles: MC1061 (rpsL, streptomycin resistant, laboratory collection), MKW278 (MG1655 ΔlacZ::cat, chloramphenicol resistant laboratory collection) and CAG18439 (MG1655 lacI::Tn10, tetracycline resistant) (28) were used for pESBL transfer experiments. SM10 λ pir (laboratory collection) was used as a donor to introduce Tn insertions and gene replacement. Klebsiella pneumoniae B5055 (O1:K2) (29) was kindly provided by Dr Jerry Pier.
The Himar1 suicide transposon delivery vector pSC189 (30) was modified to construct pYB742. In brief, the polymerase chain reaction (PCR) was used to amplify the pSC189 sequence without the bla gene and the cat gene from pKD3 (31), then the two fragments were joined by Gibson Assembly (32).
Plasmid pYB748, the M.EcoGIX::gent allele replacement vector, was constructed by Gibson Assembly using the pDM4 vector (33) along with DNA flanking the M.EcoGIX gene and the gentamicin resistance gene amplified from the O104 Δsxt::gent strain. The plasmid (pEYY12) used for deletion of the high-frequency transfer (Hft) region was also constructed by Gibson Assembly using the pDM4 vector along with DNA flanking the Hft region. Gene replacements in pESBL were carried out by conventional double crossovers with sucrose sensitivity as the counter selection in an MC1061 background (33). The EcoRI restriction modification (R-M) expression plasmid pYB753 was constructed by Gibson Assembly with pBAD18Kn vector (34) and PCR-amplified EcoRI R-M genes (from pIK172 (35), kindly provided by Dr Ichizo Kobayashi). The EcoGIII R-M expression plasmid pDM142 was kindly provided by Dr Diana Munera.
A standard plasmid (pEYY3) that contains chromosomal and plasmid DNA fragments for qPCR analyses for measurement of pESBL copy number was constructed by first amplifying narW and repZ target regions and then inserting them into pBluescript II KS+ (Agilent Tech) by Gibson Assembly.
Bacterial cells were grown in Lysogeny Broth (LB) media and antibiotics and/or inducers were used at following concentrations when appropriate: streptomycin, 200 μg/ml; carbenicillin, 50 μg/ml; tetracycline, 15 μg/ml for E. coli and 10 μg/ml for Klebsiella; chloramphenicol, 20 μg/ml; gentamicin, 5 μg/ml; kanamycin, 50 μg/ml; arabinose, 0.02%.
Oligonucleotides used to construct plasmids and strains are listed in Supplementary Table S1.
Bacterial conjugation and transfer efficiency
Equal amounts (100 μl) of overnight cultures of recipient and donor cells were washed once with LB to remove antibiotics and then mixed and spun down in Eppendorf tube. Cells were then resuspended in 100 μl of LB and placed on a 0.45-μm HAWP filter (Millipore) and placed on LB agar plate and incubated at 37°C for 5 h unless otherwise noted. The cells from the filter were then recovered in 1 ml of LB and after appropriate dilution, spread onto LB plates with appropriate antibiotics for selecting donor, recipient and exconjugants. The number of colonies obtained after overnight incubation at 37°C was counted and the number of exconjugants was divided by the number of donors to obtain transfer frequencies.
Tn insertion sequencing
We followed protocols described in (36) to create transposon insertion libraries and to carry out TnSeq. In brief, overnight cultures of SM10 λ pir/pYB742 (the Tn donor) and MC1061/pESBL were mixed and incubated for 1.5 h. Exconjugant cells with the KmR Himar1 Tn inserted somewhere in the MC1061/pESBL genome (either on the chromosome or plasmid) were recovered on large square plates as SmR CarbR KmR colonies (∼20 000/plate) that were resuspended and collected as the ‘input library’. Total DNA was extracted from a 3-ml aliquot (∼1/5) of the input library. Another aliquot of the input library (with the OD600 adjusted to match that of an overnight culture of recipient) was used as the donor in a second round of conjugation with a new recipient strain CAG18439 (TetR). Exconjugants were selected as TetR CarbR KmR colonies; this scheme selects for Tn insertions in pESBL (not the chromosome) that were still capable of transmission. Approximately 20 000 TetR CarbR KmR colonies were collected and designated as the ‘output library’. Genomic DNA of the output library was also extracted from a 3-ml aliquot of this library. Prior to sequencing, DNA was sheared by sonication, end-repaired with the Quick Blunting Kit (NEB) and then A-tailed by Taq polymerase. Adaptor DNA was ligated and two rounds of PCR were carried out to amplify DNA fragments containing the Tn and appropriate sequences for Illumina sequencing (such as P5 and P7 hybridization sequences, barcodes and spacers). The final PCR products were gel purified on a 2% agarose gel to isolate 200–500-bp fragments and sequenced on the MiSeq V2.0 platform for 65 cycles (Illumina). We prepared two independent libraries but the data analysis was done with the two data sets combined.
Twenty four colonies from the output library were subjected to PCR with himmer3out and repZup primers to isolate mutants with Tn inserted in the Hft region. Two PCR-positive clones were chosen for characterization after the exact Tn insertion site was determined by sequencing. Oligonucleotides used for TnSeq are listed in Supplementary Table S1.
TnSeq data analysis
We used the ARTIST pipeline to analyze the DNA sequences of the transposon insertion sites (37). Briefly, all reads were trimmed of adaptor sequences using CutAdapt (38) and then mapped to the pESBL-EA11 sequence using Bowtie (39). The input library contained 103 900 reads that mapped to pESBL, corresponding to disruption of 92% of all TA dinucleotides in pESBL. The output library contained 654 073 reads that mapped to 34.3% of TA sites in pESBL (Table 1).
Table 1. Summary of TnSeq.
| Input library | Output library | |
|---|---|---|
| # of mapped reads | 103 900 | 635 073 |
| %TA hits | 92.0 | 34.3 |
| % reads from Hft region | 7.6 | 94.6 |
Results from two independently constructed transposon libraries are shown. The high-frequency transfer (Hft) region encompasses 270 bp (coordinates 35394–35663) and includes 21TA sites.
The marked over-representation of reads in the output library mapping to the Hft region could not be accommodated by the analysis pipeline, and we normalized these reads down to the average of all reads from disrupted sites, without affecting reads outside the Hft. The output reads after normalization were then subjected to a sliding window and hidden Markov model (HMM)-based analysis (36,37). The output of the program classified all loci within pESBL as either ‘non-essential’, ‘essential’ (i.e. genes essential for plasmid replication or maintenance), ‘conditionally essentially’ (i.e. genes required for transfer) or ‘enriched’ (genes that promote plasmid transfer).
We utilized Con-ARTIST (37) to define genes that were conditionally essential for transfer. Briefly, the reads for each gene between the input and output libraries were compared using a Mann–Whitney U test. All insertions in genes that were significantly different (P-value <0.002) were defined as conditionally essential and then fed into an HMM. The HMM refined the likelihood that each insertion was truly conditionally essential for transfer, essential for replication/maintenance or not required under either condition. After the HMM ran to convergence, the probabilities of every insertion in the central 90% coding regions of each gene were averaged, and the genes were assigned to the following categories—required for replication/maintenance (essential), required for transfer (conditionally underrepresented) or not required for transmission—depending on which biological category had the highest probability across all statistical tests (see Table 2).
Table 2. ORFs with revised annotations of pESBL and their classification.
| Starta | Enda | Directiona,b | Locus taga | Original annotationa | Revised annotation | Non-essentialc | Essential for maintenancec | Required for transferc |
|---|---|---|---|---|---|---|---|---|
| 1 | 1477 | − | O3K_25572 | TraU protein | TraUd | 0.03 | 0.00 | 0.97 |
| 1567 | 2367 | − | O3K_25577 | TraT transfer protein | TraT | 0.00 | 0.00 | 1.00 |
| 2351 | 2539 | − | O3K_25582 | TraS transfer protein | TraS | 0.00 | 0.00 | 1.00 |
| 2603 | 3007 | − | O3K_25587 | Hypothetical protein | TraR | 0.00 | 0.00 | 1.00 |
| 3058 | 3585 | − | O3K_25592 | TraQ protein | TraQ | 0.00 | 0.00 | 1.00 |
| 3585 | 4289 | − | O3K_25597 | Hypothetical protein | TraP | 0.00 | 0.00 | 1.00 |
| 4289 | 5578 | − | O3K_25602 | Hypothetical protein | TraO | 0.00 | 0.00 | 1.00 |
| 5581 | 6564 | − | O3K_25607 | Hypothetical protein | TraN | 0.00 | 0.00 | 1.00 |
| 6575 | 7267 | − | O3K_25612 | TraM protein | TraM | 0.00 | 0.00 | 1.00 |
| 7264 | 7611 | − | O3K_25617 | F pilus assembly | TraL | 0.00 | 0.00 | 1.00 |
| 7629 | 10160 | − | NAe | SogS | 0.00 | 0.00 | 1.00 | |
| 7629 | 11393 | − | O3K_25622 | SogL protein | SogL | 0.26 | 0.00 | 0.74 |
| 11483 | 12034 | − | O3K_25627 | EDTA-resistant nuclease | Nuc | 1.00 | 0.00 | 0.00 |
| 12049 | 12339 | − | NAe | TraK | 0.03 | 0.00 | 0.97 | |
| 12336 | 13484 | − | O3K_25632 | TraJ transfer ATPase | TraJ | 0.00 | 0.00 | 1.00 |
| 13481 | 14299 | − | O3K_25637 | Lipoprotein | TraI | 0.00 | 0.00 | 1.00 |
| 14296 | 14754 | − | O3K_25642 | TraH lipoprotein | TraH | 0.02 | 0.00 | 0.98 |
| 15149 | 15733 | − | O3K_25647 | Lipopolysaccharide core heptose(II)-phosphate phosphatase | TraG | 0.79 | 0.21 | 0.00 |
| 15793 | 16995 | − | O3K_25652 | F pilus assembly | TraF | 1.00 | 0.00 | 0.00 |
| 17081 | 17905 | − | O3K_25657 | F pilus assembly | TraE | 0.93 | 0.07 | 0.00 |
| 18056 | 19210 | − | O3K_25662 | Shufflon-specific DNA recombinase | Rci | 1.00 | 0.00 | 0.00 |
| 19275 | 19562 | + | NAe | shufflon (D) | 1.00 | 0.00 | 0.00 | |
| 19604 | 19822 | + | NAe | shufflon (C’) | ||||
| 19819 | 20070 | − | NAe | shufflon (C) | ||||
| 20105 | 20350 | + | NAe | shufflon (B) | ||||
| 20231 | 20350 | + | O3K_25667 | Hypothetical protein | f | |||
| 20352 | 20603 | − | NAe | shufflon (B’) | ||||
| 20375 | 20581 | − | O3K_25672 | Hypothetical protein | f | |||
| 20623 | 20847 | + | NAe | shufflon (A’) | ||||
| 20844 | 22280 | − | O3K_25677 | Shufflon protein A | PilV (A) | 0.18 | 0.00 | 0.82 |
| 22268 | 22924 | − | O3K_25682 | PilU prepilin peptidase | PilU | 0.00 | 0.02 | 0.98 |
| 22909 | 23469 | − | O3K_25687 | Type IV prepilin cluster | PilT | 0.01 | 0.10 | 0.90 |
| 23479 | 24093 | − | O3K_25692 | Type IV prepilin cluster, prepilin | PilS | 0.09 | 0.00 | 0.92 |
| 24110 | 25195 | − | O3K_25697 | Putative transmembrane protein | PilR | 0.08 | 0.00 | 0.93 |
| 25208 | 26761 | − | O3K_25702 | Type IV prepilin cluster, ATP-binding protein | PilQ | 0.62 | 0.00 | 0.38 |
| 26772 | 27224 | − | O3K_25707 | Putative conjugative transfer PilP protein | PilP | 0.87 | 0.13 | 0.00 |
| 27211 | 28506 | − | O3K_25712 | PilO protein | PilO | 0.97 | 0.03 | 0.00 |
| 28499 | 30181 | − | O3K_25717 | PilN type IV pilus secretin | PilN | 1.00 | 0.00 | 0.00 |
| 30195 | 30632 | − | O3K_25722 | PilM type IV pilus biogenesis protein | PilM | 0.99 | 0.00 | 0.01 |
| 30632 | 31699 | − | O3K_25727 | Lipoprotein PilL | PilL | 1.00 | 0.00 | 0.00 |
| 32028 | 32621 | − | NAe | PilK | 0.50 | 0.46 | 0.04 | |
| 32746 | 33045 | − | O3K_25732 | Hypothetical protein | PilI | 0.95 | 0.00 | 0.05 |
| 33172 | 33855 | − | O3K_25737 | TraC | TraC | 0.12 | 0.00 | 0.88 |
| 34109 | 34642 | − | O3K_25742 | TraB protein | TraB | 0.44 | 0.00 | 0.56 |
| 35084 | 35371 | − | O3K_25747 | Protein TraA | TraA | 1.00 | 0.00 | 0.00 |
| 35289 | 35552 | − | O3K_25752 | Hypothetical protein | Hp1 | 1.00 | 0.00 | 0.00 |
| 36448 | 36537 | + | NAe | RepY | 0.04 | 0.65 | 0.31 | |
| 36525 | 37556 | + | O3K_25757 | Replication initiation protein | RepZ | 0.08 | 0.50 | 0.42 |
| 38626 | 39135 | − | O3K_25762 | Protein YafA | Hp2 | 0.96 | 0.04 | 0.00 |
| 39191 | 39793 | − | O3K_25767 | Hypothetical protein | Hp3 | 0.95 | 0.05 | 0.00 |
| 40146 | 41492 | + | O3K_25772 | Hypothetical protein | Hp4 | 1.00 | 0.00 | 0.00 |
| 41980 | 44454 | − | O3K_25777 | Hypothetical protein | TnpA | 1.00 | 0.00 | 0.00 |
| 44849 | 45121 | + | O3K_25782 | Hypothetical protein | Hp5 | 1.00 | 0.00 | 0.00 |
| 45168 | 46043 | − | O3K_25787 | Beta-lactamase | Bla-CTX-M-3 | 1.00 | 0.00 | 0.00 |
| 46196 | 46318 | − | O3K_25792 | Hypothetical protein | Hp6 | 0.96 | 0.04 | 0.00 |
| 46299 | 47561 | − | O3K_25797 | ISEcp1 transposase | Transposase1 | 0.44 | 0.56 | 0.00 |
| 47743 | 47961 | − | O3K_25802 | Transposase | Transposase2 | 0.98 | 0.02 | 0.00 |
| 48125 | 48682 | + | O3K_25807 | Tn3 resolvase | TnpR | 1.00 | 0.00 | 0.00 |
| 48865 | 49725 | + | O3K_25812 | Beta-lactamase TEM | Bla-TEM | 1.00 | 0.00 | 0.00 |
| 49994 | 50296 | − | O3K_25817 | Hypothetical protein | Hp7 | 0.20 | 0.80 | 0.00 |
| 50293 | 50826 | − | O3K_25822 | Type II plasmid partioning protein | ParA-like | 0.84 | 0.16 | 0.00 |
| 51123 | 52394 | − | O3K_25827 | DNA polymerase V subunit UmuC | UmuC/ImpB | 1.00 | 0.00 | 0.00 |
| 52394 | 52831 | − | O3K_25832 | DNA polymerase V subunit ImpA | ImpA | 1.00 | 0.00 | 0.00 |
| 52828 | 53076 | − | O3K_25837 | Hypothetical protein | ImpC | 1.00 | 0.00 | 0.00 |
| 53471 | 54397 | + | O3K_25842 | Hypothetical protein | Hp8 | 1.00 | 0.00 | 0.00 |
| 54782 | 55465 | + | O3K_25847 | Putative methylase | M.EcoGIX | 1.00 | 0.00 | 0.00 |
| 55466 | 55687 | + | O3K_25852 | Hypothetical protein | Hp9 | 1.00 | 0.00 | 0.00 |
| 55701 | 56135 | + | O3K_25857 | Hypothetical protein | Hp10 | 1.00 | 0.00 | 0.00 |
| 56181 | 56957 | + | O3K_25862 | YchA | Hp11 | 1.00 | 0.00 | 0.00 |
| 57371 | 57796 | + | O3K_25867 | Antirestriction protein | KlcA/ArdB | 1.00 | 0.00 | 0.00 |
| 57843 | 58265 | + | O3K_25872 | Hypothetical protein | Hp12 | 1.00 | 0.00 | 0.00 |
| 58262 | 58453 | + | O3K_25877 | Hypothetical protein | Hp13 | 1.00 | 0.00 | 0.00 |
| 59222 | 59749 | + | O3K_25882 | Single-stranded DNA-binding protein | Ssb | 0.96 | 0.02 | 0.02 |
| 59807 | 60040 | + | O3K_25887 | Hypothetical protein | Hp14 | 0.95 | 0.03 | 0.02 |
| 60099 | 62057 | + | O3K_25892 | Type II plasmid partioning protein | ParB-like | 1.00 | 0.00 | 0.00 |
| 62112 | 62546 | + | O3K_25897 | Plasmid SOS inhibition protein B | PsiB | 1.00 | 0.00 | 0.00 |
| 62543 | 63262 | + | O3K_25902 | Plasmid SOS inhibition protein A | PsiA | 1.00 | 0.00 | 0.00 |
| 63259 | 63855 | + | O3K_25907 | YgaA | Hp15 | 1.00 | 0.00 | 0.00 |
| 64317 | 64817 | + | O3K_25912 | Anti-restriction protein | ArdA | 1.00 | 0.00 | 0.00 |
| 65546 | 65980 | + | O3K_25917 | Hypothetical protein | Hp16 | 0.76 | 0.24 | 0.00 |
| 66074 | 66340 | + | O3K_25922 | Hypothetical protein | Hp17 | 1.00 | 0.00 | 0.00 |
| 66517 | 66906 | + | O3K_25927 | Hypothetical protein | Hp18 | 1.00 | 0.00 | 0.00 |
| 66903 | 67817 | + | O3K_25932 | Hypothetical protein | Hp19 | 1.00 | 0.00 | 0.00 |
| 67879 | 68085 | + | O3K_25937 | Hypothetical protein | Hp20 | 1.00 | 0.00 | 0.00 |
| 68116 | 68367 | − | O3K_25942 | Hypothetical protein | Hp21 | 1.00 | 0.00 | 0.00 |
| 68947 | 69795 | + | O3K_25947 | Hypothetical protein | Hp22 | 1.00 | 0.00 | 0.00 |
| 69882 | 70217 | − | O3K_25952 | Protein YggA | Hp23 | 0.99 | 0.00 | 0.01 |
| 70354 | 70436 | NAe | oriTg | 0.28 | 0.00 | 0.72 | ||
| 70451 | 70783 | + | O3K_25957 | Relaxosome component | NikA | 0.00 | 0.00 | 1.00 |
| 70794 | 73493 | + | O3K_25962 | Relaxase | NikB | 0.00 | 0.00 | 1.00 |
| 73530 | 75821 | − | O3K_25967 | IcmO-like type IV secretion system protein | TrbC | 0.00 | 0.00 | 1.00 |
| 75814 | 76884 | − | O3K_25972 | Hypothetical protein | TrbB | 0.00 | 0.00 | 1.00 |
| 76903 | 78111 | − | O3K_25977 | Hypothetical protein | TrbA | 0.00 | 0.00 | 1.00 |
| 78418 | 79503 | − | O3K_25982 | Fertility inhibition protein | Hp24 | 0.59 | 0.41 | 0.00 |
| 79766 | 79969 | + | NAe | PndC | 1.00 | 0.00 | 0.00 | |
| 79821 | 79976 | + | O3K_25987 | Hypothetical protein | PndA | 1.00 | 0.00 | 0.00 |
| 80048 | 80299 | − | O3K_25992 | Hypothetical protein | Hp25 | 1.00 | 0.00 | 0.00 |
| 80599 | 80895 | + | O3K_25997 | Hypothetical protein | Hp26 | 1.00 | 0.00 | 0.00 |
| 80960 | 81136 | − | O3K_26002 | Hypothetical protein | Hp27 | 0.57 | 0.42 | 0.01 |
| 81133 | 81276 | − | O3K_26007 | Hypothetical protein | Hp28 | 0.22 | 0.76 | 0.02 |
| 81319 | 81528 | − | O3K_26012 | Putative regulator protein | Hha/YmoA | 0.40 | 0.59 | 0.02 |
| 81626 | 82240 | − | O3K_26017 | Surface exclusion protein | ExcA | 0.47 | 0.29 | 0.24 |
| 82316 | 84553 | − | O3K_26022 | TraY integral membrane protein | TraY | 0.01 | 0.16 | 0.84 |
| 84581 | 85165 | − | O3K_26027 | F pilin acetylation protein | TraX | 0.00 | 0.00 | 1.00 |
| 85194 | 86396 | − | O3K_26032 | F pilus assembly | TraW | 0.00 | 0.00 | 1.00 |
| 86363 | 86977 | − | O3K_26037 | F pilus assembly | TraV | 0.00 | 0.00 | 1.00 |
| 86977 | 88544 | − | O3K_25572 | TraU protein | TraUd | 0.00 | 0.00 | 1.00 |
aThe reference genome was downloaded from NCBI with accession number NC_018659.1.
bgenes coded on the complementary strand are shown with a ‘−’.
cProbability of essentiality for replication/maintenance obtained by Con-ARTIST analysis (see the Materials and Methods section). The most probable assignment among the three categories is in bold. The ‘enriched’ category is not shown, since no genes were predicted to be enriched. Note that the unannotated Hft region is not shown in this table. The entire shufflon region, not individual genes, was used for this analysis.
dTraU is separated in two pieces because of linear presentation.
eNot annotated in original reference.
fRevised as shufflon segment shown in other rows.
goriT is not an ORF but shown in this table for clarity.
pESBL annotation
Re-annotation of the pESBL genome was performed by a BLASTP search against the NCBI plasmid database (ftp://ftp.ncbi.nlm.nih.gov/genomes/Plasmids/) and KEGG database (40). Coding DNA Sequences (CDSs) were annotated as a gene encodes a hypothetical protein when the CDSs had any of (i) E-value more than 1e−10, (ii) coverage of length less than 60% and (iii) sequence identity less than 40%, in the top hit of the BLASTP result. All the hypothetical genes were numbered for clarity. Furthermore, sogS, traK, pilK, repY and shufflon segments were added based on their identity to the R64 counterparts.
Gene expression analysis
RNA from log phase culture was extracted using the Trizol reagent (Life Tech). After DNase treatment, mRNA was converted to cDNA by Superscript III using random hexamers (Life Tech). A StepOne real time PCR system and a SYBR Green Master Mix reagent (Life Tech) and the ΔΔCt method (41) were used to measure transcript levels in mutants and the wild type. rpoB was used for the internal control. PCR primers for these experiments are listed in Supplementary Table S1 and the Student's two-tailed t-test from four independent experiments was used to analyze statistical significance.
Determination of pESBL copy number
Genomic DNA was extracted from log phase cultures with the Wizard gDNA purification kit (Promega) and subjected to quantitative PCR (qPCR) in the LightCycler 480 system with the SYBR Green I Master reagent (Roche). repZ and narW loci were used for plasmid and chromosome quantification, respectively, and pEYY3 was used for the standard curve. The primers used for these experiments are listed in Supplementary Table S1.
DNA sequences
Accession numbers for the DNA sequences analyzed in this study are NC_018659.1 (pESBL isolated from strain 2011C-3493) and AP005147.1 (R64). We also referred to the pESBL sequence from E. coli O104:H4 strain TY-2482 found in the PATRIC database (http://patricbrc.org/portal/portal/patric/Genome?cType=genome&cId=192592).
RESULTS AND DISCUSSION
pESBL is self-transmissible and stable
Although epidemiologic data suggest that pESBL is transmissible, to date there have been no reports investigating whether pESBL can be transmitted from the E. coli O104:H4 outbreak strain to other bacteria. Using a plate-based bacterial conjugation protocol, we found that pESBL can be transferred from a Δstx derivative of the E. coli O104:H4 outbreak strain to a derivative of E. coli K-12 strain MG1655 (Figure 1). Furthermore, the resulting exconjugant (MKW278/pESBL) could serve as a donor of pESBL to additional E. coli strains at frequencies of greater than 1 x 10−1 exconjugants/donor (Figure 1). Together, these observations suggest that pESBL is readily transmissible and encodes all the factors required for its high frequency transfer between E. coli strains. We also found that pESBL could be transferred from an E. coli K-12 donor to K. pneumoniae, albeit at a lower frequency than between E. coli strains (Figure 1); however, no exconjugants were detected in conjugations between MC1061/pESBL and either Pseudomonas aeruginosa or Vibrio cholerae. Thus, pESBL's host range may be relatively restricted, perhaps to the Enterobacteriaceae, consistent with its classification within IncI1 plasmids, which are known to have a relatively narrow-host-range restricted to enterobacteria (42). Besides being self-transmissible, pESBL appears to be stably maintained in its host. We were unable to isolate an E. coli O104:H4 strain that spontaneously lost pESBL after in vitro or in vivo (in rabbits) passage, despite observing loss of the outbreak strain's pAA plasmid under identical conditions (6). Furthermore, we did not detect pESBL loss from E. coli MC1061 after 25 generations of in vitro growth without antibiotic selection.
Figure 1.

Frequency of conjugative transfer of pESBL between different donor/recipient pairs. The frequency of pESBL transfer between donor strains (the indicated strains carrying pESBL) and recipient strains is expressed as exconjugants per donor. Each mating pair is tested at least three times and the means and standard deviations are shown. The limit of detection in these experiments is between 10−8 and 10−7.
Creation and utilization of a high-density transposon insertion library in pESBL
We used transposon insertion site sequencing, an approach that facilitates rapid and often comprehensive linkage of genes and phenotypes (26,27), to begin to identify the genes and sequences that contribute to pESBL's replication, stable maintenance and transmission. Himar1, a mariner-based Tn that inserts in TA dinucleotides but has little other sequence bias, was used for high-density mutagenesis of pESBL (36). Initially, we constructed a Tn-insertion library in E. coli MC1061 harboring pESBL, as a way to identify pESBL ‘essential’ genes. Such loci should be under-represented for (or lack) transposon insertions, and correspond to genes/sequences that enable pESBL's replication and/or contribute to its segregation and stability. The set of pESBL insertions present in this initial library is referred to subsequently as the ‘input library’. The input set of strains was used to donate the library of mutated pESBLs to another host, CAG18439, a tetracycline resistant E. coli K-12 strain, and resistance markers on the plasmid and on the transposon were used for selection of exconjugants. The set of pESBL insertion mutants in the CAG18439 exconjugants is referred to below as the ‘output library’. We took advantage of a recently developed analytic pipeline (37) to compare the distribution of transposon insertion sites in the input and output libraries and identify pESBL loci that are required for or modulate its transfer. Insertions in genes or intergenic sequences in pESBL that are under-represented or absent in the output library (relative to their abundance in the input library) likely correspond to loci that facilitate or are essential for pESBL transfer, and are termed ‘genes required for transfer’. In contrast, genes that are over-represented in the output library likely negatively affect plasmid transfer.
Even though the great majority of insertions generated in MC1061/pESBL did not map to the 88.5-kb pESBL, which only represents a small fraction of this strain's genome, our library of insertions included more than 100 000 reads that mapped to pESBL. The pESBL insertions in the input library covered 92% of the TA sites in this plasmid, suggesting that the library is sufficiently saturated to enable identification of genes essential for pESBL replication, maintenance and transfer. In the output library, which contained more than 600 000 reads, a far smaller fraction (34%) of the plasmid's TA sites contained insertions (Table 1), consistent with the idea that a significant portion of pESBL contributes to its transfer. In the input library, the Tn insertions were fairly evenly distributed around pESBL, although there was a small region of 270 bp that contained almost 8% of all insertions. In the output library, more than 94% of the transposon insertions mapped to this small region, suggesting that the Tn insertions in this region increase the efficiency of pESBL transfer (see below).
pESBL genes required for plasmid replication
Using a recently developed sliding window/HMM-based analytic pipeline (36) to analyze the distribution of transposon insertions, we identified regions in pESBL that were consistently underrepresented for reads in the input library, which are presumed to correspond to insertions that impair plasmid replication or segregation, and hence are selected against in the library. Because the existing pESBL annotations were incomplete, we re-annotated the pESBL genome (Table 2 and Supplementary Figure S1) and relied on this new set of annotations for all downstream transposon-insertion sequencing analyses. Only six ORFs had a sufficiently reduced frequency of insertions to suggest that they are essential for the plasmid's replication/maintenance. One of these genes, repZ, encodes the putative pESBL replication initiator, and is 95.6% identical to repZ in the IncI1 plasmids R64 and ColIb-P9. In the latter plasmid, the level of repZ expression has been found to control ColIb-P9 copy number (21,43). Insertions were also rare in repY, which is a small gene immediately upstream of repZ that regulates expression of the initiator (20,43) in R64. The means by which the other four ORFs classified as essential contribute to the replication/maintenance of pESBL is not evident. One of the four genes is annotated as a transposase, another as hha/YmoA (a putative regulator of gene expression), and two short ORFs (hp7 and hp28) encode hypothetical proteins with no homologs of known function. It is possible that one or more of these regions may not correspond to protein coding sequence, but instead represent essential DNA sequences. We also note that insertions within duplicated loci or redundant genes that participate in plasmid replication or maintenance are unlikely to be under-represented.
Genes required for pESBL transfer
As expected, many more genes (32 in total) were classed as essential for pESBL transfer than for its replication/maintenance. The majority of these genes are clustered in groups of at least five genes, so that the plasmid genome consists of domains that are essential for transfer (Figure 2, blue genes) punctuated by (sometimes large) regions of non-essential genes (see also Table 2 and Supplementary Figure S1). Genes that are dispensable for transfer include bla-CTX-M-3 and bla-TEM, which encode the plasmid's extended spectrum beta lactamases, and most of pESBL's ORFs that lack significant homologs (annotated here as hypothetical proteins; hp) (Table 2). Most of these non-essential genes do not have counterparts in R64, suggesting that this region in R64-related plasmids may consist of variable ‘cargo’ genes that inserted into the core backbone of an ancestral plasmid during the evolution of this plasmid group.
Figure 2.
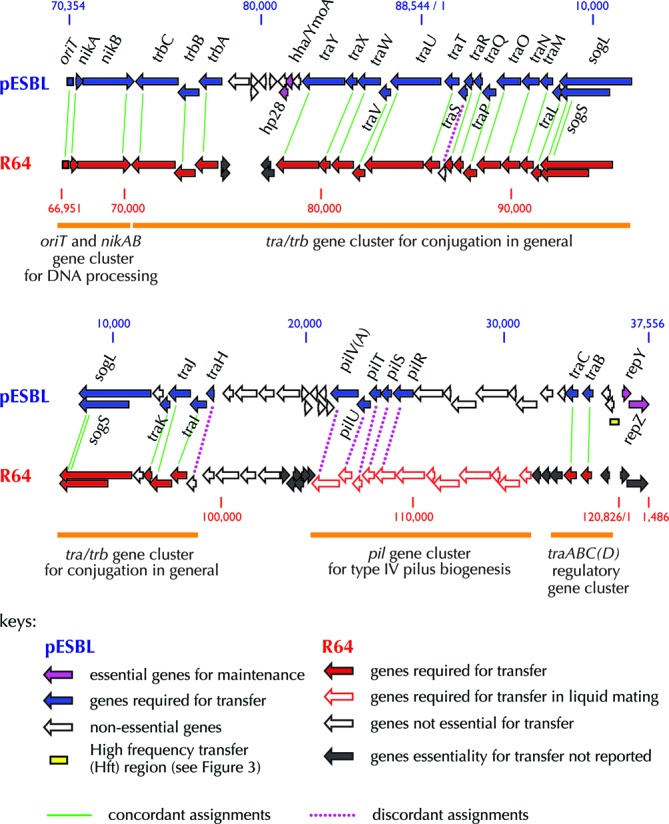
Comparison of pESBL and R64 transfer genes. Alignment of the pESBL and R64 transfer regions, depicting genes critical for pESBL transfer (as defined in this study) in blue and critical for R64 transfer on solid media shown in solid red arrows and in liquid media with open red arrows. Classifications of R64 transfer genes (based on (20) and references therein) are also indicated. Genes essential for both pESBL and R64 transfer are joined with green lines and genes that are essential for pESBL but not R64 transfer are joined with purple dotted lines. Genes required for pESBL maintenance/replication are shown in magenta. Note pESBL lacks a traD homolog and that sogL/sogS are duplicated on the top and bottom rows of the map. Additionally, the gene arrangement shown in this figure is a circular permutation of the arrangement listed in Table 2.
Notably, all 32 genes required for pESBL transfer correspond to parts of the four previously defined functional clusters within the R64 ‘transfer region’. These clusters are (i) the nikAB and oriT region for conjugative DNA processing, (ii) the tra/trb gene cluster (spanning from traH - trbC) for general conjugation functions, (iii) the pil operon for type IV pilin biogenesis and (iv) the traABCD regulatory genes ((20) and Figure 2). With a few exceptions (outlined below), the genes we classified as essential for pESBL transfer (Table 2) were homologous to R64 genes found to mediate transfer in previous non-TnSeq-based analyses (24,44–48) (Figure 2).
The pil genes, which encode the machinery for the biogenesis of a type IV pilus (45), appear to have somewhat distinct roles in R64 and pESBL. In R64, these genes are required for conjugative transfer in liquid media, but not on solid media (24). However, in pESBL, five pil genes (pilRSTUV) were essential for pESBL transfer on the solid media we used for our screen. Another difference between R64 and pESBL transfer genes concerns traS and traH. These two small genes are found in the traH-Y region of the tra/trb cluster, which is very similar in the two plasmids. In R64, only three of the 20 genes from this region—nuc, traS and traH—are not essential for transfer (24). In contrast, our analyses revealed that only nuc is dispensable for transmission of pESBL, while the remaining 19 genes are required for efficient pESBL transfer (Figure 2). The reasons for the apparent differences in roles of traH, which encodes a lipoprotein, and TraS, whose function is not known, remain to be elucidated.
Discovery of Hft region
Unexpectedly, the majority of transposon insertions in the output library mapped to a very small (270 bp) region in pESBL located between traA and repY. This region (Figure 2, yellow box, and Figure 3A), which includes only ∼0.5% of pESBL's TA dinucleotide sites, contained nearly 95% of the mapped reads in the output library (Table 1). When compared to the read abundance for flanking regions, reads in the 270-bp region are ∼100-fold enriched. To test whether Himar1 insertions in this region promote pESBL transfer, which might account for this enrichment, we isolated two independent pESBL::Tn clones (pESBL::Tn1 and pESBL::Tn2; 1 and 2 in Figure 3A) that each contain a Tn inserted in this region (dubbed Hft, for high frequency transfer) and measured their transfer frequency. As shown in Figure 3B, both plasmids exhibited ∼20-fold higher frequency of transfer compared to wild-type pESBL. Increase of transfer efficiency was also observed with these pESBL variants when K. pneumoniae was used as the recipient (Figure 3C).
Figure 3.
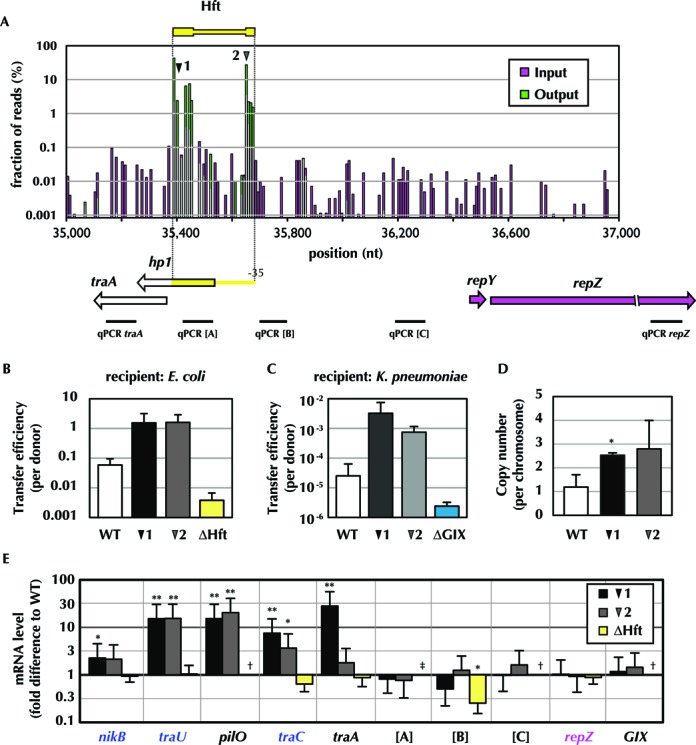
Transposon insertions in a very small region of pESBL promote its transfer. (A) Map and fraction of reads in the high-frequency transfer region and its flanking DNA. Reads from the input library are indicated with pink bars, and reads from the output library are shown with green bars. Overlap of pink and green bars yields white. The Tn insertion sites in pESBL::Tn1 and pESBL::Tn2 (1 and 2) used in the analyses below are shown with the black and gray arrowheads. DNA regions amplified in RT-qPCR analyses are also indicated. (B, C) Transfer efficiency (exconjugants per donor) of pESBL mutants into E. coli CAG18439 (B) and K. pneumoniae (C); note the result for the wild type (WT) in these panels is reiterated from Figure 1. Experiments were carried out more than three times and the mean and standard deviations are shown. (D) Copy number of wild type (WT) pESBL, pESBL::Tn1 and pESBL::Tn2 measured by qPCR. (E) Transcript abundance of the indicated genes and regions from pESBL::Tn1 and pESBL::Tn2 and the deletion mutant (ΔHft) relative to wild-type levels. The results represent the mean and standard deviation from three independent experiments; **P < 0.01 and *P < 0.05. † not tested, ‡ not measured as ΔHft lacks corresponding sequence. The mean and standard deviations of three biological replicates are shown; *P < 0.05.
Interestingly, insertion in the Hft region was also enriched in the input library, although to a lesser extent. In part this appears to reflect an ∼2× increase in the copy numbers of pESBL::Tn1 and pESBL::Tn2 relative to wild-type pESBL (Figure 3D); it might also reflect plasmid transmission among donor strains, although this has not been assessed, and might be prevented by surface exclusion. The elevated frequency of Hft insertions among donor strains, coupled with the observed ∼20× increase in transmission of pESBL::Tn1 and pESBL::Tn2, likely accounts for the ∼100× enrichment of such mutants in the output library. It is also possible that elevated plasmid copy number enhances the probability of plasmid transfer; however, we think it is unlikely that this accounts for the full 20× increase in transmission of pESBL::Tn1 and pESBL::Tn2.
From a genetic perspective, insertions in the Hft region could heighten pESBL transfer either by disrupting the activity of a negative regulator or by promoting the activity of a positive regulator. To begin to distinguish between these two possibilities, we assessed the transfer frequency of pESBL lacking the Hft region. We found that deletion of the entire Hft region (ΔHft) reduced the frequency of pESBL transfer by ∼10× (Figure 3B), rather than increasing it as the transposons do, suggesting that the transposons do not simply inactivate a negative regulatory gene or locus within Hft (e.g. the putative ORF hp1). The 10× reduction in transfer frequency is consistent with the idea that the Hft locus contains or encodes an activator of transfer, although it is not clear how Tn insertions in this locus might augment its activity. Polar effects caused by readthrough from the kanamycin resistance cassette in the transposon do not appear to account for enhanced transfer, since we did not observe enrichment of a particular transposon orientation within the Hft insertions in the output library compared to the input library.
To further characterize the effects of Hft insertions, we compared expression of transfer-related genes from pESBL::Tn1 and pESBL::Tn2, wild-type pESBL and ΔHft. Notably, pESBL::Tn1 and pESBL::Tn2 were associated with higher expression of several genes required for transfer, including nikB, traU and traC, which is a putative activator of transfer gene expression (47) (Figure 3E). With pESBL::Tn1, increased expression of traA, which is not required for transfer but is adjacent to the Hft locus, was also observed, whereas traA expression from pESBL::Tn2 did not significantly differ from wild-type pESBL. This result raises the possibility that the two insertion mutants do not act in entirely the same fashion to increase pESBL transmission. Notably, we also observed that neither mutation altered expression of the replication gene repZ or of sequences between the insertion sites and repZ (Figure 3E, sites A, B, C and repZ). These findings suggest that the increases in expression of nikB, traU and traC are not a non-specific result of increased plasmid copy number, but may instead be restricted to transfer-related genes. Additionally, they somewhat surprisingly suggest that the increase in plasmid copy number is not dependent upon increased production of the pESBL replicator, RepZ. Finally, we observed that none of the pESBL transcripts assayed were elevated for the ΔHft mutant, a difference in phenotype that provides further evidence that the transposon insertions do not simply result in loss of Hft function.
Careful inspection of the distribution of insertions within the short (270 bp) Hft region revealed that the enriched reads in the output library were not equally distributed across this region. Instead, the enriched reads mapped to its ends, and bracket a middle zone with TA sites that were not enriched in the output library (Figure 3A). This distribution provides additional support for the idea that the insertions may not all act in the same fashion and raises the possibility that the Hft region encodes more than a single activity, e.g. it mediates both activation and repression, and either activity can be modulated by transposon insertion. Such complexity would complicate interpretation of the effects of deleting the Hft region, and therefore add a caveat to conclusions drawn from study of the ΔHft mutant. Although the full significance of this bimodal distributions of insertions remains to be explored, these observations illustrate the tremendous resolving power of transposon insertion site sequencing for interrogating functions encoded in unannotated DNA sequences. Collectively, our findings suggest that the means by which the Hft region controls pESBL transfer, and probably transfer of other R64 related plasmids where this region is well conserved, will be quite complicated and an intriguing area for future investigation.
Genes that modulate pESBL host range
Genes required for plasmid replication, maintenance and transfer should be readily identifiable in all TnSeq analyses of conjugative plasmids. In contrast, identification of genes that contribute to plasmid establishment in new host cells is likely to be more variable, as it is modulated by features of the recipient cells as well as of the plasmid. For example, pESBL contains sequence homologous to the ‘shufflon’ region of R64, which functions (along with the Rci recombinase) as a site-specific inversion system to generate diversity in the lipopolysaccharide (LPS) binding, C-terminal region of the PilV adhesin, thereby expanding the plasmid's host range (49). However, the importance of a shufflon should depend on the nature of the potential recipient's LPS. In pESBL, the apparently intact (Supplementary Figure S2) but previously unannotated shufflon sequence likely encodes a functional system, since the two reported pESBL DNA sequences differ in the configurations of their respective shufflon regions (Supplementary Figure S2). Our TnSeq-based analyses indicate that the pESBL shufflon region is not required for transfer of pESBL to E. coli K-12 strain CAG18439; however, it is possible that analyses with a range of recipients might reveal a role for the shufflon region in expanding pESBL's host range.
Plasmid establishment in a new host can also be influenced by host factors designed to counter invasion by foreign DNA, as well as by plasmid mechanisms for countering such defenses, such as restriction enzymes and their associated methyltransferases (MTase). The pESBL locus M.EcoGIX encodes an unusual single-strand DNA MTase activity with no conventional nucleotide recognition sequence (23), and homologs of this MTase can be found in many plasmids, e.g. YfbB of R64 (94% identity) and YfeA of F (91% identity) and in some chromosomes as well (Supplementary Table S2). Our transposon insertion sequencing analyses revealed that M.EcoGIX is not required for transmission of pESBL to E. coli CAG18439; however, CAG18439 does not encode a Type II restriction enzyme that targets sequences that might be methylated by M.EcoGIX. We hypothesized that introduction of DNA restriction endonucleases into recipient cells might reveal a role for M.EcoGIX during transmission, and to explore this possibility, we compared transmission of wild-type pESBL and pESBL lacking M.EcoGIX (ΔGIX) to a variety of hosts. Notably, deletion of M.EcoGIX does not alter the stability of pESBL within E. coli MC1061, and growth of MC1061 cells containing ΔGIX was equivalent to that of the strain containing the wild-type plasmid (Supplementary Figure S3). As expected based on our TnSeq results, transmission of ΔGIX to CAG18439 was not impaired relative to transmission of pESBL. In contrast, when R-M systems (e.g. the recently described R-M EcoGIII (23) or the standard endonuclease EcoRI and its associated MTase) were present in recipient cells but not in donors, transmission of both wild-type and ΔGIX plasmids was reduced, but transfer of ΔGIX was significantly lower (as much as 1000×) than that of wild-type pESBL (Figure 4). This difference was abolished when the EcoGIII or EcoRI R-M systems were also present in donor cells, presumably because all potential restriction sites were methylated and hence protected in the donor strains prior to conjugation (Figure 4). Collectively, our data suggest that the non-specific adenine MTase M.EcoGIX provides pESBL with some defense against restriction endonucleases that might otherwise hamper its transmission to new hosts. This defense does not appear to protect 100% of transmitted DNA, perhaps because M.EcoGIX does not appear to be a potent MTase (23); however, it likely can counter a broader range of endonucleases than do sequence-specific MTases. This may account for the ∼9-fold reduction in transfer efficiency to K. pneumoniae of ΔGIX pESBL compared to wild-type pESBL (Figure 3C), although we do not know the precise restriction system content of the K. pneumoniae strain utilized in this experiment.
Figure 4.
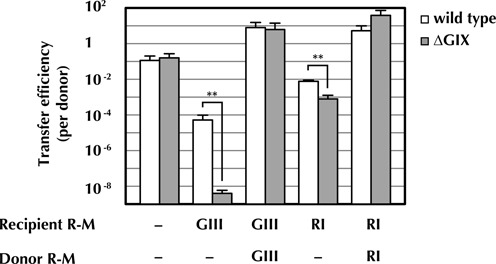
M.EcoGIX can promote pESBL transfer to strains expressing restriction modification systems. Transfer efficiency (exconjugants per donor) of wild-type pESBL (white bars) and a pESBL M.EcoGIX deletion mutant (ΔGIX) (gray bars) from and to E. coli MC1061 harboring plasmid expressing the indicated R-M system (GIII, EcoGIII and RI, EcoRI). The experiment was carried out three times and the mean and standard deviations are shown. Student's two-tailed t-test was used to compare the values between wild type and ΔGIX with log10 values, **P < 0.01.
Conclusions/perspectives
Although IncI1 plasmids such as R64 have been the subject of many outstanding studies over the past few decades, our findings highlight that there is still much to be learned about this and other plasmid families. In particular, our work demonstrates the power of the TnSeq approach to enable rapid and comprehensive analyses of the genes and sequences that enable plasmid replication, stability and transmission. The resolving power of this approach is illustrated by our discovery of a small 270-bp region in pESBL that modulates its transmission. This previously unrecognized Hft region is conserved among several IncI, IncIγ and IncFII plasmids, including R64, R621a and pRK1, and likely governs plasmid transfer in these systems as well. The relative ease of TnSeq-based analyses should facilitate high-throughput analyses of several features of plasmid biology that have not been feasible using gene-by-gene approaches. For example, TnSeq-based discovery of plasmid factors that counter host defenses against acquisition of foreign DNA should be possible employing panels of recipient strains, rather than a single strain, as we used here. Similarly, studies like to those reported here could employ a variety of donor cell backgrounds, in order to investigate how the host cell's genotype modulates plasmid phenotypes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Drs Ichizo Kobayashi, Jerry Pier and Diana Munera for plasmids and strain, and Mahnaz Sabeti Azad for her experimental assistance. We also thank members from Waldor and Yamaichi laboratories for discussions and comments on the manuscript.
FUNDING
National Institute of Allergy and Infectious Diseases [R37 AI-042347]; Howard Hughes Medical Institute. Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Carattoli A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013;303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Juhas M. Horizontal gene transfer in human pathogens. Crit. Rev. Microbiol. 2013. doi:10.3109/1040841X.2013.804031. [DOI] [PubMed]
- 3.Scheutz F., Nielsen E.M., Frimodt-Møller J., Boisen N., Morabito S., Tozzoli R., Nataro J.P., Caprioli A. Characteristics of the enteroaggregative Shiga toxin/verotoxin-producing Escherichia coli O104:H4 strain causing the outbreak of haemolytic uraemic syndrome in Germany, May to June 2011. Euro. Surveill. 2011;16:pii19889. doi: 10.2807/ese.16.24.19889-en. [DOI] [PubMed] [Google Scholar]
- 4.Frank C., Werber D., Cramer J.P., Askar M., Faber M., an der Heiden M., Bernard H., Fruth A., Prager R., Spode A., et al. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N. Engl. J. Med. 2011;365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 5.Bielaszewska M., Mellmann A., Zhang W., Köck R., Fruth A., Bauwens A., Peters G., Karch H. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 2011;11:671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- 6.Munera D., Ritchie J.M., Hatzios S.K., Bronson R., Fang G., Schadt E.E., Davis B.M., Waldor M.K. Autotransporters but not pAA are critical for rabbit colonization by Shiga toxin-producing Escherichia coli O104:H4. Nat. Commun. 2014;5:3080. doi: 10.1038/ncomms4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grad Y.H., Lipsitch M., Feldgarden M., Arachchi H.M., Cerqueira G.C., Fitzgerald M., Godfrey P., Haas B.J., Murphy C.I., Russ C., et al. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc. Natl Acad. Sci. U.S.A. 2012;109:3065–3070. doi: 10.1073/pnas.1121491109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasko D.A., Webster D.R., Sahl J.W., Bashir A., Boisen N., Scheutz F., Paxinos E.E., Sebra R., Chin C.S., Iliopoulos D., et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 2011;365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohde H., Qin J., Cui Y., Li D., Loman N.J., Hentschke M., Chen W., Pu F., Peng Y., Li J., et al. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N. Engl. J. Med. 2011;365:718–724. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- 10.Mayer C.L., Leibowitz C.S., Kurosawa S., Stearns-Kurosawa D.J. Shiga toxins and the pathophysiology of hemolytic uremic syndrome in humans and animals. Toxins. 2012;4:1261–1287. doi: 10.3390/toxins4111261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghafourian S., Sadeghifard N., Soheili S., Sekawi Z. Extended spectrum beta-lactamases: definition, classification and epidemiology. Curr. Issues Mol. Biol. 2014;17:11–22. [PubMed] [Google Scholar]
- 12.Ahmed S.A., Awosika J., Baldwin C., Bishop-Lilly K.A., Biswas B., Broomall S., Chain P.S., Chertkov O., Chokoshvili O., Coyne S., et al. Genomic comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including shiga toxin encoding phage stx2. PLoS One. 2012;7:e48228. doi: 10.1371/journal.pone.0048228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brzuszkiewicz E., Thürmer A., Schuldes J., Leimbach A., Liesegang H., Meyer F.D., Boelter J., Petersen H., Gottschalk G., Daniel R. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC) Arch. Microbiol. 2011;193:883–891. doi: 10.1007/s00203-011-0725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grad Y.H., Godfrey P., Cerquiera G.C., Mariani-Kurkdjian P., Gouali M., Bingen E., Shea T.P., Haas B.J., Griggs A., Young S., et al. Comparative genomics of recent Shiga toxin-producing Escherichia coli O104:H4: short-term evolution of an emerging pathogen. MBio. 2013;4:e00452–e00412. doi: 10.1128/mBio.00452-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christie P.J., Whitaker N., González-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta. 2014;1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh P.K., Meijer W.J. Diverse regulatory circuits for transfer of conjugative elements. FEMS Microbiol. Lett. 2014;358:119–128. doi: 10.1111/1574-6968.12526. [DOI] [PubMed] [Google Scholar]
- 17.Smet A., Van Nieuwerburgh F., Vandekerckhove T.T., Martel A., Deforce D., Butaye P., Haesebrouck F. Complete nucleotide sequence of CTX-M-15-plasmids from clinical Escherichia coli isolates: insertional events of transposons and insertion sequences. PLoS One. 2010;5:e11202. doi: 10.1371/journal.pone.0011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer J., Rodríguez I., Baumann B., Guiral E., Beutin L., Schroeter A., Kaesbohrer A., Pfeifer Y., Helmuth R., Guerra B. blaCTX-M-15-carrying Escherichia coli and Salmonella isolates from livestock and food in Germany. J. Antimicrob. Chemother. 2014;69:2951–2958. doi: 10.1093/jac/dku270. [DOI] [PubMed] [Google Scholar]
- 19.García-Fernández A., Chiaretto G., Bertini A., Villa L., Fortini D., Ricci A., Carattoli A. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J. Antimicrob. Chemother. 2008;61:1229–1233. doi: 10.1093/jac/dkn131. [DOI] [PubMed] [Google Scholar]
- 20.Sampei G., Furuya N., Tachibana K., Saitou Y., Suzuki T., Mizobuchi K., Komano T. Complete genome sequence of the incompatibility group I1 plasmid R64. Plasmid. 2010;64:92–103. doi: 10.1016/j.plasmid.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Asano K., Mizobuchi K. Copy number control of IncIalpha plasmid ColIb-P9 by competition between pseudoknot formation and antisense RNA binding at a specific RNA site. EMBO J. 1998;17:5201–5213. doi: 10.1093/emboj/17.17.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nekrasov S.V., Agafonova O.V., Belogurova N.G., Delver E.P., Belogurov A.A. Plasmid-encoded antirestriction protein ArdA can discriminate between type I methyltransferase and complete restriction-modification system. J. Mol. Biol. 2007;365:284–297. doi: 10.1016/j.jmb.2006.09.087. [DOI] [PubMed] [Google Scholar]
- 23.Fang G., Munera D., Friedman D.I., Mandlik A., Chao M.C., Banerjee O., Feng Z., Losic B., Mahajan M.C., Jabado O.J., et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 2012;30:1232–1239. doi: 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komano T., Yoshida T., Narahara K., Furuya N. The transfer region of IncI1 plasmid R64: similarities between R64 tra and legionella icm/dot genes. Mol. Microbiol. 2000;35:1348–1359. doi: 10.1046/j.1365-2958.2000.01769.x. [DOI] [PubMed] [Google Scholar]
- 25.Rees C.E., Bradley D.E., Wilkins B.M. Organization and regulation of the conjugation genes of IncI1 plasmid colIb-P9. Plasmid. 1987;18:223–236. doi: 10.1016/0147-619x(87)90065-5. [DOI] [PubMed] [Google Scholar]
- 26.van Opijnen T., Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 2013;11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barquist L., Boinett C.J., Cain A.K. Approaches to querying bacterial genomes with transposon-insertion sequencing. RNA Biol. 2013;10:1161–1169. doi: 10.4161/rna.24765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singer M., Baker T.A., Schnitzler G., Deischel S.M., Goel M., Dove W., Jaacks K.J., Grossman A.D., Erickson J.W., Gross C.A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Domenico P., Salo R.J., Cross A.S., Cunha B.A. Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumoniae. Infect. Immun. 1994;62:4495–4499. doi: 10.1128/iai.62.10.4495-4499.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang S.L., Rubin E.J. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene. 2002;296:179–185. doi: 10.1016/s0378-1119(02)00856-9. [DOI] [PubMed] [Google Scholar]
- 31.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 33.Milton D.L., O'Toole R., Horstedt P., Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guzman L.M., Belin D., Carson M.J., Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kusano K., Naito T., Handa N., Kobayashi I. Restriction-modification systems as genomic parasites in competition for specific sequences. Proc. Natl Acad. Sci. U.S.A. 1995;92:11095–11099. doi: 10.1073/pnas.92.24.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao M.C., Pritchard J.R., Zhang Y.J., Rubin E.J., Livny J., Davis B.M., Waldor M.K. High-resolution definition of the Vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nucleic Acids Res. 2013;41:9033–9048. doi: 10.1093/nar/gkt654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pritchard J.R., Chao M.C., Abel S., Davis B.M., Zhang Y.J., Baronowski K., Rubin E.J., Waldor M.K. ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet. 2014;10:e1004782. doi: 10.1371/journal.pgen.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. [Google Scholar]
- 39.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Johnson T.J., Shepard S.M., Rivet B., Danzeisen J.L., Carattoli A. Comparative genomics and phylogeny of the IncI1 plasmids: a common plasmid type among porcine enterotoxigenic Escherichia coli. Plasmid. 2011;66:144–151. doi: 10.1016/j.plasmid.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 43.Asano K., Mizobuchi K. An RNA pseudoknot as the molecular switch for translation of the repZ gene encoding the replication initiator of IncIalpha plasmid ColIb-P9. J. Biol. Chem. 1998;273:11815–11825. doi: 10.1074/jbc.273.19.11815. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida T., Kim S.R., Komano T. Twelve pil genes are required for biogenesis of the R64 thin pilus. J. Bacteriol. 1999;181:2038–2043. doi: 10.1128/jb.181.7.2038-2043.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S.R., Komano T. The plasmid R64 thin pilus identified as a type IV pilus. J. Bacteriol. 1997;179:3594–3603. doi: 10.1128/jb.179.11.3594-3603.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furuya N., Komano T. Nucleotide sequence and characterization of the trbABC region of the IncI1 Plasmid R64: existence of the pnd gene for plasmid maintenance within the transfer region. J. Bacteriol. 1996;178:1491–1497. doi: 10.1128/jb.178.6.1491-1497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim S.R., Funayama N., Komano T. Nucleotide sequence and characterization of the traABCD region of IncI1 plasmid R64. J. Bacteriol. 1993;175:5035–5042. doi: 10.1128/jb.175.16.5035-5042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komano T., Funayama N., Kim S.R., Nisioka T. Transfer region of IncI1 plasmid R64 and role of shufflon in R64 transfer. J. Bacteriol. 1990;172:2230–2235. doi: 10.1128/jb.172.5.2230-2235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komano T. Shufflons: multiple inversion systems and integrons. Annu. Rev. Genet. 1999;33:171–191. doi: 10.1146/annurev.genet.33.1.171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


