Abstract
As a champion of small RNA research for two decades, Caenorhabditis elegans has revealed the essential Argonaute CSR-1 to play key nuclear roles in modulating chromatin, chromosome segregation and germline gene expression via 22G-small RNAs. Despite CSR-1 being preserved among diverse nematodes, the conservation and divergence in function of the targets of small RNA pathways remains poorly resolved. Here we apply comparative functional genomic analysis between C. elegans and Caenorhabditis briggsae to characterize the CSR-1 pathway, its targets and their evolution. C. briggsae CSR-1-associated small RNAs that we identified by immunoprecipitation-small RNA sequencing overlap with 22G-RNAs depleted in cbr-csr-1 RNAi-treated worms. By comparing 22G-RNAs and target genes between species, we defined a set of CSR-1 target genes with conserved germline expression, enrichment in operons and more slowly evolving coding sequences than other genes, along with a small group of evolutionarily labile targets. We demonstrate that the association of CSR-1 with chromatin is preserved, and show that depletion of cbr-csr-1 leads to chromosome segregation defects and embryonic lethality. This first comparative characterization of a small RNA pathway in Caenorhabditis establishes a conserved nuclear role for CSR-1 and highlights its key role in germline gene regulation across multiple animal species.
INTRODUCTION
RNA interference (RNAi) and related small interfering RNA (siRNA) pathways entail a potent means of regulating gene expression. Such pathways are characterized by Argonaute proteins (AGOs) and small RNAs, in which the small RNAs are complementary to target transcripts and guide the AGO to the target to elicit regulatory effects ranging from transcriptional silencing to transcript degradation or translational inhibition (1). Caenorhabditis elegans has been a champion of small RNA research since the initial discoveries of RNAi and microRNA pathways (2–4). From this early foundation, small RNA biology in the worm has developed to encompass at least four distinct types of small RNAs, including microRNA, piRNA/21U-RNA and two endogenous siRNA pathways (22G-RNA and 26G-RNA) that interact with 27 Argonaute proteins (5). These diverse pathways play key gene regulatory roles in development, differentiation and genome surveillance, and many of these functions are conserved in humans.
Despite the depth of accumulated knowledge of the functions and targets of small RNA pathways in C. elegans, understanding of their conservation and functional divergence between species remains more limited. The morphological and life history similarities between C. elegans and Caenorhabditis briggsae provide a powerful comparative system to examine the function and evolution of small RNA pathways and their gene targets. C. briggsae shares many model organism virtues with C. elegans, including a genome assembly of exceptional quality, molecular genetic tools and growing inclusion by researchers for comparative genomic and developmental studies (6–9). These two species diverged tens of millions of years (MY) ago, with sequence divergence comparable to the distance between mice and humans, despite extensive conservation of protein coding gene content between them (6,10–11). C. briggsae retains the four major classes of small RNAs found in C. elegans, but the functions of these pathways have yet to be explored (12,13). C. briggsae and C. elegans also possess an overlapping complement of Argonaute proteins (Supplementary Figure S1), including homologs of key C. elegans Argonautes such as CSR-1, WAGO-1, ALG-1 and -2, and PRG-1, thus providing a useful context to compare functional overlap and specificity of small RNA pathways over an intermediate evolutionary timescale (12,14–15).
To begin to evaluate the function and evolution of small RNA pathways, here we focus on the Argonaute CSR-1 (Chromosome Segregation and RNAi Deficient) in C. briggsae, facilitated by the cross-utility of antibodies for C. elegans CSR-1 (16,17). In both the male and hermaphrodite germlines of C. elegans, CSR-1 interacts with endogenous small RNAs called 22G-RNAs that target, via base-complementary, the majority of germline-expressed protein coding genes (in C. elegans, ∼5600 genes in males and ∼4200 genes in hermaphrodites) (16,18–19). Notably, the 22G-RNAs associated with CSR-1 entail a subset of the total 22G-RNA class. The other major subset of 22G-RNAs associates with WAGO-1 (Worm Argonaute 1) and, in contrast to CSR-1, this pathway targets repetitive elements, pseudogenes and intergenic regions, along with some protein coding genes (18). The biogenesis of both types of 22G-RNAs requires the activity of several additional factors, including DRH-3 (Dicer-Related Helicase 3, a DEAD box helicase), EKL-1 (Enhancer of KSR-1 Lethality 1, a dual-tudor domain protein) and an RNA-dependent RNA polymerase (RdRP) (16,18). In the CSR-1 pathway, the RdRP EGO-1 (Enhancer of GLP-1) is utilized, while in the WAGO-1 pathway, both EGO-1 and a paralog, RRF-1 (RNA-Dependent RNA Polymerase Family-1), act in a partially redundant manner (16,18). The poly-uridyl transferase, CDE-1 (Co-suppression DEfective-1), also functions in the CSR-1 pathway and may influence the stability of CSR-1 22G-RNAs by uridylating them at the 3′ end (20).
CSR-1 and the other known components of this pathway are essential in C. elegans, and loss of csr-1 leads to mitotic chromosome segregation defects in the embryo, defects in spermatogenesis and oogenesis due in part to aberrant meiotic chromatin organization and alterations in histone mRNA biogenesis (16,20–26). CSR-1 mediates chromosome-related functions in the nucleus, as it has been shown to associate with the genomic loci of its gene targets by chromatin immunoprecipitation in adult males, hermaphrodites and embryos (16,19). Why and how CSR-1 targets these particular germline genes remains an open question. Nonetheless, recent studies indicate that CSR-1 plays a rare positive role to promote the expression of both somatic and germline genes (19,21,27–29). In the germline, this positive activity acts in opposition to the other small RNA pathways, including the piRNA pathway, that perform germline genome surveillance and silence invading nucleic acids (19,27–28).
Here we use comparative functional genomics to characterize the Argonaute CSR-1 in C. briggsae to understand the evolution of this small RNA pathway and its targets. We find that C. briggsae CSR-1 (CBR-CSR-1) has a conserved, nuclear role in regulating germline-expressed protein coding genes in Caenorhabditis. CSR-1 target genes are largely preserved between species and are enriched in operons relative to other germline-expressed genes that are not CSR-1 targets. Despite the general signature of evolutionary constraint on CSR-1 targets, we identified a class of target genes that switched Argonaute pathways between species and that also evolve rapidly at the sequence level. Argonaute pathway switching points to a novel means by which small RNA systems could alter regulatory network architecture.
MATERIALS AND METHODS
Worm strains and husbandry
C. briggsae strains AF16 (reference strain) and JU1018 (mfIs42 [Cel-sid-2 + Cel-myo-2::DsRed]) (30), Caenorhabditis remanei SB146, Caenorhabditis brenneri PB2801, C. elegans Bristol N2 were obtained from the Caenorhabditis Genetics Center (CGC). Worms were cultured according to widely used protocols (31). Nomenclature is as described by: http://www.wormbase.org/about/userguide/nomenclature. Orthologous genes or proteins are designated by the prefix cbr/CBR for C. briggsae and cel/CEL for C. elegans.
Protein preparation, immunoprecipitation and western blotting
Protein lysate preparation and immunoprecipitations were performed as described in (16), and greater detail is provided in the Supplementary material.
RNAi assays
RNAi experiments were conducted as previously described using strain JU1018 (4,32). The largest exon of each gene was amplified with T7 promoter sequences at the ends of each primer, then each polymerase chain reaction (PCR) product was inserted into pCR2.1 using a TOPO TA® cloning kit (Life Technologies). Primer sequences can be found in the Supplementary material. RNAi plasmids were transformed into ht115 bacteria.
Quantitative Reverse Transcription PCR (qRT-PCR)
RNA and cDNA were prepared from adult C. briggsae hermaphrodites as previously described (16). qRT-PCR was performed as described previously (16) with minor adjustments. qRT-PCR was performed on the StepOnePlus™ Real-time PCR System (Life Technologies), using default thermocycling conditions. Reactions were composed of 7.5-μl Fast SYBR® Green Master Mix (Life Technologies), 0.6 μl of 10-μM stock for each primer, 2-μl diluted cDNA and 4.3-μl dH20. Each reaction was performed in technical triplicates. Primer sequences are provided in the Supplementary material.
Brood size and viability assays
Synchronous populations of young adult empty vector (L4440) and csr-1 RNAi-treated hermaphrodites (JU1018) were single-picked onto Nematode Growth Medium (NGM) plates. Each worm was transferred to a fresh plate every day and the number of embryos laid (total brood size) and that subsequently hatched (viable brood size) were counted. For each condition, at least 10 hermaphrodites were assayed.
Immunostaining and Immunofluorescence Microscopy
Samples were prepared and images for Figure 3 were collected as described in (16). Images in Figure 4 were collected using a Nikon TiE inverted microscope and Nikon C2 confocal system, with a 60X oil immersion lens. Antibodies used were: rabbit anti-CSR-1 (16), rabbit anti-HCP-3 (Novus Biologicals), mouse anti-Tubulin (Sigma, clone DM1A) or mouse anti-nuclear pore proteins (Abcam, MAb414). Secondary antibodies were used as described in (16).
Figure 3.
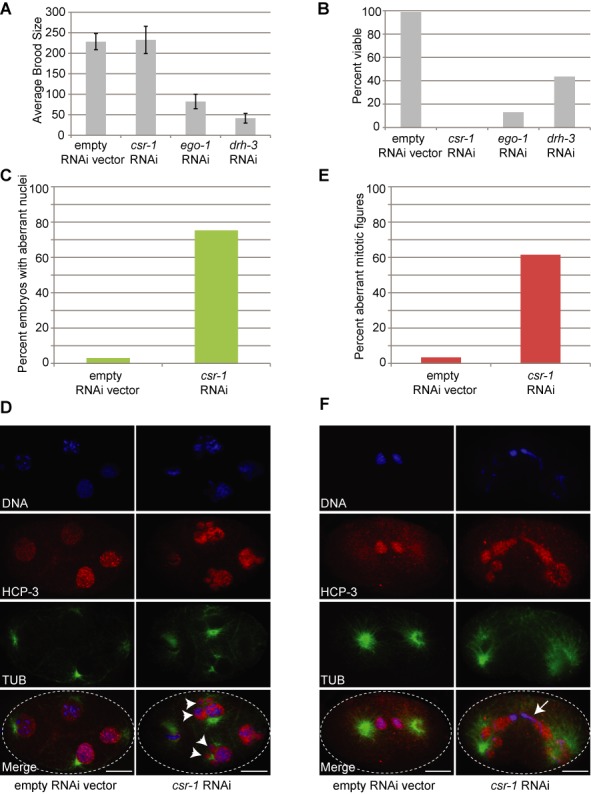
Loss of csr-1 in C. briggsae leads to defects in embryonic chromosome segregation. (A) RNAi knockdown of cbr-csr-1, cbr-drh-3 and cbr-ego-1 in strain JU1018 leads to defects in brood size. The average brood size (total number of eggs laid) is decreased when cbr-ego-1 and cbr-drh-3 are knocked down, while the total brood size remains the same as worms fed dsRNA against an empty vector when csr-1 RNAi is administered. Error bars represent standard deviation, and at least 10 adult hermaphrodites were used for each assay. (B) RNAi knockdown of cbr-csr-1, cbr-drh-3 and cbr-ego-1 in strain JU1018 leads to defects in embryo viability. None of the embryos laid by cbr-csr-1 RNAi worms are viable, while only a small proportion of the embryos laid by cbr-ego-1 and cbr-drh-3 RNAi worms are viable. Viable progeny are calculated as the number of embryos that hatch into L1 larvae relative to the total number of embryos laid. (C) RNAi of cbr-csr-1 leads to defects in chromosome segregation. JU1018 C. briggsae were fed dsRNA against an empty vector or cbr-csr-1 from the L1 stage. Those fed empty vector dsRNA gave rise to embryos with a very small proportion of aberrant nuclei (<3%) (aberrant nuclei are multi-lobed, abnormally shaped or sized, see (D) right column), while those worms fed dsRNA against cbr-csr-1 show >75% of embryos with at least one aberrant nucleus. (D) Representative images of embryos used in the quantification in (C) are shown. Embryos were stained with DAPI (blue), anti-CEL-Histone HCP-3 (red) and anti-tubulin (green). Arrowheads indicate features of aberrant nuclei (mis-shapen and multi-lobed). (E) RNAi to cbr-csr-1 in strain JU1018 also leads to a large proportion of lagging chromosomes in anaphase/telophase and aberrant mitotic figures (shown in (F)). (F) Representative images of embryos used in quantification for (E) are shown. Embryos were stained with DAPI (blue), anti-CEL-Histone HCP-3 (red) and anti-alpha-tubulin (green). The arrow indicates abnormally trailing DNA after a previous mitotic division.
Figure 4.
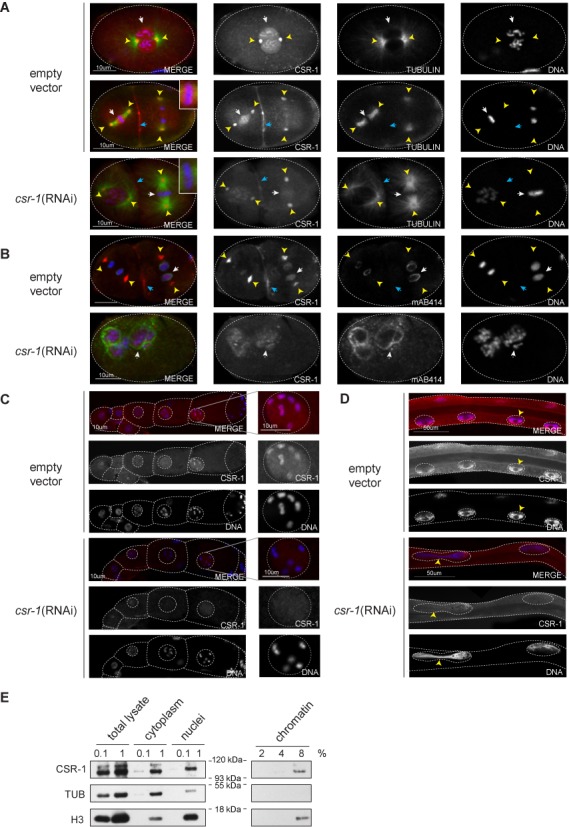
CBR-CSR-1 localizes to chromosomes and centrosomes in multiple stages of C. briggsae development. (A) CBR-CSR-1 localizes to mitotic chromatin (white arrows, prophase and metaphase stages are shown), centrosomes (yellow arrowheads) and the cleavage furrow (blue arrowheads) in C. briggsae embryos. Strain JU1018 was fed dsRNA against empty vector or cbr-csr-1 in (A), (B), (C) and (D). Embryos were stained with DAPI (blue), anti-CEL-CSR-1 (red) and anti-alpha tubulin (green). (B) CBR-CSR-1 shows reduced association with chromatin in anaphase and telophase (white arrow), but remains associated with centrosomes (yellow arrowheads) and the cleavage furrow (blue arrowheads). Embryos were stained with DAPI (blue), anti-CEL-CSR-1 (red) and MAb414, which recognizes nuclear pore complex proteins (green). (C) CBR-CSR-1 localizes to chromatin in C. briggsae oocytes. The germlines of mature adult wild-type and cbr-csr-1 RNAi worms were dissected and stained with DAPI (blue) and anti-CEL-CSR-1 (red). CSR-1 localization to chromatin is reduced in cbr-csr-1 RNAi worms. Single oocyte nuclei are shown as indicated. (D) CBR-CSR-1 localizes to chromatin in C. briggsae gut nuclei. Mature adult wild-type and cbr-csr-1 RNAi worms were stained with DAPI (blue) and anti-CEL-CSR-1 (red). CBR-CSR-1 localization to chromatin is reduced in cbr-csr-1 RNAi worms. Note that cbr-csr-1 RNAi worms display incomplete division of nuclei, consistent with defects in chromosome segregation (arrow heads). (E) Chromatin fractionation assay conducted in C. briggsae embryos confirms the association of CBR-CSR-1 with chromatin (strain AF16). Anti-alpha-tubulin was used as a cytoplasmic control and anti-histone H3 was used as a chromatin control.
Subcellular fractionation and chromatin isolation
Fractionation was performed as described in (16,33), with modification. Further details are described in the Supplementary material.
Small RNA cloning and Illumina sequencing
The C. briggsae CSR-1 immunoprecipitate (IP) and Input (cloned from gravid adult hermaphrodite AF16 worms), csr-1 RNAi and RNAi empty vector (cloned from JU1018 gravid adult hermaphrodite worms) libraries were created by the Tobacco Acid Pyrophosphatase (TAP) Cloning Method, and cloning was performed as described (16,18). Previous data sets re-analyzed: CSR-1 IP versus input in C. elegans (16); csr-1(tm892) mutant and control DA1316 (avr-14 (ad1302) I; avr-15(ad1051) glc-1)pk54)V) strain of C. elegans (16); glp-4(bn2), drh-3(ne4253) and WAGO-1 IP (From N2 gravid adults) as described in (18).
Small RNA and RNA-seq data analysis
Small RNA analysis was performed using custom Shell and Perl (5.10.0) scripts, and is described in detail in the Supplementary material. The SRA accession number for all RNA sequencing samples is SRP021463, including the CBR-CSR-1 IP (SRR833245), C. briggsae Input (SRR1043066), cbr-csr-1 RNAi (SRR1044359), empty RNAi vector (SRR1044399).
Construction of Argonaute phylogenetic tree
Multiple sequence alignments were conducted using T-coffee (34) and bootstrap values were generated using ClustalX (Neighbor-Joining) with 111 random seeds and 1000 replicates. The trees were then visualized using Fig Tree.
RESULTS
The C. elegans Argonaute CSR-1 is conserved in C. briggsae
The ability of CSR-1 to target most germline-expressed protein coding genes in C. elegans led us to test for its conserved functions in other Caenorhabditis species (16,19). We identified CSR-1 homologs in 10 related Clade III and Clade V nematode species (Figure 1A). Using a previously characterized rabbit polyclonal antibody that we generated against a conserved peptide of CEL-CSR-1 (Figure 1B), we tested for cross-reactivity in four Caenorhabditis species (16). We found that the antibody recognized a protein of the predicted size of CSR-1 in all species examined (∼120 kDa for the predominant long isoform, and ∼100 kDa for the short isoform), but immunoprecipitated C. briggsae CSR-1 most robustly (Figure 1C). We validated the specificity of the antibody in C. briggsae by performing RNAi against cbr-csr-1 and observed a significant depletion of the protein recognized by the antibody relative to the empty vector RNAi control (Figure 1D). With this antibody and ability to knockdown cbr-csr-1 in hand, we next explored the functions of the CSR-1 pathway in C. briggsae.
Figure 1.
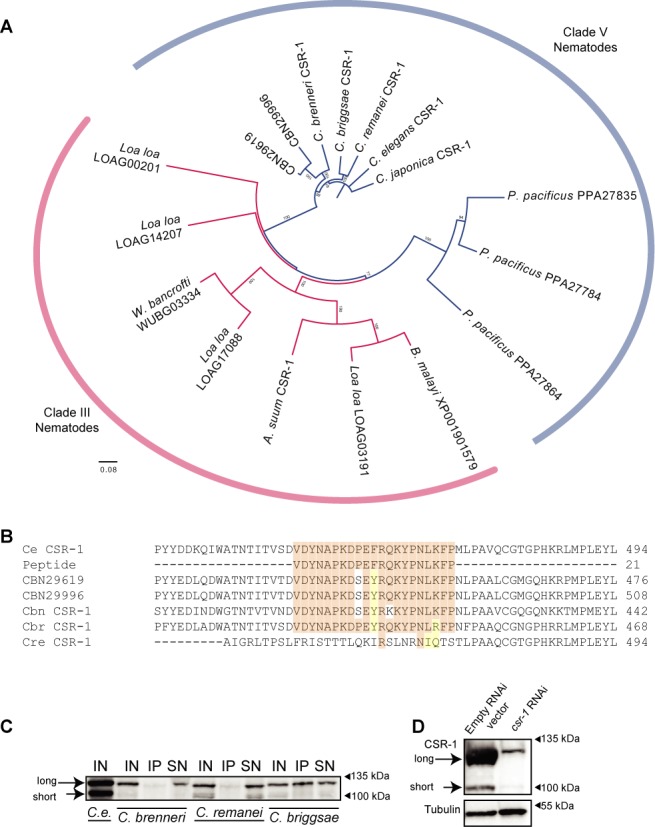
An antibody for CEL-CSR-1 recognizes orthologs in various nematode species. (A) A phylogenetic tree indicating the relationship between CSR-1 proteins in various Clade III and Clade V nematode species. Numbers on the branches report the level of confidence by bootstrap analysis (1000 bootstrap replicates; bootstrap values less than 70% were excluded from the figure). (B) An alignment of the CSR-1 protein sequence and peptide used to generate the C. elegans CSR-1 rabbit polyclonal antibody described in (16). Orange indicates conservation of the residue present in C. elegans; yellow indicates a conservative change in the residue; white indicates that the residue is not conserved. (C) Immunoprecipitation and western blotting of the CSR-1 protein in adult hermaphrodites of various Caenorhabditis species (C. briggsae strain AF16, C. remanei strain SB146, C. brenneri strain PB2801, C. elegans strain Bristol N2). IN = input protein (100 μg), IP = Immunoprecipitated material, SN = 10% of supernatant from IP. IP and WB were performed with the CEL-CSR-1 antibody described in (16). Size markers are as shown (right), and long and short refer to the two isoforms of CSR-1 (16). (D) RNAi of cbr-csr-1 demonstrates the specificity of the anti-CEL-CSR-1. RNAi was performed by feeding bacteria expressing double stranded RNA (dsRNA) of an ‘empty’ control plasmid vector (called L4440, left) or against cbr-csr-1 (right) to the RNAi-competent C. briggsae strain JU1018. (100 μg of protein purified from gravid adult hermaphrodites was loaded per lane, blots were probed with anti-alpha-tubulin, bottom; and anti-CEL-CSR-1, top). Size markers as shown (right), and long and short refer to the two isoforms of CSR-1 (16). (For cbr-csr-1 mRNA levels in worms treated with cbr-csr-1 RNAi, see Supplementary Figure S11.)
The C. briggsae CSR-1 pathway is essential for viability and mitotic chromosome segregation
We next characterized gene expression across C. briggsae development for the known components of the C. elegans CSR-1 pathway, including drh-3, ego-1, ekl-1 and cde-1 (16,18,20,26,35). In C. elegans, all CSR-1 pathway members display similar mutant phenotypes (sterility, embryonic lethality and changes in chromatin), developmental expression profiles and localization patterns, with highest expression in the germline and embryos (16,18,20,22,36). We found that in C. briggsae, all components were expressed at low to moderate levels throughout development, but were enriched in embryos and in stages possessing a mature germline (Figure 2A and B). This developmental expression profile is consistent with the expression pattern of the CSR-1 pathway expression in C. elegans, implicating this pathway as also being important for C. briggsae germline and embryonic development.
Figure 2.
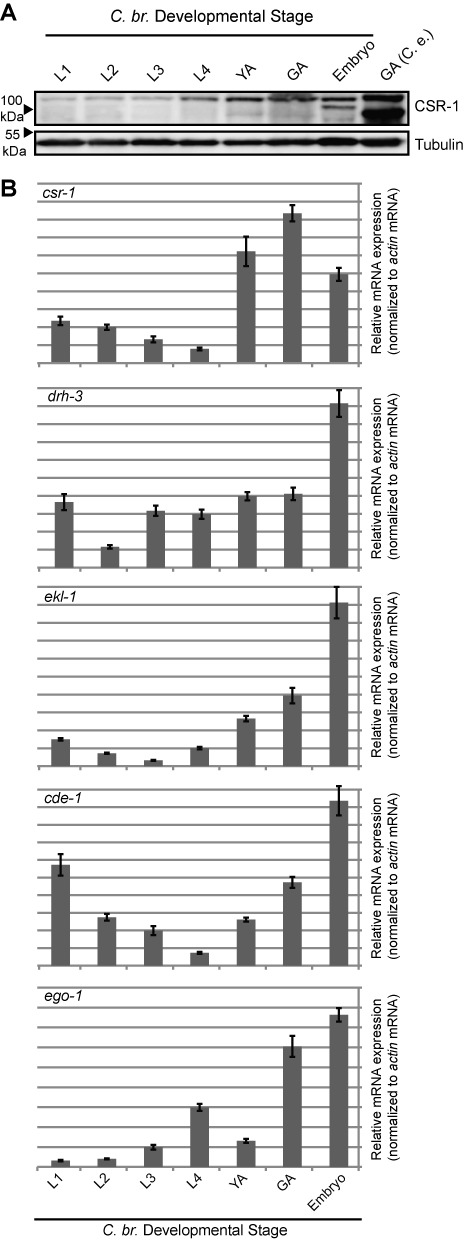
CBR-CSR-1 pathway members are highly expressed during reproductive stages and are required for embryonic viability. (A) Developmentally staged wild-type AF16 C. briggsae samples were subjected to western blotting for CBR-CSR-1 protein. Tubulin is shown as a loading control. L1, L2, L3 and L4 are larval stages, YA = young adult hermaphrodites lacking embryos, GA = mature adult hermaphrodites possessing embryos, Embryo = mixed stage embryo, GA (C.e.) = C. elegans mature adult hermaphrodite protein as a positive control. (B) Quantitative real-time PCR analysis of the mRNA expression of CSR-1 pathway components throughout wild-type development (strain AF16). Levels are expressed as arbitrary values, relative to actin mRNA, developmental stages are as in (A), error bars represent standard deviation of triplicate experiments.
We next assessed the role(s) of the CSR-1 pathway in C. briggsae development by testing the effect of RNAi knockdown of cbr-csr-1, cbr-drh-3 and cbr-ego-1 on brood size and viability of progeny (Figure 3A and B). We observed that cbr-csr-1 knockdown resulted in no reduction in the number of embryos laid (brood size; Figure 3A), but led to 100% inviable embryos (Figure 3B). In comparison, knockdown of cbr-drh-3 or cbr-ego-1 led to a dramatic reduction in the total number of embryos laid (Figure 3A), with only 15 and 43% of the ego-1 and drh-3 RNAi embryos surviving, respectively (Figure 3B). These phenotypes indicate that the CSR-1 pathway plays key developmental roles in the germline and embryo to make it essential for proper development in C. briggsae.
To explore why embryos died upon loss of the CSR-1 pathway, we examined chromosome segregation in embryos produced after cbr-csr-1 RNAi (Figure 3C–F). Embryos arrested at variable stages in early embryogenesis, with the vast majority of embryos possessing <100 cells (Figures 3D and F, and 4A and B). Approximately 75% of the embryos had at least one nucleus with an aberrant nuclear appearance, including abnormally shaped and positioned nuclei and aberrant masses of DNA distinct from nuclei (Figure 3C and D). Likewise, >60% of embryos undergoing mitosis exhibited aberrant mitotic figures, including lagging anaphase chromosomes and fragmented chromosomes (Figure 3E and F). These results indicate that the embryonic lethality of C. briggsae embryos upon knockdown of cbr-csr-1 is likely due to chromosome segregation abnormalities resulting in aneuploidy.
C. briggsae CSR-1 associates with chromatin
Could CBR-CSR-1 play a role in regulating chromatin? We examined the subcellular localization of CBR-CSR-1 in embryos and found that it localizes to the nucleus, mitotic chromosomes and centrosomes from the first mitotic cell division onward (Figure 4A and B). Interestingly, we also observed localization of CBR-CSR-1 to the cleavage furrow during embryonic cell divisions, a pattern that has not been described for C. elegans (Figure 4A and B). However, we did not detect CBR-CSR-1 granules in the cytoplasm of embryonic germ cells. Such granules would be indicative of localization to P granules as we have observed in C. elegans. We note that antibodies for C. elegans P granule markers, including PGL-1, did not recognize C. briggsae proteins, either (data not shown) (16,37). The localization of P granule proteins in C. briggsae has not been thoroughly explored, although homologs of many C. elegans P granule components, including DEPS-1, PGL-1, GLH-1 and GLH-4, are present in the C. briggsae genome (38). The knockdown of CBR-CSR-1 was significant (Supplementary Figure S2), despite CBR-CSR-1 not being completely ablated by RNAi (Figures 1A–D and 4D), thus indicating that the antibody specifically recognizes CSR-1 in C. briggsae and that these staining patterns are reflective of CSR-1 activities in C. briggsae.
In adult hermaphrodites, CBR-CSR-1 was enriched in both germline and somatic nuclei (Figure 4C and D). We observed CBR-CSR-1 in association with oocyte nuclei and enriched on chromosomes in oocytes (Figure 4C). Again, we did not reliably observe CSR-1 localization to perinuclear and cytoplasmic granules, which would be indicative of P granules. CBR-CSR-1 was also enriched in somatic gut nuclei, further supporting a nuclear role for this Argonaute (Figure 4D).
To validate the interaction of C. briggsae CSR-1 with chromatin, we performed subcellular fractionation experiments on embryos to separate cytoplasm, nuclei and chromatin (16). We probed these fractions for CBR- CSR-1 and found that the protein was present in the cytoplasm and nuclei, and was associated with chromatin in a manner comparable to the nuclear control, Histone H3 (Figure 4E and Supplementary Figure S3). The immunolocalization and cellular fractionation data (i) clearly demonstrate that CBR-CSR-1 associates with chromatin in the germline and soma, (ii) suggest a role for CBR-CSR-1 in regulating chromatin and (iii) support a role for this Argonaute in the transcriptional regulation of target genes.
CSR-1 associates with 22G-RNAs to target protein coding genes in C. briggsae
The capability of our C. elegans CSR-1 antibody to IP C. briggsae CSR-1 complexes allowed us to identify the small RNAs associated with CBR-CSR-1, and thus their target genes. Likewise, we were able to determine the effect of loss of CBR-CSR-1 on small RNA populations, owing to the successful knockdown of CSR-1 by RNAi in C. briggsae (Figures 1D and 4). Thus, we sequenced small RNA libraries from mature adult hermaphrodites possessing embryos (gravid adults), obtaining on average 3.8 million genome matching reads per each library (CBR-CSR-1 IP, Input total small RNAs, cbr-csr-1 RNAi and empty vector RNAi; excluding reads that mapped to known non-coding RNAs such as rRNAs, snoRNAs, etc.; Supplementary Table S1).
We first examined the size and first nucleotide bias for each of the libraries (Figure 5A) and then classified the small RNAs from each library by the annotations of the genomic locations to which they mapped (Figure 5B): piRNAs/21U-RNAs, miRNAs, mRNAs (sense and antisense separately), pseudogenes and repetitive elements. The small RNAs that mapped to regions of the C. briggsae genome in which no gene models have been predicted are labeled as unannotated. Because the C. briggsae genome is not as well refined as the C. elegans genome, we observed a higher fraction of unannotated small RNAs in C. briggsae.
Figure 5.
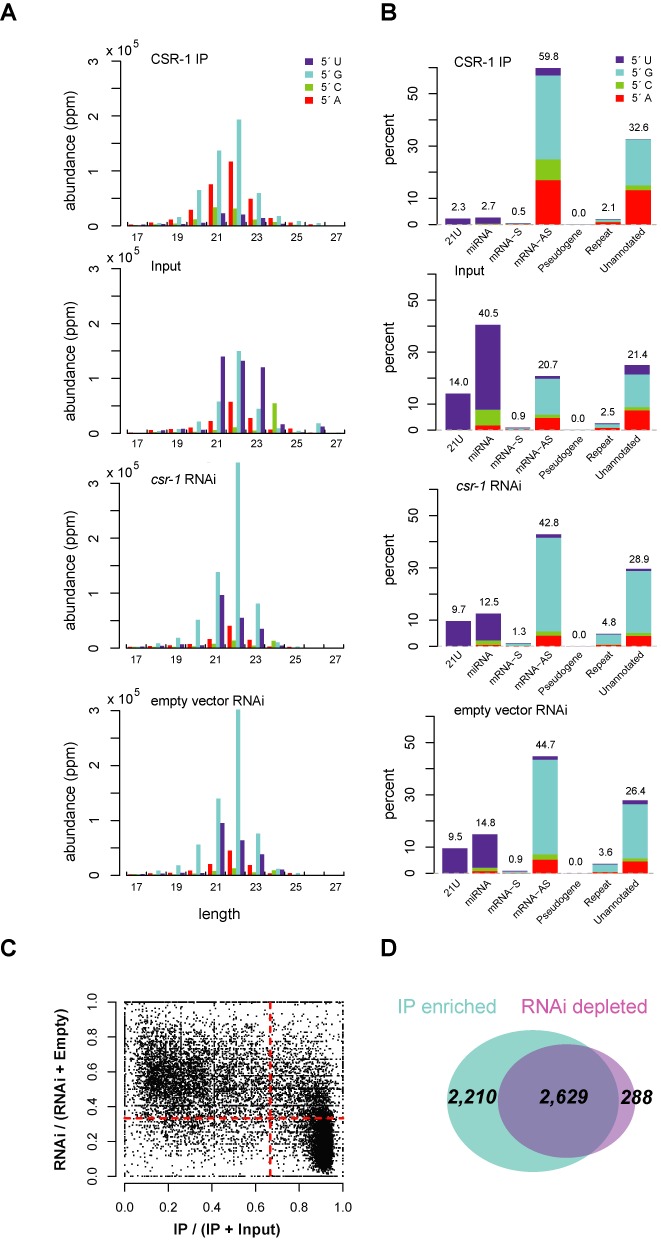
Analysis of small RNAs in C. briggsae. (A) Length and first nucleotide distribution of small RNAs in CBR-CSR-1 IP, C. briggsae input, cbr-csr-1 RNAi and empty vector RNAi. The bar graphs show the length and first nucleotide distributions of small RNAs that are mapped to the C. briggsae genome or exon–exon junctions of annotated C. briggsae genes. Small RNA abundance is measured in parts per million (ppm). (B) Proportions of reads that map to 21U-RNAs, miRNAs, sense (S) mRNAs, antisense (AS) mRNAs, pseudogenes, repeat elements and the unannotated portion of the genome. The stacked bar graphs display the first nucleotide distribution of small RNAs in each class. (C) Comparison of enrichment in CBR-CSR-1 IP versus input and cbr-csr-1 RNAi depletion versus empty vector RNAi. Each point in the scatter plot corresponds to a protein coding gene, for which depletion of small RNAs in cbr-csr-1 RNAi libraries is plotted against enrichment in the CBR-CSR-1 IP libraries. Depletion in cbr-csr-1 RNAi is defined as the number of small RNA reads in the cbr-csr-1 RNAi sample divided by the sum of small RNA reads in the cbr-csr-1 RNAi and empty vector RNAi samples. Enrichment in CBR-CSR-1 IP is defined as the number of small RNA reads in the CBR-CSR-1 IP sample divided by the sum of small RNA reads in the CBR-CSR-1 IP and input samples, with a minimum of 10 ppm. The vertical red line indicates a 2-fold enrichment in the CSR-1 IP, while the horizontal one represents a 2-fold depletion in csr-1 RNAi, thus genes in the bottom right quadrant are depleted in csr-1 RNAi and enriched in the CSR-1 IP. (D) The Venn diagram shows the overlap between the genes with ≥2-fold CBR-CSR-1 IP enrichment and the genes with ≥2-fold cbr-csr-1 RNAi depletion.
In comparison with the input library, the CBR-CSR-1 IP library is strongly enriched for 22G-RNAs and depleted for other types of small RNAs (Figure 5A) (16). Among CBR-CSR-1-bound small RNAs, 59.8% map antisense to mRNAs, while only 0.5% map sense to mRNAs and 7.1% map to 21U-RNAs, miRNAs, pseudogenes or repeat elements collectively (Figure 5B). We suspect that much of the 32.6% of small RNAs in the unannotated category also map antisense to mRNAs, but refining this set will require improved genome annotation. Overall, we found that 4839 genes are the targets of CSR-1 and its associated small RNAs in C. briggsae, as defined by 2-fold or greater enrichment in the CBR-CSR-1 IP relative to input, with a minimum 10 ppm (parts per million) (Supplementary Table S2).
We also observed an increase in small RNAs with a 5′ adenine in the CBR-CSR-1 IP library, with the most abundant length as 22 nucleotides (Figure 5A). Notably, our previously published CEL-CSR-1 IP small RNA data also possessed a small fraction of this 22A-RNA species, but we did not specifically examine it at the time (see Supplementary Figure S6 in (16)). Our previous analysis also demonstrated the 22A-RNAs are characterized by a 5′ triphosphate, suggesting similar biochemical characteristics to 22G-RNAs (see Figure 2 in (18)). We examined the 22A-RNA species further in this study and found that the targets of the 22A-RNAs are well correlated with the targets of the 22G-RNAs (Supplementary Figure S4) and that both types of RNAs possess a similar sequence motif surrounding the first nucleotide (Supplementary Figure S5). Finally, both 22G-RNAs and 22A-RNAs are depleted in cel-drh-3 mutants, suggesting a common mode of biogenesis (Supplementary Figure S6) (18). These findings all point to the 22A-RNAs representing a bona fide small RNA species. At the present, however, we do not have evidence to suggest that the 22A-RNAs function any differently than the 22G-RNAs, thus for further analyses, we have focused on the small RNAs enriched in the CBR-CSR-1 IP collectively (including both 22G- and 22A-RNAs), and continue to refer to the group as 22G-RNAs, as they are the more abundant and best characterized small RNA species.
We next asked which small RNAs were depleted upon loss of cbr-csr-1. Our initial examination of the small RNA populations in the cbr-csr-1 RNAi versus control samples showed little difference in the overall proportion of 22G-RNAs (Figure 5A and B, bottom two panels), but closer inspection revealed differences between the samples in the distribution of 22G-RNA subsets. Although cbr-csr-1 knock down led to only a minor decrease in the percentage of small RNAs that were antisense to mRNAs (44.7% in the empty vector RNAi control library and 42.8% in the cbr-csr-1 RNAi library; Figure 5B), the percentage of small RNAs anti-sense to the subset of mRNAs that were CSR-1 targets was much decreased (25.69% in the empty vector RNAi control library versus 13.44% in the cbr-csr-1 RNAi library). Conversely, the cbr-csr-1 RNAi sample contained 1.34% more WAGO-1 small RNAs than the empty vector RNAi control sample (for conserved gene targets with 1:1 orthologs between C. elegans and C. briggsae), as well as ∼12% more small RNAs from non-CSR-1, non-WAGO-1 target genes.
We observed a large overlap between genes depleted of small RNAs in cbr-csr-1 RNAi with genes enriched for small RNAs in the CBR-CSR-1 IP (Figure 5C and D and Supplementary Table S2). Among the 4839 genes enriched in small RNAs in the CBR-CSR-1 IP, the majority (54%) of them were depleted of small RNAs in the cbr-csr-1 RNAi sample. This value is consistent with observations in C. elegans, for which about half of the CEL-CSR-1 target genes are depleted of small RNAs when cel-csr-1 is mutated. The converse is more pronounced, such that 90% of the genes depleted of small RNAs in the cbr-csr-1 RNAi sample were enriched in small RNAs in the CBR-CSR-1 IP. The overlap in these two data sets defines a high confidence set of genes targeted by CSR-1 in C. briggsae. However, it is important to note that RNAi does not fully deplete CBR-CSR-1 (Figure 1D), thus the reduction of small RNAs is likely to be underestimated. Therefore, we define the CBR-CSR-1 target genes as those 4839 genes enriched for small RNAs in the CBR-CSR-1 IP sample, consistent with our previous analysis (16).
Finally, we tested whether there was a role for the 3′ uridyl-transferase CDE-1 in uridylating small RNAs associated with CBR-CSR-1 (20). We observed that CSR-1-associated small RNAs possessing a 3′ uridine tail in C. briggsae were enriched by 3.5-fold relative to input (Supplementary Figure S7) (16). These data suggest that uridylation could stabilize CSR-1 bound small RNAs. Alternatively, uridylation may comprise one step in the turnover of the small RNAs, as data from C. elegans indicate that CSR-1 complexes are more robustly occupied by small RNAs in cde-1 mutants (20).
CSR-1 small RNAs target conserved genes in C. briggsae and C. elegans
Given the conserved functional features and small RNAs that we documented for CSR-1 in C. elegans and C. briggsae, we next tested for evolutionary conservation of CSR-1 gene targets between the two species. Using our set of 4839 CBR-CSR-1 targets and a comparable set of 4932 CEL-CSR-1 targets (obtained by re-analyzing our previously published C. elegans data (16) in the same manner as the new C. briggsae data), we observed that the vast majority of CSR-1 target genes had 1:1 orthologs in the genomes of C. briggsae and C. elegans (Supplementary Table S2). Specifically, 86.3% of CBR-CSR-1 targets had 1:1 orthologs in C. elegans and, among them, 90.4% were also CEL-CSR-1 targets (Figure 6A). Reciprocally, 88.1% of C. elegans targets had 1:1 orthologs in C. briggsae, of which 87% are also targeted by CBR-CSR-1 (Figure 6A). Our results are consistent with a study by Shi et al. in which the authors reported that ∼85% of the previously defined CEL-CSR-1 targets (16) had 1:1 orthologs in C. briggsae, C. remanei or C. brenneri (12). Our CSR-1 IP analysis further confirms experimentally that the orthologs of CSR-1 targets in one species were bona fide CSR-1 targets in the other species. Thus, these observations support the idea that genes are more likely to be retained in genomes if they are stronger CSR-1 targets.
Figure 6.
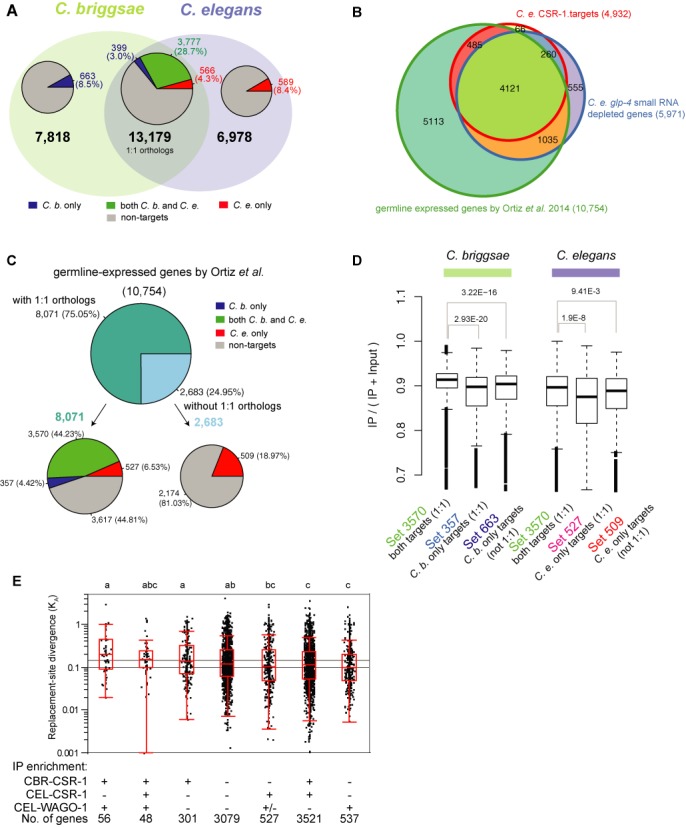
Conservation of the CSR-1 pathway in C. briggsae and C. elegans. (A) Comparison of the protein coding genes that are CSR-1 targets in C. briggsae and C. elegans. There are 13 179 total genes that exist as one-to-one (1:1) orthologs between C. briggsae and C. elegans; 7818 C. briggsae genes that do not have 1:1 orthologs in C. elegans; and 6978 C. elegans genes that do not have 1:1 orthologs in C. briggsae. The pie charts display the percentages of CSR-1 targets in these three groups. Among the 13 179 1:1 orthologs, 3777 genes are CSR-1 targets in both species (green), while 399 genes are CSR-1 targets in C. briggsae only (blue) and 566 are CSR-1 targets in C. elegans only (red). Non-target genes are shown in gray. (B) Overlap between germline-expressed, germline-specific and CEL-CSR-1 target genes. The Venn diagram displays the overlap among the 4932 CEL-CSR-1 targets, 5971 genes depleted of small RNAs in C. elegans glp-4(bn2) mutants and the Ortiz et al. set of 10 754 germline-expressed genes. (C) Comparison of the protein coding genes that are CSR-1 targets in C. briggsae and C. elegans within the Ortiz et al. 10 754 germline-expressed genes in C. elegans. There are 8071 C. elegans genes that have 1:1 orthologs, and the remaining 2683 genes do not have 1:1 orthologs in C. briggsae. Among the 8071 1:1 orthologs, 3570 genes are CSR-1 targets in both species (green), while 357 genes are CSR-1 targets in C. briggsae only (blue) and 509 are CSR-1 targets in C. elegans only (red). Non-targets are shown in gray. (D) CSR-1 IP enrichment of various gene groups. Among the 10 754 Ortiz et al. germline-expressed genes, the CBR-CSR-1 targets conserved in C. elegans (set 3570) are more highly enriched in the CBR-CSR-1 IP than the non-conserved CBR-CSR-1 targets with 1:1 C. elegans orthologs (set 357, Wilcoxon rank sum test P-value = 2.93E−20), and the CBR-CSR-1 targets without 1:1 orthologs (set 663, P-value = 3.22E−16). Likewise, the CEL-CSR-1 targets conserved in C. briggsae (set 3570) are more highly enriched in the C. elegans CSR-1 IP than the non-conserved CEL-CSR-1 targets with 1:1 C. briggsae orthologs (set 527, P-value = 1.9E−8), and the CEL-CSR-1 targets without 1:1 orthologs (set 509, P-value = 9.41E−3). (E) A plot of the KA values for 1:1 orthologous genes depicts rates of molecular evolution for each functional category. Each gene is depicted as a single dot, with higher values indicating more rapid sequence evolution at replacement sites. The box overlays indicate median and interquartile range, with points beyond whiskers being potential outliers. The gene sets differ significantly from each other (one-way ANOVA on log-transformed values, F6,8082 = 10.65, P < 0.0001); gene sets sharing the same letter above the box are not significantly different (Tukey's post-hoc test). Twenty genes with KA = 0 are not shown due to the log scale.
To investigate whether a high level of conservation was a property specific to CSR-1 targets or a general property of genes expressed in the germline, we analyzed the set of 10 754 genes recently defined by Ortiz et al. using RNA-seq on dissected C. elegans gonads (39). The Ortiz et al. data set of germline-expressed genes almost entirely encompasses the set of 5971 germline-specific genes we identified previously (18) (Figure 6B). These germline-specific genes are defined by 2-fold or greater depletion of small RNAs in glp-4(bn2) mutants (glp-4(bn2) is a C. elegans strain in which the germline precursor cells do not proliferate at a restrictive temperature), and analyzed in a manner consistent with our current analysis (18,40). Furthermore, the Ortiz et al. data set contains all but 326 of 4932 CEL-CSR-1 targets (Figure 6B), with the 326 genes likely being CSR-1 targets in the soma. Thus we proceeded with our analyses using the Ortiz et al. data set as germline-expressed genes.
Notably, only 75.05% of the germline-expressed genes have 1:1 orthologs. This is a much lower percentage than CSR-1 targets (88.9% of the germline-expressed CEL-CSR-1 targets have 1:1 orthologs), yet it is a higher percentage than the genome-wide average (65.4% of C. elegans genes have 1:1 orthologs) (Figure 6C). Furthermore, germline-expressed genes that are preserved as CSR-1 targets in both species are disproportionately enriched in both of our CSR-1 IP samples than are species-specific CSR-1 targets (Figure 6D). Indeed, our analysis supports the notion that CSR-1 targets are more evolutionarily conserved than germline-expressed genes in general.
To assess the conservation of CSR-1 target genes in a quantitative manner, we computed the rate of replacement site substitution (KA) between 1:1 orthologs of C. elegans and C. briggsae (41,42). KA provides a metric of molecular evolution for evaluating selective pressure on the protein sequence of target genes. To perform this analysis, we partitioned genes into seven mutually exclusive categories defined by whether they represent the germline-expressed targets of CEL-CSR-1, CBR-CSR-1 or CEL-WAGO-1 (Figure 6E). We find that the germline-expressed genes that are targeted by the CSR-1 or WAGO-1 small RNA pathways (the two ‘c’ groups in Figure 6E) are evolving at a significantly slower rate than those that are not the targets of these small RNA pathways (the ‘ab’ group in Figure 6E), using one-way ANOVA on log-transformed values (F6,8082 = 10.65, P < 0.0001) and Tukey's post-hoc test to compare between categories. These relationships also hold true when germline-specific transcripts from the cel-glp-4(bn2) data set are analyzed (Supplementary Figure S8), demonstrating that CSR-1 targets are evolving more slowly than other germline genes.
Evidence for 22G-RNA pathway specificity and switching between species
The 22G-RNAs of C. elegans are mainly divided between the CSR-1 pathway and the WAGO-1 pathway (16,18). Therefore, we sought to determine whether any of the CBR-CSR-1 targets that were not CEL-CSR-1 targets could be targets of the WAGO-1 pathway in C. elegans. A mosaic plot that partitions our small RNA data between CEL-CSR-1, CBR-CSR-1 and CEL-WAGO-1 pathways for 1:1 ortholog pairs illustrates the extensive overlap of germline-expressed CSR-1 targets in both C. elegans and C. briggsae, and distinguishes these genes from germline-expressed genes that are not CSR-1 targets in either species (Figure 7A). Much higher percentages of genes that are CSR-1 targets in both species are depleted of small RNAs in the cbr-csr-1 RNAi sample and in cel-csr-1 mutants in comparison to germline-expressed genes that are not CSR-1 targets in either species (60% of conserved CSR-1 targets are depleted of 22G-RNAs in cbr-csr-1 RNAi versus 1% of non-target germline-expressed genes; 21% of conserved CSR-1 targets are depleted of 22G-RNAs in cel-csr-1 mutants versus 8% of non-target germline-expressed genes). In sharp contrast, the converse is true for the enrichment in the CEL-WAGO-1 IP (1% of conserved CSR-1 targets overlap with CEL-WAGO-1 targets versus 15% of non-targets overlap with CEL-WAGO-1 targets).
Figure 7.

CSR-1 22G-RNA Pathway Specificity and Genomic Organization. (A) A mosaic plot parses pathway divisions for CEL-CSR-1 and CBR-CSR-1 target genes. The 8071 1:1 orthologs out of 10 754 Ortiz et al. germline-expressed genes are divided into four groups in the mosaic plot, according to whether they are CSR-1 targets in both species (3570), only one species (357 for C. briggsae, 527 in C. elegans), or neither (3617). Three pie charts in each quadrant illustrate the fractions of genes in each group that show depleted small RNAs in cbr-csr-1 RNAi (pink), depleted small RNAs in a cel-csr-1 mutant (purple) or are WAGO-1 targets in C. elegans (blue). (B) The Venn diagram compares the overlap among the subsets of the 357 non-conserved CBR-CSR-1 targets that are depleted of small RNAs in cbr-csr-1 RNAi (pink); depleted small RNAs in a cel-csr-1 mutant (but not enriched in the CEL-CSR-1 IP, purple); and are WAGO-1 targets in C. elegans (blue). (C) The stacked bar graph shows the distribution of 1:1 orthologs from various categories of genes that are found within operons compared with those that are not in operons. Y-axis indicates proportion of total genes (total number of genes in operons = 2690; total number of genes not in operons = 5381).
Among the 527 C. elegans-specific, germline-expressed CSR-1 targets, 20% are depleted for small RNAs in the cel-csr-1 mutants (Figure 7A). Supporting the idea that these are truly C. elegans-specific CSR-1 targets, we observe 10-fold fewer of them being depleted for small RNAs in the cbr-csr-1 RNAi sample in C. briggsae (2%) and 5-fold fewer genes are enriched in CEL-WAGO-1 IP (4%). Perhaps more unexpected, nearly one-third of the 357 C. briggsae-specific CSR-1 targets are depleted of small RNAs in each of the samples derived from cbr-csr-1 RNAi or cel-csr-1 mutants (Figure 7A and B). The overlap of 37 genes in these two subsets suggests that many of these genes are likely to be CSR-1 targets in both species, but fall below the detection threshold in C. elegans. Interestingly, 56 of the 357 C. briggsae-specific CSR-1 targets (16%) are enriched in small RNAs in the CEL-WAGO-1 IP, and 45 of these 56 genes are not depleted in small RNAs in cel-csr-1 mutants, strongly suggesting that some of the C. briggsae-specific gene targets of CSR-1 have diverged in their pathway affiliation between species, making use of the WAGO-1 pathway in C. elegans (Figure 7B). Notably, we also found that this labile set of germline-expressed genes targeted by CSR-1 in C. briggsae and WAGO-1 in C. elegans evolves most rapidly in our KA analysis (the first ‘a’ group in Figure 6E). Together, these data point to functional conservation in targeting by the CSR-1 pathway and elevated rates of evolution in these intriguing targets that appear to have switched pathways.
Genomic organization of CSR-1 targets
Because the CEL-CSR-1 pathway was previously implicated in genome-wide chromatin organization (16,19,21,29), we next asked how the CBR-CSR-1 targets are organized throughout the genome. The locations of the 3777 CSR-1 target genes common to both species’ genomes are distributed roughly evenly across both genomes, except that the densities are lower on chromosome X than on the autosomes for both species (Supplementary Figure S9). This observation correlates with a dearth of germline-expressed protein coding genes on the X chromosome in both species (see below) (43). One additional striking aspect CSR-1 target gene organization across the genome is that these genes are strongly enriched in operons. While 33% (2690/8071) of germline-expressed genes with 1:1 orthologs are found in operons, 56% (1971/3522) of the conserved CSR-1 target genes are found in operons, and only 9% of each group of germline-expressed non-target genes (282/3080) and CEL-WAGO-1 target genes (50/537) occur in operons, respectively (Figure7C and Supplementary Figure S10). Examined from a slightly different perspective, there is an over two-fold enrichment of conserved CSR-1 target genes in operons: of the 1:1 germline-expressed genes found in operons, 73%(1971/2690)are conserved CSR-1 targets, while only 28.8% (1551/5381) of non-operon genes are conserved CSR-1 targets. Operons are positively linked to germline gene expression (44) and we hypothesize that this organization may enable coordinated transcriptional regulation and expression of CSR-1 targets in the germline.
Functional insights into the conserved CSR-1 22G-RNA pathway
Several recent studies have shown that C. elegans CSR-1 small RNAs target nearly all germline protein coding genes to license their germline expression (16,19,27–28). Our analysis thus far has clearly demonstrated that nearly all of the conserved CSR-1 targets are expressed in the germline (Figure 6C). By further utilizing the Ortiz et al. data (39), we next assessed whether CSR-1 targets were expressed in the female germline (oogenesis), male germline (spermatogenesis) or in both germlines (gender neutral). We found that the largest group of conserved targets was gender neutral targets (76.7%, 2897/3777), followed by oocyte-specific targets (16.23%, 613/3777) and sperm-specific targets (1.6%, 60/3777) (Figure 8A). Here it is important to note that we likely did not capture all of the sperm specific CSR-1 targets simply because our samples were isolated from adult hermaphrodites, a developmental stage at which spermatogenesis is not ongoing. Indeed, Conine et al. identified an additional ∼1800 CEL-CSR-1 targets from isolated male samples that were not previously identified in our hermaphrodite libraries (16,19). These data indicate that CSR-1 targets conserved ‘gender neutral’ genes that are expressed broadly in both male and female germlines, along with a smaller subset of gender-specific germline genes. These results implicate CSR-1 in a conserved regulatory role during gametogenesis of both sperm and oocytes.
Figure 8.
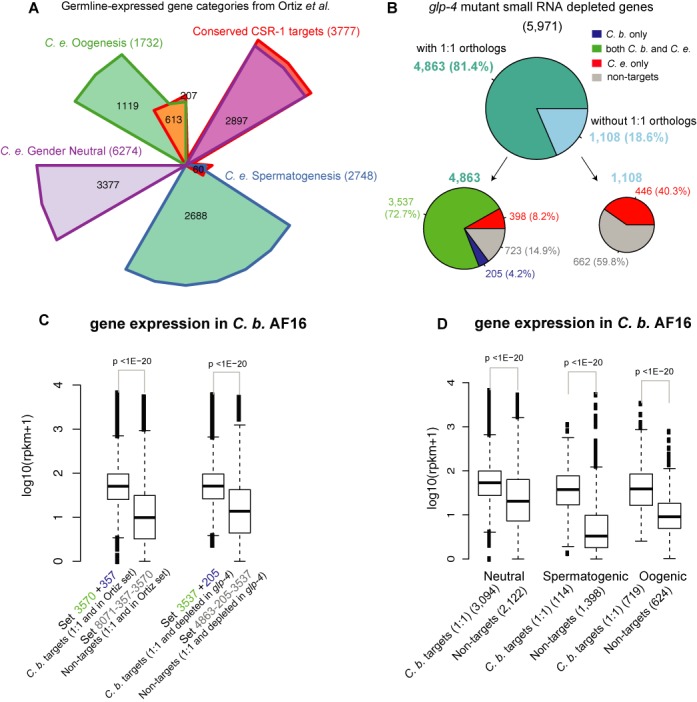
Conservation of function of CSR-1 targets in C. briggsae and C. elegans. (A) Comparison of the protein-coding genes between the conserved CSR-1 targets and the Ortiz et al. germline-expressed genes, which were further divided into three categories: female germline (oogenesis), male germline (spermatogenesis) and both germlines (gender neutral). The Venn diagram indicates that 2897 (76.7%) of the 3777 conserved CSR-1 targets are gender neutral targets. (B) The 5971 C. elegans genes depleted of small RNAs in the cel-glp-4(bn2) mutant are divided into two groups according to whether they have 1:1 C. briggsae orthologs, shown in the upper pie chart. The two bottom pie charts show the proportions in the two groups according to whether the genes are CSR-1 targets in both species, in C. briggsae only, in C. elegans only or in neither. (C) The CBR-CSR-1 targets with 1:1 orthologs (set 3570+357 defined in Figure 6C) within the Ortiz et al. set of germline-expressed genes are expressed in higher levels in mature adult hermaphrodites of the wild-type C. briggsae reference strain (AF16) than the C. briggsae genes that have 1:1 orthologs within the Ortiz et al. set but are not CSR-1 targets (Wilcoxon P-value <1E−20, left set). The observation also holds when comparing the targets with non-targets within the genes depleted of small RNAs in the cel-glp-4(bn2) mutant (right set). Expression levels were calculated as reads per kilobases of transcript in a sequencing depth of 1 million (RPKM), and the y-axis is the log10 transform of the average RPKM (added with a pseudo count 1) from the RNA-seq data of three AF16 biological replicates. (D) Among the Ortiz et al. set of 10 754 germline-expressed genes in C. elegans, there are 8071 genes with 1:1 orthologs in C. briggsae, which can be divided into three categories: gender neutral, spermatogenic and oogenic. The CBR-CSR-1 targets are expressed at higher levels in mature adult hermaphrodites of the wild-type C. briggsae reference strain AF16 than the non CSR-1 targets within each category (Wilcoxon P-value <1E−20). The y-axis is the same as the one in (C).
Beyond a role for CSR-1 in regulating germline-expressed genes, we observed that, in fact, 88.9% of the CEL-CSR-1 targets are germline-specific in nature. Among the 5971 germline-specific genes identified using the cel-glp-4(bn2) mutant, 4863 (81.4%) have 1:1 orthologs in C. briggsae, in contrast to the 75.05% of 1:1 orthologs for germline-expressed genes defined by Ortiz et al. (39). CSR-1 targets in both species comprise 72.7% of these 4863 germline-specific genes with 1:1 orthologs, with an additional 8.2% being CSR-1 targets in C. elegans only and 4.2% being CSR-1 targets in C. briggsae only (Figure 8B). In contrast, of the C. elegans germline-specific genes lacking 1:1 orthologs, only 40.3% are CSR-1 targets in C. elegans. Thus, CSR-1 small RNAs preferentially target germline-specific genes that are present in both species, suggesting that like CEL-CSR-1, CBR-CSR-1 could promote the expression of these target genes in the germline.
Next, to gain additional insight into the functions of CSR-1 target genes, we performed gene ontology (GO) enrichment analysis using the DAVID algorithm (45). Due to the incomplete annotations for C. briggsae genes, we performed GO enrichment analysis only for CSR-1 target genes in C. elegans, separated into four groups according to their preservation in C. briggsae. The 3777 1:1 orthologs that are CSR-1 targets in both species are strongly enriched in a large number of functional annotation clusters in comparison with the entire set of 10 754 germline-expressed genes, with the three most significant clusters being nematode larval development (false discovery rate or FDR = 2.11E−42), reproductive developmental process (FDR = 5.18E−25) and positive regulation of growth rate (FDR = 2.10E−23) (Supplementary Table S3). These results are consistent with a role for CSR-1 in regulating conserved germline genes that are important for gametogenesis.
To determine the effect of loss of CSR-1 on its target transcripts, we used qRT-PCR to measure the steady state levels of CSR-1 target genes in C. briggsae mature adult hermaphrodites fed RNAi for cbr-csr-1 or for a control empty vector. In this experiment, we found that the CSR-1 target genes did not significantly change in their steady state levels (Supplementary Figure S11). These data are consistent with observations in C. elegans that steady state target mRNA levels for the majority of CSR-1 targets are not significantly altered upon loss of csr-1 (16). However, a more recent survey using GRO-seq (Global Run-On high-throughput sequencing) demonstrated that the nascent transcript levels of a subset of CSR-1 target genes are decreased when csr-1 or drh-3 are compromised (29), thus a more sensitive sequencing method may shed further light on this question.
Finally, we reasoned that if CSR-1 plays a broad role in promoting the expression of its targets, CSR-1 target mRNA levels would likely be higher than non-target germline genes. Thus, we used RNA-seq to analyze mRNA expression levels in adult wild-type C. briggsae hermaphrodites. We focused our analysis on the C. briggsae 1:1 orthologs of the Ortiz et al. set of germline-expressed C. elegans genes (defined in Figure 6C) (39). This analysis demonstrated that CBR-CSR-1 targets are significantly more highly expressed than germline-expressed genes that are not CSR-1 targets (Wilcoxon rank sum test P-value <1E−20; Figure 8C, left set). CBR-CSR-1 targets are also more highly expressed than other germline-specific genes (P-value <1E−20; Figure 8C, right set), and our results were consistent when we examined the sperm, oocyte and gender neutral germline-expressed genes for CBR-CSR-1 targets versus non-targets (Figure 8D). These results also held true for CEL-CSR-1 targets (Supplementary Figure S12). As a confirmation of the RNA-seq data, qRT-PCR demonstrated a consistent trend of higher mRNA expression levels of genes in the species for which they were the targets of CSR-1, whereas non-target genes were variably expressed in both species (data not shown). These data point to mRNA expression patterns or mRNA levels as a possible means of triggering small RNA production, and are consistent with a role for CSR-1 in promoting germline gene expression in multiple species (19,27–29).
DISCUSSION
The C. elegans Argonaute CSR-1 has been the focus of intense interest due to its essential nature and functions in the nucleus, which are distinguishing features among nematode Argonautes (17). Here we employed a comparative functional genomic approach to characterize the roles of CSR-1 in two distinct nematode species, C. elegans and C. briggsae, which are evolutionally separated by tens of millions of years (6,10). Our analyses were enabled in part by the ability of a C. elegans antibody to recognize a C. briggsae protein. Despite the deepening understanding of the CSR-1 pathway in C. elegans, little was known about the conservation and divergence of this pathway and its targets through direct functional analysis of other species.
We demonstrated that the core activity of the CSR-1 pathway is conserved in C. briggsae. As is true in C. elegans (16), the C. briggsae ortholog of CSR-1 is essential for animal viability, such that loss of cbr-csr-1 leads to chromosome segregation defects. Likewise, other components of the CSR-1 pathway in C. briggsae are present and expressed in a similar developmental pattern and at least two of these components, ego-1 and drh-3, are also essential (16). The enrichment of uridylated small RNAs in the CBR-CSR-1 IP sample also implicates C. briggsae CDE-1 in the pathway (20). Furthermore, CBR-CSR-1 localizes to chromosomes and associates with chromatin, indicating a conserved nuclear role for this pathway, perhaps at the level of transcriptional control and chromatin organization for CBR-CSR-1.
The association of CBR-CSR-1 with 22G-RNAs that target germline-expressed genes is another conserved feature of this pathway. The germline-expressed and germline-specific protein coding genes targeted by CSR-1/22G-RNA complexes are preserved as 1:1 orthologs between the two species to a greater extent than expected from the genome-wide average for protein coding genes. This observation could be interpreted to mean that CSR-1 simply targets more highly conserved genes, as indeed, gene orthologs targeted by CSR-1 in both species evolve especially slowly at the sequence level. Alternatively, these data could point to a role for CSR-1 in promoting patterns of germline gene expression and the retention of essential genes throughout evolution. Notably, and again consistent with a role for CSR-1 in promoting germline gene expression, the highest confidence CSR-1 targets are those with the highest expression levels.
Our comparative functional genomic study has uncovered two previously unappreciated features of the CSR-1 pathway. First is the enrichment of a 22A-RNA species associated with CSR-1. Although it is currently unclear whether these small RNAs behave distinctly from canonical 22G-RNAs, they do collectively target the same set of germline-expressed genes and bear similar biogenesis characteristics to the 22G-RNAs. Second, more than half of CSR-1 pathway targets occur in operons, twice the incidence for other genes in the genome. The link between operons and germline gene expression in C. elegans has been made previously (44), and is consistent with CSR-1 targeting germline-expressed genes and only a minority of CSR-1 targets being X-linked. However, the functional implication of CSR-1 targeting operon genes is not entirely clear. It is possible that targeting clusters of genes that are coordinately expressed within the genome may provide CSR-1 with a more effective means of regulating chromatin and transcription of the targets. It is also possible that CSR-1's regulation of the local chromatin environment might facilitate recruitment by translocation of additional germline-expressed genes into operons (46).
Despite the overall strong conservation for the CSR-1 pathway between C. elegans and C. briggsae, our analysis revealed several intriguing disparities. The CSR-1 protein has been preserved among the diverse Clade V nematode species, which includes C. elegans and C. briggsae (14,15). Homologs of CSR-1 can also be identified in Clade III, which includes human parasitic nematodes such as Loa loa and Ascaris lumbricoides (14,15). Indeed, small RNAs of the 22G-RNA class have been identified by sequencing studies in Ascaris suum, although those associated with CSR-1 have not yet been isolated (47). Due to its essential nature and nuclear role in chromosome organization, it is somewhat surprising that CSR-1 is not preserved more universally in additional nematode species. In fact, several other WAGO-class Argonautes are more highly represented in Clades III–V, such as HRDE-1/WAGO-9 and a currently uncharacterized Argonaute that appears to be closely related to CSR-1, C04F12.1 (14,15). Thus, it is possible that in more distantly related organisms, the essential functions of CSR-1 are assumed by other WAGOs, including C04F12.1. As experimental techniques such as genome editing become more widely utilized, additional nematode species will become increasingly tractable for cross-species examination of the functional roles of CSR-1 and other Argonautes.
The question of how particular transcripts are directed to one pathway or another remains somewhat mysterious. Our analysis of small RNA populations indicates that 16% of the genes targeted by CSR-1 in C. briggsae only are the targets of WAGO-1 in C. elegans. In contrast, only 4% of C. elegans-specific CSR-1 targets are also targets of WAGO-1. This comparative genomics approach has highlighted the possibility that the pathway fidelity of some genes may be more labile than others, permitting the evolution of pathway ‘switching,’ whereby different Argonautes regulate a target gene's expression in different species. Moreover, this subset of genes that appears to have undergone pathway switching evolves nearly twice as fast at the sequence level as do genes that are targeted by CSR-1 in both C. elegans and C. briggsae. Clearly these labile targets provide intriguing candidates for understanding the evolution of regulatory network architecture in general, as well as for understanding in greater molecular detail how targets are chosen. In particular, should these targets share any consistent sequence elements, mRNA localization patterns or other characteristics, then they could provide key insights into which features of a specific transcript enable it to be targeted by one pathway over another, and how such features differ from those of conserved targets. Heterologous expression studies are also likely to reveal the molecular rules dictating the shunting of mRNAs into each pathway.
Our comparative genetic and genomic approach has revealed both conserved and species-specific features of the CSR-1 22G-RNA pathway. Collectively, we find evidence for a conserved nuclear role of CSR-1 in promoting germline gene expression, such that CSR-1 may be capable of modulating chromatin and licensing germline gene expression in multiple species. As genome scale techniques and genome editing tools come into wider use, further comparative genomic studies that integrate function and evolution on a wide array of species will empower a comprehensive understanding of small RNA biology across nematodes.
ACCESSION NUMBER
All sequencing data have been deposited in the SRA, accession number SRP021463.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank Dr. Alexander W. Ensminger, Vivian Cheung, Michelle A. Francisco, Aldis Krizus and Christopher J. Wedeles for critical reading of this manuscript. Some strains for this study were provided by the Caenorhabditis Genetics Center (which is supported by the NIH Office of Research Infrastructure Programs [P40 O010440]).
Footnotes
These authors contributed equally to the paper as first authors.
Present address: Jie Wang, Department of Biochemistry, University at Buffalo, and NY State Center of Excellence in Bioinformatics & Life Sciences Buffalo 14203, NY, USA.
FUNDING
Ontario Graduate Scholarship [to M.W.]; University of Toronto Open Fellowship [to M.W.]; Canada Research Chair in Small RNA Biology [J.M.C.]; NSERC [RGPIN-418 to J.M.C.]. Funding for open access charge: NSERC [RGPIN-418 to J.M.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ketting R.F. The many faces of RNAi. Dev. Cell. 2011;20:148–161. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 4.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 5.Grishok A. Biology and mechanisms of short RNAs in Caenorhabditis elegans. Adv. Genet. 2013;83:1–69. doi: 10.1016/B978-0-12-407675-4.00001-8. [DOI] [PubMed] [Google Scholar]
- 6.Stein L.D., Bao Z., Blasiar D., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta B.P., Sternberg P.W. The draft genome sequence of the nematode Caenorhabditis briggsae, a companion to C. elegans. Genome Biol. 2003;4:238.1-238.4. doi: 10.1186/gb-2003-4-12-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta B.P., Johnsen R., Chen N. Genomics and biology of the nematode Caenorhabditis briggsae. WormBook. 2007:1–16. doi: 10.1895/wormbook.1.136.1. doi/10.1895/wormbook.1.136.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koboldt D.C., Staisch J., Thillainathan B., Haines K., Baird S.E., Chamberlin H.M., Haag E.S., Miller R.D., Gupta B.P. A toolkit for rapid gene mapping in the nematode Caenorhabditis briggsae. BMC Genomics. 2010;11:236.1–236.16. doi: 10.1186/1471-2164-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutter A.D. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol. Biol. Evol. 2008;25:778–786. doi: 10.1093/molbev/msn024. [DOI] [PubMed] [Google Scholar]
- 11.Hillier L.W., Miller R.D., Baird S.E., Chinwalla A., Fulton L.A., Koboldt D.C., Waterston R.H. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 2007;5:e167. doi: 10.1371/journal.pbio.0050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Z., Montgomery T.A., Qi Y., Ruvkun G. High-throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes. Genome Res. 2013;23:497–508. doi: 10.1101/gr.149112.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Wit E., Linsen S.E., Cuppen E., Berezikov E. Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res. 2009;19:2064–2074. doi: 10.1101/gr.093781.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalzell J.J., McVeigh P., Warnock N.D., Mitreva M., Bird D.M., Abad P., Fleming C.C., Day T.A., Mousley A., Marks N.J., et al. RNAi effector diversity in nematodes. PLoS Negl. Trop. Dis. 2011;5:e1176. doi: 10.1371/journal.pntd.0001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buck A.H., Blaxter M. Functional diversification of Argonautes in nematodes: an expanding universe. Biochem. Soc. Trans. 2013;41:881–886. doi: 10.1042/BST20130086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claycomb J.M., Batista P.J., Pang K.M., Gu W., Vasale J.J., van Wolfswinkel J.C., Chaves D.A., Shirayama M., Mitani S., Ketting R.F., et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wedeles C.J., Wu M.Z., Claycomb J.M. A multitasking Argonaute: exploring the many facets of C. elegans CSR-1. Chromosome Res. 2013;21:573–586. doi: 10.1007/s10577-013-9383-7. [DOI] [PubMed] [Google Scholar]
- 18.Gu W., Shirayama M., Conte D.J., Vasale J., Batista P.J., Claycomb J.M., Moresco J.J., Youngman E.M., Keys J., Stoltz M.J., et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conine C.C., Moresco J.J., Gu W., Shirayama M., Conte D.J., Yates J.R.r., Mello C.C. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell. 2013;155:1532–1544. doi: 10.1016/j.cell.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Wolfswinkel J.C., Claycomb J.M., Batista P.J., Mello C.C., Berezikov E., Ketting R.F. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 21.Hall S.E., Chirn G.W., Lau N.C., Sengupta P. RNAi pathways contribute to developmental history-dependent phenotypic plasticity in C. elegans. RNA. 2013;19:306–319. doi: 10.1261/rna.036418.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smardon A., Spoerke J.M., Stacey S.C., Klein M.E., Mackin N., Maine E.M. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 23.Yigit E., Batista P.J., Bei Y., Pang K.M., Chen C.C., Tolia N.H., Joshua-Tor L., Mitani S., Simard M.J., Mello C.C. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 24.Duchaine T.F., Wohlschlegel J.A., Kennedy S., Bei Y., Conte D.J., Pang K., Brownell D.R., Harding S., Mitani S., Ruvkun G., et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 25.Avgousti D.C., Palani S., Sherman Y., Grishok A. CSR-1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J. 2012;31:3821–3832. doi: 10.1038/emboj.2012.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.She X., Xu X., Fedotov A., Kelly W.G., Maine E.M. Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 2009;5:e1000624. doi: 10.1371/journal.pgen.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seth M., Shirayama M., Gu W., Ishidate T., Conte D.J., Mello C.C. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell. 2013;27:656–663. doi: 10.1016/j.devcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wedeles C.J., Wu M.Z., Claycomb J.M. Protection of germline gene expression by the C. elegans argonaute CSR-1. Dev. Cell. 2013;27:664–671. doi: 10.1016/j.devcel.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 29.Cecere G., Hoersch S., O'Keeffe S., Sachidanandam R., Grishok A. Global effects of the CSR-1 RNA interference pathway on the transcriptional landscape. Nat. Struct. Mol. Biol. 2014;21:358–365. doi: 10.1038/nsmb.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nuez I., Felix M.A. Evolution of susceptibility to ingested double-stranded RNAs in Caenorhabditis nematodes. PLoS One. 2012;7:e29811. doi: 10.1371/journal.pone.0029811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Timmons L., Court D.L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 33.Chu D.S., Liu H., Nix P., Wu T.F., Ralston E.J., Yates J.R.r., Meyer B.J. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature. 2006;443:101–105. doi: 10.1038/nature05050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 35.Rocheleau C.E., Cullison K., Huang K., Bernstein Y., Spilker A.C., Sundaram M.V. The Caenorhabditis elegans ekl (enhancer of ksr-1 lethality) genes include putative components of a germline small RNA pathway. Genetics. 2008;178:1431–1443. doi: 10.1534/genetics.107.084608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vought V.E., Ohmachi M., Lee M.H., Maine E.M. EGO-1, a putative RNA-directed RNA polymerase, promotes germline proliferation in parallel with GLP-1/notch signaling and regulates the spatial organization of nuclear pore complexes and germline P granules in Caenorhabditis elegans. Genetics. 2005;170:1121–1132. doi: 10.1534/genetics.105.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strome S., Wood W.B. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- 38.Bezares-Calderon L.A., Becerra A., Salinas L.S., Maldonado E., Navarro R.E. Bioinformatic analysis of P granule-related proteins: insights into germ granule evolution in nematodes. Dev. Genes Evol. 2010;220:41–52. doi: 10.1007/s00427-010-0327-3. [DOI] [PubMed] [Google Scholar]
- 39.Ortiz M.A., Noble D., Sorokin E.P., Kimble J. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 (Bethesda) 2014;4:1765–1772. doi: 10.1534/g3.114.012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beanan M.J., Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992;116:755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- 41.Wang D., Zhang Y., Zhang Z., Zhu J., Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z., Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 43.Reinke V., Gil I.S., Ward S., Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- 44.Reinke V., Cutter A.D. Germline expression influences operon organization in the Caenorhabditis elegans genome. Genetics. 2009;181:1219–1228. doi: 10.1534/genetics.108.099283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 46.Cutter A.D., Agrawal A.F. The evolutionary dynamics of operon distributions in eukaryote genomes. Genetics. 2010;185:685–693. doi: 10.1534/genetics.110.115766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J., Czech B., Crunk A., Wallace A., Mitreva M., Hannon G.J., Davis R.E. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 2011;21:1462–1477. doi: 10.1101/gr.121426.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


