Abstract
The 5′ untranslated region of hepatitis C virus (HCV) genomic RNA contains an internal ribosome entry site (IRES) element, composed of domains II–IV, which is required for cap-independent translation initiation. Little information on the 3D structure of the whole functional HCV IRES is still available. Here, we use atomic force microscopy to visualize the HCV IRES conformation in its natural sequence context, which includes the upstream domain I and the essential, downstream domains V and VI. The 574 nt-long molecule analyzed underwent an unexpected, Mg2+-induced switch between two alternative conformations: from ‘open’, elongated morphologies at 0–2 mM Mg2+ concentration to a ‘closed’, comma-shaped conformation at 4–6 mM Mg2+. This sharp transition, confirmed by gel-shift analysis and partial RNase T1 cleavage, was hindered by the microRNA miR-122. The comma-shaped IRES-574 molecules visualized at 4–6 mM Mg2+ in the absence of miR-122 showed two arms. Our data support that the first arm would contain domain III, while the second one would be composed of domains (I–II)+(V–VI) thanks to a long-range RNA interaction between the I-II spacer and the basal region of domain VI. This reinforces the previously described structural continuity between the HCV IRES and its flanking domains I, V and VI.
INTRODUCTION
Hepatitis C virus (HCV) is the major etiological agent of chronic liver disease. There is no HCV vaccine and the traditional treatment based on a combination of alpha-interferon (IFN) and ribavirin (RBV) failed in about half of the patients. The need for new, alternative therapeutic approaches has encouraged the exploration of HCV life cycle as well as the development of direct-acting antiviral agents that have substantially increased sustained virologic response, what suggests that IFN-free regimens could lead to HCV eradication (1). With this aim, a thorough study of the native structure of the HCV genomic RNA is currently required, since certain structural/functional RNA elements, in particular those present at the 5′ and 3′ untranslated regions (UTR), are promising targets for antiviral therapy (2,3).
The 5′ UTR of HCV is highly conserved among all the viral genotypes and contains an internal ribosome entry site (IRES) element that drives cap-independent initiation of translation of the viral polyprotein (4,5). The minimum sequence required for HCV IRES activity spans nucleotides (nts) 39–371 of the viral genome (6,7), and contains domains II to IV followed by the first 27 nts of the core coding sequence (4). Its secondary structure has been proposed using in silico RNA folding, covariation sequence analysis and biochemical methods (4,6,8–12) (Figure 1). The structures of individual HCV IRES domains or subdomains have been studied using techniques with atomic resolution (such as X-ray diffraction (XRD) and nuclear magnetic resonance (NMR)) (5,13–16), while electron microscopy (EM) has been used to visualize the structure of HCV IRES in its free form (17) and bound to the ribosome (18). A model of the HCV IRES structure in solution, based on small-angle X-ray scattering (SAXS) in combination with molecular dynamics simulations, has been also published (19).
Figure 1.
HCV IRES in its natural sequence context. (A) Schematic representation of the 9.6-Kb-long, ssRNA HCV genome. The secondary structures of the 5′ UTR and 3′ UTR are schematized. The structural (gray background) and non-structural (white background) proteins within the viral polyprotein are boxed. The regions corresponding to the IRES-418, IRES-574 and E2/p7/NS2 transcripts are shown. (B) Sequence and secondary structure of the 5′ UTR of HCV, previously deduced by different bioinformatic, biochemical and biophysical methods. Nucleotides are numbered every 50 residues, and domains/subdomains are labeled from I to VI. HCV IRES domains II to IV are boxed. The last residues of the IRES-418 and IRES-574 molecules used in this work are marked with arrows. Dashed lines show the complementary sequences at the I-II spacer (nts 24–38) and the basal region of domain VI (nts 428–442), while the two miR-122-binding sites are marked with solid gray lines.
HCV IRES is flanked upstream by a short stem-loop termed domain I, which is connected with domain II through a 23-nt-long ssRNA spacer region. Additionally, two downstream, structured domains termed V and VI (spanning nts 388–510 of the viral genome and located within the core coding sequence (20)), are essential for HCV viability (21) through a still unknown RNA structure-based mechanism. Phylogenetic (22), functional (23) and structural (24) studies have shown that a 15-nt-long sequence at the I-II spacer region can anneal with a complementary sequence at the basal region of domain VI (Figure 1), thus promoting a ‘closed’ conformation of the HCV IRES. This long-range RNA interaction is destabilized upon binding of the most abundant liver-specific microRNA, miR-122, which also interacts with the I-II spacer region by binding to two seed sites located 8 or 9 nts apart, depending on the viral isolate (25,26), and promotes the switch to an ‘open’ IRES conformation (27). However, the transition from ‘open’ to ‘closed’ (or vice versa) HCV IRES conformations has not been described in the absence of any effector RNA or protein molecule.
At the functional level, miR-122 binding has been associated to several effects including the increase of HCV RNA stability (28,29), reduction of the 5′ decay rates of the viral genome in infected cells (30) and stimulation of IRES-mediated translation (31). However, the distinct effects of miR-122 on translation observed in vitro and in cell culture (32,33) make it difficult to associate different HCV IRES conformations with miR-122 activities and to assess the role of miR-122 in the viral life cycle (34,35). This encourages the acquisition of additional structural and functional data on the interaction of miR-122 with HCV IRES in different experimental conditions.
RNA folding depends on the concentration of cations around the negatively charged phosphate backbone. Due to its high charge density, Mg2+ ions stabilize RNA tertiary structure more efficiently than other divalent or monovalent cations (36), thus being essential for the functional folding of large RNAs (37,38). The Mg2+ concentration required for optimal HCV IRES activity in translation-competent extracts ranges from 1 to 2.5 mM (39). HCV IRES can promote translation initiation at 2.5 mM Mg2+ without requiring the complete set of initiation factors, and it is able to drive initiation factor-independent translation at 5 mM Mg2+ (40). Although the Mg2+-dependent secondary structure of the minimal HCV IRES (domains II–IV) has already been investigated (10), new technological approaches are needed to visualize the effect of Mg2+ on the tertiary fold of the HCV IRES in its natural sequence context.
Atomic force microscopy (AFM) is a powerful nanotechnology-based tool for the structural analysis of a wide range of biological entities. It provides a 3D surface profile of the imaged sample without requiring any staining or coating, thus being less destructive and disruptive than EM. The nanometer resolution of this technique is optimal for the visualization of nucleic acid molecules (41–45), including RNA and RNA-protein complexes (46–49). AFM is increasingly being used in virology (50–53) and low-resolution images of the whole HCV genomic RNA have been published (54,55). However, AFM has not yet been applied to the structural characterization of either viral 5′ UTR regions or viral/cellular IRES elements. Here, we analyze by AFM the Mg2+-induced RNA folding of the HCV IRES in two constructions that contain either domains I-IV or I-VI. AFM results, in combination with gel-shift analysis and partial RNase T1 digestion, have allowed us to unveil a sharp magnesium-induced switch from an ‘open’ to a ‘closed’ conformation of the HCV IRES in its natural sequence context (domains I-VI), this transition being partially hindered by the presence of miR-122 in the medium.
MATERIALS AND METHODS
RNA synthesis
HCV RNA transcripts were obtained from the plasmid pN(1–4728) that contains the subgenomic region of HCV genotype 1b (GenBank accession number AF139594) spanning nts 1–4728, preceded by the T7 promoter (56). The IRES-418 construction (nts 1–418, IRES domains I-IV) and the E2/p7/NS2 fragment (nts 2478–3107) were obtained by polymerase chain reaction (PCR) amplification of pN(1–4728). The forward primer 1b/2a-5′-T71(+) (5′-TAATACGACTCACTATAGGACCCGCCCCGATTGGGGGCGA-3′, where the underlined sequence corresponds to the T7 RNA polymerase promoter), and the reverse primer 1b-C393(-) (5′-CCGGAATTCCCCGGGAACTTGACGTCCTGTGGGC-3′) were used for the amplification of IRES-418, while 1b-NS2T7–2487(+) (5′-TAATACGACTCACTATAGGGAGTATGTCGTGTTGCTT-3′) and 1b-NS2–3087(-) (5′-AGCGCGTACGAAGTACGGCA-3′) (11) allowed the PCR amplification of E2/p7/NS2. All high pressure liquid chromatography (HPLC) grade primers were purchased to IBA GmbH. In turn, the IRES-574 fragment (nts 1–574, IRES domains I-VI) was obtained by digestion of pN(1–4728) with the restriction enzyme BlpI (NEB). All the DNA fragments were purified by QIAquick Gel Extraction Kit (Qiagen) and eluted in water. These DNA templates (0.5–1 μg) were in vitro transcribed using AmpliScribe T7 RNA polymerase (Epicentre) at 37ºC for 4 h. After DNase I treatment, RNA was purified by precipitation with 2.5 M ammonium acetate and 2.5 volumes of absolute ethanol, and finally resuspended in diethylpyrocarbonate (DEPC)-treated water. The integrity of the RNA transcripts was checked in a denaturing 4% polyacrylamide gel, and the concentration of the transcribed RNA was determined by absorbance at 260 nm.
Control RNA molecule for AFM experiments
The dsRNA molecule used as a control for measuring the height of the A-RNA duplex adsorbed onto mica surfaces modified with (3-aminopropyl)triethoxysilane (APTES) was the 4579-bp-long genomic RNA of the totivirus L-A (57) (kindly provided by Dr. Rosa Esteban, Instituto de Microbiología Bioquímica, CSIC/USAL, Spain), with a theoretical length of 1236 nm.
RNA folding in different Mg2+-containing buffers
RNA transcripts were resuspended at 2–3 nM concentration in folding buffer (100 mM NaCl and 100 mM HEPES pH 7.5) either magnesium-free or supplemented with 2, 4, 6 or 10 mM MgCl2. The RNAs were heated at 90ºC for 5 min, cooled on ice for 5 min and incubated at 37ºC for 20 min. The use of other renaturation protocols did not affect the results. Control experiments were performed by incubating IRES-574 in 80% formamide at 90ºC for 5 min, and cooling on ice for 5 min. A folding buffer in which sodium cations were replaced by ammonium ones (100 mM NH4Cl, 100 mM HEPES pH 7.5 and 10 mM MgCl2) was also used.
MicroRNA-122 annealing assay
The miR-122 RNA sequence (5′-UGGAGUGUGACAAUGGUGUUUGU-3′) was synthesized and HPLC-purified by IBA GmbH. HCV IRES-574 was folded under different concentrations of Mg2+ as described above, incubated (at a concentration of 3 nM) with 15 nM miR-122 at 37°C for 20 min, and the mixture was successively transferred to ice.
RNA adsorption on mica surfaces
We used APTES-modified mica (58), a method that drives a tight enough immobilization of RNA via electrostatic interactions without damaging RNA molecules. This approach avoided the use of Mg2+ or other divalent cations as a bridge to immobilize the negatively charged RNA (59), since they would interfere with our envisaged study. Freshly cleaved muscovite mica Hi-Grade V2 (Monocomp Instrumentation) was treated with a 0.1% solution of APTES (SIGMA-Aldrich) for 15 min, washed with 2-propanol, rinsed with ultrapure, DEPC-treated milliQ water and dried at 37ºC. Then, 30 μl of the RNA sample at 2–3 nM concentration was deposited onto the treated mica disc and incubated in a humidity chamber for 20 min at room temperature (22 ± 2ºC). The RNA-containing mica discs were gently washed with ultrapure, DEPC-treated milliQ water and finally air-dried. A minimum of three independent samples of each HCV IRES preparation (IRES-418, IRES-574 and IRES-574/miR-122 complex) at each buffer composition was imaged.
AFM imaging
Samples were imaged in air, at room temperature, with two commercial AFM microscopes: PicoSPM (Agilent) and Nanoscope IIIa (Veeco). Their performance was equivalent in the analyzed samples, as assessed after measuring different control preparations of either IRES-574 molecules or IRES-574/miR-122 complexes in parallel using both microscopes (data not shown). Acoustic tapping mode AFM was performed using silicon cantilevers (Nanosensors and Bruker) with spring constants of 0.5–9.5 N/m and resonance frequency in the 50–80 KHz range. Two different kinds of tips, with nominal curvature radius of 2 and 10 nm, were used to obtain different levels of resolution during the image acquisition. The set-points used were kept relatively low, in the 0.3–0.6 V range, in order to use soft imaging conditions. The images (from 512 × 512 to 2048 × 2048 pixels) were recorded at a scan rate of 1 line per second and processed using WSxM v3.1 software (Nanotec) (60). The average length of the RNA molecules was measured using the same software. In the case of branched shapes, the length of every imaged arm of the molecule was measured, and the total length was computed. The number of analyzed IRES molecules in each buffer condition was at least 100 for IRES-574 samples, and at least 50 for IRES-574/miR-122 complexes. The angle between the arms of the selected IRES-574 molecules was measured with Image-J software (61).
Statistical analysis
The non-parametric Kolmogorov–Smirnov test was used to check the data sets for normality. A non-parametric Mann–Whitney test was applied to IRES-574 at different Mg2+ concentration. The independent samples t-test was also applied to the IRES-574, IRES-574/miR-122 and E2/p7/NS2 samples incubated under different ionic conditions.
RNA analysis by native gel electrophoresis
Purified RNA transcripts were internally radiolabeled with [α-32P]-GTP (specific radioactivity of 107 dpm/μg) as described (62). After folding under different concentrations of Mg2+, the IRES-574 molecule was left on ice for at least 10 min and resuspended at a final 3 nM concentration (3 nM IRES-574 and 15 nM IRES for IRES-574/miR-122 complexes) in loading buffer (previously described in (27)) containing 60% glycerol, 1× TMN, 0.4 μg/μl yeast tRNA (Ambion), 0.4% (w/v) xylene cyanol and 0.4% (w/v) bromophenol blue. The samples were loaded into 0.8-mm thick, non-denaturing 6% polyacrylamide (acrylamide:bis-acrylamide ratio 1:19) gels containing 50 mM Tris-Ac pH 8.3 and 10 mM NH4(OAc). The gels were run at 12 mA (constant intensity) for 24 h at 4ºC, and autoradiographed. Product bands were scanned in a Phosphorimager (Storm 820, GE) and quantified with Image Quant 5.2 software (GE). The percentage of formation of each conformer X was calculated as follows: % conformer X = (signal conformer X / Σ signals of all conformers) × 100. Mean values and standard deviations were calculated from three independent experiments.
RNase T1 and RNase III cleavage assays
Previous studies ((27) and data not shown) showed that the optimal conditions for RNase T1 (Calbiochem) partial digestion of the IRES-574 molecule were 0.001 μg/μl of enzyme (concentrations assayed: 0.01, 0.001, 0.0001, 0.00001 μg/μl), in a 20-min reaction at 37ºC. [32P]pCp 3′-end labeled IRES-574 RNA (27) was resuspended in each of the following control conditions: in water and kept on ice for 2 h, to check the RNA integrity (sample ‘I’); in standard folding buffer supplemented with 2 mM ethylenediaminetetraacetic acid (EDTA) (samples ‘0*’ mM Mg2+) and in folding buffer without Mg2+ (samples ‘0′ mM Mg2+), after preheating to 90ºC and slowly cooling to room temperature. RNase T1-digested samples were previously resuspended in the corresponding folding buffer (in the presence of 0–10 mM Mg2+), pre-heated to 90ºC for 1 min and cooled to room temperature. Note that 300 cpm of labeled and digested RNA were loaded in each lane. Adequate band separation was achieved when bromophenol blue dye reached 80% of the length of a 7M urea-containing denaturing 4% polyacrylamide gel. IRES-574 molecules incubated with 0.0001 μg/μl RNase T1 under denaturing conditions were used as an electrophoretic mobility control (sample ‘D’). Reference bands were identified from previous studies (10,27). RNase T1 digestion bands that decrease their intensity with increasing Mg2+ concentration, consistently detected in different gels and exposure times, were identified.
Commercial Escherichia coli RNase III (Ambion), a nuclease specific for dsRNA, was used to cleave the internally labeled (24) HCV-574 molecule. Digestion was performed on renatured HCV RNA under ‘secondary conditions of cleavage’: RNA substrate was pre-heated at 90°C for 1 min, before the addition of reaction buffer [10 mM HEPES–KOH, pH 7.5, 100 mM NH4(OAc) and 10 mM Mg(OAc)2] and cooled to room temperature. Cleavage reactions were performed at 37°C for 1 h in a volume of 10 μl, in the presence of 20 U RNasin, 0.0005 U/μl (final concentration) of RNase III and 2 μg/μl of yeast tRNA. Note that 300 cpm of digested RNA were loaded in the corresponding gel lane. Digestion products corresponding to fragments spanning nts 1–439 and 27–574 of the IRES-574 molecule, previously characterized by direct RNA fingerprinting analysis and other classical RNA sequencing techniques (24,27), were used as size markers.
RESULTS
AFM analysis of HCV IRES in denaturing conditions
A systematic AFM study of HCV IRES molecules adsorbed onto APTES-treated mica surfaces (58) (Supplementary Figure S1) was performed using an RNA transcript encompassing nts 1–574 (domains I–VI) of HCV genome, termed IRES-574 (Figure 1). The theoretical length of the unfolded IRES-574 molecule is 155 nm, assuming that each ribonucleotide unit is 0.27-nm long in an A-RNA helical conformation with a pitch of 3.0 nm and 11 nts per turn. The AFM analysis of the IRES-574 in 80% formamide (Supplementary Figure S2) evidenced that part of its secondary structure is highly stable under harsh denaturing conditions, since the imaged molecules showed 65–80% of the theoretical molecular length. This observation agrees with a previous report on the preservation of secondary structure elements of the hepatitis delta virus ribozyme in 95% formamide (63).
The measured height of the IRES-574 molecule in formamide varied between 1.3 and 2.2 nm (Supplementary Figure S2D). To experimentally check the height of a control linear dsRNA molecule adsorbed onto mica-APTES, we chose the whole genomic RNA of the totivirus L-A (Supplementary Figure S3A). The height of this molecule was 1.2–1.4 nm (Supplementary Figure S3B). Therefore, the height range of the imaged IRES-574 molecule in formamide might correspond to dsRNA regions lying on the surface, together with other elements of non-denatured secondary structure in different orientations.
AFM analysis of the Mg2+-induced IRES-574 folding process
To analyze the influence of Mg2+ on IRES-574 folding, different preparations of the molecule in buffers containing 100 mM Na+ supplemented with rising Mg2+ concentrations (0, 2, 4, 6 and 10 mM) were visualized by AFM. Tapping mode AFM images of more than 100 molecules under each ionic condition showed that the increase of Mg2+ concentration induced conformational changes in IRES-574, while molecular aggregation or dimerization was not observed. Supplementary Figure S4 shows a selection of 12 individual molecules imaged at each Mg2+ concentration, and Figure 2 summarizes the observed effect of Mg2+ on the conformational rearrangement of the molecule.
Figure 2.
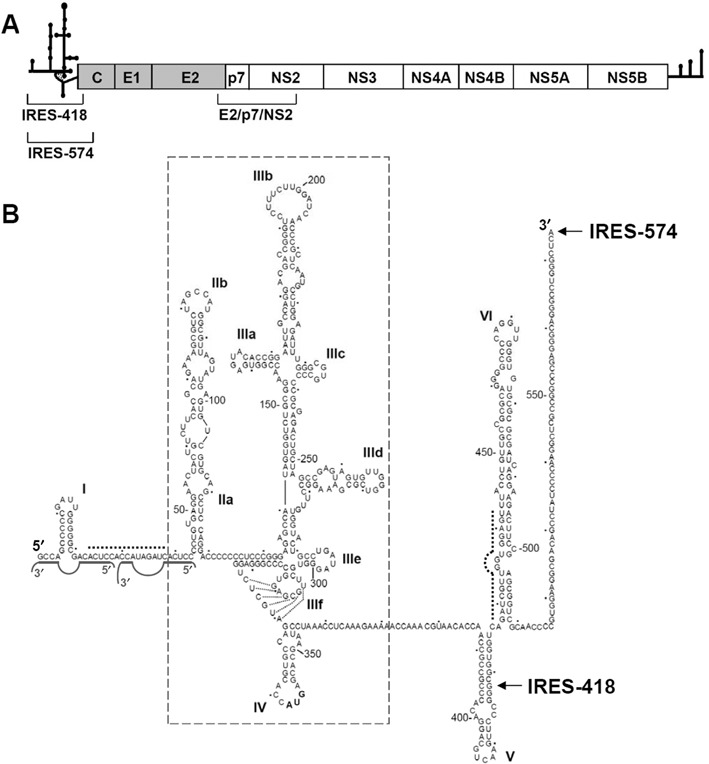
AFM images of IRES-574 molecules in a folding buffer supplemented with increasing Mg2+ concentration. IRES-574 RNA molecules were incubated in a buffer containing 100 mM Na+, either without Mg2+ (panel A) or supplemented with 2, 4, 6 and 10 mM Mg2+ (panels B, C, D and E, respectively). In all panels, top row contains large AFM images of molecules deposited onto a pre-treated mica-APTES surface (550 × 550 nm, scale bar of 110 nm), medium row shows a zoom area containing a selected molecule (100 × 100 nm image, scale bar of 20 nm), and the bottom row depicts a 3D representation of the selected molecule, showing its measured length. The nominal curvature radius of the AFM tips was 2 nm. Overall, this figure shows the Mg2+-induced IRES-574 folding process and reveals a major conformational switch when Mg2+ concentration increases from 2 to 4 mM.
IRES-574 molecules incubated in a buffer lacking Mg2+ ions showed an elongated, zigzag shape with one to three bumps (Figure 2A and Supplementary Figure S4A), likely corresponding to regions that preserved their secondary structure in the absence of Mg2+, in agreement with previous data on minimal HCV IRES (10) and also consistent with a recent analysis of the Mg2+-dependent folding of picornavirus IRES (64). At 2 mM Mg2+, the imaged shapes showed a large variability even within the same preparation: a majority of elongated morphologies similar to those found at 0 mM Mg2+ (Figure 2B) coexisted with some heterogeneous, more compact molecules with 2–4 arms protruding from a central axis (Supplementary Figure S4B) that could inform about certain Mg2+-driven tertiary interactions (36,38). A neat increase in the homogeneity and compactness of the imaged molecules was found in the presence of 4 mM Mg2+ (Figure 2C and Supplementary Figure S4C). This ionic condition promoted the unexpected formation of compact, comma-shaped molecules in which two arms could be distinguished: a bulky one and a flatter one. Both arms were 15–20 nm long, and the angle between them ranged between 110º and 150º, with a maximum in the interval 117º–127º (Supplementary Figure S5). Similar angles could also be measured in the fraction of non-elongated molecules imaged at 2 mM Mg2+ and even in some molecules at 0 mM Mg2+ (see examples in Supplementary Figure S4A and B). The trend toward a further RNA compaction was maintained in the presence of 6 mM Mg2+ (Figure 2D and Supplementary Figure S4D), with morphologies analogous to those found at 4 mM Mg2+ together with some more compact, elliptical-shaped molecules. Finally, in the presence of 10 mM Mg2+ only compact molecules with a highly uniform elliptical shape were observed (Figure 2E and Supplementary Figure S4E). Overall, this analysis evidenced that the main structural switch in the IRES-574 conformation occurred between 2 and 4 mM Mg2+ concentration.
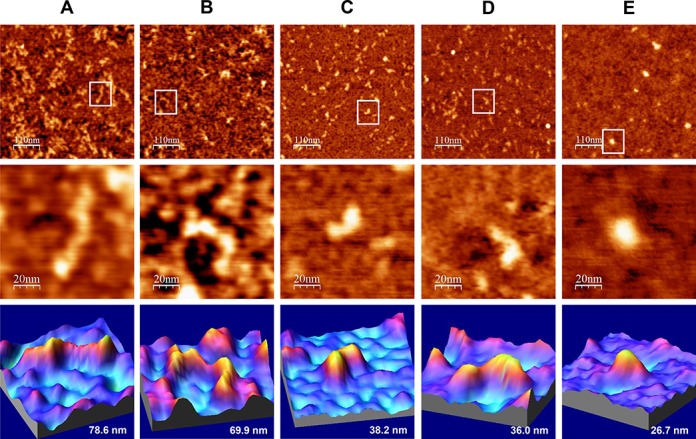
To quantitatively study the Mg2+-induced folding process, the average end-to-end distance of 100 IRES-574 molecules was measured for each Mg2+ concentration (Figure 3). At 0 mM Mg2+, molecules with lengths in the range of 25–135 nm were imaged (Figure 3A), most of them being in the interval of 60–80 nm (Figure 3B). Their median length was 63 nm, about 40% of the theoretical length of the molecule. This confirmed that a significant number of RNA secondary structure elements are preserved in the presence of 100 mM Na+. IRES-574 molecules under 2 mM Mg2+ also showed a broad length distribution between 25 and 120 nm, the majority of them being in the intervals of 40–60 and 60–80 nm. Their median length was 61 nm. A clear shift in the length distribution was found at 4 mM Mg2+: the molecules were imaged in a narrower interval of 14–93 nm, with a clear peak in the interval of 20–40 nm and a computed median length of 28 nm. Thus, a greater than 2-fold reduction in the median length was produced when Mg2+ concentration increased from 2 to 4 mM, in agreement with the qualitative data derived from Figure 2 and Supplementary Figure S4. In the presence of 6 mM Mg2+, the length distribution of the IRES-574 molecules decreased to 12–70 nm while their median length was slightly higher (32 nm). Finally, the additional compaction of the imaged molecules at 10 mM Mg2+ was reflected in a very narrow length distribution of the molecules (14–50 nm), concentrated in the interval of 20–40 nm and showing a median length of 25 nm. Along this process, the only statistically significant reductions of the average lengths occurred in the transitions 2–4 and 6–10 mM Mg2+ (Figure 3A).
Figure 3.
Length distribution of IRES-574 molecules at different Mg2+ concentrations. (A) Length distribution (computed over 100 molecules) of the IRES-574 molecule in folding buffer supplemented with 0, 2, 4, 6 and 10 mM Mg2+ concentration is depicted in a box plot. Boxes represent 25–75 percentile range, vertical lines span 10–90 percentile range and horizontal bar within the box represent the median. Outlier values are depicted as circles (mild outliers) and asterisks (extreme outliers). Statistical P-values corresponding to the average length of RNA molecules for 0 versus 2, 2 versus 4, 4 versus 6 and 6 versus 10 Mg2+ concentration are shown: statistically significant differences (P < 0,001) are marked in bold. (B) Distribution of molecular length versus Mg2+ concentration, computed for 100 IRES-574 molecules at each buffer composition. Box: intervals of molecular length (nm). (C) Length distribution (computed over 50 molecules) of the IRES-574/miR-122 complex at 0, 2, 4, 6 and 10 mM Mg2+ concentration depicted in a box plot, as detailed in panel A. (D) Distribution of molecular length versus Mg2+ concentration, computed for 50 IRES-574/miR-122 complexes in each condition. Box: intervals of molecular length (nm).
As a control experiment, we analyzed the eventual influence of Mg2+ concentration on the folding of a coding region of the HCV genome with a length similar to that of IRES-574: the E2/p7/NS2 junction, spanning nts 2487–3087 of the viral genome (Figure 1A). The results depicted in Supplementary Figure S6 showed that the HCV IRES element, whose biological function relies on its tridimensional structure, is much more dependent on the Mg2+ concentration than a coding region of the same length within the same viral genome. We were also interested in analyzing whether two monovalent cations, Na+ and NH4+, might show a differential effect on the folding of IRES-574 molecule in the presence of 10 mM Mg2+. The molecules incubated in a buffer containing 100 mM NH4+ (equivalent to that previously used in RNase III cleavage experiments that supported a ‘closed’ conformation of the HCV IRES including domains I–VI (24)) showed elliptical morphologies that were less compact that those folded in the presence of 100 mM Na+ (Supplementary Figure S7). This observation agrees with previous results evidencing that the lower charge density of ammonium with respect to sodium makes the former cation less efficient than the latter one in promoting RNA folding (36,65).
AFM analysis of a 418-nt-long HCV IRES transcript in the presence of 4 mM Mg2+
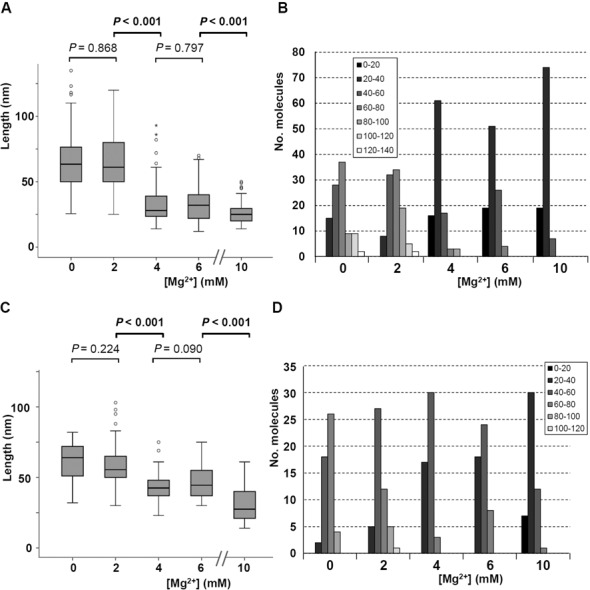
We also analyzed by AFM an RNA molecule spanning nts 1–418 of HCV genome (molecule termed IRES-418). It contained the IRES domains II–IV flanked upstream by domain I and, downstream, by the first 76 nts of the core coding region (Figure 1). Most of the imaged IRES-418 molecules in the presence of 4 mM Mg2+ showed elongated conformations together with branched shapes where three arms of different lengths protruded from a central axis, some of which exhibited a characteristic ‘λ-like’ shape (Figure 4).
Figure 4.
AFM images of IRES-418 molecules in a folding buffer containing 4 mM Mg2+. (A) Large AFM image (1000 × 1000 nm) of molecules deposited onto a mica-APTES surface. (B) AFM images (100 × 100 nm) of three selected molecules (top row) and their corresponding 3D representations from two view angles (medium and bottom rows). The nominal curvature radius of the AFM tips was 2 nm.
Monitoring the Mg2+-induced folding of IRES-574 by gel-shift
To deepen into the conformational rearrangements imaged by AFM in the IRES-574 molecules, two complementary techniques for RNA structural analysis were used: gel-shift and ssRNA-specific digestions. Native gel electrophoresis allowed the identification of only four major bands, corresponding to conformers termed C1 to C4 (Figure 5A). Among them, C1 and C2 were minority under all the buffer conditions assayed, while the fast-migrating conformers C3 and C4 were the most abundant. The band intensity was quantified in three independent experiments, and the ratio of each conformer was graphically represented (Figure 5B). In the absence of Mg2+ (both with and without added EDTA, see Materials and Methods) the most intense band was C3 (mean of 71.7% and 76.1% in lanes 2 and 3 of Figure 5A, respectively), followed by C4 (18.3% and 14.8%, respectively). Thus, the use of EDTA to remove the residual Mg2+ ions eventually trapped into the IRES-574 molecule showed a very limited effect in our experimental conditions. In turn, when the folding buffer contained 2 mM Mg2+ a reduction of the amount of C3 was observed concomitantly to the increase of C4, their relative intensities equalizing (45.7% and 46.3%, respectively). The increase of Mg2+ up to 4 mM completed the observed shift between C3 and C4 (rates of 25.3% and 66.3%, respectively). A further increase of Mg2+ up to 6 mM produced slight variations (21.5% of C3 and 69.4% of C4) while the dominance of the most compact conformer was again reinforced at 10 mM Mg2+ (19.9% of C3 and 71.0% of C4).
Figure 5.
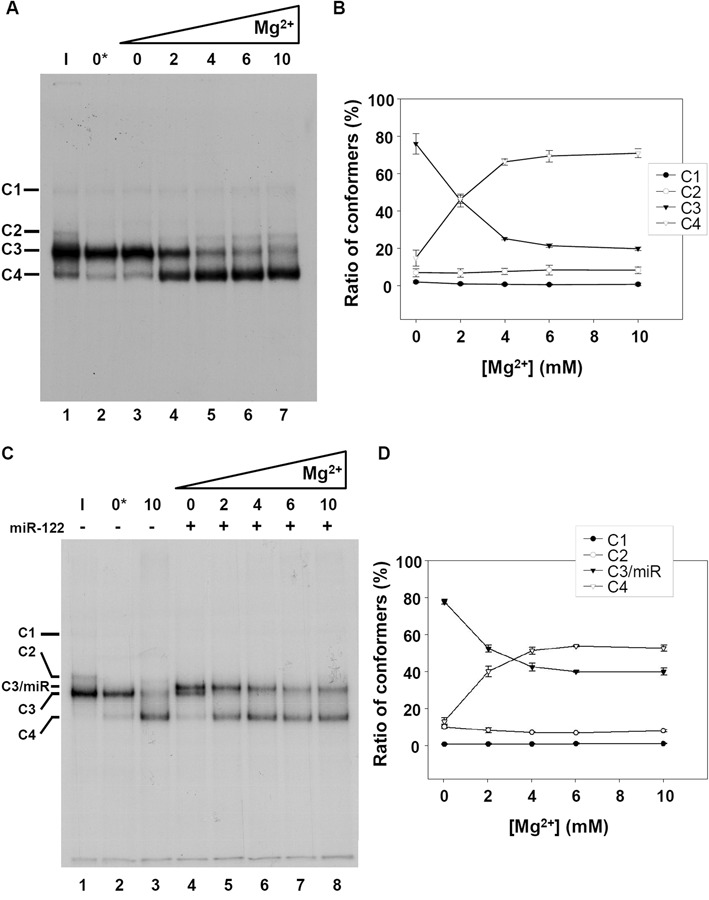
Native gel electrophoresis of the HCV IRES-574 at different Mg2+ concentrations. (A) Autoradiography of a non-denaturing 6% polyacrylamide gel. Internally labeled IRES-574 RNA was resuspended at 3 nM in each of the following conditions: in water and kept on ice for 10 min, as a control of intact RNA (sample ‘I’, lane 1); in standard folding buffer supplemented with 2 mM EDTA (sample ‘0*’ mM Mg2+, lane 2); in folding buffer alone (sample ‘0′ mM Mg2+, lane 3); in folding buffer containing Mg2+ at a final concentration of 2, 4, 6 and 10 mM (lanes 4–7). Resuspended RNA in lanes 2–7 was pre-heated to 90ºC for 1 min and slowly cooled down to room temperature. Four conformers, labeled as C1 to C4, are detected at different ratios in all the lanes. (B) Quantification of the percentage of formation of each conformer at 0, 2, 4, 6 and 10 mM Mg2+ in three independent experiments analogous to that showed in panel A (lanes 3–7). Mean values and standard deviations are shown. (C) Same as panel A showing the effect of the interaction of miR-122 with IRES-574: ice-incubated control IRES-574 RNA (sample ‘I’, lane 1); IRES-574 alone in buffer supplemented with EDTA (sample ‘0*’ mM Mg2+, lane 2); IRES-574 in folding buffer containing Mg2+ at 10 mM concentration (lane 3); IRES-574 (at 3 nM concentration) incubated with miR-122 (at 15 nM concentration) in folding buffer either lacking Mg2+ (lane 4) or in the presence of increasing concentrations of Mg2+ from 2 to 10 mM (lanes 5–8). Five conformers (labeled as C1, C2, C3/miR, C3 and C4) were observed in total. (D) Quantification of the percentage of formation of conformers C1, C2, C3/miR and C4 at 0–10 mM Mg2+ in three independent experiments analogous to that showed in panel C (lanes 4–8). Mean values and standard deviations are shown.
Analysis of the folding process of IRES-574 by partial RNase T1 cleavage
Partial RNase T1 cleavage assays were carried out on IRES-574 molecules under different Mg2+ concentrations. Nuclease T1 cuts after G nts in ssRNA regions, being a sensitive method to detect global rearrangements in RNA structure that introduce steric hindrance. Since the Mg2+-dependent IRES structure has been previously analyzed for domains II and III in the context of a 332-nt-long molecule (10), we focused our study on the effect of Mg2+ on the 574-long transcript that also included domains I, IV, V and VI. The partial digestion of IRES-574 molecules (Figure 6) showed a defined cleavage pattern in the absence of added divalent ions, which changed as a result of the presence of increasing Mg2+ concentrations. The samples without Mg2+ (lanes 4 and 5 of Figure 6A) showed that the cleavage yield of the EDTA-treated molecule (lane 4) was slightly lower than that of the sample without EDTA (lane 5). Thus, it is likely that the total absence of Mg2+ ions in the EDTA-containing buffer impaired the activity of the nuclease. Upon addition of Mg2+, the overall reactivity of IRES-574 decreased (lanes 6–9), thus reflecting a progressive Mg2+-induced compaction of the molecule, in agreement with previous results (10). However, several regions of the molecule, in particular, certain hairpin loops, remained accessible to the nuclease.
Figure 6.
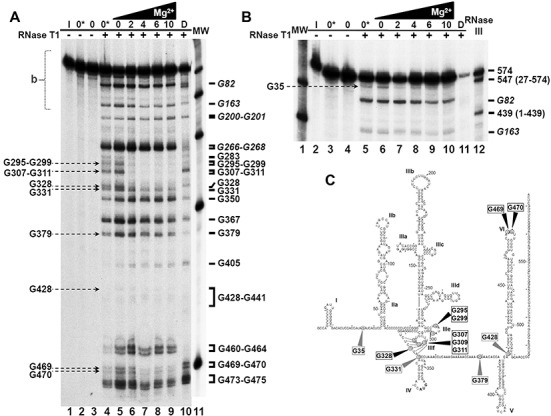
Mg2+-induced protection from RNase T1 cleavage. (A) Partial RNase T1 cleavage of 3′-end labeled IRES-574 resolved in a denaturing gel. Undigested control samples (lanes 1–3) are described in the Materials and Methods section. Lanes 4–9 correspond to samples resuspended in folding buffer either without Mg2+ (lanes 4 and 5) or supplemented with 2, 4, 6 and 10 mM Mg2+ (lanes 6–9, respectively), renatured and digested by RNase T1. Electrophoretic mobility control: sample ‘D’ (see Materials and Methods), lane 10. Molecular weight (100–500 nt) markers (‘MW’) corresponding to a less exposed gel were added as lane 11. Reference bands are shown on the right of the gel. Bands that decrease their intensity with increasing Mg2+ are identified by arrows on the left. (B) A longer electrophoretic run used to resolve the upper region marked with ‘b’ in panel A. MW markers are shown in lane 1. Lanes 2–11 correspond to lanes 1–10 in panel A. Control RNase III digestion fragments spanning nts 1–439 and 27–574 of the IRES-574 molecule are shown in lane 12. Reference positions 82 and 163 (10) are also marked. The band corresponding to nt G35, which decreases its intensity with increasing Mg2+ concentration, is identified on the left. (C) Secondary structure of the HCV IRES-574 molecule, showing the positions of the RNase T1 cleavage sites corresponding to the bands identified in panels A and B. Nucleotides that either decrease their intensity or virtually disappear with increasing Mg2+ concentration are marked with grey and black arrowheads, respectively.
We chose the electrophoretic bands that could be used as a reference to identify the nts whose reactivity to RNase T1 cleavage was affected by the Mg2+ concentration. Among them, positions 82, 163, 200–201 and 266–268 corresponded to the highly accessible Gs located at the apical loops of subdomains IIb, IIIa, IIIb and IIId, respectively (10), while reactive positions between nts 350 and 475 were detected in other previous work (27) (Figure 6A). Additional reference bands were identified as the eventually accessible Gs between positions 268 and 350: 283, 295, 299, 307, 309, 311, 328 and 331 (the latter G was previously shown to be a reactive nucleotide despite being paired in the secondary structure models (10)). This allowed us to identify the main Gs that changed their reactivity to RNase T1 as a result of the Mg2+-driven compaction of the IRES-574 molecule. Additionally, a longer electrophoretic run allowed resolving the upper part of the gel showed at Figure 6A, corresponding to the 5′-end of the HCV IRES (Figure 6B).
The first observation was that the region encompassing nts 379–460 showed a very low reactivity to RNase T1 even in the absence of Mg2+, which is indicative of the high stability of the RNA stems present at domains V and VI, as well as the protection of two Gs (at positions 435 and 436) whose accessibility might have been predicted from the secondary structure of the HCV IRES (Figure 1). This exemplifies that in the absence of added Mg2+ certain elements of secondary structure are stabilized by the monovalent cations present in the buffer, as imaged by AFM. On the contrary, certain nts in the analyzed region (including G350, G367, G460-G464 and G473-G475) remain highly reactive to RNase T1 digestion in folding buffers containing Mg2+ concentrations in the range 0–10 mM.
The conformational changes induced by Mg2+ (lanes 5–9 of Figure 6A, and lanes 6–10 of Figure 6B) were accompanied by a decrease in the intensity of at least nine RNase T1 bands. This effect was detected when Mg2+ concentration increased from 0 to 2 mM (bands G295-G299, G307-G309-G311, G328, G428, G469 and G470 in Figure 6A; G35 in Figure 6B) and from 2 to 4 mM (G331 and G379, Figure 6A). Among them, the most pronounced protections from RNase T1 cleavage was found in bands G295-G299, G307-G309-G311, G328, G469 and G470. In all the positions showing Mg2+-dependent reactivity, the band intensities were similar at 4, 6 and 10 mM Mg2+, what suggests that the main conformational rearrangement of the IRES-574 molecule is already completed at 4 mM Mg2+, in agreement with our previous AFM and gel-shift results. The nts whose reactivity to RNase T1 is affected by Mg2+ are located at the spacer region between domains I and II (G35), the IIIef/IV pseudo-knot (G295-G299, G307-G309-G311 and G328), the basal region of domain IV (G331), the spacer between domains IV and V (G379), the basis of domain VI (G428) and the apical loop of this domain (G469 and G470), as depicted in Figure 6C.
Effect of miR-122 on the Mg2+-induced conformational switch of IRES-574
The microRNA miR-122 interacts with two tandem binding sites in the I-II spacer region of the HCV IRES (25,26) (Figure 1) and induce switching of the IRES element in its natural sequence context (domains I–VI) between two alternative conformers: from a quick migrating form associated with a ‘closed’ conformation to a slow migrating one corresponding to an ‘open’ conformation) (27). Thus, the possibility that the conformational effect of Mg2+ on IRES-574 folding was inversely related to that of miR-122 deserved a detailed analysis by AFM and gel-shift. First, IRES-574 at 3 nM concentration was titrated using native gel electrophoresis with a range of miR-122 concentrations (from 0.5 to 150 nM) in the folding buffer containing 6 mM Mg2+. As a 15 nM concentration of miR-122 was found to initiate the linear phase of the saturation curve from ‘closed’ to ‘open’ conformations (data not shown), such conditions (corresponding to an IRES-574:miR-122 ratio of 1:5) were used for subsequent AFM and gel-shift experiments.
The visualization by AFM of IRES-574/miR-122 complexes in buffers containing 100 mM Na+ and 0–10 mM Mg2+ was performed as described above. As a control experiment, the miR-122 molecule alone (whose 23-nt-long sequence corresponds to a theoretical length of 6.2 nm) was imaged at 15 nM concentration under different ionic conditions, showing a uniform and elliptical shape of 10–14 nm in length (Supplementary Figure S8). Therefore, unbound miR-122 and IRES-574 molecules were clearly distinguishable in the analyzed AFM images (see examples at Figure 7E and Supplementary Figure S9), while neither dimerization nor molecular aggregation were observed.
Figure 7.
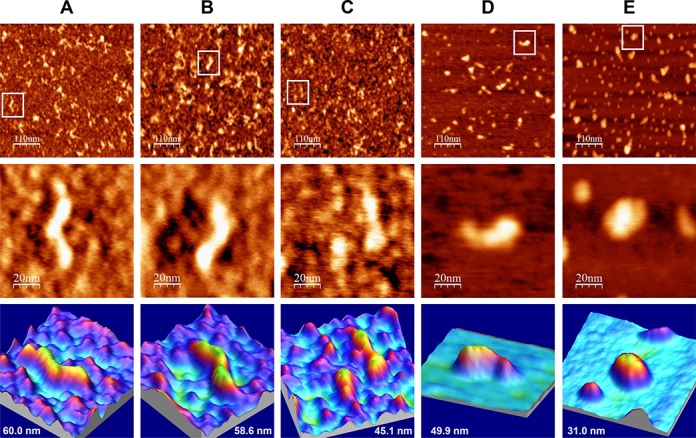
AFM images of IRES-574/miR-122 complexes in a folding buffer supplemented with increasing Mg2+ concentration. IRES-574 RNA molecules were folded in a buffer containing 100 mM Na+, either without Mg2+ (panel A) or supplemented with 2, 4, 6 and 10 mM Mg2+ (panels B, C, D and E, respectively) and incubated with miR-122 (see Materials and Methods). Large AFM images (550 × 550 nm, top row), zoom areas containing a selected molecule (100 × 100 nm images, medium row) and 3D representations of the selected molecule showing its measured length (bottom row) are shown. The medium and bottom row at panel C show an open and a closed IRES-574 conformer coexisting at 4 mM Mg2+, the depicted length corresponding to the open one. Two miR-122 molecules are imaged at panel E (10 mM Mg2+) together with a compact IRES-574 conformer. The nominal curvature radius of the AFM tips was 2 nm.
AFM images of IRES-574/miR-122 complexes in the folding buffer lacking Mg2+ ions showed a majority of elongated and angular shapes similar to those previously found in the absence of miR-122 (Figure 7A and Supplementary Figure S9A). Their median length (computed over 50 IRES-574 molecules) was 64 nm (Figure 3C), and most of the imaged molecules were in the length interval of 60–80 nm (Figure 3D). Elongated and zigzaging molecules were also imaged at 2 mM Mg2+ (Figure 7B and Supplementary Figure S9B), with a median length of 56 nm (Figure 3C) and the majority of them being 40–60 nm long (Figure 3D). In contrast to the observation in the absence of miR-122, the complex IRES-574/miR-122 at 4 mM Mg2+ did not show uniform, comma-shaped images, but a heterogeneous distribution of partially elongated and branched shapes together with a fraction of more compact ones (Figure 7C and Supplementary Figure S9C). The majority of their molecular lengths belonged to the interval of 40–60 nm, with a computed median value of 43 nm (Figure 3C and D). Thus, the decrease in the median length associated to the transition from 2 to 4 mM Mg2+ was 13 nm (23% of the length at 2 mM), while the previously observed shortening in the absence of miR-122 (Figure 3A) was 33 nm (54% of the length at 2 mM). However, both decreases showed statistically significant differences at the level of the average molecular length. The coexistence of partially elongated and compact shapes was also imaged at 6 mM Mg2+ (Figure 7D and Supplementary Figure S9D): most of the molecular lengths were still found in the interval of 40–60 nm, with a median value of 45 nm (Figure 3C and D). Among the molecules observed in the presence of 2, 4 and 6 mM Mg2+, examples with 3 to 4 arms, analogous to those found in the absence of miR-122 at 2 mM Mg2+, were imaged, as well as additional morphologies including triangles, F- and C-shaped molecules (Supplementary Figure S9B–D). Finally, the maximum Mg2+ concentration assayed (10 mM) promoted a clear and statistically supported compaction of the imaged molecules to a median length of 28 nm (Figure 3C and D), with a majority of 20–40 nm long structures (Figure 3D) showing triangular, elliptical and round shapes (Figure 7E and Supplementary Figure S9E).
The effect on IRES-574 conformation of increasing Mg2+ concentration from 0 to 10 mM in the presence of miR-122 was also analyzed by gel-shift (Figure 5C and D) as described above, and the results were compared to those previously obtained in the absence of miR-122 (Figure 5A and B). In the absence of miR-122 and Mg2+ (lane 2 of Figure 5C, equivalent to lane 2 of Figure 5A) a single band, corresponding to the slow migrating form (conformer C3), was observed. The addition of miR-122 (lane 4 of Figure 5C), displaced more than one half of the total signal to a slightly slower migrating band, which corresponded to the IRES-574/miR-122 complex (conformer termed ‘C3/miR’), leaving a residual band with an electrophoretic mobility equivalent to that of the unbound IRES-574 molecule (C3). The presence of 2 mM Mg2+ in the folding buffer (lane 5 of Figure 5C) virtually eliminated the fraction of unbound IRES-574, thus showing that the divalent ion promoted the annealing of miR-122 (present in a 5:1 excess) with the available IRES-574 molecules. The relative band intensity of the conformer C3/miR (quantified in three independent experiments and graphically represented in Figure 5D) showed a mean value of 52.1% of the total signal. Also, a fast migrating band (conformer C4) increased its mean ratio from 12.6% in the absence of Mg2+ to 39.7% at 2 mM Mg2+. Thus, the Mg2+-driven promotion of a ‘closed’ conformation of IRES-574 occurred in a smaller fraction of molecules with respect to the previous observation in the absence of miR-122 (see Figure 5B). The further addition of Mg2+ increased the mean ratio of C4 versus C3/miR conformers to 51.0%:42.1%, 53.4%:39.6% and 52.3%:39.5% at 4, 6 and 10 mM Mg2+, respectively (Figure 5D). Therefore, two main differences were observed with respect to the situation found in the absence of miR-122 (compare Figure 5B and D): (i) when miR-122 was present in the medium, the Mg2+ concentration at which the two major conformers (C3/miR and C4) reached an equal representation was 3 mM, a value higher than that for the equilibrium between C3 and C4 in the absence of miR-122 (2 mM Mg2+); (ii) miR-122 affected the equilibrium between ‘open’ and ‘closed’ conformations at 4–10 mM Mg2+ in such a way that the ‘closed’ conformer was still predominant over the ‘open’ one, though to a lesser extent than in the absence of the microRNA (roughly a 5:4 ratio versus a 7:2 ratio).
DISCUSSION
In this work, we used AFM for visualizing the Mg2+-induced conformational rearrangement of a 574-nt-long RNA molecule that includes the HCV IRES element together with its flanking domains I, V and VI. The quantitative analysis of the molecular lengths measured at each Mg2+ concentration revealed a two-regime behavior in the IRES-574 folding. A sharp conformational switch was monitored from a heterogeneous distribution of elongated shapes at 0–2 mM Mg2+ to a uniform population of compact structures at 4–6–10 mM Mg2+. It is reasonable to assume that the alternative conformations imaged at 2 mM Mg2+ reflect the relative movements of rigid parts (defined by stable secondary structures) instead of the presence of disordered regions, in agreement with previous analysis of the IRES domains II–IV at 2.5 mM Mg2+ (19). Thus, a fraction of the imaged shapes at 2 mM Mg2+ could allow the identification of some of the folded, individual IRES domains, while the morphologies imaged at 4 and 6 mM Mg2+ would eventually reveal long-range RNA interactions among the preformed domains. In our system, 2 mM Mg2+ could represent the divalent cation concentration at which some compact or ‘collapsed’ RNA intermediate conformations appear before the proper tertiary structure is acquired, by analogy to certain group I introns where the transition from collapsed RNA conformations to their functional tertiary structures occurs at 2–3 mM Mg2+ (66).
Gel-shift analysis confirmed the existence of a switch between two major, alternative conformations of the IRES-574 molecule. The stepwise increase of Mg2+ concentration from 0 to 4 mM completely shifted the ratio between such RNA conformers toward the most compact one, both of them being equally represented at 2 mM Mg2+. A previous study showed that the HCV IRES containing domains II–IV folded into a unique, extended structure with certain degree of flexibility at 2.5 mM Mg2+ (10). This fact suggests that the presence of two major conformers in the molecule IRES-574 at 2 mM Mg2+ is likely due to the conformational flexibility allowed by the flanking domains I, V and VI.
The AFM analysis of the shorter transcript IRES-418 (containing domains I–IV) showed that this molecule can adopt three-arms morphologies at 4 mM Mg2+, including λ-shaped conformations. These molecular shapes are consistent with those derived from a previous TEM analysis (17) of the HCV IRES (nts 1–380) at 5 mM Mg2+, and from a SAXS-based model (19) of a shorter version of the HCV IRES (nts 39–371) at 2.5 mM Mg2+. Both approaches showed molecules with three main arms protruding from a central axis, what has been interpreted as a structure based on a flexible pseudo-knot that constitutes a hinge between IRES domains II, III and IV (14). The comparison of our AFM images with such previous data allowed us to hypothesize that the IRES domains II–IV might be assigned as depicted in Figure 8A. Hence, the bended arm corresponding to the upper part of the λ-like imaged shapes (showing a characteristic angular morphology previously revealed by NMR (13), also identified in a SAXS-based model of the HCV IRES (19) and in the cryo-EM map of the HCV IRES bound to the 40S ribosomal subunit (18)) would correspond to domain II. In turn, the bulky and straight arm would contain the apical part of domain III (subdomains IIIabc (16)). The comparison of our data with the SAXS-based model (19) also suggests that the third, shorter arm imaged by AFM could be assigned to either subdomain IIId or domain IV plus the core-coding sequence fragment. The putative domain I cannot be identified by AFM in any of the molecules, however, it is assumed to be placed next to the basal region of domain II.
Figure 8.
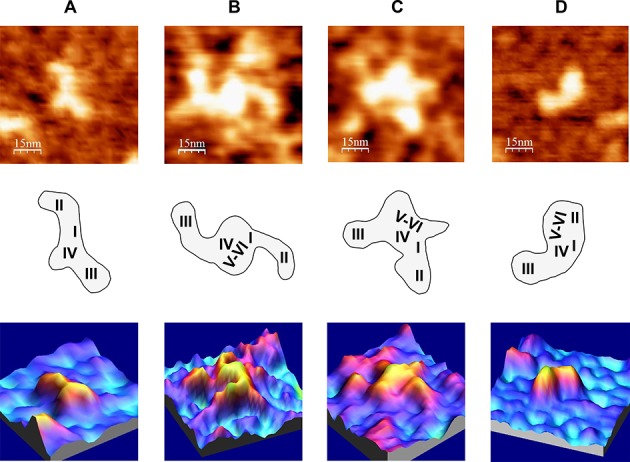
AFM images of HCV IRES-containing RNA molecules under different ionic conditions, in the absence of miR-122. (A) IRES-418 molecule at 4 mM Mg2+. (B) and (C) Two examples of IRES-574 molecules at 2 mM Mg2+. (D) IRES-574 molecule at 4 mM Mg2+. AFM images (75 × 75 nm) of the selected molecule (top row), schematic representations of the molecule sowing the suggested assignment of IRES domains (medium row) and 3D representations of the selected molecules (bottom row) are shown. The nominal curvature radius of the AFM tips was 2 nm.
Although it is evident that further experiments will be needed to unambiguously identify individual IRES domains in the molecules imaged by AFM, the three-arms conformation of the IRES-418 at 4 mM Mg2+ helped us interpret a fraction of the AFM images of the IRES-574 molecule at 2 mM Mg2+, in which three or four arms have been imaged (two representative examples are shown in Figure 8B and C). According to our model, domains II and III could be distinguished, while the downstream domains V-VI might either be placed in the central region of the molecule close to the pseudo-knot (Figure 8B) or (thanks to the flexibility provided by the ssRNA spacer between domains IV and V) partially protrude from such central region as one or two extra arms (Figure 8C). In turn, the compact two-arms, comma-shaped morphology showed by most of the IRES-574 molecules at 4 and 6 mM Mg2+ (Figure 8D) would suggest the presence of a long-range interaction of the downstream domains V-VI with either domains I-II or domain III. The comparison of the images in Figure 8 and Supplementary Figure S4 with previously published data (17,19) would favor the former possibility. Therefore, we hypothesize that, in the IRES-574 molecule, the downstream domains V (likely excluding its apical region, which is also present in the IRES-418 molecule, see Figure 1) and/or VI could interact with the arm composed of domains I-II provided that Mg2+ concentration is at least 2 mM, this interaction being favored at 4 mM Mg2+.
In order to test this hypothesis, a complementary analysis of the IRES-574 molecule was performed by RNase T1 probing. Our results showed that the main transition in the observed conformational rearrangements was produced between 0 and 4 mM Mg2+ concentration: the intensity of at least nine RNase T1 bands (corresponding to 12 reactive Gs) decreased, and such a protection was already evident at 2 mM Mg2+ in seven bands (10 Gs). The protected positions at 2 mM Mg2+ included six Gs directly involved in the formation of the IIIef/IV pseudo-knot (14). The requirement of at least 0.25 mM Mg2+ for the establishment of this key tertiary interaction has been shown in the HCV IRES containing domains II–IV (10). Therefore, it seems evident that the formation of the central pseudo-knot is not affected by the presence of the flanking domains I, V and VI. Among the remaining six protected Gs, two of them (G35 and G428) are placed at the I-II spacer region and at the basis of domain VI, respectively. Notably, they lie at both strands of a previously proposed long-range annealing motif (involving sequences at nts 24–38 and 428–442, Figure 1) that could promote a ‘closed’ conformation of HCV IRES in its natural sequence context (22–24). This fact suggested that such a long-range RNA interaction could be involved in the Mg2+-induced conformational switch reported in IRES-574.
The conformational rearrangement of the IRES-574 molecule produced between 0 and 4 mM Mg2+ (both conformers being in equilibrium at 2 mM Mg2+) could rely, first, on the formation of the IIIef/IV pseudo-knot, which affects the central region of the IRES element including the basis of domain IV. Concomitantly, this tertiary interaction would allow the long-range RNA interaction involving domains I-II and domains V-VI. In the model we are envisaging, such interactions could be responsible for the main conformers visualized by AFM and quantified by gel-shift analysis: (i) elongated molecules at 0 mM Mg2+, where only intra-domain, stable secondary structure elements are present; (ii) molecules with heterogeneous and angular shapes at 2 mM Mg2+, including topologies with three arms, likely corresponding to domains I-II, III and V-VI, which protrude from a central region defined by the IIIef/IV pseudo-knot; (iii) homogeneous, comma-shaped molecules at 4–6 mM Mg2+ showing two arms, the first of them being likely composed of the interacting domains (I-II) + (V-VI) and the second one containing domain III alone.
The angle formed between the two IRES-574 arms at 4 mM Mg2+ showed a maximum in the interval of 117º–127º. Previous studies indicated that the dominant motion in HCV IRES is the in-plane movement of domains II and III toward and away from each other (19). Thus, it is reasonable to assume that, while in the IRES molecule than contains domains II–IV imaged by EM (17) the maximum angle between domains II and III lies in the interval 100º–110º, the presence in IRES-574 of a bulky arm composed of the interacting domains (I-II) + (V-VI) displaces the maximum in the distribution of the angles formed with domain III toward higher values.
The AFM and gel-shift analyses performed in the presence of miR-122 supported the involvement of the miR-122-binding site at the I-II spacer region in the reported molecular switch. AFM images showed that, under Mg2+ concentrations of 0 and 10 mM Mg2+, the distribution of molecular lengths and shapes was similar in IRES-574 molecules and in IRES-574/miR-122 complexes. However, the sharp conformational switch imaged in IRES-574 molecules when Mg2+ concentration increased from 2 to 4 mM was not evident in the presence of miR-122, since the IRES-574/miR-122 complex folded in heterogeneous shapes of different lengths in the interval of 2–6 mM Mg2+ concentration. The results of gel-shift analyses were in agreement with the equilibrium between ‘open’ and ‘closed’ conformations of the IRES-574/miR-122 complex in buffers containing Mg2+ concentrations higher than 2 mM.
Previously, a transition from ‘closed’ to ‘open’ IRES-574 conformation was demonstrated in the presence of miR-122 (27). Here, we show that this transition can also be promoted by Mg2+, though in the opposite direction: while miR-122 favours the ‘open’ form, the increasing Mg2+ displaces the equilibrium toward the ‘closed’ conformer. Remarkably, both effectors have been described to affect HCV IRES-dependent initiation of translation in vitro: the presence of miR-122 stimulates the process (31), while the increase of Mg2+ concentration up to 5 mM allows IRES-dependent translation initiation in the absence of initiation factors (40). Therefore, in the frame of the many regulated functions in which the interactions between miR-122 and HCV IRES may participate (32–35), the data reported here point toward the existence of a bidirectional conformational switch stimulated (in opposite directions) by two effectors of very different nature.
Gel-shift analysis showed that ‘open’ and ‘closed’ conformations of IRES-574 are in equilibrium at 2 mM Mg2+ in the absence of miR-122, and at 3 mM Mg2+ when the micro-RNA is present in the medium. These values are higher than the closely regulated 0.5–1 mM concentration (0.59 mM in rat hepatocytes) of free Mg2+ in the eukaryotic cytoplasm (67–69), where positively charged RNA-binding proteins and other effectors also modulate the functional structure of HCV IRES. However, such a 2–3 mM Mg2+ concentrations correspond to the 2.5 mM value at which HCV IRES can promote the initiation of translation in the absence of the complete set of initiation factors (40) and define the upper limit of the Mg2+ concentration that allows IRES activity in translation-competent extracts (2.5 mM (39)). Therefore, the Mg2+-induced ‘open’/’closed’ switch reported here could be of relevance for IRES-dependent translation initiation in vitro under different experimental settings. Our results also support that the complementary sequences present at the I-II spacer and at the basal region of domain VI are prone to establish a long-range RNA interaction provided that the ionic or biochemical environment of the folded HCV 5′ UTR allows a compatible tertiary structure. The structural data reported here provide further insights on the conformational plasticity of HCV IRES in its genomic context (22–24) and, as a result, encourage the development of novel HCV inhibitors based on small molecules targeting either the IRES regions essential for translation initiation (2,3) or the liver-specific microRNAs that act as enhancers of viral replication (70).
In summary, here we report for the first time the use of AFM to analyze the 3D structure of an IRES element. We have shown that, in its natural sequence context, HCV IRES can undergo a Mg2+-induced molecular switch between two alternative conformations, such a structural rearrangement being hindered by the presence of miR-122 in the medium. Our data are compatible with those previously reported for HCV IRES domains II–IV based on chemical and enzymatic probing, EM and SAXS-derived models. Additionally, our results shed light on the 3D conformation of HCV IRES in its natural sequence context, thus reinforcing the previously suggested structural continuity between the domain I, the minimal HCV IRES element and (despite the apparent functional discontinuity between coding and non-coding sequences) the essential domains V and VI within the core coding region.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank Dr. Victor M. González and Dr. Francisco J. López de Saro for preliminary analysis of the RNA conformation of IRES-574 molecules, and María Fernández-Algar for excellent technical assistance.
Footnotes
Miguel Moreno and Ascensión Ariza-Mateos contributed equally to the paper as second authors.
FUNDING
Spanish Ministerio de Ciencia e Innovación and Ministerio de Economía y Competitividad [BIO2010–20696 and BIO2013–47228-R to C.B.; MAT2011–26534 to J.A.M.-G.; BFU2010–11612-E to J.G.; FIS2012–38866-C05–05 to L.V.]; Spanish National Research Council [CSIC-200920I040 to C.B.]; Comunidad de Madrid [NANOAVANSENS S2013/MIT-3029 to L.V.]; FEDER funds from the European Union. CIBERehd is funded by the Instituto de Salud Carlos III. JAE-Doc postdoctoral fellowship from the CSIC Program Junta de Ampliación de Estudios, co-funded by the European Science Foundation [to M.M.]. Funding for open access charge: Spanish National Research Council (CSIC).
Conflict of interest statement. None declared.
REFERENCES
- 1.Pawlotsky J.-M. Treatment of chronic hepatitis C: current and future. Curr. Topics Microbiol. Immunol. 2013;369:321–342. doi: 10.1007/978-3-642-27340-7_13. [DOI] [PubMed] [Google Scholar]
- 2.Davis D.R., Seth P.P. Therapeutic targeting of HCV internal ribosomal entry site RNA. Antiviral Chem. Chemother. 2011;21:117–128. doi: 10.3851/IMP1693. [DOI] [PubMed] [Google Scholar]
- 3.Romero-Lopez C., Berzal-Herranz A. Unmasking the information encoded as structural motifs of viral RNA genomes: a potential antiviral target. Rev. Med. Virol. 2013;23:340–354. doi: 10.1002/rmv.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda M., Ping L.H., Rijnbrand R.C.A., Amphlett E., Clarke B., Rowlands D., Lemon S.M. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 5.Fraser C.S., Doudna J.A. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Micro. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 6.Wang C., Sarnow P., Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honda M., Beard M.R., Ping L.H., Lemon S.M. A phylogenetically conserved stem-loop structure at the 5 ‘ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang P.S., Elazar M., Pham E.A., Glenn J.S. Simplified RNA secondary structure mapping by automation of SHAPE data analysis. Nucleic Acids Res. 2011;39:e151. doi: 10.1093/nar/gkr773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown E.A., Zhang H.C., Ping L.H., Lemon S.M. Secondary structure of the 5′ nontranslated regions of hepatitis-c virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieft J.S., Zhou K., Jubin R., Murray M.G., Lau J.Y.N., Doudna J.A. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 11.Martell M., Briones C., de Vicente A., Piron M., Esteban J.I., Esteban R., Guardia J., Gomez J. Structural analysis of hepatitis C RNA genome using DNA microarrays. Nucleic Acids Res. 2004;32:e90. doi: 10.1093/nar/gnh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero-Lopez C., Barroso-delJesus A., Garcia-Sacristan A., Briones C., Berzal-Herranz A. The folding of the hepatitis C virus internal ribosome entry site depends on the 3 ‘-end of the viral genome. Nucleic Acids Res. 2012;40:11697–11713. doi: 10.1093/nar/gks927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukavsky P.J., Kim I., Otto G.A., Puglisi J.D. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Mol. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 14.Berry K.E., Waghray S., Mortimer S.A., Bai Y., Doudna J.A. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19:1456–1466. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieft J.S., Zhou K.H., Grech A., Jubin R., Doudna J.A. Crystal structure of an RNA tertiary domain essential to HCVIRES-mediated translation initiation. Nat. Struct. Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 16.Collier A.J., Gallego J., Klinck R., Cole P.T., Harris S.J., Harrison G.P., Aboul-ela F., Varani G., Walker S. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat. Struct. Mol. Biol. 2002;9:375–380. doi: 10.1038/nsb785. [DOI] [PubMed] [Google Scholar]
- 17.Beales L.P., Rowlands D.J., Holzenburg A. The internal ribosome entry site (IRES) of hepatitis C virus visualized by electron microscopy. RNA. 2001;7:661–670. doi: 10.1017/s1355838201001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spahn C.M.T., Kieft J.S., Grassucci R.A., Penczek P.A., Zhou K.H., Doudna J.A., Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 19.Perard J., Leyrat C., Baudin F., Drouet E., Jamin M. Structure of the full-length HCV IRES in solution. Nat. Commun. 2013;4:1612. doi: 10.1038/ncomms2611. [DOI] [PubMed] [Google Scholar]
- 20.Tuplin A., Evans D.J., Simmonds P. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J. Gen. Virol. 2004;85:3037–3047. doi: 10.1099/vir.0.80141-0. [DOI] [PubMed] [Google Scholar]
- 21.McMullan L.K., Grakoui A., Evans M.J., Mihalik K., Puig M., Branch A.D., Feinstone S.M., Rice C.M. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2879–2884. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honda M., Rijnbrand R., Abell G., Kim D.S., Lemon S.M. Natural variation in translational activities of the 5 ‘ nontranslated RNAs of hepatitis C virus genotypes 1a and 1b: Evidence for a long-range RNA-RNA interaction outside of the internal ribosomal entry site. J. Virol. 1999;73:4941–4951. doi: 10.1128/jvi.73.6.4941-4951.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y.K., Lee S.H., Kim C.S., Seol S.K., Jang S.K. Long-range RNA-RNA interaction between the 5 ‘ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA-Publ. RNA Soc. 2003;9:599–606. doi: 10.1261/rna.2185603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beguiristain N., Robertson H.D., Gomez J. RNase III cleavage demonstrates a long range RNA: RNA duplex element flanking the hepatitis C virus internal ribosome entry site. Nucleic Acids Res. 2005;33:5250–5261. doi: 10.1093/nar/gki822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jopling C.L., Yi M.K., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 26.Jopling C.L., Schuetz S., Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diaz-Toledano R., Ariza-Mateos A., Birk A., Martinez-Garcia B., Gomez J. In vitro characterization of a miR-122-sensitive double-helical switch element in the 5 ‘ region of hepatitis C virus RNA. Nucleic Acids Res. 2009;37:5498–5510. doi: 10.1093/nar/gkp553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machlin E.S., Sarnow P., Sagan S.M. Masking the 5 ‘ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimakami T., Yamane D., Welsch C., Hensley L., Jangra R.K., Lemon S.M. Base pairing between hepatitis C virus RNA and microRNA 122 3 ‘ of its seed sequence is essential for genome stabilization and production of infectious virus. J. Virol. 2012;86:7372–7383. doi: 10.1128/JVI.00513-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Masaki T., Yamane D., McGivern D.R., Lemon S.M. Competing and noncompeting activities of miR-122 and the 5 ‘ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1881–1886. doi: 10.1073/pnas.1213515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henke J.I., Goergen D., Zheng J., Song Y., Schuettler C.G., Fehr C., Juenemann C., Niepmann M. MicroRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goergen D., Niepmann M. Stimulation of Hepatitis C Virus RNA translation by microRNA-122 occurs under different conditions in vivo and in vitro. Virus Res. 2012;167:343–352. doi: 10.1016/j.virusres.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Pineiro D., Martinez-Salas E. RNA structural elements of hepatitis C virus controlling viral RNA translation and the implications for viral pathogenesis. Viruses-Basel. 2012;4:2233–2250. doi: 10.3390/v4102233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson J.A., Huys A. miR-122 promotion of the hepatitis C virus life cycle: sound in the silence. RNA. 2013;4:665–676. doi: 10.1002/wrna.1186. [DOI] [PubMed] [Google Scholar]
- 35.Li Y., Masaki T., Lemon S.M. miR-122 and the Hepatitis C RNA genome: more than just stability. RNA Biol. 2013;10:919–923. doi: 10.4161/rna.25137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Draper D.E. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auffinger P., Grover N., Westhof E. Metal ion binding to RNA. Metal Ions Life Sci. 2011;9:1–35. [PubMed] [Google Scholar]
- 38.Bowman J.C., Lenz T.K., Hud N.V., Williams L.D. Cations in charge: magnesium ions in RNA folding and catalysis. Curr. Opin. Struct. Biol. 2012;22:262–272. doi: 10.1016/j.sbi.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Shenvi C.L., Dong K.C., Friedman E.M., Hanson J.A., Cate J.H.D. Accessibility of 18S rRNA in human 40S subunits and 80S ribosomes at physiological magnesium ion concentrations - implications for the study of ribosome dynamics. RNA. 2005;11:1898–1908. doi: 10.1261/rna.2192805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lancaster A.M., Jan E., Sarnow P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA. 2006;12:894–902. doi: 10.1261/rna.2342306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bustamante C., Keller D. Scanning force microscopy in biology. Phys. Today. 1995;48:32–38. [Google Scholar]
- 42.Hansma H.G., Kasuya K., Oroudjev E. Atomic force microscopy imaging and pulling of nucleic acids. Curr. Opin. Struct. Biol. 2004;14:380–385. doi: 10.1016/j.sbi.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Hansma H.G., Revenko I., Kim K., Laney D.E. Atomic force microscopy of long and short double-stranded, single-stranded and triple-stranded nucleic acids. Nucleic Acids Res. 1996;24:713–720. doi: 10.1093/nar/24.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Briones C., Mateo-Marti E., Gomez-Navarro C., Parro V., Roman E., Martin-Gago J.A. Ordered self-assembled monolayers of peptide nucleic acids with DNA recognition capability. Phys. Rev. Lett. 2004;93:208103. doi: 10.1103/PhysRevLett.93.208103. [DOI] [PubMed] [Google Scholar]
- 45.Neaves K.J., Huppert J.L., Henderson R.M., Edwardson J.M. Direct visualization of G-quadruplexes in DNA using atomic force microscopy. Nucleic Acids Res. 2009;37:6269–6275. doi: 10.1093/nar/gkp679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wells S.E., Hillner P.E., Vale R.D., Sachs A.B. Circularization of mRNA by Eukaryotic Translation Initiation Factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 47.Hansma H.G., Golan R., Hsieh W., Daubendiek S.L., Kool E.T. Polymerase activities and RNA structures in the atomic force microscope. J. Struct. Biol. 1999;127:240–247. doi: 10.1006/jsbi.1999.4170. [DOI] [PubMed] [Google Scholar]
- 48.Hansma H.G., Oroudjev E., Baudrey S., Jaeger L. TectoRNA and ‘kissing-loop’ RNA: atomic force microscopy of self-assembling RNA structures. J. Micros. 2003;212:273–279. doi: 10.1111/j.1365-2818.2003.01276.x. [DOI] [PubMed] [Google Scholar]
- 49.Basame S., Wai-lun Li P., Howard G., Branciforte D., Keller D., Martin S.L. Spatial assembly and RNA binding stoichiometry of a LINE-1 protein essential for retrotransposition. J. Mol. Biol. 2006;357:351–357. doi: 10.1016/j.jmb.2005.12.063. [DOI] [PubMed] [Google Scholar]
- 50.Giro A., Bergia A., Zuccheri G., Bink H.H., Pleij C.W., Samorì B. Single molecule studies of RNA secondary structure: AFM of TYMV viral RNA. Micros. Res. Tech. 2004;65:235–245. doi: 10.1002/jemt.20123. [DOI] [PubMed] [Google Scholar]
- 51.Alvarez D.E., Lodeiro M.F., Luduena S.J., Pietrasanta L.I., Gamarnik A.V. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuznetsov Y.G., Daijogo S., Zhou J., Semler B.L., McPherson A. Atomic force microscopy analysis of icosahedral virus RNA. J. Mol. Biol. 2005;347:41–52. doi: 10.1016/j.jmb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 53.Kuznetsov Y.G., Dowell J.J., Gavira J.A., Ng J.D., McPherson A. Biophysical and atomic force microscopy characterization of the RNA from satellite tobacco mosaic virus. Nucleic Acids Res. 2010;38:8284–8294. doi: 10.1093/nar/gkq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sagan S.M., Nasheri N., Luebbert C., Pezacki J.P. The efficacy of siRNAs against hepatitis C virus is strongly influenced by structure and target site accessibility. Chem. Biol. 2010;17:515–527. doi: 10.1016/j.chembiol.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 55.Gilmore J.L., Aizaki H., Yoshida A., Deguchi K., Kumeta M., Junghof J., Wakita T., Takeyasu K. Nanoimaging of ssRNA: genome architecture of the hepatitis C virus revealed by atomic force microscopy. J. Nanomed. Nanotechol. 2014:010. S5. [Google Scholar]
- 56.Beard M.R., Abell G., Honda M., Carroll A., Gartland M., Clarke B., Suzuki K., Lanford R., Sangar D.V., Lemon S.M. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology. 1999;30:316–324. doi: 10.1002/hep.510300137. [DOI] [PubMed] [Google Scholar]
- 57.Icho T., Wickner R.B. The double-stranded-RNA genome of yeast virus L-A encodes its own putative RNA-polymerase by fusing 2 open reading frames. J. Biol. Chem. 1989;264:6716–6723. [PubMed] [Google Scholar]
- 58.Lyubchenko Y., Shlyakhtenko L., Harrington R., Oden P., Lindsay S. Atomic force microscopy of long DNA - Imaging in air and under water. Proc. Natl. Acad. Sci. U.S.A. 1993;90:2137–2140. doi: 10.1073/pnas.90.6.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng H., Zhang K., Libera J.A., de la Cruz M.O., Bedzyk M.J. Polynucleotide adsorption to negatively charged surfaces in divalent salt solutions. Biophys. J. 2006;90:1164–1174. doi: 10.1529/biophysj.105.070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horcas I., Fernandez R., Gomez-Rodriguez J.M., Colchero J., Gomez-Herrero J., Baro A.M. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instr. 2007;78:013705. doi: 10.1063/1.2432410. [DOI] [PubMed] [Google Scholar]
- 61.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ariza-Mateos A., Prieto-Vega S., Diaz-Toledano R., Birk A., Szeto H., Mena I., Berzal-Herranz A., Gomez J. RNA self-cleavage activated by ultraviolet light-induced oxidation. Nucleic Acids Res. 2012;40:1748–1766. doi: 10.1093/nar/gkr822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duhamel J., Liu D.M., Evilia C., Fleysh N., Dinter-Gottlieb G., Lu P. Secondary structure content of the HDV ribozyme in 95% formamide. Nucleic Acids Res. 1996;24:3911–3917. doi: 10.1093/nar/24.20.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lozano G., Fernandez N., Martinez-Salas E. Magnesium-dependent folding of a picornavirus IRES element modulates RNA conformation and eIF4G interaction. FEBS J. 2014;281:3685–3700. doi: 10.1111/febs.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen S.J. Annual Review of Biophysics. Vol. 37. Palo Alto: Annual Reviews; 2008. pp. 197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woodson S.A. Structure and assembly of group I introns. Curr. Opin. Struct. Biol. 2005;15:324–330. doi: 10.1016/j.sbi.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 67.Raju B., Murphy E., Levy L.A., Hall R.D., London R.E. A fluorescent indicator for measuring cytosolic free magnesium. Am. J. Physiol. 1989;256:C540-C548. doi: 10.1152/ajpcell.1989.256.3.C540. [DOI] [PubMed] [Google Scholar]
- 68.Gaussin V., Gailly P., Gillis J.M., Hue L. Fructose-induced increase in intracellular free Mg2+ ion concentration in rat hepatocytes: Relation with the enzymes of glycogen metabolism. Biochem. J. 1997;326:823–827. doi: 10.1042/bj3260823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Günther T. Concentration, compartmentation and metabolic function of intracellular free Mg2+ Magnes. Res. 2007;19:225–236. [PubMed] [Google Scholar]
- 70.Lindow M., Kauppinen S. Discovering the first microRNA-targeted drug. J. Cell Biol. 2012;199:407–412. doi: 10.1083/jcb.201208082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


