Abstract
Using sedimentation and cryo electron tomography techniques, the conformations of eukaryotic polyribosomes formed in a long-term cell-free translation system were analyzed over all the active system lifetime (20–30 translation rounds during 6–8 h in wheat germ extract at 25°C). Three distinct types of the conformations were observed: (i) circular polyribosomes, varying from ring-shaped forms to circles collapsed into double rows, (ii) linear polyribosomes, tending to acquire planar zigzag-like forms and (iii) densely packed 3D helices. At the start, during the first two rounds of translation mostly the circular (ring-shaped and double-row) polyribosomes and the linear (free-shaped and zigzag-like) polyribosomes were formed (‘juvenile phase’). The progressive loading of the polyribosomes with translating ribosomes induced the opening of the circular polyribosomes and the transformation of a major part of the linear polyribosomes into the dense 3D helices (‘transitional phase’). After 2 h from the beginning (about 8–10 rounds of translation) this compact form of polyribosomes became predominant, whereas the circular and linear polyribosome fractions together contained less than half of polysomal ribosomes (‘steady-state phase’). The latter proportions did not change for several hours. Functional tests showed a reduced translational activity in the fraction of the 3D helical polyribosomes.
INTRODUCTION
Dissociated ribosomal particles initiate translation at the initiation codon within the 5′-terminal region of mRNA. Upon the initiation, the translating ribosome moves along the mRNA chain toward the 3′-end of mRNA, thus vacating the initiation site for a next ribosome. In this way a group of ribosomes moving one after another and translating the same mRNA chain is formed. Such a group is called polyribosome or polysome.
A number of early electron microscopy (EM) studies demonstrated the circular (ring-shaped) array of ribosomes in eukaryotic polyribosomes (1–4). Later, the EM studies of larger polyribosomes (i.e. the polyribosomes formed on longer mRNAs) showed that they often look like two parallel rows of ribosomes (‘double-row polyribosomes’). These images were interpreted as collapsed circles, when two anti-parallel halves of the circles are laying side-by-side, with retention of the circular topology of their mRNA (5–7). The studies of exchange of polysomal ribosomes with the pool of free ribosomal particles during many rounds of translation showed a slow rate of such an exchange and gave evidence in favor of preferential reinitiation of terminating ribosomes without leaving mRNA, thus suggesting the possibility of a circular translation of mRNA by ribosomes (7–10).
At the same time, in parallel with the reports on circularity of eukaryotic polyribosomes numerous reports about helical conformations of eukaryotic polyribosomes appeared (11–19). Sometimes zigzag-like conformations of polyribosomes, and especially the conformations of planar zigzags with linear topology of mRNA, were observed and discussed (19,20). Thus, the question has arisen about the relations between eukaryotic polyribosomes of different morphological types, and especially between the polyribosomes with circular topology of their mRNA, on one hand, and the helical polyribosomes with linear topology of mRNA, on the other.
In our previous work (21) we studied the formation of eukaryotic polyribosomes in a cell-free translation system during the first rounds of translation and demonstrated that the circular polyribosomes were a characteristic feature of the juvenile phase of the polyribosome formation. In the present work, we took advantage of the long-term cell-free translation system (22) in the continuous exchange cell-free (CECF) version (23,24), which allowed us to follow the changes of conformations of eukaryotic polyribosomes over long time. Using sedimentation and cryo electron tomography (cryo-ET) analyses at different stages of polyribosome formation and transformations, it has been demonstrated that the eukaryotic polyribosomes formed in the long-term cell-free translation system undergo significant structural changes and pass several discrete stages (juvenile, transitional and steady-state phases) in the course of their lifetime, thus raising the issue of the ‘ontogenesis’ of eukaryotic polyribosomes.
MATERIALS AND METHODS
The following mRNA constructs were used for translation and formation of polyribosomes in a cell-free system based on wheat germ extract (WGE):
Capped mRNA with poly(A) tail, consisting of (i) the cap structure; (ii) the 5′-untranslated region (5′-UTR) of rabbit β-globin mRNA; (iii) the 225-nt sequence encoding for streptavidin- and calmodulin-binding peptides (serves as the N-terminal tag for the protein, encoded by the following sequence; abbreviated as ‘s’ and ‘c’, respectively); (iv) the following 750-nt sequence encoding for green fluorescent protein (GFP); (v) the 40-nt 3′-UTR (3′-UTR(N)40); (vi) approximately 100 nt poly(A) tail (A100). The construct as a whole is designated Cap-5′UTRβ-globin-scGFP-3′UTR(N)40-(A)100.
Uncapped non-polyadenylated mRNA consisting of (i) the 5′-UTR of obelin mRNA; (ii) the 750-nt GFP-encoding sequence; (iii) the 3′-UTR of tobacco mosaic virus (TMV) RNA. The construct as a whole is designated 5′UTRObe-GFP-3′UTRTMV.
Uncapped non-polyadenylated mRNA consisting of (i) the 5′-UTR of the obelin-encoding mRNA; (ii) the firefly luciferase-encoding sequence (1650 nt); (iii) the 3′-UTR of TMV RNA. The construct as a whole is designated 5′UTRObe-Luc-3′UTRTMV.
Uncapped non-polyadenylated mRNA consisting of (i) the 5′-UTR of TMV RNA (also called omega sequence); (ii) the firefly luciferase-encoding sequence (1650 nt); (iii) the 3′-UTR of TMV RNA. The construct as a whole designated 5′UTROmega-Luc-3′UTRTMV.
Capped mRNA without poly(A) tail, consisting of (i) the cap structure; (ii) the 5′-UTR of rabbit β-globin mRNA; (iii) the N-tagged GFP-encoding sequence (975 nt); (iv) the non-specific 180 nt 3′-UTR derived from plasmid non-coding sequence. The construct as a whole designated Cap-5′UTRβGlobin-scGFP-3′UTR(N)180).
Preparation of the above-listed mRNA constructs was described in our previous publications (21,25; see also (7) for the luciferase construct 3). Capped mRNAs were prepared by T7-polymerase in vitro transcription with 3′OMe-m7G(5′)ppp(5′)G cap-analog, the poly(A)-tail was added post-transcriptionally in reaction with poly(A)-polymerase, as described in (21).
Cell-free translation and sedimentation analysis
Translation of the mRNAs was performed in a CECF system based on WGE (Roche Diagnostics, Penzberg, Germany) according to the protocol published in (24). All possible precautions to minimize mRNA degradation during long-term in vitro translation runs were undertaken, and the integrity of mRNA 3′-end in polyribosomes was checked as described in Supplementary Data. The polyribosomes formed during the active lifetime of the translation system (usually 360–480 min) were analyzed, as described earlier in detail (7).
Analysis of translation activities of polyribosome fractions
The polyribosomes were preformed by translation of 5′UTROmega-Luc-3′UTRTMV mRNA (construct 4) in the CECF system during 2 h at 25°C; then the translation reaction was switched over to batch format, radioactive amino acids ([14C]Ser and [14C]Phe) were added (up to 40 and 60 μM, respectively), and the translation reaction continued at 25°C for 1 h more. Aliquots were taken at 0, 4, 8, 16 and 24 min after addition of the radioactive amino acids and subjected to zonal centrifugation in 15–50% sucrose gradient using SW-41 rotor at 37 000 revolutions per minute (rpm) during 2 h at 4°C. The incorporation of the radioactive amino acids into nascent polypeptides (trichloroacetic acid-insoluble product) was determined in fractions of the sucrose gradient.
Cryo-ET analysis
Samples for cryo-ET analysis were taken during translation and immediately diluted 5-fold with ice-cold buffer A (25 mM HEPES-KOH pH 7.6, 0.5 mM Mg(OAc)2, 85 mM KOAc, 0.01 mg/ml cycloheximide). After dilution, 3 μl samples were applied to carbon grids using the Vitrobot apparatus (FEI, Eindhoven, Netherlands) and plunged into liquid ethane pre-cooled with liquid nitrogen (21). Cryo-ET data were recorded on Polara F30 FEG instrument (FEI, Eindhoven, Netherlands) run at 150 kV acceleration voltage and a nominal underfocus of Δz = 3 μm. Tilt series were recorded between −66° and +66°, with a tilt angle increment of 1°–3°; the cumulative electron dose did not exceed 60 e−/Å2. Data acquisition was carried out using a 4096 × 4096 CCD Eagle camera (FEI, Eindhoven, Netherlands). The images alignment and tomogram reconstruction were performed by using the ETOMO (IMOD software, Boulder Laboratory for 3-D EM, University of Colorado, USA) (26) and Inspect3D (FEI, Eindhoven, Netherlands) software. Subtomograms of the individual polysomal ribosomes images were extracted and subjected to maximum likelihood based iterative alignment and averaging using XMIPP software package (27). Positioning of the subtomogram average back into the tomogram was done in IMAGIC (Image Science Software GmbH, Germany) and UCSF Chimera software (28).
The averaged ribosome structure with estimated resolution of 4–5 nm was obtained for each individual tomogram separately in the same way as described in our previous papers (21,29). The reconstruction was based only on the cryo-ET data obtained in this work; no external reference structure was exploited. In total, 64 tomograms were processed and 9764 ribosomes in 1274 polyribosomes were analyzed by subtomogram averaging.
Tracing of mRNA paths within reconstructed polyribosomes
The tracing of the mRNA path was based on the mutual orientation of neighboring ribosomes within reconstructed polyribosomes, taking into account that the positions of the exit and entry sites for mRNA on ribosomes are well known (see (21) for details). The mRNA path through ribosomes is marked with arrowheads in all figures illustrating the polyribosome structures. In this way the direction of mRNA chain through the polysomal ribosomes could be outlined and thus the overall path of the mRNA within a polyribosome could be traced. This approach allowed us to clearly distinguish between circular and linear topologies of polysomal mRNAs, as well as to reveal 3D helical elements in ‘curls’ inside intermediate (transitional) conformations.
RESULTS
Sedimentation study of polyribosome formation and transformation in a multi-round cell-free translation system
In our cell-free translation experiments the process was initiated by addition of only one species of mRNA. Either mRNA encoding for GFP (GFP-mRNA, 750 nt coding sequence (construct 2 in the above list), or scGFP-mRNA, 975 nt coding sequence when the protein was tagged by streptavidin (s) and calmodulin binding peptide (c) (constructs 1 and 5), or mRNA encoding for firefly luciferase (Luc-mRNA), 1650 nt coding sequence (constructs 3 and 4), were used. The duration of one round of translation, or transit time (estimated as described earlier, (7,30)) was found to be 12 min for Luc-mRNA and about 7–8 min for GFP-mRNAs. The period of active translation in our system varied from 6 to 8 h, so that no less than 30 rounds of translation passed during the lifetime of the system.
Figure 1, panel A, presents the time course of protein synthesis in the long-term cell-free translation system based on WGE and programmed with 975-nt N-tagged GFP-encoding mRNA (construct 1), and panel B shows the sedimentation distribution of polyribosomes formed on the same scGFP-mRNA during the period when some visible changes in sedimentation profile were observed. The first two rounds of translation (15 min, ‘juvenile phase’) were characterized by formation of the typical polyribosome profile where most polysomal ribosomes were more or less equally distributed between trisomes and octasomes, with minority of larger polyribosomes, up to dodecasomes. After four rounds of translation (ca. 30 min, ‘transitional phase’) the distribution pattern persisted in general, but the fraction of the larger polyribosomes (from octasomes to dodecasomes) became increased. After 8–9 rounds of translation (ca. 60 min) a fast-sedimenting fraction appeared ahead of the typical polysomal peaks. The vertical line marking the position of dodecasomes indicates that this level of occupancy of polyribosomes by translating ribosomes—about 80 nucleotides of the coding sequence per translating ribosome (abbreviated as nts/RS = ca. 80)—should be considered as critical: the higher occupancy provokes the conformational transition of the polyribosomes into a more compact form (namely, into the dense 3D helical conformation, see below). Indeed, as seen in Figure 1, panel B, a rapidly moving hump was revealed in the post-dodecasome zone of the sedimentation profile. As follows from summation of our results, it is this conformational transition of the overloaded polyribosomes that contributed to the increase of their sedimentation velocity. The direct consequence was the appearance of the rapidly moving hump, which is well-defined at the sedimentation profile of the polyribosome zone, especially after 60-min translation.
Figure 1.
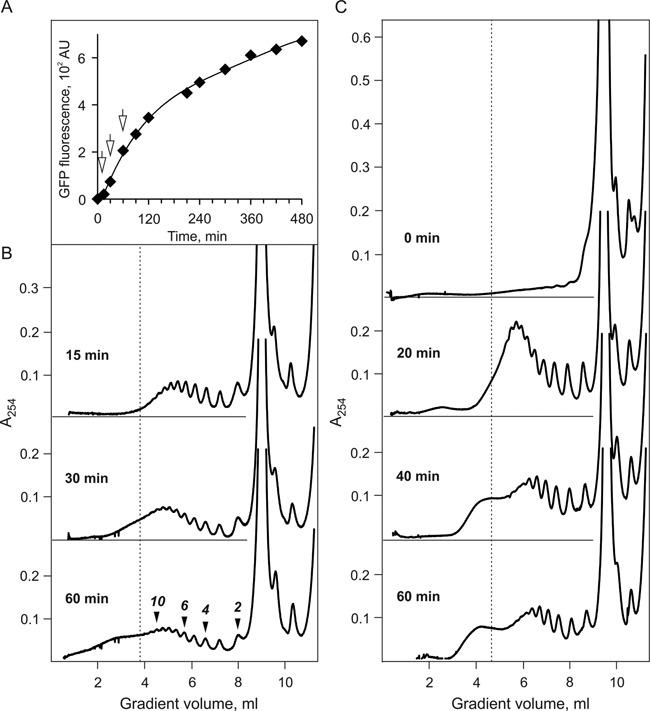
Time course of protein synthesis (A) and polyribosomes formation (B and C) in the long-term cell-free translation system (CECF) programmed with 975-nt N-tagged GFP-encoding mRNA (A and B), or with 1650-nt luciferase-encoding mRNA (C) (see Materials and Methods, constructs 1 and 4, respectively). (A) Synthesis of GFP during 8 h of translation at 25°C. Accumulation of the protein during translation was recorded by measurement of the fluorescence intensity at 510 nm (with excitation at 395 nm). The arrows indicate the moments (15, 30 and 60 min) when the 25 μl samples of the translation mixture were taken for the sedimentation analysis. (B) Sucrose gradient sedimentation analysis of the polyribosomes formed in the cell-free translation system shown in (A): sedimentation profiles of polyribosomes formed after 15, 30 and 60 min translation (approximately corresponding to 2, 4 and 8 rounds of translation*, respectively). Zonal centrifugation was performed as follows: the 25 μl aliquots of the translation mixture were layered on the 15–45% sucrose gradient in 20 mM HEPES-KOH pH7.6, 5 mM magnesium acetate, 100 mM potassium acetate, 0.01 mg/ml cycloheximide buffer and centrifuged for 2 h at 37 000 rpm and 4°C in SW41 rotor. Sedimentation profiles were recorded by continuous measuring optical density at 254 nm in flow cell of Uvicord SII monitor during gradient fractionation. The vertical line on the sedimentation diagrams marks the position of dodecasome (12 translating ribosomes on 975 nt coding sequence, i.e. 80 nt per ribosome). (C) Time course of polyribosomes formation after 0, 20, 40 and 60 min of translation in CECF-system programmed with 1650 nt luciferase-encoding mRNA (construct 4). Sedimentation analysis of polyribosomes was performed in the same way as in (B) using 15–50% sucrose gradient. [*One round of translation is the time required for a ribosome to pass the full length of mRNA including initiation and termination.]
A similar dynamics of the sedimentation profile of polyribosomes was demonstrated in the case of translation of a longer mRNA, namely, Luc-mRNA, construct 3 (Figure 1, panel C). In this case the rapidly moving hump characteristic for the conformational transition of polyribosomes upon attainment of a certain level of occupancy by translating ribosomes was found even more pronounced and convincing.
Reconstructed averaged structure of polysomal ribosomes
In the process of cryo-ET analysis the averaged ribosome structure was reconstructed for every tomogram and then used for solving the polyribosome structure within the tomograms (see Materials and Methods section and (21,29). The typical averaged ribosome structure with the resolution of 4–5 nm is shown in Figure 2. The characteristic structural domains of the ribosome, such as the head and the body of the 40S subunit and P1/P2 stalk of 60S subunit, can be clearly seen. The mRNA entry and exit sites are marked by dotted lines with red arrowheads.
Figure 2.
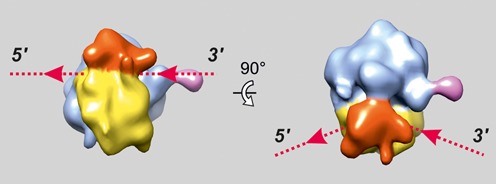
Structure of the eukaryotic (wheat) 80S ribosome obtained by averaging of the subtomograms resulted from cryo-ET analysis of polyribosomes within single tomogram. Here and in the Figures 3–6 the following colors are used: the head of the 40S ribosomal subunit is red, the body of the 40S subunit is yellow, the 60S subunit is blue and P1/P2 stalk is pink. The entry and exit sites of mRNA on the ribosome are indicated with dotted lines and arrowheads.
Cryo-ET analysis of the juvenile phase of polyribosome formation: from ring-shaped to double-row polyribosomes
In our previous experiments (21), where translation of GFP-encoding mRNAs (750 and 975 nt coding sequences) was initiated in a cell-free system and lasted for two translation rounds (15 min, ‘juvenile phase’), the formation of circular polyribosomes consisting mostly of 4–8 ribosomes per circle was shown, and the circularity was confirmed by tracing of the mRNA path.
Here, we demonstrate that in the course of further active work of the cell-free translation system the changes in size and forms of the circular polyribosomes occurred. First of all, the ring-shaped polyribosomes became larger, accommodating up to 11–12 ribosomes (Figure 3B), and some of them begin to collapse into the double-row polyribosomes (Figure 3B, the upper part of the polysomal cycle), where two halves of the cycle became juxtaposed and arrayed side-by-side with anti-parallel paths of the two halves of their mRNA chain (see below). Thus, the resultant double-row polyribosomes retain the circular topology of their mRNAs.
Figure 3.
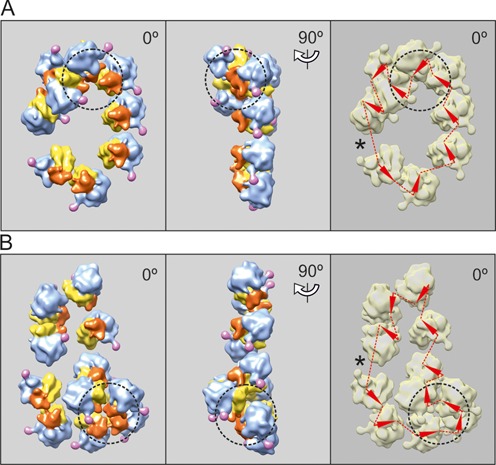
Cryo-ET reconstruction of circular polyribosomes. Nonasome (A) and undecasome (B) formed on uncapped non-adenylated GFP-encoding mRNA, 750-nt coding sequence (construct 2, see Materials and Methods). The ribosomes involved in 3D bulges (see the text) are inscribed into a dashed-line cycle. The undecasome (B) is an example of a double-row polyribosome with anti-parallel paths of the mRNA chain (collapsed circle). Here and in the following Figures 4–6, the left-hand panels present the vertical projections (elevation views) of the polyribosome reconstruction images. The middle panels show the side projections of the same polyribosome reconstruction. The right-hand panels present the deduced path of mRNA, the arrowheads being markers of the direction of mRNA path through the ribosomes. The asterisk indicates the presumed site of the junction of the 5′ and 3′ ends of mRNA, suggested by the gap and/or irregularity in orientation of neighboring ribosomes.
It should be also mentioned that the overloading of the polysomal circles with translating ribosomes, when the occupancy of 9–11 ribosomes per mRNA of this size (975 nt coding sequence, i.e. nts/RS = 100 ± 10) had been attained, led to the formation of characteristic compact bulges on the planar circles and the double rows. Examples of such high-loaded polysomal ring-shaped or collapsing circles with bulges are shown in Figure 3A and B. It is noteworthy that the ribosomes in the bulges were usually arrayed in a compact ‘curl’, similar to a helix turn of the 3D helical polyribosomes (see Discussion).
Linearization of circular mRNA and formation of planar zigzag-like polyribosomes
The continued loading of circular polyribosomes, resulting in the increase of polysome length and the appearance of the bulges (‘curls’), was shown to correlate with the process of progressive de-circularization (circle opening). Probably the bulges cause a certain tension in circular conformation and thus favor the de-circularization process. As a result, the circular polyribosomes may transform into string-shaped polyribosomes with linear topology of mRNA. The polysomal strings displayed the tendency to acquire the planar zigzag-like forms (Figure 4A and B), sometimes with the bulge (‘curl’) inside or in the vicinity of the 3′-end of the string (Figure 4B, see the lower part of the string). When the occupancy of mRNA by translating ribosomes achieved the level of about 100 nucleotides per ribosome, the linearization of all polyribosomes with circular topology of their mRNA became complete, and the linear polyribosomes were transformed further into the densely folded 3D helices with linear topology of mRNA. Possibly the preformed 3D bulges (‘curls’) could be the helical embryos promoting the conformational transition (see above).
Figure 4.
Cryo-ET reconstruction of topologically linear, zigzag-like polyribosomes. (A) Nonasome formed on uncapped non-polyadenylated GFP-encoding mRNA, 750-nt coding sequence (construct 2, see Materials and Methods). (B) Tridecasome formed on capped polyadenylated N-tagged GFP-encoding mRNA, 975-nt coding sequence (construct 1, see Materials and Methods). The nonasome (A) is an example of pseudo-double-row polyribosome with zigzag-like path of mRNA. The tridecasome in panel B contains a 3D bulge at its 3′-end (marked by a dashed-line cycle) with structural parameters similar to those of the turn of the dense 3D helical polyribosomes (see Discussion in the text).
However, under our experimental conditions only about half of the circular polyribosomes were subjected to the overloading with translating ribosomes and to the following linearization during translation. The other half of the circular polyribosome population kept the balance between initiation and termination rates and avoided the overloading, so that some part of the total polyribosome population continued to be circular up to the end of the active lifetime of the translation system (see below).
It must be noted that along with the circular polyribosomes, the linear (non-circular) free-form polyribosomes were also assembled and present in a cell-free translation system from the beginning. (It cannot be excluded that the presence of such a high proportion of the non-circular polyribosomes at the start of translation may be an artifact caused by some conditions of the cell-free system; for instance, the deficit of some factors required for mRNA circularization may occur in the diluted cytosol.) Anyhow, these two conformation types—circles and linear forms—were present in comparable amounts and covered together about 80% polyribosomes during the first two rounds of translation. The linear polyribosomes were also progressively loaded with translating ribosomes, and at a high level of the occupancy the local 3D bulges (‘curls’) with parameters of the 4-fold helical turn were formed (see, e.g. Figure 4B). The incubation mixture after 60 min translation contains all forms of polyribosomes discussed here, including circular, linear and helical formations (see Supplementary Figure S1).
The confusing circumstance is that the topologically linear polyribosomes in the planar zigzag-like configuration (Figure 4), are seen as two parallel rows of ribosomal particles, when observed in the vertical projection using classic EM methods, and thus such a pseudo-double-row polyribosome can be hardly distinguished from the true double-rows of topologically circular polyribosomes (see Figure 5). The analysis of mRNA pathways within polyribosomes based on cryo-ET 3D reconstructions allowed solving the problem. In Figure 5 the two reconstructed short hexasomes look very similar (cf. left-hand images in Figure 5A and B), but the tracing of mRNA path shows that the upper is a true double-row polyribosome with circular path of mRNA (Figure 5A, right-hand panel), whereas the lower is a pseudo-double-row with linear topology of mRNA chain (Figure 5B, right-hand panel).
Figure 5.
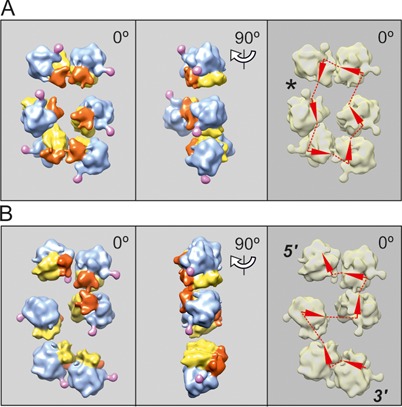
Cryo-ET reconstruction of two topologically different hexasomes formed on capped polyadenylated N-tagged GFP-encoding mRNA, 975-nt coding sequence (construct 1, see Materials and Methods) after 15 min translation. (A) The hexasome with circular mRNA path, visible as short double-row polyribosome. (B) The hexasome with topologically linear, zigzag-like mRNA path, also visible as short polyribosome with two parallel rows of ribosomes (‘pseudo-double-row polysome’).
Compaction of overloaded polyribosomes: formation of dense 3D helices
In studies of formation and transformations of eukaryotic polyribosomes during prolonged incubation, when they continued replenishing with ribosomes, the analysis of the sedimentation profiles revealed the anomalous behavior of overloaded polyribosomes: instead of the expected increase of the sedimentation coefficients, more or less proportionally to the number of ribosomes added to polyribosomes, a heavy sedimentation shift was recorded that was inadequate to the contribution of acquired ribosomes. As a result, a separate heavier zone appeared and moved ahead of the main polysomal zone (see above, Figure 1). It is seen that after formation of dodecasomes the following additions of ribosomes resulted in anomalously increased sedimentation rates of the overloaded polyribosomes and the appearance of a more or less discrete ‘super-heavy’ zone. Formation of such a discrete zone of overloaded polyribosomes was found to be even more clearly exhibited on sedimentation profiles with longer mRNAs, such as firefly luciferase-encoding mRNA (1650 nts coding sequence) shown in Figure 1C. In this case the appearance of the ‘super-heavy’ zone began after the formation of polyribosomes accommodating more than 20 translating ribosomes (i.e. icosasomes). Such a behavior suggested that the overloading of polyribosomes induced a cooperative conformational transition of polysomal structures resulting in their overall compaction.
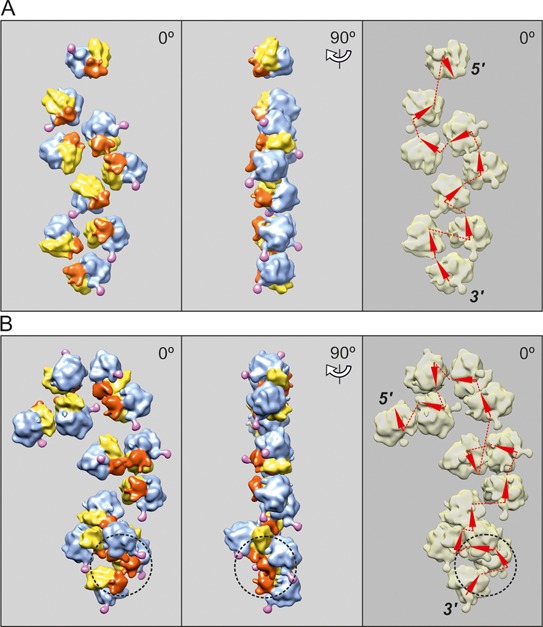
The cryo-ET analysis fully confirms these conclusions. It was found that when the occupancy of mRNA coding sequence exceeded 80 nucleotides per translating ribosome and approached 60 nucleotides per ribosome, the polyribosomes of all types, including those with the circular topology (ring-shaped and double rows) and the linear topology of their mRNAs (zigzag-like and pseudo-double rows), began to undergo a cooperative conformational transition into the densely packed 4-fold 3D helices. The reconstructed helical polyribosomes and the scheme of the mRNA path of the helix are shown in Figure 6. The arrangement of ribosomes in the helical polyribosome viewed from its end clearly demonstrated the 4-fold pattern of ribosome packing. The detailed molecular structure of a dense helical polyribosome with a number of specific inter-ribosomal contacts is described by us elsewhere (29).
Figure 6.
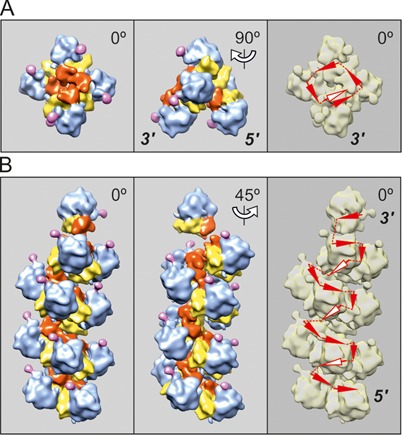
Cryo-ET reconstruction of 3D helical polyribosomes formed on capped polyadenylated N-tagged GFP-encoding mRNA, 975-nt coding sequence (construct 1, see Materials and Methods) after 30 min translation (A) and 120 min translation (B). (A) Pentasome folded into tetrad with one additional ribosome; the tetrad resembles the turn of the 4-fold helix as the typical conformation of the dense helical polyribosomes that appear at the late stages of the lifetime of translation (see below). (B) Tetradecasome folded into the dense 4-fold left-handed helix (see the text). The arrowheads indicating the mRNA path, which are pertinent to the ribosomes on the rear (invisible) side of helical polysomes, are marked by white color.
Proportions of different polysomal conformations and their changes in the course of the long-term translation
The cryo-ET structural analysis of the polyribosomes was performed at several time points during all the lifetime of the CECF translation systems. Three mRNA constructs (1, 2 and 5, see Materials and Methods) were used for the analyses.
Figure 7 shows a typical example of the dynamics of changes in proportions of the different polysomal conformations during 6 h incubation of the translation systems programmed with capped polyadenylated scGFP-encoding mRNA (construct 1, see Materials and Methods section). Under conditions of our experiment (WGE, 25°C) during the first 15 min after the translation start the young polyribosome population consisted predominantly of circular (ring-shaped and double-row) and linear (mostly zigzag-shaped) polyribosomes. Although helical tetrasomes (‘curls’) could be also observed, they were considered circular because of the proximity of their 3′ and 5′ ends.
Figure 7.
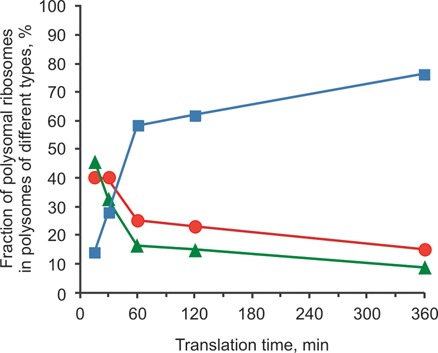
Dynamics of conformational changes of polyribosomes during multi-round translation in a long-term cell-free translation system programmed with the capped polyadenylated N-tagged GFP-encoding mRNA, 975 nt coding sequence (construct 1, see Materials and Methods): the distribution of polysomal ribosomes between different types of polyribosomes—circular (red circles), linear (green triangles) and 3D helical (blue squares)—depending on the time passed after translation start. The percentages of ribosomes in the polysomes of each type are plotted for each time point, the total number of polysomal ribosomes at this point being accepted as 100% (based on the data of Supplementary Table S1).
After 60 min the fraction of the 3D helical polyribosomes prevailed over polyribosomes of the two other types: more than a half of polysomal ribosomes belonged to the 3D helical polyribosomes, whereas the circular and the linear formations each comprised less than a quarter of polysomal ribosomes. During the next hour (120 min after start) this tendency persisted, so that the dense 3D helix fraction became strongly predominant, possessing up to two-thirds of all polysomal ribosomes. The circular and the linear polyribosomes were found in minority containing about 15–20% of all polysomal ribosomes each; this situation continued during further translation.
Thus, after 2 h of activity and conformational transitions (‘transitional phase’) the translation system entered into a well-balanced phase, so that no significant changes of polyribosome types and their proportions occurred any more during the next 4 h (steady-state).
It should be noted that similar dynamics of the structural transformations of polyribosomes during a long-term translation were observed in the cases of the other mRNA constructs used for polyribosome formation and studied in our work (see Supplementary Data, Table S1 and Figure S2).
Because the mRNA degradation, especially cleavage of the 3′-end including the stop-codon, may be the cause of ribosome stalling, we checked the integrity of 3′-end of the polysomal mRNA by controlling the change of the length of poly(A) tail (initially about 100 nt) in polysomal [32P]mRNA during translation. We have found that the length of poly(A) tail gradually shortened in the course of translation, but was still long enough (about 30 nt) in the major part of polysomes after 6 h of reaction (Supplementary Figure S4). Hence, the 3′-terminal degradation under conditions of translation process did not affect yet the stop-codon region of the polysomal mRNA. Small fraction of polysomal mRNA was found deadenylated, while retaining about 40 nucleotides of 3′UTR including the stop-codon. Notably, the extent of degradation was similar in all the polysomes fractions, no enrichment of the longer polysomes by mRNA with degraded 3′-end was found.
The occupancy of polyribosomes and its change in the course of the long-term translation
Sedimentation patterns and EM analyses of the samples taken from incubation mixtures of our cell-free system showed that approximately one-third of the total ribosome population was involved in translating polyribosomes. At the same time the main tendency observed in the active systems providing the long-term multi-round translation process was the growth of translating polyribosomes. Typically, this growth is revealed as one-directional elongation of polyribosomes; in other words, the polyribosomes grow via one-by-one addition of ribosomal particles at the 5′ side of mRNA chain. There are all grounds to believe that the growth is realized by using the conventional translation initiation mechanism. This implies that in the growing polyribosomes the rate of translation initiation exceeds the rate of translation termination. It is not clear what the contribution of free ribosomes to the polysome growth is, and whether the terminating ribosomes can be preferentially involved in the polysome growth process. In any case, the number of translating ribosomes on mRNA could grow due to threading both free ribosomes and reinitiating ribosomal particles after termination on mRNA of the existing polyribosomes. In such a way the occupancy of polyribosomes by translating ribosomes could increase during the translation process up to a certain level. In our experiments it was found that in the polyribosomes of both the circular and the linear types the occupancy of mRNA by translating ribosomes varied within a wide range, mostly from 200 to 80 nucleotides per ribosome (Figure 8), and thus it was far from saturation (ca. 30–40 nucleotides per ribosome). It seems evident that such a rarefaction of translating ribosomes along mRNA strand should exclude direct permanent contacts between the neighboring ribosomes.
Figure 8.
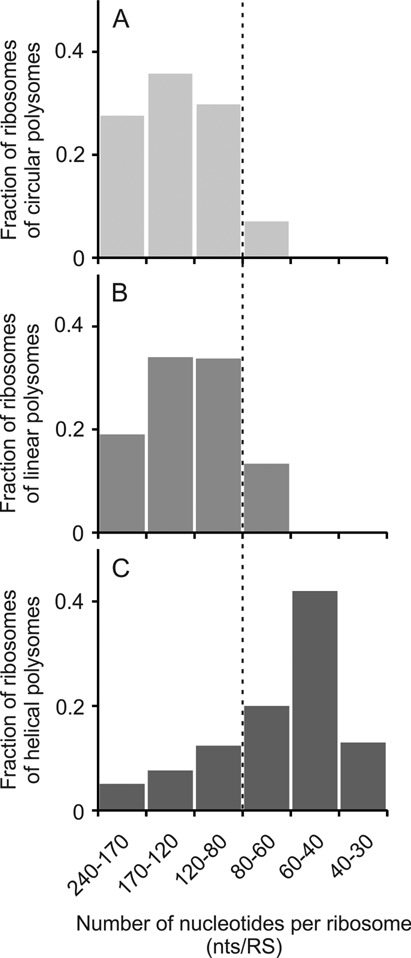
Dependence of proportions of three main types of polysomal conformations—circular (A), linear including zigzag-like (B) and 3D helical (C)—on the occupancy of the coding region of mRNA by translating ribosomes (number of nucleotides per ribosome, nts/RS ratio).
Quite a different situation was observed in the case of the 3D helical polyribosomes. In most cases the dense 3D helices were characterized by a high occupancy level, such as nts/RS = 60 (±20). Therefore, inter-ribosomal contacts appear to become inevitable in the dense helical polyribosomes. As to the local sites of high occupancy in the circular and linear polyribosomes, it is likely that they could arise at the sites of occasional local crowding of ribosomes on mRNA chain described above as bulges or ‘curls’ (see Figures 3 and 4).
As seen in Figure 8, when the polyribosome occupancy attained nts/RS = 80, the fraction of the dense 3D helices began to prevail, being accompanied by decreasing the circular polyribosome fraction. At the occupancy of nts/RS = 60 both circular and linear (zigzag-like) polyribosomes did not exist anymore, and all polyribosomes became densely packed 3D helices.
In order to check the functional (translation) activity of the compact 3D polyribosomes we made use of the phenomenon demonstrated in Figure 1 where polyribosomes with the occupancy of nts/RS = 80 and less were abruptly (cooperatively) transformed into compact, rapidly sedimenting polyribosomes. These proved to be densely packed 3D helices revealed as a partially separated ‘super-heavy’ zone of the sedimentation profile. The polyribosomes formed on luciferase-encoding mRNA (1650 nt coding sequence) during 120 min translation were pulse labeled with [14C] amino acid, and then subjected to sedimentation fractionation; the radioactivity incorporation into the nascent polypeptides of the polyribosomes with different occupancy was analyzed in gradient fractions (see Materials and Methods section). The results are presented in Supplementary Figure S3 (see also (29)). It proved that the translation activity, judging by amino acid incorporation, is significantly lower in the fractions of the ‘super-heavy’ zone comprising the densely packed 3D helical polyribosomes, as compared with the highly active polyribosomes of a lighter zone.
DISCUSSION
In the case of prokaryotes, where transcription and translation processes are not spatially separated within the cell, the initiation of translation typically occurs at the emergent 5′-section of a nascent mRNA chain which is still being elongated by RNA polymerase (the so-called ‘transcription-translation coupling’, see (31,32)). This predetermines the linear topology of arising prokaryotic polyribosomes. The densely packed helical polyribosomes of bacterial cells recently studied by cryo-EM and cryo-ET techniques (33) may be considered as a final stage of transformation of the loose linear polyribosomes into dense 3D helical polyribosomes.
On the contrary, in eukaryotes the polyribosomes are formed in the cytoplasm after the completion of the synthesis of mRNA chains and their processing in the nucleus. Thus, the formation of eukaryotic polyribosomes begins and proceeds in the cytoplasm on completed polyribonucleotide chains associated with a number of accompanying cytoplasmic proteins (see, e.g. (34,35)) including some translation initiation factors. It is the situation that is modeled in a cell-free translation system when a mature, ready for translation mRNA is added to the cytoplasmic extract. As commonly accepted, the formation of eukaryotic polyribosomes in the cytoplasmic milieu starts from the non-covalent circularization of mRNA and the appearance of ring-shaped oligosomes and polyribosomes. The primary appearance of the ring-shaped polyribosomes and the circular topology of their mRNA have been confirmed by our cryo-ET observations (see (21)). This stage of the eukaryotic polyribosome formation seems to be relatively short-lived and can be considered as a juvenile phase of the polysomal lifetime.
The biological role of circularization of the newly formed (juvenile) polyribosomes is still the subject of discussions. Two of our recent observations may be related to this problem. First, it seems that the circular polyribosomes enable their ribosomes to reinitiate translation immediately after termination on the same mRNA (7; see also (9,10)). Second, just after the first round of translation the phenomenon of significant acceleration of translational activity of the juvenile polyribosomes occurs (36). These observations together imply that the circularization of newly formed polyribosomes may be required for fast loading of polysomal mRNA with translating ribosomes: the queue of the ribosomes that have initiated translation at the 5′-end of the mRNA (de novo initiation) are moving further one after another along the mRNA chain becomes supplemented after the first round of translation with the moving queue of the ribosomes terminating at 3′-end and immediately reinitiating at 5′-region of the same mRNA (post-termination re-initiation). As a result, the circular polyribosome can be very quickly filled with translating ribosomes thus displaying translation acceleration during the first rounds of translation.
Along with the circular polyribosomes, certain amount of the polyribosomes with linear mRNA topology is also formed from the beginning. When the string-like polyribosomes grow, they tend to form planar zigzags which look like two parallel rows of ribosomes. These are pseudo-double-row (double-row-like) polyribosomes with the linear topology of mRNA, in contrast to true double-row polyribosomes with the circular topology of mRNA. As shown in our previous cryo-ET studies (25), the double-row polyribosomes with the circular topology of their mRNA and anti-parallel paths of their ribosome rows, on one part, and the pseudo-double-row polyribosomes with the zigzag-like path of the ribosomes and hence the linear topology of mRNA, on the other part, can be distinguished in the case of the vitrified (flash-frozen) polyribosomes. An alternative approach was the selective labeling of 3′ and 5′ ends of mRNA with subsequent analysis of the location of the markers at double-labeled polyribosomes (25). It proved that both the double-row polyribosomes (with circular topology of mRNA) and the pseudo-double-row polyribosomes (the zigzag-like forms with linear topology of mRNA) may co-exist in the same translation mixture. This fact can be the source of errors in interpretations of EM images (including in the cellular context) and the corresponding conclusions about polyribosome conformations. That is why a number of previous EM reports where the double-row-like polyribosomes were interpreted as circular (5–7) cannot be considered as unconditional demonstrations of the circularity.
Being replenished with translating ribosomes, the growing ring-shaped polyribosomes can be modified into double-row polyribosomes with anti-parallel rows of ribosomes, keeping the circular topology of their mRNAs. When observed with cryo-EM or cryo-ET, the vitrified double-row polyribosomes are seen as two parallel, more or less separated rows of ribosomes, whereas the classic negative staining EM methodology where contrasting and drying procedures are involved gives the misleading impression of two side-by-side stuck rows. At the same time the later stages of the polysomal lifetime are accompanied also by de-circularization of previously circular polyribosomes, so that they transform into topologically linear forms with the tendency to fold into the planar zigzag-like formations.
The careful investigation of the mRNA paths in polyribosomes had led to an important discovery of anomalous bulges within the polyribosomes of both circular and linear types (see Figures 3 and 4B). It seems that the bulges resulted from the local overloading of some sections of mRNA by translating ribosomes (local clotting), which induced a more compact packing of the polysomal ribosomes at the corresponding segment of the polyribosome. A compact packing of polysomal ribosomes is known to be characteristic of the 3D helical polyribosomes (29). Indeed, the character of mutual arrangement of ribosomes within such a bulge bears a strong resemblance to a turn of the 3D helical polyribosome (see Figure 6, 90° projection). It seems that the appearance of the bulges (‘curls’) in circular or linear polyribosomes evidences a critical state of the polysomal structure. In circular type polyribosomes these bulges may cause some tension within the polysomal circle, thus promoting the opening of the circular polyribosomes. In linear polyribosomes the bulges may be considered to be the ‘embryos’ or nucleation points for transformation of the polyribosomes into the dense 3D helices.
The study of dynamics of conformational transitions of polyribosomes in the course of all active lifetime of the translation system (Figure 7, see also Supplemetary Table S1 and Figure S2) evidenced that the most demonstrative events in polyribosome transformations occurred soon after the beginning, during the first 2 h after the start. If the first two rounds of translation during 15 min led to primary formation of circular and linear polyribosomes (‘juvenile phase’), the following 15 min period was characterized by the appearance of the dense 3D polyribosome fraction, with simultaneous decrease of the two other fractions. During the next half an hour the 3D helical polyribosomes became a predominant fraction, whereas the circular and linear polysome fractions were found in the minority (‘transitional phase’). After 1–2 h of the conformational transitions the situation was stabilized and the system passed into the steady-state phase with the major fraction of the 3D polyribosomes. It is demonstrative that the three successive phases of existence and conformational changes of the polyribosomes are well reflected in the slope of the translation rate curve (Figure 1A): during the first hour the maximal slope of the translation rate curve is observed (juvenile phase); then, at around the 60th min of translation some curve inflection occurs, and the somewhat decreased translation level follows until the 120th min (transitional phase); after the 120th min the most significant decrease of the translation rate can be noted (steady-state phase). The same sequence of events was found in the cases of all other tested mRNAs.
Progressive loading of polyribosomes with translating ribosomes seems to be the main factor leading to the changes of polysomal conformations. At the same time, there is a significant variety in the occupancy of the polyribosomes by translating ribosomes, from 240 to 80 nucleotides per ribosome, both for the circular conformation type polyribosomes and the polyribosomes with linear topology of mRNA. This range is in agreement with the average occupancy found in vivo (0.64 ± 0.31 ribosomes per 100 nt, or 156 ± 50 nts/RS) (37). However, the loading to the occupancy of up to 60 ± 20 nucleotides per ribosome results in abrupt transformation of polyribosomes into dense, highly ordered 3D helices with restrained activity. It seems that in polyribosomes of both circular and linear types the ribosomal particles are deprived of permanent contacts with each other and any functional coordination with each other. On the other hand, inter-ribosomal contacts and functional coordination are evident in the compact 3D polysomes (29). In other words, the compact 3D helical polyribosomes might be responsible for a special level of translational regulation in dormant cells. Indeed, the dense 3D helices may be down-regulated to zero, but retaining vital capacity. The fraction of 3D polysomes in the steady-state phase varied from 50% to 90% in the cell-free systems programmed with different mRNAs in this work (see Supplementary Figure S2); it may vary in the in vivo situation as well depending on the cell state. The helices with densely packed ribosomes were observed in dormant cells, such as plant zygotes (17), cysts of Protozoa (Entamoeba invadens) (38–41), mouse oocytes (42), as well as in hypothermic chick embryo cells (43), some drug-inhibited cells (44), and also in the case of host cells infected by viruses (18). Along with this, however, the dense 3D helical polyribosomes were recently demonstrated to be present in active human living cells (19).
Recently, the evidence is provided that mRNA may be transported in synapses in the form of stably paused (stalled) polyribosomes, which upon proper signal can start immediate translation, thus bypassing the rate-limiting steps of translation initiation and polyribosome formation (45). We believe that the dense 3D helical polyribosomes may be the formations where the high occupancy by ribosomes and their regular arrangement provide conditions for the negative self-regulation of their activity via direct specific contacts between active centers of neighboring ribosomes along the helix. Such a reversible self-regulation might explain temporary pausing and stalling phenomena in protein synthesis and the low-active states of eukaryotic polyribosomes without their irreversible inactivation and degradation. In our parallel cryo-ET work on the structure of the 3D helical polyribosomes (29) the new results about specific contacts between ribosomes within the dense 4-fold left-handed 3D polyribosome helices have been obtained. We believe that this work may stimulate searching for novel molecular mechanisms of the protein synthesis regulation at the polysomal level.
The conformations of polyribosomes have been shown to be in direct dependence on the level of occupancy of mRNA by translating ribosomes, whereas the occupancy is the result of the balance between the consecutive loading of mRNA chains with new initiating ribosomes and the release of terminating ribosomes. Thus, the observed changes of the conformations imply that in the course of multi-round translation a number of regulatory mechanisms of translational control at the polysomal level are realized. The results have also indicated that the different conformations of the eukaryotic polyribosomes observed represent the sequential phases of a natural process, which may be termed ‘ontogenesis of polyribosomes’, starting from the juvenile phase of circular polyribosomes, then passing through some structural transformations (the transitional phase) and coming into the steady-state phase with the 3D helical polyribosomes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors are very grateful to V.I. Agol, V.A. Kolb and K.S. Vassilenko for critically reading the manuscript and fruitful discussions, and also to A. Kommer for invaluable help in the preparation of the manuscript.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
FUNDING
Program on Molecular and Cell Biology of the Russian Academy of Sciences; Russian Foundation for Basic Research [09–04–01726, 13–04–40213-N]; European Research Council (ERC Starting Grant); French Infrastructure for Integrated Structural Biology (FRISBI) [ANR-10-INSB-05–01]; Instruct as part of the European Strategy Forum on Research Infrastructures (ESFRI). ARCUS (Alsace/ Russia-Ukraine) [to Z.A.A.]; EMBO [ASTF 388.00–2009 to Z.A.A.]. Alsace Region; FRM; IBiSA; INSERM; CNRS; Association pour la Recherche sur le Cancer (ARC). Funding for open access charge: Program on Molecular and Cell Biology of the Russian Academy of Sciences; Russian Foundation for Basic Research [13-04-40213-N].
Conflict of interest statement. None declared.
REFERENCES
- 1.Mathias A.P., Williamson R., Huxley H.E., Page S. Occurrence and function of polysomes in rabbit reticulocytes. J. Mol. Biol. 1964;9:154–167. doi: 10.1016/s0022-2836(64)80097-8. [DOI] [PubMed] [Google Scholar]
- 2.Dallner G., Siekevitz P., Palade G.E. Biogenesis of endoplasmic reticulum membranes. I. Structural and chemical differentiation in developing rat hepatocyte. J. Cell Biol. 1966;30:73–96. doi: 10.1083/jcb.30.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shelton E., Kuff E.L. Substructure and configuration of ribosomes isolated from mammalian cells. J. Mol. Biol. 1966;22:23–31. [Google Scholar]
- 4.Christensen A.K., Kahn L.E., Bourne C.M. Circular polysomes predominate on the rough endoplasmic reticulum of somatotropes and mammotropes in the rat anterior pituitary. Am. J. Anat. 1987;178:1–10. doi: 10.1002/aja.1001780102. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida T., Wakiyama M., Yazaki K., Miura K. Transmission electron and atomic force microscopic observation of polysomes on carbon-coated grids prepared by surface spreading. J. Electron Microsc. (Japan) 1997;46:503–506. doi: 10.1093/oxfordjournals.jmicro.a023550. [DOI] [PubMed] [Google Scholar]
- 6.Madin K., Sawasaki T., Kamura N., Takai K., Ogasawara T., Yazaki K., Takei T., Miura K.-I., Endo Y. Formation of circular polyribosomes in wheat germ protein synthesis system. FEBS Lett. 2004;562:155–159. doi: 10.1016/S0014-5793(04)00221-2. [DOI] [PubMed] [Google Scholar]
- 7.Kopeina G.S., Afonina Z.A., Gromova K.V., Shirokov V.A., Vasiliev V.D., Spirin A.S. Step-wise formation of eukaryotic double-row polyribosomes and circular translation of polysomal mRNA. Nucleic Acids Res. 2008;36:2476–2488. doi: 10.1093/nar/gkm1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Philipps G.R. Haemoglobin synthesis and polysomes in intact reticulocytes. Nature. 1965;205:53–56. doi: 10.1038/205567a0. [DOI] [PubMed] [Google Scholar]
- 9.Adamson S.D., Howard G.A., Herbert E. The ribosome cycle in a reconstituted cell-free system from reticulocytes. Cold Spring Harb. Symp. Quant. Biol. 1969;34:547–554. doi: 10.1101/sqb.1969.034.01.062. [DOI] [PubMed] [Google Scholar]
- 10.Baglioni C., Vesco C., Jacobs-Lorena M. The role of ribosomal subunits in mammalian cells. Cold Spring Harb. Symp. Quant. Biol. 1969;34:555–565. doi: 10.1101/sqb.1969.034.01.063. [DOI] [PubMed] [Google Scholar]
- 11.Behnke O. Helical arrangement of ribosomes in the cytoplasm of differentiating cells of the small intestine of rat fetuses. Exp. Cell Res. 1963;30:597–598. [Google Scholar]
- 12.Waddington C.H., Perry M.M. Helical arrangement of ribosomes in differentiating muscle cells. Exp. Cell Res. 1963;30:599–600. doi: 10.1016/0014-4827(63)90339-2. [DOI] [PubMed] [Google Scholar]
- 13.Echlin P. An apparent helical arrangement of ribosomes in developing pollen mother cells of Ipomoea purpurea (L.) J. Cell Biol. 1964;24:150–153. doi: 10.1083/jcb.24.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfuderer P., Cammarano P., Holladay D.R., Novelli G.D. A helical polysome model. Biochim. Biophys. Acta. 1965;109:595–606. doi: 10.1016/0926-6585(65)90186-x. [DOI] [PubMed] [Google Scholar]
- 15.Weiss P., Grover N.B. Helical array of polyribosomes. Proc. Natl. Acad. Sci. U.S.A. 1968;59:763–768. doi: 10.1073/pnas.59.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wooding F.B. Ribosome helices in mature cells. J. Ultrastruct. Res. 1968;24:157–164. doi: 10.1016/s0022-5320(68)80025-5. [DOI] [PubMed] [Google Scholar]
- 17.Jensen W.A. Cotton embryogenesis. J. Cell Biol. 1968;36:403–406. doi: 10.1083/jcb.36.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djaczenko W., Benedetto A., Pezzi R. Formation of helical polyribosomes in poliovirus-infected cells of the 37 RC line. J. Cell Biol. 1970;45:173–177. doi: 10.1083/jcb.45.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandt F., Carlson L.-A., Hartl U., Baumeister W., Grünewald K. The three-dimensional organization of polyribosomes in intact human cells. Mol. Cell. 2010;39:560–569. doi: 10.1016/j.molcel.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Martin K.A., Miller O.L. Polysome structure in sea urchin eggs and embryos: an electron microscopic analysis. Dev. Biol. 1983;98:338–348. doi: 10.1016/0012-1606(83)90364-0. [DOI] [PubMed] [Google Scholar]
- 21.Afonina Z.A., Myasnikov A.G., Shirokov V.A., Klaholz B. P., Spirin A.S. Formation of circular polyribosomes on eukaryotic mRNA without cap-structure and poly(A)-tail: A cryo electron tomography study. Nucleic Acids Res. 2014;42:9461–9469. doi: 10.1093/nar/gku599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spirin A.S., Baranov V.I., Ryabova L.A., Ovodov S.Yu., Alakhov Yu.B. A continuous cell-free translation system capable of producing polypeptides in high yield. Science. 1988;242:1162–1164. doi: 10.1126/science.3055301. [DOI] [PubMed] [Google Scholar]
- 23.Spirin A.S. High-throughput cell-free systems for synthesis of functionally active proteins. Trends Biotech. 2004;22:538–545. doi: 10.1016/j.tibtech.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Shirokov V.A., Kommer A.A., Kolb V.A., Spirin A.S. Continuous-exchange protein-synthesizing systems. In: Grandi G, editor. Methods in Molecular Biology, ‘In vitro Transcription and Translation Protocols’. Vol. 375. Totowa, NJ: Humana Press Inc; 2007. pp. 19–55. [DOI] [PubMed] [Google Scholar]
- 25.Afonina Z.A., Myasnikov A.G., Khabibullina N.F., Belorusova A.Yu., Ménétret J.-F., Vasiliev V.D., Klaholz B.P., Shirokov V.A., Spirin A.S. Topology of mRNA chain in isolated eukaryotic double-row polyribosomes. Biochemistry (Moscow) 2013;78:445–454. doi: 10.1134/S0006297913050027. [DOI] [PubMed] [Google Scholar]
- 26.Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 27.Sorzano C.O.S., Marabini R., Velazquez-Muriel J., Bilbao-Castro J.R., Scheres S.H.W., Carazo J.M., Pascual-Montano A. XMIPP: a new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.Myasnikov A.G., Afonina Zh.A., Ménétret J.-F., Shirokov V.A., Spirin A.S., Klaholz B.P. The molecular structure of the left-handed supra-molecular helix of eukaryotic polyribosomes. Nat. Comm. 2014;5:5294. doi: 10.1038/ncomms6294. [DOI] [PubMed] [Google Scholar]
- 30.Vassilenko K.S., Alekhina O.M., Dmitriev S.E., Shatsky I.N., Spirin A.S. Unidirectional constant rate motion of the ribosomal scanning particle during eukaryotic translation initiation. Nucleic Acids Res. 2011;39:5555–5567. doi: 10.1093/nar/gkr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller O.L., Hamkalo B.A., Thomas C.A. Visualization of bacterial genes in action. Science. 1970;169:392–395. doi: 10.1126/science.169.3943.392. [DOI] [PubMed] [Google Scholar]
- 32.Gowrishankar J., Harinarayanan R. Why is transcription coupled to translation in bacteria. Mol. Microbiol. 2004;54:598–603. doi: 10.1111/j.1365-2958.2004.04289.x. [DOI] [PubMed] [Google Scholar]
- 33.Brandt F., Etchells S.A., Ortiz J.O., Elcock A.H., Hartl U., Baumeister W. The native 3D organization of bacterial polysomes. Cell. 2009;136:261–271. doi: 10.1016/j.cell.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 34.Spirin A.S., Ajtkhozhin M.A. Informosomes and polyribosome-associated proteins in eukaryotes. TIBS. 1985;10:162–165. [Google Scholar]
- 35.Ryazanov A.G., Ovchinnikov L.P., Spirin A.S. Development of structural organization of protein-synthesizing machinery from prokaryotes to eukaryotes. BioSystems. 1987;20:275–288. doi: 10.1016/0303-2647(87)90035-9. [DOI] [PubMed] [Google Scholar]
- 36.Alekhina O.M., Vassilenko K.S., Spirin A.S. Translation of non-capped mRNAs in a eukaryotic cell-free system: acceleration of initiation rate in the course of polysome formation. Nucleic Acids Res. 2007;35:6547–6559. doi: 10.1093/nar/gkm725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arava Y., Wang Y., Storey J.D., Liu C.L., Brown P.O., Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgan R.S., Uzman B.G. Nature of the packing of ribosomes within chromatoid bodies. Science. 1966;152:214–216. doi: 10.1126/science.152.3719.214. [DOI] [PubMed] [Google Scholar]
- 39.Morgan R.S. Structure of ribosomes of chromatoid bodies: three-dimensional Fourier synthesis at low resolution. Science. 1968;162:670–671. doi: 10.1126/science.162.3854.670. [DOI] [PubMed] [Google Scholar]
- 40.Baker D.C., Swales L.S. Comparison of trophozoite helical polysomes with cyst ribosomogen microcrystals in axenic Entamoeba sp. Cell Diff. 1972;1:307–315. doi: 10.1016/0045-6039(72)90006-1. [DOI] [PubMed] [Google Scholar]
- 41.Lake J.A., Slayter H.S. Three-dimensional structure of the chromatoid body helix of Entamoeba invadens. J. Mol. Biol. 1972;66:271–282. doi: 10.1016/0022-2836(72)90478-0. [DOI] [PubMed] [Google Scholar]
- 42.Burkholder G.D., Comings D.E., Okada T.A. A storage form of ribosomes in mouse oocytes. Exp. Cell Res. 1971;69:361–371. doi: 10.1016/0014-4827(71)90236-9. [DOI] [PubMed] [Google Scholar]
- 43.Byers B. Structure and formation of ribosome crystals in hypothermic chick embryo cells. J. Mol. Biol. 1967;26:155–167. doi: 10.1016/0022-2836(67)90288-4. [DOI] [PubMed] [Google Scholar]
- 44.Kusamrarn T., Sobhon P., Bailey G.B. The mechanism of formation of inhibitor-induced ribosome helices in Entamoeba invadens. J. Cell Biol. 1975;65:529–539. doi: 10.1083/jcb.65.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graber T.E., Hébert-Seropean S., Khoutorsky A., David A., Yewdell J.W., Lacaille J.-C., Sossin W.S. Reactivation of stalled polyribosomes in synaptic plasticity. Proc. Natl. Acad. Sci. U.S.A. 2013;110:16205–16210. doi: 10.1073/pnas.1307747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


