Abstract
Translation fidelity and efficiency require multiple ribosomal (r)RNA modifications that are mostly mediated by small nucleolar (sno)RNPs during ribosome production. Overlapping basepairing of snoRNAs with pre-rRNAs often necessitates sequential and efficient association and dissociation of the snoRNPs, however, how such hierarchy is established has remained unknown so far. Here, we identify several late-acting snoRNAs that bind pre-40S particles in human cells and show that their association and function in pre-40S complexes is regulated by the RNA helicase DDX21. We map DDX21 crosslinking sites on pre-rRNAs and show their overlap with the basepairing sites of the affected snoRNAs. While DDX21 activity is required for recruitment of the late-acting snoRNAs SNORD56 and SNORD68, earlier snoRNAs are not affected by DDX21 depletion. Together, these observations provide an understanding of the timing and ordered hierarchy of snoRNP action in pre-40S maturation and reveal a novel mode of regulation of snoRNP function by an RNA helicase in human cells.
INTRODUCTION
The modification of multiple types of eukaryotic RNAs is key for their biogenesis, structure and function. Ribosomal (r)RNAs represent one of the most highly modified classes of RNA and the numerous modifications introduced during ribosome production cluster in functionally important regions of the ribosome, such as the decoding centre, intersubunit bridge and peptidyl transferase centre (1). Here, the modifications have been proposed to be necessary for the efficiency and fidelity of translation.
The biogenesis of ribosomes is initiated by nucleolar transcription of a primary transcript (47S pre-rRNA in humans; 35S pre-rRNA in yeast), which contains the sequences of the 18S, 5.8S and 28S (human) or 25S (yeast) rRNAs (reviewed in (2,3)). Ribosome assembly starts co-transcriptionally and is closely coordinated with pre-rRNA processing, modification and folding by a multitude of cofactors. Many of the more than 200 protein cofactors required for ribosome assembly are recruited as pre-assembled subcomplexes that have hardly been studied in human cells. In yeast, the small subunit processome, which mediates the initial maturation and pre-rRNA processing steps on the nascent transcript, is assembled from individual proteins and several subcomplexes, such as the U3 snoRNP, tUTP/UTP-A, bUTP/UTP-B and UTP-C complexes (reviewed in (4)). The initial processing steps of the pre-rRNAs lead to the separation of the biogenesis pathways of the small (SSU, 40S) and large ribosomal subunits (LSU, 60S), followed by their further assembly in the nucleus, nuclear export and final maturation steps in the cytoplasm (2,3).
Almost all rRNA modifications are mediated by small nucleolar RNA-protein complexes (snoRNPs; reviewed in (5)). These contain a snoRNA component that basepairs with the pre-rRNA and thereby guides modification of the target residue by the enzymatic subunit of the snoRNP. While box C/D snoRNPs introduce 2′-O-methylations of the ribose, box H/ACA snoRNPs mediate the conversion of uridine to pseudouridine (6,7). The snoRNAs, in particular those of the box C/D class, possess significant complementarity to their target pre-rRNA and form extensive basepairing interactions (8). Coupled with the high density of rRNA modifications, this results in overlapping pre-rRNA interaction sites for many snoRNAs, indicating that they act sequentially. However, it has been shown in yeast that almost all rRNA modifications are introduced co-transcriptionally (9), implying rapid action and exchange of snoRNPs that modify neighbouring sites. It is not yet known, whether such snoRNPs act stochastically or in a defined sequence and how a ‘snoRNA hierarchy’ might be established. Insight into how the timing of snoRNA association and snoRNP action may be regulated is currently lacking.
Besides their modification functions, basepairing interactions between snoRNAs and the nascent pre-rRNA transcript are likely to help maintain the open conformation of early pre-ribosomal complexes, allowing access of ribosomal proteins and numerous ribosome biogenesis cofactors. The sequential removal of snoRNAs from the pre-rRNA might further regulate the folding and structural compaction of pre-ribosomal complexes. Interestingly, several RNA helicases have been shown in yeast to be required for the release of specific snoRNAs from pre-ribosomes (10–13). In mammalian cells, however, only the RNA helicase DDX51 has, so far, been implicated in the release of a snoRNA, U8/SNORD118 (14), while the roles of multiple other helicases proposed to function in ribosome synthesis have remained elusive (reviewed in (15,16)). It has also been suggested that RNA helicases structurally remodel pre-ribosomal complexes during their biogenesis, but this has yet to be demonstrated.
Here, we identify the RNA helicase DDX21 as a novel component of the human UTP-B complex and show that DDX21 is found in early and late pre-ribosomal intermediates. Using UV crosslinking and analysis of cDNA (CRAC), we reveal several binding sites of DDX21 on pre-rRNAs. Intriguingly, DDX21 is required for specific recruitment and function of several late-acting box C/D snoRNPs, which basepair at the DDX21 crosslinking sites in pre-40S complexes and mediate post-transcriptional methylation of 18S rRNA precursors. We further identify snoRNAs with different timing in ribosome biogenesis including several that are only recruited to pre-40S complexes, providing evidence that snoRNP modifications can occur at various stages during ribosome biogenesis.
MATERIALS AND METHODS
Cell culture and RNAi
Stable HEK293 cell lines enabling tetracycline-inducible expression of fusions of PWP2, DDX21 or NOP2 with C-terminal His-PreScission protease site-2xFlag (Flag) tags were generated using the Flp-In T-REx 293 Expression System (Invitrogen). For the expression of RNAi-resistant DDX21 five silent mutations were introduced into the siRNA target site. For expression of the DDX21SAT mutant, point mutations were introduced to convert S375 to L and A376 to E. 30 nM of siRNA duplexes (DDX21, GGGAAAUCACCCUGAAAGGTT; PWP2, GGAAGUUUGUUGGCAACCUTT, UGACAGAGUUUGGCAACCUTT, GGAGCUGGACAAGAUUACATT) were transfected using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer's recommendations and cells were harvested after 72 h. Note, the three siRNAs targeting PWP2 were used in combination to produce data presented after verification that each individual siRNA gave the same phenotype.
Pulldown experiments and complex analysis
HEK293 stable cell lines were treated with 1 μg/ml doxycycline to induce expression of DDX21-Flag, NOP2-Flag and PWP2-Flag for 24 h before cells were harvested in lysis buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 8, 150 mM KCl, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 0.1 mM dithiothreitol (DTT), 1 μg/ml Leupeptin/Pepstatin, 1 μg/ml Aprotinin, 1 mM phenylmethylsulfonyl fluoride (PMSF), Complete Mini protease inhibitor cocktail (Roche)) and lysed by sonication before addition of 0.2% Triton X-100, 10% glycerol and 1.5 mM MgCl2. The resultant lysate was cleared by centrifugation and incubated with anti-FLAG M2 magnetic beads (Sigma-Aldrich). After washing, Flag-tagged proteins were eluted using 250 μg/ml Flag peptide (Sigma-Aldrich) in 10 mM HEPES pH 8, 300 mM KCl and 1.5 mM MgCl2, 0.05% nonidet P-40 supplemented with 0.5 mM PMSF and Complete Mini protease inhibitor cocktail by shaking at 12°C for 30 min. Co-immunoprecipitated proteins were detected by mass spectrometry. In addition, co-immunoprecipitated proteins were precipitated using methanol:chloroform and analysed alongside non-concentrated samples of the cleared lysate, by Western blotting using specific antibodies as described in the Supplementary Materials and Methods. Association of DDX21 with pre-ribosomal complexes was determined by glycerol gradient (10-40%) separation (17) of whole cell extracts prepared from control cells or those depleted of PWP2 by RNAi followed by Western blot analysis using an antibody against the endogenous DDX21 protein.
UV crosslinking and mapping of binding sites
The UV Crosslinking and Analysis of cDNA (CRAC) method used previously (11,18) was adapted for use on human cells. Expression of His-PreScission protease site-2xFlag (Flag) tagged proteins was induced in HEK293 cell lines by addition of 1 μg/ml doxycycline for 24 h. Cell culture medium was removed, cells were washed in phosphate-buffered saline (PBS) and ultraviolet (UV) crosslinking was performed with cells in petri dishes in a Stratalinker (Agilent). Cells were lysed by sonication and complexes were first isolated using anti-Flag magnetic beads under native conditions and eluted using PreScission protease, before RNase trimming using RNace-IT. A second denaturing purification step on Ni-NTA was performed in the presence of 6 M guanidium-HCl. 5′ and 3′ linkers were ligated and the libraries generated by reverse transcriptase-polymerase chain reaction (RT-PCR) were sent for Illumina deep sequencing. Reads were mapped on the human transcriptome using a 23 nucleotide cut-off as previously described (11,18). High peaks observed in control samples are likely due to over-represention of non-specific sequences after PCR amplification. For mapping of peaks on the 2D structures of the rRNAs (19) and the 3D structure of the human ribosome (20), the highest peak was set to 100% and peaks above 20% were mapped in a colour gradient from yellow (20%) to red (100%) using Python scripts (Version 2.7). 3D ribosome structure analysis was performed using Pymol.
Pre-rRNA analysis
RNA extracted from siRNA treated cells using Trizol (Qiagen) was separated on a 1.2% agarose-glyoxal gel and transferred to nitrocellulose membrane. Northern blotting was performed using 32P-labelled oligonucleotide probes hybridizing to the 5′ end of internal transcribed spacer (ITS) 1 and to ITS2 (5′-CCTCGCCCTCCGGGCTCCGTTAATGATC-3′ and 5′-CTGCGAGGGAACCCCCAGCCGCGCA-3′, respectively). Pre-rRNA levels were quantified using ImageQuant software and normalized to the amount of actin mRNA (probe 5′-AGGGATAGCACAGCCTGGATAGCAAC-3′).
Sucrose gradient density centrifugation and snoRNA detection
Extracts from cells either treated with non-target siRNAs or those targeting DDX21 or PWP2 were separated on 12 ml 10-45% sucrose density gradients (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, 10/45% sucrose) by centrifugation for 16 h at 23,500 rpm in an Sw40Ti rotor. Gradients were then fractionated and RNA extracted from each fraction was analysed by separation on a 10% denaturing polyacrylamide gel followed by Northern blotting using probes hybridizing to specific snoRNAs ((10); see Supplementary Materials and Methods for details of the probes used). The intensity of signals for each snoRNA was quantified in all fractions, added and set as 100% (total) snoRNA signal and the distribution of each snoRNA in the fractions was calculated as percentage of total signal.
rRNA modification analysis
SNORD68-directed methylation of U428 of the 18S rRNA sequence was assessed using site-specific RNase H cleavage directed by a chimeric, modified RNA/DNA oligonucleotide (5′- mCmGmGdAdAdTdCmGmAmAmCmCmCmUmUmAmUmU-3′; dN, DNA base; mN, 2′-O-methylated RNA) as previously described (8,21). High concentrations of oligonucleotides or RNase H can lead to non-specific interactions cleavages in long RNAs. While unspecific cleavage persisted in the case of C517 modification (SNORD56) analysis, the assays could be optimized for U428 (SNORD68) and were therefore performed using conditions that ensured site-specific cleavage but which resulted in incomplete cleavage of pre-rRNAs. In brief, the oligonucleotide was annealed to 4μg total RNA and samples were incubated with RNase H for 30 min at 37°C. Reactions were stopped by addition of EDTA to a concentration of 0.2 mM and phenol:chloroform:isoamylalcohol extraction of the RNA. RNA was separated on a 1.2% agarose–glyoxal gel and transferred for Northern blotting using a probe hybridizing to the 5′ end of ITS1.
Additional methods can be found in the Supplementary Data online.
RESULTS
DDX21 and NOP2 are components of the human UTP-B complex
An interesting feature of ribosome biogenesis in yeast is the hierarchical association of pre-assembled subcomplexes to form both the early small subunit (SSU) processome and later intermediates. To shed light on the mode of cofactor recruitment in human cells, we performed pulldown experiments using extracts prepared from HEK293 cell lines stably expressing a Flag-tagged version of the human orthologue of yeast Periodic tryptophan protein 2 (Pwp2), a component of the UTP-B complex (22,23), and a control cell line expressing only the Flag-tag. Co-precipitated proteins were identified by mass spectrometry (MS) and Western blotting (Figure 1A and Supplementary Table S1). Using these two methods, comparison to the Flag-tag control revealed that PWP2-Flag specifically enriched the human orthologues of all the yeast UTP-B proteins, Utp21 (WDR36), Dip2/Utp13 (TBL3), Utp12 (UTP12), Utp18 (UTP18) and Utp6 (UTP6). Our MS analysis indicates that under these conditions, the UTP-B subcomplex, rather than pre-ribosomal complexes, is precipitated. The role of these orthologues of the yeast UTP-B components in ribosome biogenesis in human cells is supported by their recruitment to pre-ribosomal particles (17) and RNAi experiments, which reveal defects in pre-rRNA processing (24,25).
Figure 1.
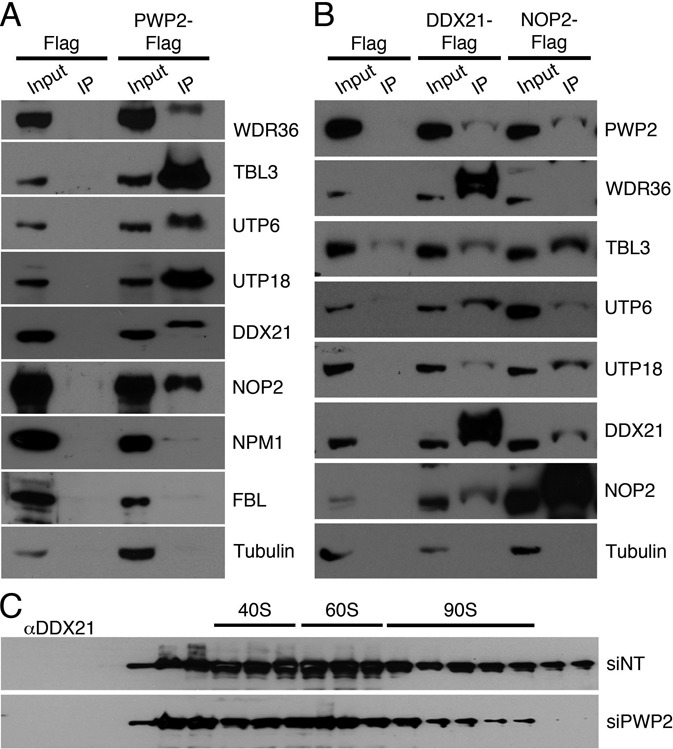
DDX21 and NOP2 are novel components of the UTP-B complex in human cells. (A) Extracts from HEK293 cells expressing Flag-tagged PWP2 or only the Flag-tag were incubated with anti-Flag magnetic beads, eluates were precipitated and analysed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using antibodies against human orthologues of the yeast UTP-B proteins, DDX21, NOP2 and controls. Note, proteins in eluate fractions often migrate more slowly than those in the input samples due to different protein precipitation methods. (B) Eluates of pulldowns using DDX21-Flag, NOP2-Flag or Flag tag alone were analysed as in (A). Note, WDR36 was found to co-precipitate with Nop2-Flag in other experiments performed and identified by mass spectrometry. (C) Extracts from cells treated with non-target siRNAs (siNT) or those targeting PWP2 (siPWP2) were separated by glycerol gradient centrifugation and the distribution of DDX21 was determined by Western blotting using an antibody against the endogenous protein.
Interestingly, in addition to the orthologues of the yeast UTP-B components, the nucleolar RNA helicase DDX21 and the putative methyltransferase NOP2 were also identified by these analyses (Figure 1A and Supplementary Table S1). Yeast Nop2 has recently been shown to mediate m5C methylation of position 2870 of the 25S rRNA (26), while the human protein has, so far, been shown to be required for biogenesis of the large ribosomal subunit (27). DDX21, which does not have a yeast orthologue (Supplementary Figure S1), is overexpressed in various cancers and is implicated in viral infection (see, e.g. (28,29)). DDX21 functions in ribosome biogenesis and was suggested to link c-Jun-activated transcription to ribosome assembly in human cells (30), but its molecular function has remained unclear so far. The highly abundant nucleolar proteins Fibrillarin (FBL) and Nucleophosmin (NPM1) were also identified by MS analysis, but along with a control, Tubulin, were not significantly or repeatedly detectable by Western blotting (Figure 1A).
To determine whether DDX21 and NOP2 interact with PWP2 individually or rather associate with a human UTP-B complex, we performed reverse pulldowns using DDX21-Flag or NOP2-Flag. Both proteins co-precipitated each other as well as the conserved core UTP-B factors (Figure 1B), indicating that DDX21 and NOP2 are UTP-B components in human cells.
The identification of DDX21 as an additional UTP-B component in human cells raised the question of which pre-ribosomal complexes DDX21 is associated with. Density gradient centrifugation followed by Western blotting revealed that DDX21 co-migrates with early 90S pre-ribosomal complexes as well as later pre-60S and pre-40S particles (Figure 1C). Interestingly, we found that in cells depleted of the core UTP-B component PWP2, the amount of DDX21 co-sedimenting with early, 90S pre-ribosomal complexes significantly decreased while its accumulation in complexes co-migrating with 40S or 60S subunits was unaffected. This may suggest that DDX21 is recruited to early complexes together with the UTP-B complex but that it could also associate independently of UTP-B later during ribosome synthesis.
DDX21 interacts with both ribosomal subunits
The UV Crosslinking and Analysis of cDNA (CRAC) approach (18,31) has been used very successfully to determine binding sites of several proteins, among them the yeast RNA helicases Prp43 and Rok1, on cellular RNAs (11,32). To identify interaction sites of the novel UTP-B components on pre-rRNAs, we adapted the CRAC protocol for use in human cells (see ‘Materials and Methods’ section) and employed it on the DDX21-Flag and NOP2-Flag cell lines together with the control cell line expressing only the Flag-tag. Under these conditions stable interactions between RNA and NOP2-Flag could not be detected, however, DDX21-Flag crosslinked to RNA in the CRAC experiments and mapping of the reads obtained by Illumina deep sequencing on the human transcriptome identified specific crosslinking sites in rRNAs from both ribosomal subunits (Figure 2A). DDX21 crosslinked to two major sites within the 18S rRNA sequence that correspond to helices (H)12/13 and H15 (Figure 2A and B; (19)) and to H67 of the 28S rRNA secondary structure. We then mapped the specific crosslinking sites on the 3D structure of the human ribosome (20) and found that the sites in the 18S rRNA sequence cluster together in 3D (Figure 2C), suggesting that the two major peaks in the 18S rRNA might be derived from a single DDX21 binding site. Interestingly, the identified DDX21 crosslinking site in the 18S rRNA sequence overlaps with the binding site of the ribosomal protein RPS9, with which DDX21 has previously been shown to interact (33).
Figure 2.
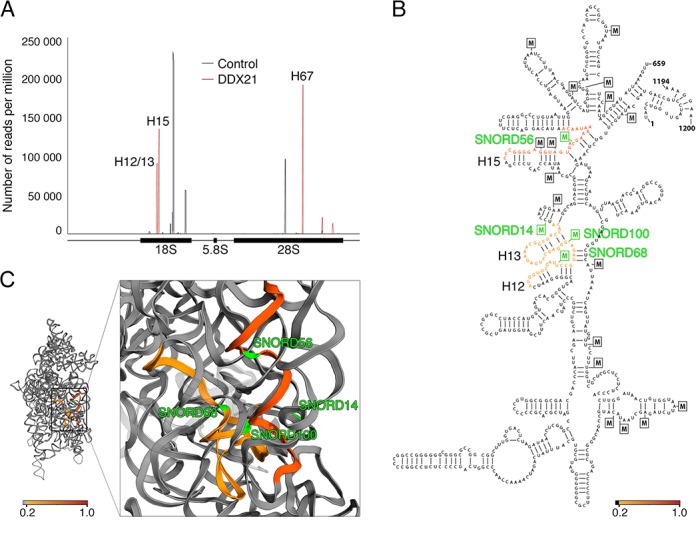
DDX21 crosslinks to ribosomal RNAs. (A) HEK293 cells expressing either the Flag-tag (control) or Flag-tagged DDX21 (DDX21) were UV crosslinked in vivo, covalently linked RNAs were purified via the tagged protein, trimmed and ligated to linkers, followed by RT-PCR and Illumina deep sequencing. Sequence reads were aligned with the human transcriptome and the number of reads per nucleotide is plotted for the control (grey) and for DDX21 (red) on the 47S rDNA sequence. The positions of the mature 18S, 5.8S and 28S rRNAs is shown below. Helices (H) to which DDX21 peaks were mapped are indicated. (B) DDX21 crosslinking sites from (A) were mapped on to the 2D structure of the 5′ domain of the 18S rRNA. Colours indicate peak height in a gradient from yellow (20%) to red (100% of highest peak). Boxed Ms-2′-O-methylations. (C) DDX21 crosslinking sites were mapped on the 18S rRNA in the 3D structure of the SSU with a magnified view shown on the right; colouring is as in (B). The modification sites targeted by SNORD68, SNORD56, SNORD100, SNORD14 are marked in green. Note that helix 21 lies in front of the DDX21 crosslinking site in the mature ribosome.
Consistent with the crosslinking of DDX21 to the SSU and LSU rRNAs, RNAi against DDX21 affected the biogenesis of both ribosomal subunits. Analysis of pre-rRNA levels by Northern blotting following depletion of DDX21 revealed accumulation of the early 47/45S pre-rRNAs and decreased levels of the late SSU and LSU pre-rRNAs 21S/18SE and 12S, respectively (Figure 3A-C). RNAi-mediated depletion of PWP2 affected production of the late SSU precursors to a similar extent as DDX21 depletion. However, while DDX21 depletion lead to accumulation of the 30S pre-rRNA, decreasing the levels of PWP2 caused accumulation of the aberrant 30SL5′ pre-rRNA, indicating an additional defect in the initial pre-rRNA cleavage at the A’ site (Figure 3A-C). Furthermore, co-expression of a Flag-tagged, RNAi-resistant version of wild-type DDX21 rescued the SSU pre-rRNA processing defects caused by depletion of the endogenous protein (Figure 3D). To determine if the activity of DDX21, or merely the presence of the protein, is required for SSU maturation an RNAi-resistant, inactive form of DDX21 was used. S375L and A376E mutations within motif III (DDX21SAT) have previously been shown to reduce the ATPase activity DDX21 (34). Interestingly, expression of this protein in cells depleted of endogenous DDX21 did not restore normal pre-rRNA processing (Figure 3D). Together, these data suggest that, although DDX21 is not required alongside other UTP-B complex components for A’ cleavage, its activity is required for downstream steps in SSU biogenesis.
Figure 3.
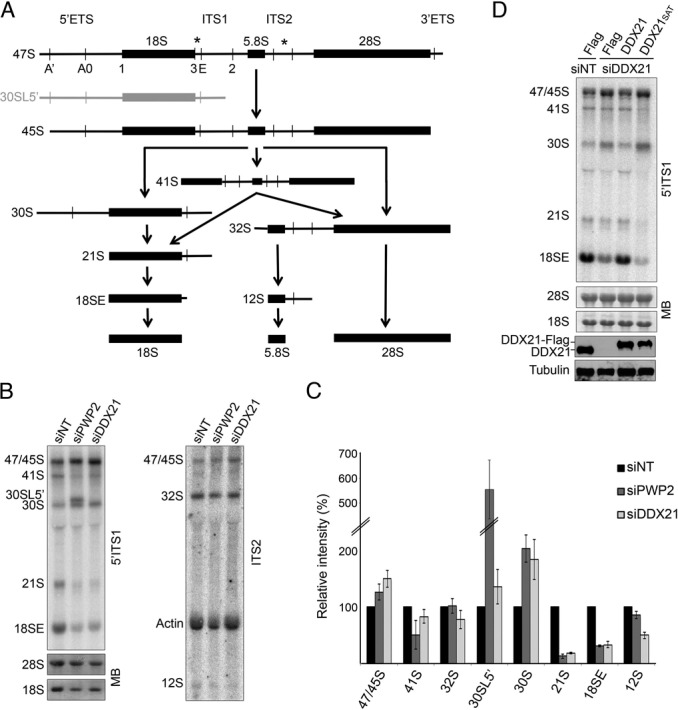
The activity of DDX21 is required for 18S rRNA production. (A) Schematic outline of pre-ribosomal (r)RNA processing in human cells. The 47S primary transcript containing the 18S, 5.8S and 28S mature rRNAs (thick lines) and external (ETS) and internal (ITS) transcribed spacers (thin lines) is shown with the positions of cleavage sites and probes (asterisk) used in Northern blotting indicated. The aberrant pre-rRNA 30SL5′, which accumulates when A’ cleavage is inhibited, is shown in grey. (B) HEK293 cells were transfected with non-target siRNAs (siNT) or those targeting DDX21 (siDDX21) or PWP2 (siPWP2). After 72 h cells were harvested, RNA was extracted, separated by agarose–glyoxal gel electrophoresis and analysed by Northern blotting using probes hybridizing in ITS1 and ITS2 of the pre-rRNA transcript and to the actin mRNA. RNA was visualized using a phosphorimager and mature rRNAs were detected by methylene blue staining (MB). (C) The levels of pre-rRNA intermediates were quantified, normalized to actin mRNA levels and are given relative to the non-target siRNA control. Data are represented as a mean of three independent experiments ± SEM. (D) HEK293 cells expressing the Flag-tag (Flag) or Flag-tagged, RNAi-resistant wild-type (DDX21) or a helicase-inactive form of DDX21 (DDX21SAT), were transfected with either non-target siRNAs (siNT) or siRNAs targeting the endogenous DDX21 mRNA (siDDX21) as indicated. The levels of tagged and endogenous DDX21 were determined by Western blotting and pre-rRNA levels were analysed by Northern blotting using a probe hybridizing to the 5′ end of ITS1.
DDX21 is required for the association of snoRNAs with pre-ribosomal complexes
A key function of RNA helicases in ribosome biogenesis is the release of snoRNAs from pre-ribosomes. Since several snoRNP modification sites are found within, or close to, the DDX21 pre-rRNA crosslinking sites, we investigated whether depletion of DDX21 affected the levels of such snoRNAs in pre-ribosomal complexes. For the SSU, we selected several box C/D snoRNAs, SNORD100 (HBII-439), SNORD68 (HBII-202), SNORD56 (U56), SNORD14 (U14) that guide modifications at, or close to, the DDX21 crosslinking sites in the 18S rRNA sequence (Figure 2B; (35)). In addition, for the LSU we analysed SNORD88, which directs a modification in H67 within the DDX21 crosslinking site in the 28S rRNA sequence (Supplementary Figure S2A). Soluble extracts from control cells or cells depleted of DDX21 by RNAi were separated by sucrose-gradient density centrifugation and the distribution of the selected snoRNAs in free snoRNPs and in pre-ribosomal complexes was analysed by Northern blotting. The SSU snoRNAs SNORD14 and SNORD100 and the LSU snoRNA SNORD88 were detected with free snoRNPs and co-migrated with early, 90S complexes. Depletion of DDX21 did not affect the distribution or total levels of these snoRNAs (Figure 4A, Supplementary Figure S2B-D). In control cells, both SNORD56 and SNORD68 were detected with free snoRNPs and, interestingly, in pre-40S complexes. However, while RNAi against the 40S biogenesis factor PWP2 only resulted in a minor reduction in pre-40S associated SNORD68 and SNORD56 (∼35%), depletion of DDX21 led to a significant reduction of SNORD68 and SNORD56 (approximately 85%) in pre-40S complexes (Figure 4B-D). This suggests a specific reduction in SNORD68 and SNORD56 recruitment to pre-40S particles upon knockdown of DDX21 and demonstrates that this effect does not arise due to a general defect in SSU biogenesis. Furthermore, expression of Flag-tagged, wild-type DDX21 in cells depleted of endogenous DDX21 rescued the SNORD68 and SNORD56 recruitment defects, while expression of inactive DDX21SAT did not (Figure 4B-D). Together, these data reveal that DDX21 is actively required for pre-ribosomal access of SNORD68 and SNORD56, which basepair with 18S rRNA sequences that overlap with the DDX21 crosslinking sites.
Figure 4.
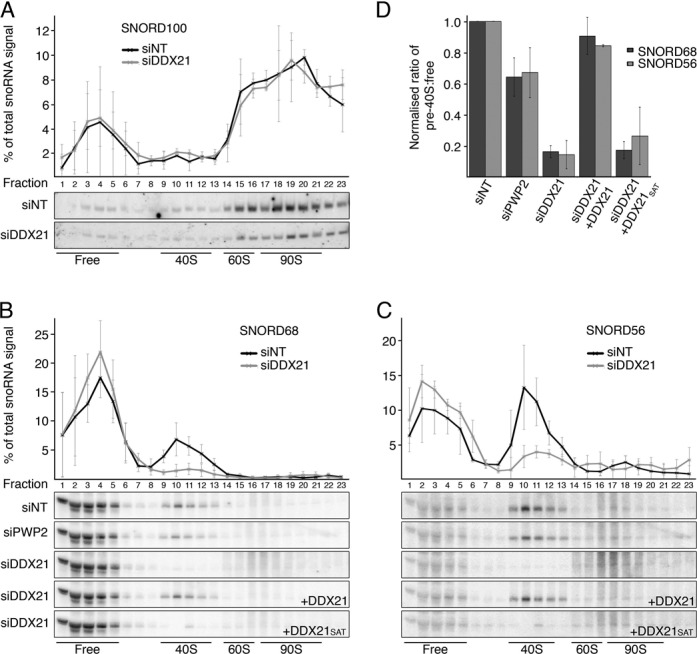
DDX21 is required for access of the late-acting SNORD56 and SNORD68 to pre-40S intermediates. (A-C) Extracts from cells transfected with siRNAs targeting DDX21 (siDDX21), with or without expression of RNAi-resistant wild-type DDX21 or inactive DDX21 (DDX21SAT), as well as extracts from cells treated with non-target siRNAs (siNT) or those targeting PWP2 (siPWP2), were separated by sucrose gradient density centrifugation. RNA was isolated from gradient fractions and analysed by denaturing polyacrylamide gel electrophoresis followed by Northern blotting for the snoRNAs SNORD100 (A), SNORD68 (B) and SNORD56 (C). Quantifications of three independent experiments are shown above the Northern blots for each fraction as percentage of the total snoRNA signal (data are presented as mean ± SEM); fractions containing free snoRNPs and ribosomal complexes are indicated below. (D) The ratio of SNORD68 and SNORD56 signals in pre-40S-containing fractions to those containing free snoRNPs in (B) and (C) was calculated, normalized to the ratio in the corresponding sample transfected with non-target siRNAs and plotted as mean ± SEM.
The activity of DDX21 is required for rRNA modification
The function of SNORD68 is to guide the 2′-O-methylation of residue U428 of the 18S rRNA sequence. The decreased association of SNORD68 with pre-ribosomes observed upon depletion on DDX21 would be expected to be mirrored by a decrease in the extent of modification of its target residue. We therefore used site-specific RNase H cleavage directed by chimeric 2′-O-methyl RNA/DNA oligonucleotides spanning U428 to monitor the extent of SNORD68-guided rRNA modification in pre-rRNAs (Figure 5A; see ‘Materials and Methods’ section). Incubation of control RNA with RNase H resulted in a mild accumulation of a cleaved fragment of the late 18SE pre-rRNA, implying that this site is incompletely modified in vivo (Supplementary Figure S3). Consistent with our gradient analysis, depletion of DDX21, or expression of only the inactive DDX21SAT (Figure 3C; (34)), led to a significant increase in the relative accumulation of this cleavage fragment compared to the 18SE pre-rRNA, indicating decreased modification of the SNORD68 target residue upon depletion of DDX21 (Figure 5B and Supplementary Figure S3).
Figure 5.
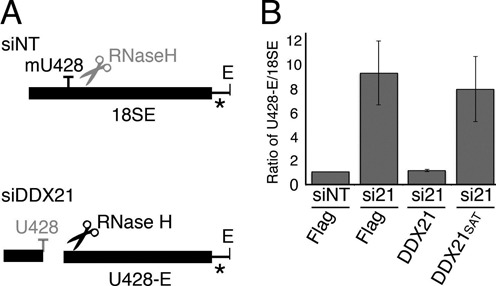
DDX21 is a non-snoRNP protein required for rRNA modification. (A) Schematic representation of the site-specific RNase H cleavage assay used, where unmodified (siDDX21, lower panel) 18SE pre-rRNA can be cleaved by RNase H while modified (siNT; upper panel) 18SE pre-rRNA is not. Asterisk marks the position of the probe used in Northern blotting. (B) Cells stably expressing RNAi-resistant, Flag-tagged DDX21, DDX21SAT or only the FLAG-tag were transfected with non-target siRNAs (siNT) or those targeting DDX21 (siDDX21) as indicated. RNA was extracted and analysed by site-directed RNase H cleavage to monitor rRNA methylation of U428 in the 18S rRNA. The ratio of RNase H cleaved fragment (U428-E):18SE pre-rRNA in three independent experiments was calculated (data are presented as mean ± SEM).
snoRNAs associate with both early and late pre-ribosomal complexes
Based on work performed in the yeast Saccharomyces cerevisiae, snoRNPs have generally been assumed to be recruited very early in the ribosome biogenesis pathway and to mediate pre-rRNA modification in a co-transcriptional manner ((9); and references therein). Consistent with this model, we found that human SNORD100 and SNORD14 were associated with early 90S complexes (Figure 4A and Supplementary Figure S2C). Intriguingly, however, other snoRNAs tested were found in later pre-40S complexes implying that they function at later stages of ribosome biogenesis. To investigate this further, we used Northern blotting and analysed the pre-ribosomal association of additional human snoRNAs that basepair within the DDX21 crosslinking site and at various, spatially distinct regions of the 18S rRNA sequence (Supplementary Figure S4). Our data show that most snoRNAs (including SNORD100, SNORD14 and SNORD70 (HBII-234), which basepair close to the DDX21 crosslinking site, as well as SNORD20 (U20) that basepairs with the distant H44 of the 18S rRNA sequence) were distributed between the free snoRNP pool (gradients fractions 1-5) and early, 90S complexes (fractions 17-21, Figure 6A and B). Interestingly, two additional snoRNAs, SNORD27 and SNORD25, were found in these fractions, but they also accumulated in later, pre-40S complexes (fractions 9-12), suggesting that they remain associated with pre-ribosomes after pre-rRNA cleavages that separate the pre-SSU and pre-LSU complexes. This is in contrast to SNORD68 and SNORD56, which were not detected in early, 90S complexes, but instead were only recruited to late pre-40S complexes (Figures 4B and C and 6A and B). Additionally, we examined SNORD13 (U13), which has previously been suggested to be involved in the final maturation of the 3′ end of 18S rRNA (36), and found that this snoRNA was primarily detected co-migrating with pre-40S complexes (Figure 6A and B). As SNORD27, SNORD25 and SNORD13 were found in later pre-40S complexes we investigated whether their association with these complexes was regulated by DDX21. This is particularly relevant for SNORD27, which guides a modification close to the DDX21 crosslinking site. However, for all three snoRNAs, no changes in their gradient profiles were detected upon depletion of DDX21 (Supplementary Figure S5). Together, our data therefore identify three distinct sets of snoRNAs based on the timing of their pre-ribosome association: (i) those that function in early ribosome biogenesis and dissociate before separation of the subunits (SNORD14, SNORD100, SNORD20, SNORD70), (ii) snoRNAs that associate with early pre-ribosomes but maintain interactions until later stages of ribosome maturation (SNORD27, SNORD25) and 3) those that are only recruited to later pre-ribosomal complexes (SNORD68, SNORD56, SNORD13; Figures 6A and B and 7). Furthermore, DDX21 is specifically required for the pre-40S association of late-acting snoRNAs that basepair close to its pre-ribosomal crosslinking site in the small subunit (SNORD68, SNORD56).
Figure 6.
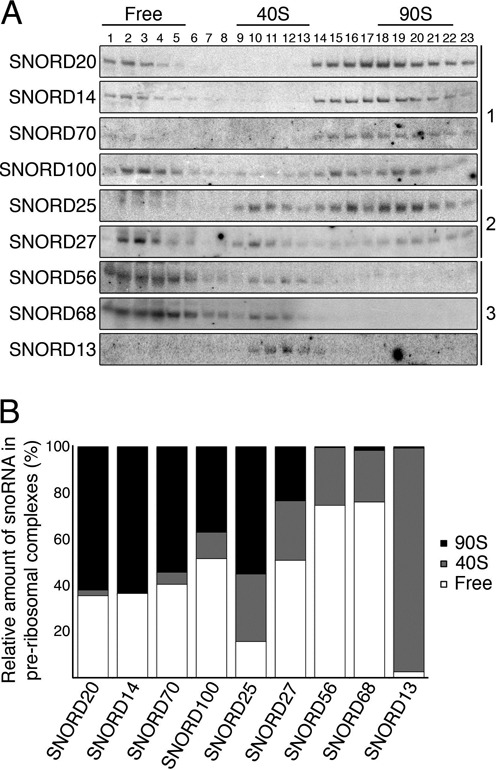
snoRNAs associate with pre-ribosomal complexes at different stages during ribosome maturation. (A) Cell lysates were separated by sucrose-gradient density centrifugation and RNA isolated from each fraction was analysed by Northern blotting using probes hybridizing to the snoRNAs indicated. Fractions containing free snoRNPs and ribosomal complexes are indicated above. snoRNAs can be grouped into 90S snoRNAs (group 1, see right), those present in both 90S and pre-40S particles (group 2) and pre-40S snoRNAs (group 3). (B) Quantification of the relative amounts of each snoRNA in fractions containing free snoRNPs (fractions 1-5), pre-40S (fractions 9-12) and 90S (fractions 17-21) complexes expressed as a percentage of the total amount of each snoRNA.
Figure 7.
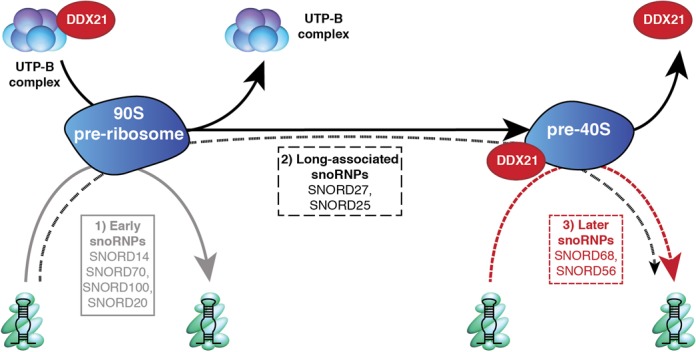
Schematic overview of snoRNA timing and DDX21 function in human cells. snoRNPs can be divided into three sets: (1) those that function in early pre-ribosomal complexes (grey), (2) those that are found associated with early and later pre-ribosomal particles (black) and (3) later snoRNAs (red), such as SNORD68 and SNORD56, whose association with pre-40S complexes is regulated by DDX21.
DISCUSSION
Early ribosome synthesis in yeast is characterized by the modular assembly of the pre-ribosomal intermediates from pre-existing subcomplexes. While the inventory of human cofactors has recently begun to be explored (22,24,37,38), the mode and timing of cofactor recruitment has largely remained unknown. Here, we show that the UTP-B complex, a subcomplex of the SSU processome in yeast, is conserved but contains additional components in human cells, including the RNA helicase DDX21. Our findings support the model of modular assembly also for human pre-ribosomal intermediates, where it is likely that this aids the coordinated and efficient recruitment of the numerous cofactors involved in ribosome assembly, pre-rRNA processing and modification.
The modification of rRNAs is a key step in their maturation and is largely mediated by a host of snoRNPs that are guided to their target sites by snoRNA-pre-rRNA basepairing. Given the high density of modifications, particularly in human rRNAs, the question of how the numerous snoRNA-pre-rRNA interactions are coordinated is highly pertinent. In yeast, most snoRNA-guided rRNA modifications in the 18S and 25S rRNA sequences are introduced co-transcriptionally and snoRNPs are thought to act very early in the pathway (9). In contrast, lone-standing RNA methyltransferases, such as Dim1 and Emg1 mostly act later in ribosome biogenesis and, in the case of Emg1, even require prior modification of the modification site by a snoRNP (39–41). The close proximity of modified nucleotides, especially in human cells, and the extensive base-pairing interactions that box C/D snoRNAs form with their pre-rRNA targets means that multiple snoRNAs have overlapping binding sites, necessitating their sequential action. It is not yet clear how the temporal action of snoRNPs is regulated but, based on the observation that most snoRNPs modify co-transcriptionally, it is possible to imagine that one snoRNA outcompetes or rapidly displaces another snoRNA. Interestingly, we find here that in human cells snoRNAs associate with both 90S and also pre-40S complexes, suggesting that rRNA modifications occur over a much broader timeframe than previously anticipated. Our analysis of a number of snoRNAs that guide modification in SSU intermediates allows their classification into early snoRNAs only present in 90S complexes, snoRNAs found in both 90S and pre-40S intermediates and late snoRNAs, which specifically bind pre-40S complexes (Figures 6 and 7). This clearly indicates that several human snoRNPs analysed here follow a defined hierarchy in their pre-ribosomal association.
Our analyses of DDX21 have revealed that this RNA helicase actively regulates the pre-ribosomal access of the late-acting snoRNAs SNORD56 and SNORD68, which basepair at the DDX21 crosslinking sites in pre-40S complexes. This is in contrast to several yeast RNA helicases, and the only other mammalian RNA helicase for which a pre-ribosomal function is described, as these enzymes are implicated in the release of snoRNAs from pre-ribosomes. However, in yeast the recruitment of the snoRNAs snR64 and snR67 was previously shown to be affected by depletion of Prp43 (11,42). Since RNA annealing functions have recently been described for helicases, DDX21 might actively participate in establishing snoRNA-pre-rRNA interactions. However, in this case, since direct interactions between DDX21 and any snoRNAs that guide modifications in the SSU were not detected in our CRAC analysis, DDX21 might rather function by remodelling pre-rRNA secondary structures or by inducing the release of protein cofactors from the snoRNA basepairing sites to allow the late snoRNAs to gain access to their target sites. In these scenarios, DDX21 would likely not, or only transiently, contact the snoRNAs directly. Consistent with this model, depletion of DDX21 did not affect the distribution of snoRNAs that associate with early pre-ribosomal complexes, such as SNORD100, SNORD14 and SNORD27, but only influenced snoRNAs that are recruited later. Interestingly, the basepairing site of the late-acting SNORD68 overlaps with the pre-rRNA interaction sites of the early-acting SNORD100 (Figure 6 and Supplementary Figure S4), which functions independently of DDX21. Instead, DDX21 likely enables the access of SNORD68 after SNORD100 has already been released from pre-ribosomal intermediates. Furthermore, both SNORD68 and SNORD100 (as well as SNORD56 and SNORD13) form extended basepairing interactions with the pre-rRNA (Supplementary Figures S4, S5 and S8). However, since depletion of DDX21 only affects association of SNORD68 (and SNORD56) with pre-ribosomes it is unlikely that DDX21 generally functions in helping establish these additional snoRNA–pre-rRNA contacts.
It is likely that late-acting snoRNPs represent exceptions as they will almost certainly require the co-ordinated action of an RNA helicase to enable them to gain access to their basepairing sites after the pre-ribosome has undergone significant folding during the early phases of ribosome synthesis. Our results therefore suggest that, together with DDX21, SNORD56 and SNORD68 might constitute a late modification module in human SSU biogenesis. This notion is supported by the fact that both modification by SNORD56 and SNORD68, as well as the DDX21 helicase (see Supplementary Figure S1), seem conserved in most eukaryotes while neither the rRNA modifications, nor the helicase, are found in yeast. It is tempting to speculate, therefore, that DDX21, at least for its function in SSU biogenesis, might have co-evolved with its late snoRNA targets to facilitate their recruitment onto pre-ribosomal intermediates. It will be interesting to see whether future analyses will reveal additional late-acting snoRNPs and partner helicases, especially in the dense clusters of modification sites of the large ribosomal subunit.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We would like to thank Lukas Brüning for comments on the manuscript.
Footnotes
The authors wish it to be known that, in their opinion, the first three authors should be regarded as joint First Authors.
Present addresses:
Matthias S. Leisegang and Carmen Doebele, Medical Faculty, Goethe University, 60596 Frankfurt, Germany.
Ana S. Ramírez, Institute of Molecular Biology and Biophysics, ETH, 8093 Zürich, Switzerland.
FUNDING
Deutsche Forschungsgemeinschaft [BO 3442/1-1 to M.T.B. and SFB 902 to E.S.]; Göttingen University Medical Department [to M.T.B.]; Dorothea Schlözer and Alexander von Humboldt postdoctoral fellowships [to K.E.S.]; Cluster of Excellence Frankfurt [to M.T.B. and E.S.]; Wellcome Trust [to N.J.W.]; Biodiversity and Climate Research Centre Frankfurt [BIK-F to I.E.]. Funding for open access charge: Göttingen University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Decatur W.A., Fournier M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 2.Henras A.K., Soudet J., Gerus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woolford J.L., Jr, Baserga S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phipps K.R., Charette J., Baserga S.J. The small subunit processome in ribosome biogenesis-progress and prospects. Wiley Interdiscip. Rev. RNA. 2011;2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watkins N.J., Bohnsack M.T. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA. 2012;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 6.Ganot P., Bortolin M.L., Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 7.Kiss-Laszlo Z., Henry Y., Bachellerie J.P., Caizergues-Ferrer M., Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 8.van Nues R.W., Granneman S., Kudla G., Sloan K.E., Chicken M., Tollervey D., Watkins N.J. Box C/D snoRNP catalysed methylation is aided by additional pre-rRNA base-pairing. EMBO J. 2011;30:2420–2430. doi: 10.1038/emboj.2011.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kos M., Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohnsack M.T., Kos M., Tollervey D. Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep. 2008;9:1230–1236. doi: 10.1038/embor.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohnsack M.T., Martin R., Granneman S., Ruprecht M., Schleiff E., Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kos M., Tollervey D. The putative RNA helicase Dbp4p Is required for release of the U14 snoRNA from preribosomes in Saccharomyces cerevisiae. Mol. Cell. 2005;20:53–64. doi: 10.1016/j.molcel.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 13.Liang X.H., Fournier M.J. The helicase Has1p is required for snoRNA release from pre-rRNA. Mol. Cell. Biol. 2006;26:7437–7450. doi: 10.1128/MCB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srivastava L., Lapik Y.R., Wang M., Pestov D.G. Mammalian DEAD box protein Ddx51 acts in 3′ end maturation of 28S rRNA by promoting the release of U8 snoRNA. Mol. Cell. Biol. 2010;30:2947–2956. doi: 10.1128/MCB.00226-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin R., Straub A.U., Doebele C., Bohnsack M.T. DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 2013;10:4–18. doi: 10.4161/rna.21879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez-Galan O., Garcia-Gomez J.J., de la Cruz J. Yeast and human RNA helicases involved in ribosome biogenesis: current status and perspectives. Biochim. Biophys. Acta. 2013;1829:775–790. doi: 10.1016/j.bbagrm.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Turner A.J., Knox A.A., Prieto J.L., McStay B., Watkins N.J. A novel small-subunit processome assembly intermediate that contains the U3 snoRNAP, nucleolin, RRP5 and DBP4. Mol.Cell. Biol. 2009;29:3007–3017. doi: 10.1128/MCB.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bohnsack M.T., Tollervey D., Granneman S. Identification of RNA helicase target sites by UV cross-linking and analysis of cDNA. Methods Enzym. 2012;511:275–288. doi: 10.1016/B978-0-12-396546-2.00013-9. [DOI] [PubMed] [Google Scholar]
- 19.Petrov A.S., Bernier C.R., Hershkovits E., Xue Y., Waterbury C.C., Hsiao C., Stepanov V.G., Gaucher E.A., Grover M.A., Harvey S.C., et al. Secondary structure and domain architecture of the 23S and 5S rRNAs. Nucleic Acids Res. 2013;41:7522–7535. doi: 10.1093/nar/gkt513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anger A.M., Armache J.P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 21.Yu Y.T., Shu M.D., Steitz J.A. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA. 1997;3:324–331. [PMC free article] [PubMed] [Google Scholar]
- 22.Ebersberger I., Simm S., Leisegang M.S., Schmitzberger P., Mirus O., von Haeseler A., Bohnsack M.T., Schleiff E. The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res. 2014;42:1509–1523. doi: 10.1093/nar/gkt1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dosil M., Bustelo X.R. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J. Biol. Chem. 2004;279:37385–37397. doi: 10.1074/jbc.M404909200. [DOI] [PubMed] [Google Scholar]
- 24.Tafforeau L., Zorbas C., Langhendries J.L., Mullineux S.T., Stamatopoulou V., Mullier R., Wacheul L., Lafontaine D.L. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell. 2013;51:539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Sloan K.E., Bohnsack M.T., Schneider C., Watkins N.J. The roles of SSU processome components and surveillance factors in the initial processing of human ribosomal RNA. RNA. 2014;20:540–550. doi: 10.1261/rna.043471.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma S., Yang J., Watzinger P., Kotter P., Entian K.D. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013;41:9062–9076. doi: 10.1093/nar/gkt679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sloan K.E., Bohnsack M.T., Watkins N.J. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 2013;5:237–247. doi: 10.1016/j.celrep.2013.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonzheim I., Irmler M., Klier-Richter M., Steinhilber J., Anastasov N., Schafer S., Adam P., Beckers J., Raffeld M., Fend F., et al. Identification of C/EBPbeta target genes in ALK+ anaplastic large cell lymphoma (ALCL) by gene expression profiling and chromatin immunoprecipitation. PLoS One. 2013;8:e64544. doi: 10.1371/journal.pone.0064544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yasuda-Inoue M., Kuroki M., Ariumi Y. Distinct DDX DEAD-box RNA helicases cooperate to modulate the HIV-1 Rev function. Biochem. Biophys. Res. Commun. 2013;434:803–808. doi: 10.1016/j.bbrc.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmstrom T.H., Mialon A., Kallio M., Nymalm Y., Mannermaa L., Holm T., Johansson H., Black E., Gillespie D., Salminen T.A., et al. c-Jun supports ribosomal RNA processing and nucleolar localization of RNA helicase DDX21. J. Biol. Chem. 2008;283:7046–7053. doi: 10.1074/jbc.M709613200. [DOI] [PubMed] [Google Scholar]
- 31.Granneman S., Kudla G., Petfalski E., Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9613–9618. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin R., Hackert P., Ruprecht M., Simm S., Brüning L., Mirus O., Sloan K.E., Kudla G., Schleiff E., Bohnsack M.T. A pre-ribosomal RNA interaction network involving snoRNAs and the Rok1 helicase. RNA. 2014;20:1173–82. doi: 10.1261/rna.044669.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lachapelle S., Gagne J.P., Garand C., Desbiens M., Coulombe Y., Bohr V.A., Hendzel M.J., Masson J.Y., Poirier G.G., Lebel M. Proteome-wide identification of WRN-interacting proteins in untreated and nuclease-treated samples. J. Prot. Res. 2011;10:1216–1227. doi: 10.1021/pr100990s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valdez B.C., Henning D., Perumal K., Busch H. RNA-unwinding and RNA-folding activities of RNA helicase II/Gu–two activities in separate domains of the same protein. Eur. J. Biochem. 1997;250:800–807. doi: 10.1111/j.1432-1033.1997.00800.x. [DOI] [PubMed] [Google Scholar]
- 35.Lestrade L., Weber M.J. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:158–162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavaille J., Hadjiolov A.A., Bachellerie J.P. Processing of mammalian rRNA precursors at the 3′ end of 18S rRNA. Identification of cis-acting signals suggests the involvement of U13 small nucleolar RNA. Eur. J. Biochem. 1996;242:206–213. doi: 10.1111/j.1432-1033.1996.0206r.x. [DOI] [PubMed] [Google Scholar]
- 37.Wild T., Horvath P., Wyler E., Widmann B., Badertscher L., Zemp I., Kozak K., Csucs G., Lund E., Kutay U. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol. 2010;8:e1000522. doi: 10.1371/journal.pbio.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sloan K.E., Mattjssen S., Lebaron S., Tollervey D., Pruijn G.J., Watkins N.W. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J. Cell Biol. 2013;200:577–588. doi: 10.1083/jcb.201207131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lafontaine D., Vandenhaute J., Tollervey D. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev. 1995;9:2470–2481. doi: 10.1101/gad.9.20.2470. [DOI] [PubMed] [Google Scholar]
- 40.Wurm J.P., Meyer B., Bahr U., Held M., Frolow O., Kotter P., Engels J.W., Heckel A., Karas M., Entian K.D., et al. The ribosome assembly factor Nep1 responsible for Bowen-Conradi syndrome is a pseudouridine-N1-specific methyltransferase. Nucleic Acids Res. 2010;38:2387–2398. doi: 10.1093/nar/gkp1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer B., Wurm J.P., Kötter P., Leisegang M.S., Schilling V., Buchhaupt M., Held M., Bahr U., Karas M., Heckel A., et al. The protein mutated in Bowen-Conradi Syndrome, Nep1 (Emg1), is required for a unique modification in 18S rRNA. Nucleic Acids Res. 2011;39:1526–1537. doi: 10.1093/nar/gkq931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leeds N.B., Small E.C., HIley S.L., Hughes T.R., Staley J.P. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol. Cell. Biol. 2006;26:513–522. doi: 10.1128/MCB.26.2.513-522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


