Abstract
CRISPR-Cas systems have shown tremendous promise as heterologous tools for genome editing and transcriptional regulation. Because these RNA-directed immune systems are found in most prokaryotes, an opportunity exists to harness the endogenous systems as convenient tools in these organisms. Here, we report that the Type I-E CRISPR-Cas system in Escherichia coli can be co-opted for programmable transcriptional repression. We found that deletion of the signature cas3 gene converted this immune system into a programmable gene regulator capable of reversible gene silencing of heterologous and endogenous genes. Targeting promoter regions yielded the strongest repression, whereas targeting coding regions showed consistent strand bias. Furthermore, multi-targeting CRISPR arrays could generate complex phenotypes. This strategy offers a simple approach to convert many endogenous Type I systems into transcriptional regulators, thereby expanding the available toolkit for CRISPR-mediated genetic control while creating new opportunities for genome-wide screens and pathway engineering.
INTRODUCTION
CRISPR (clustered regularly interspaced short palindromic repeat)-Cas (CRISPR-associated) systems provide prokaryotes with adaptive immunity against foreign invaders (1,2). Recognition of these invaders is conducted by CRISPR RNAs (crRNAs) that direct Cas proteins to cleave complementary nucleic acid sequences. The crRNAs are processed from transcribed arrays of identical repeats and intervening target-specific spacers, conferring immunity against multiple unique sequences. To manage the diversity of proteins associated with CRISPR-Cas systems, three general types have been defined each with a collection of subtypes (3). Apart from their particular suite of Cas proteins, these types can be distinguished by the molecular target, the mode of target recognition and the mechanism of crRNA processing. For instance, Type I systems target DNA with a protospacer-adjacent motif (PAM) flanking the 3′ end of the target sequence and rely on the Cas proteins for crRNA processing (4,5), whereas Type II systems target DNA with a PAM flanking the 5′ end of the target sequence and rely on a tracrRNA and RNase III for crRNA processing (6,7). Of the three types, Type I systems are the most prevalent in both bacteria and archaea (3).
A defining feature of CRISPR-Cas systems is that crRNAs can be readily designed to guide the specific targeting of virtually any sequence. Aside from genome editing and DNA imaging, this capability has opened widespread opportunities in programmable gene regulation (8–11). Toward this goal, Type II systems were recently engineered to bind but not cleave target DNA through point mutations in the endonucleolytic domains of the signature cas9 gene (12). Designed crRNAs directed the resulting catalytically dead Cas9 (dCas9) protein to bind specific promoters and coding regions, thereby modulating the recruitment of or extension by RNA polymerase (13–17). Separately, Type III-B systems, the only CRISPR-Cas systems known to naturally target RNA (18), can be directed to cleave chromosomal mRNAs (19,20). While these efforts demonstrated the capacity of Type II and III systems for gene regulation, what remains unexplored is the capacity of Type I systems to exhibit this same phenomenon. If confirmed, this prevalent type could be harnessed to advance genetic control with CRISPR-Cas systems and provide insights into their potential roles as natural gene regulators.
Type I CRISPR-Cas systems generally involve two protein elements for DNA targeting: Cascade and Cas3. Cascade, a multimeric complex of three to six different Cas proteins, is responsible for processing CRISPR arrays (4) and for binding target DNA sequences through PAM and protospacer recognition (4,21–23). Cas3, the signature protein of Type I systems, is responsible for cleaving and degrading target DNA (21,24–26). Recent biochemical studies of different Type I subtypes revealed that Cascade is a stable complex that recruits Cas3 only after DNA binding (21–23,25,27–29). Based on these insights, we hypothesized that removal of Cas3 from an endogenous CRISPR-Cas system would allow Cascade to tightly bind target DNA sequences without subsequent degradation. As a result, designed CRISPR arrays would be sufficient to direct targeted DNA binding, thereby blocking RNA polymerase recruitment or extension (13,17). Using the Type I-E CRISPR-Cas system in Escherichia coli K-12 as a model, we found that deleting the cas3 gene from the E. coli genome allowed the targeted and multiplexed regulation of gene expression using the endogenous CRISPR-Cas system. CRISPR arrays could also be generated for the coordinated silencing of multiple endogenous genes and the generation of complex phenotypes.
MATERIALS AND METHODS
Strains and plasmid construction
See Supplementary Table S1 for a list of all E. coli K-12 strains used in this work. To generate BW25113 Δcas3::cat and MG1655 Δcas3::cat, the cat resistance cassette was polymerase chain reaction (PCR)-amplified from the pKD3 plasmid (30) using oligonucleotides that append the synthetic constitutive promoter J23119 (BBa_J23119 in the registry for standard biological parts; www.partsregistry.org) (J23119-pKD3.for, J23119-pKD3.rev). Following a second PCR amplification to introduce homology arms (HR-cas3.for, HR-cas3.rev), the resulting PCR product was recombineered into NM500 by mini-λ-mediated recombination (31). The insertion replaced the native cas3 gene and the native promoter for the Cascade operon with the cat cassette and the J23119 promoter. Successful recombination was verified by sequencing. P1 transduction was then used to transfer the cat cassette and the synthetic promoter into BW25113 and into MG1655. Successful transduction was verified by PCR. To generate BW25113 Δcas3, the cat cassette from BW25113 Δcas3::cat was excised using the pCP20 plasmid as described previously (32). To generate NM500 cas3+, the cat resistance cassette was PCR-amplified from the pKD3 plasmid using oligonucleotides that append the constitutive promoter J23119 (J23119-pKD3.for, HR-casA.rev). Following a second PCR amplification to introduce homology arms (HR-cas3.for, HR-casA.rev), the resulting PCR product was recombined into NM500. This NM500 cas3+ strain replaces the native promoter for the Cascade operon with a constitutive promoter while retaining the native cas3 gene. To generate BW25113 ΔCRISPR-Cas::cat, the cat resistance cassette was PCR-amplified from the pKD3 plasmid (HR-CRISPR.for, HR-cas3.rev) and recombineered into NM500, followed by P1 transduction into BW25113. This BW25113 ΔCRISPR-Cas eliminates the entire CRISPR locus as well as cas3, the Cascade operon and the CRISPR1 locus.
See Supplementary Table S2 for a list of all plasmids used in this work. The green fluorescent protein (GFP) reporter plasmids were based on the pUA66 plasmid (low-copy sc101 origin-of-replication) (33) and are reported in previous work (34). To construct the arabinose-inducible pcrRNA.ind plasmid (medium-copy pBR322 origin-of-replication), oligonucleotides were designed to encode a single repeat and a synthetic rho-independent terminator (BBa_B1006 in the registry for standard biological parts) (pcrRNA.ind.for, pcrRNA.ind.rev). These oligonucleotides were annealed, 5′ phosphorylated using polynucleotide kinase (PNK) and ligated into the pBAD18 plasmid digested with KpnI-HF and HindIII-HF. To construct the constitutive pcrRNA.con plasmid, oligonucleotides encoding the synthetic constitutive promoter J23119 (pcrRNA.con.for, pcrRNA.con.rev) were annealed, 5′ phosphorylated with PNK and ligated into the pcrRNA.ind plasmid digested with NsiI and NheI. The insertion replaced the araC gene and ParaB promoter with the synthetic constitutive promoter. To insert new repeat–spacer pairs into pcrRNA.con or pcrRNA.ind, oligonucleotides encoding the palindromic repeat and crRNA spacers were annealed, 5′ phosphorylated with PNK and ligated into either plasmid digested with KpnI and XhoI. See Supplementary Figure S2 for an illustration of the cloning scheme.
All plasmid cloning was verified by sequencing. See Supplementary Table S3 for a list of all oligonucleotides used in this work. All oligonucleotides were chemically synthesized by IDT. All enzymes were purchased from NEB.
Growth conditions
All strains were cultured in 14-ml round-bottom polypropylene tubes at 37°C and 250 RPM in up to 5 ml of LB medium (10-g/l tryptone, 5-g/l yeast extract, 10-g/l NaCl) or M9 minimal medium (1X M9 salts, 2 mM MgSO4, 0.1 mM CaCl2, 10 μg/ml thiamine) containing the indicated combination of 0.4% glycerol, 0.2% indicated sugar and 0.2% casamino acids. All strains were plated on LB agar (LB medium with 1.2% agar) in 100 × 15-mm polystyrene petri dishes. To maintain any plasmids, cells were cultured in liquid medium or on agar plates containing appropriate antibiotics at the following concentration: 50 μg/ml of ampicillin, 34 μg/ml of chloramphenicol and 50 μg/ml of kanamycin.
Spacer design
See Supplementary Table S4 for a list of all of protospacers targeted in this work. Protospacers were selected by identifying a PAM (CTT, CCT, CAT, CTC located at the 3′ end of the target sequence) for the Type I-E system in E. coli (23). Note that only CTT and CCT were used in this work based on our previous experience with these PAM sequences (35). The 32 nucleotides immediately downstream of the PAM were then used as the spacer. The cloning scheme required changing the final two nucleotides of the spacer to TC (Supplementary Figure S2), which is not expected to impact crRNA activity (5).
Transformation assays
The transformation assay as shown in Supplementary Figure S4B was conducted similar to previous work (35). Briefly, E. coli BW25113 Δcas3::cat or NM500 cas3+ cells harboring pUA66-lacZ were cultured overnight in LB medium. Cultures were back-diluted 1:25 into 25 ml of LB medium in 125-ml Erlenmeyer flasks and grown to an ABS600 of 0.6–0.8, which was quantified using a Nanodrop 2000c spectrophotometer (Thermo Scientific). The cells were then washed in ice-cold 10% glycerol and concentrated by a factor of ∼100. A total of 50 μl of the concentrated cells were transformed with 50 ng of plasmid DNA using a MicroPulser electroporator (Bio-Rad). Transformed cells were recovered in 500-μl SOC medium for 1 h at 37°C. After the recovery period, the cells were diluted by factors of 104–106 and 250 μl of the dilution was plated on LB agar with appropriate antibiotics and inducers.
Flow cytometry analysis
Cells grown overnight in M9 minimal medium containing 0.2% casamino acids and 0.4% glycerol were back-diluted to an ABS600 of 0.01 into M9 minimal medium with the specified combination of 0.1 mM of Isopropyl β-D-1-thiogalactopyranoside (IPTG) and 0.2% of the indicated inducing sugar. Upon reaching an ABS600 of ∼0.2 after ∼3–4 h of growth, the cultures were diluted 1:100 in 1X phosphate buffered saline and run on an Accuri C6 Flow Cytometer (Becton Dickinson) equipped with CFlow plate sampler, a 488-nm laser and a 530 ± 15-nm bandpass filter. Events reflecting cells were gated based on forward scatter (FSC-H) and side scatter (SSC-H) with respective lower cutoffs of 11 500 and 600 to reduce the measurement of particulates. The gate was set using E. coli cells stained with the DRAQ5 dye (Thermo Scientific). The fluorescence of the gated cells was then measured in FL1-H. At least 20 000 events were analyzed for each sample.
For the reversibility experiments, cells were grown overnight in M9 minimal medium containing 0.2% casamino acids, 0.4% glycerol and 0.1 mM IPTG, with or without 0.2% L-arabinose. Overnight cultures were pelleted and resuspended twice in M9 minimal media with 0.2% casamino acids, 0.4% glycerol and 0.1 mM IPTG to remove any residual L-arabinose. The washed cultures were then back-diluted to an ABS600 of ∼0.001 in 30 ml of the same medium without or with 0.2% L-arabinose, respectively. Every hour, 800 μl of culture was withdrawn for flow cytometry analysis and measurement of the ABS600.
Doubling-time measurements
Cells were grown overnight in M9 minimal medium with 0.4% glycerol. The overnight cultures were pelleted and resuspended twice in M9 minimal medium with no carbon source. The washed cultures were then back-diluted to an ABS600 of ∼0.001 into 25 ml of M9 minimal medium containing 0.2% of the indicated sugar in 125-ml Erlenmeyer flasks. Every 30 min, 800 μl of culture was withdrawn for measurement of the ABS600.
Quantitative real-time PCR
Cells were grown overnight in M9 minimal medium containing 0.2% casamino acids and 0.4% glycerol. Overnight cultures were back-diluted 1:250 in M9 minimal medium containing 0.2% casamino acids, 0.4% glycerol and 0.2% of the indicated sugar. Once cultures reached an ABS600 of ∼0.4, total RNA was isolated as reported previously (36) followed by treatment with DNase I. cDNAs were generated from 2 μg of the resulting RNA using random primers and SuperScript III reverse-transcriptase (Invitrogen) followed by treatment with RNase H. Quantitative PCR was conducted on cDNA samples using SYBR Green (Bio-Rad) and the gene-specific primers (X-qPCR.fwd/rev, where X is the target gene) listed in Supplementary Table S3. cDNAs were run on a Mastercycler ep realplex2 real-time PCR system (Eppendorf) according to the manufacturer's instructions. For the PCR runs, each cDNA was heated to 95°C for 2 min followed by 50 cycles of a 15-s denaturing step at 95°C, a 15-s annealing step at 55°C and a 30-s extension step at 72°C. At the end of the run, a melt curve was generated to ensure the absence of non-specific products. Relative quantitation of gene expression was calculated using the ΔCt method.
Growth assays
Cells were inoculated into M9 minimal medium containing 0.4% glycerol and grown overnight. After 24 h, cells were pelleted and resuspended in 2 ml of M9 minimal medium with no carbon source two times to remove glycerol as a possible source of growth. The washed cultures were then back-diluted to an ABS600 of 0.001 into 2 ml of M9 minimal medium containing 0.2% of the indicated sugar(s). Finally, the cultures were grown for 24 h until the ABS600 was measured.
RESULTS
Targeted gene repression following deletion of cas3
To explore the capacity of Type I systems for gene regulation, we employed the Type I-E CRISPR-Cas system in E. coli K-12 (Supplementary Figure S1), the best characterized Type I system in a genetically tractable bacterium (4–5,21,23). Because the operon encoding Cascade (cse1-cse2-cas7-cas5-cas6e) is strongly repressed under normal growth conditions (37,38), we replaced cas3 and the native cse1 promoter with a constitutive promoter in one round of homologous recombination (Figure 1). The resulting strain (BW25113 Δcas3::cat) was transformed with a medium-copy plasmid encoding L-arabinose-inducible single-spacer arrays (Supplementary Figure S2) and a low-copy reporter plasmid encoding the green fluorescent protein (gfp) gene downstream of the lacZ promoter (pUA66-lacZ; Supplementary Table S5). The spacers were designed to target 10 locations in the promoter and gfp coding region as well as two locations far upstream of the promoter (Figure 2A and Supplementary Table S4). Using flow cytometry analysis, the fluorescence of individual cells was then measured following induction of GFP and crRNA expression.
Figure 1.
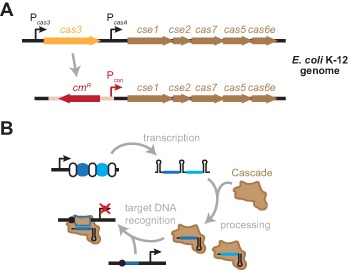
Repurposing the Type I-E CRISPR-Cas system in E. coli K-12 for programmable gene repression. (A) Conversion of the Type I-E CRISPR-Cas system into a programmable repressor. The deletion of cas3 and insertion of a constitutive promoter upstream of the Cascade operon allows crRNA-directed DNA binding without cleavage. (B) Putative mechanism of crRNA-directed gene repression. Cascade processes the transcribed CRISPR array into individual crRNAs. The Cascade–crRNA complex then binds target DNA sequences (blue line) flanked by a PAM (black circle), leading to transcriptional repression.
Figure 2.
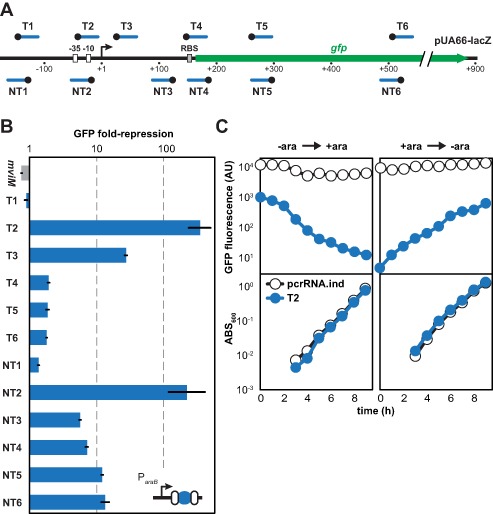
RNA-mediated transcriptional repression with the repurposed Type I-E CRISPR-Cas system in E. coli K-12. (A) Targeted silencing of plasmid-based GFP expression. The gfp gene is under the control of the lacZ promoter in the low-copy plasmid pUA66-lacZ. Each spacer sequence (blue line) and PAM (black circle) match the closest strand of the protospacer. RBS, ribosome-binding site. (B) Location-dependent and strand-dependent repression of GFP expression. BW25113 Δcas3::cat harboring the medium-copy pUA66-lacZ and the indicated single-spacer plasmid were subjected to flow cytometry analysis following induction with IPTG and L-arabinose. The non-targeting mviM spacer serves as a negative control. Repression is calculated as the ratio of the autofluorescence-subtracted fluorescence for the inducible no-spacer plasmid (pcrRNA.ind) and each single-spacer plasmid. See Supplementary Figure S3A for representative histograms from the flow cytometry analysis. (C) Reversibility of gene silencing. BW25113 Δcas3::cat cells harboring pUA66-lacZ and either the no-spacer plasmid (pcrRNA.ind, white circles) or the T2 single-spacer plasmid (T2, blue circles) either were pre-induced with only IPTG and switched to both IPTG and L-arabinose (left) or were pre-induced with both IPTG and L-arabinose and switched to only IPTG (right). Following the addition or removal of L-arabinose at t = 0, the autofluorescence-subtracted fluorescence for individual cells and the turbidity of the culture were followed over time. GFP fluorescence was ∼10-fold lower for cells with the targeting plasmid versus the spacer-free plasmid, which we attribute to leaky expression from the ParaB promoter under these growth conditions.
In comparison to the spacer-free plasmid (pCRISPR.ind; Supplementary Figure S2), we observed ranging extents of repression that depended on which region of pUA66-lacZ was targeted (Figure 2B). Targeting either strand of the promoter region strongly reduced GFP fluorescence (∼200-fold). Targeting the transcribed region moderately reduced GFP fluorescence, but only when targeting anywhere along the non-template strand or in the vicinity of the RNA polymerase footprint on the template strand (39). Interestingly, the strand bias observed when targeting the template versus non-template strand mirrored that observed for dCas9 in bacteria (13,17). As expected, targeting upstream of the promoter region negligibly reduced fluorescence. In all cases, the extent of gene silencing was uniform across the entire bacterial population (Supplementary Figure S3). Importantly, GFP levels were similar for the no-spacer plasmid and a plasmid encoding a spacer targeting the mviM gene in Salmonella enterica (Figure 2B), ruling out potential differences due to the assembly of Cascade. We also found that GFP silencing was reversible based on the change in fluorescence following addition or removal of L-arabinose (Figure 2C). The associated dynamics can be attributed to the stability of GFP similar to previous work (13).
We next performed a series of control experiments to assess the impact of deleting cas3 and constitutively expressing the Cascade operon. We first measured GFP fluorescence in the original wild-type strain in which the Cascade operon was tightly repressed and cas3 was still present (BW25113) and in a strain in which cas3 and the Cascade operon were both deleted (BW25113 ΔCRISPR-Cas::cat). The fluorescence levels were similar regardless of whether a targeting or non-targeting spacer was used (Supplementary Figure S4A), indicating that Cascade must be present for gene silencing. Next, to assess the impact on DNA integrity, we measured the transformation efficiencies for targeting and non-targeting plasmids in strains with Cascade constitutively expressed and cas3 present (NM500 cas3+) or absent (BW25113 Δcas3::cat). Surprisingly, we observed similar transformation efficiencies for the targeting and non-targeting plasmids even when cas3 was present (Supplementary Figure S4B), suggesting that Cas3 is poorly expressed or inactive in this particular strain. As further support, the strain with cas3 present could still strongly silence GFP (Supplementary Figure S4C). Finally, to gauge the impact of the resistance cassette, we excised the cassette used to delete cas3 and measured gene silencing. The resulting strain (BW25113 Δcas3) and the original strain (BW25113 Δcas3::cat) exhibited similar silencing efficiencies (Supplementary Figure S4C), indicating a negligible impact of the resistance cassette.
Impact of array length and spacer position
One beneficial feature of Cascade is that it can process multiple crRNAs from a single-spacer array. However, little is known about how the composition of natural or synthetic multi-spacer arrays quantitatively impacts individual targets. To evaluate the impact of array length, we generated arrays with one promoter-targeting spacer (T2) followed by zero to three non-targeting spacers (mviM) (Figure 3). Flow cytometry analysis revealed a gradual decrease in silencing efficiency with each additional spacer. We speculate that this decrease may be due to non-targeting spacers diluting available Cascade complexes for targeting crRNAs, as observed with other RNA-based systems (40). In support of this assertion, the single-spacer array and an array of four targeting spacers exhibited statistically indistinguishable extents of silencing (two-tailed t-test, t(4) = 1.05, p = 0.35) (Figure 3). To evaluate the impact of spacer position, we generated arrays with different permutations of one targeting and three non-targeting spacers (Figure 3). With the exception of a targeting spacer in the first position of the four-spacer array, the extent of gfp silencing was similar regardless of spacer position (one-way ANOVA, F(2,6) = 0.15, p = 0.86). These results suggest that longer arrays can reduce the potency of individual spacers, whereas the exact location of a spacer within an array has a lesser contribution to the potency of silencing.
Figure 3.
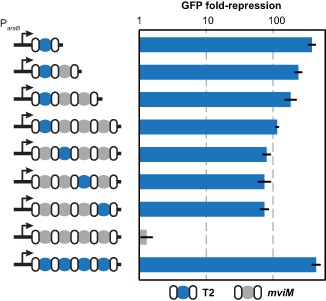
Impact of array length and spacer location on silencing efficiency. BW25113 Δcas3 cells harboring pUA66-lacZ and the indicated inducible CRISPR array plasmid were subjected to flow cytometry analysis following induction with IPTG and L-arabinose. Repression is calculated as the ratio of the autofluorescence-subtracted fluorescence for the inducible no-spacer plasmid (pcrRNA.ind) and each multi-spacer plasmid. Repeats, white ovals; T2 spacers, blue circles; non-targeting spacers matching the S. enterica mviM gene, gray circles. See Supplementary Figure S3B for representative histograms from the flow cytometry analysis. Values represent geometric mean and SEM from independent experiments staring with three separate colonies.
Multiplexed repression of endogenous genes
As a complement to targeting heterologous genes such as gfp, we explored the ability of spacers to regulate endogenous targets. We focused on operons involved in the catabolism of the sugars L-arabinose (araBAD), L-rhamnose (rhaBAD), D-xylose (xylAB) and D-lactose (lacZYA) (Figure 4A) because these operons are well characterized and are required for growth on their cognate sugar (41–44). As the araBAD, rhaBAD and lacZYA operons are disrupted in BW25113, we imported the cas3 deletion and synthetic promoter into another strain of E. coli K-12 (MG1655 Δcas3::cat). We also placed each single-spacer array under the control of the strong, constitutive promoter J23119 to circumvent the need for L-arabinose as an inducer (Supplementary Figure S2).
Figure 4.
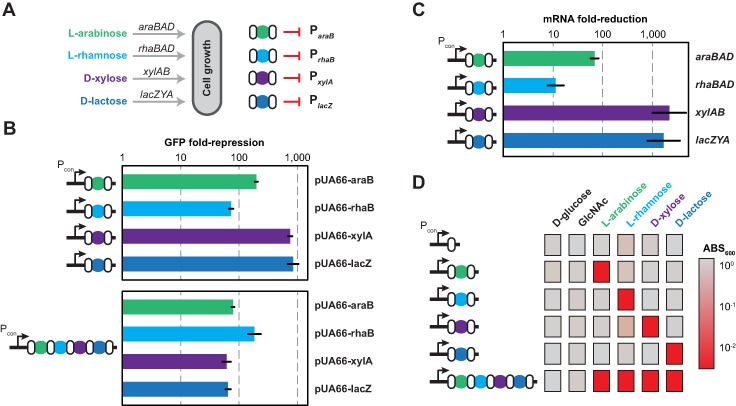
Targeted repression of endogenous genes and pathways. (A) Targeting operons responsible for sugar catabolism. Spacers were designed to target the promoter of each catabolic operon required for growth on its cognate sugar. (B) Repression of promoter activity. Each promoter was cloned upstream of the gfp gene in pUA66. The resulting plasmids were then tested in MG1655 Δcas3::cat cells harboring the corresponding single-spacer plasmid (top) or multi-spacer plasmid (bottom) by flow cytometry analysis following promoter induction with the cognate sugar. Repression is calculated as the ratio of the autofluorescence-subtracted fluorescence for the constitutive no-spacer plasmid (pcrRNA.con) and each single-spacer or multi-spacer plasmid. Values represent geometric mean and SEM from independent experiments with three colonies. See Supplementary Figure S3C for representative histograms from the flow cytometry analysis. (C) Repression of endogenous genes. MG1655 Δcas3::cat cells harboring the indicated single-spacer plasmid were harvested for total RNA following induction with the cognate sugar and subjected to qRT-PCR analysis. Repression is calculated as the ratio of the relative mRNA levels from the no-spacer plasmid (pcrRNA.con) and the indicated single-spacer plasmid. Values represent the geometric mean and SEM for quadruple technical replicates. (D) Targeted suppression of growth. MG1655 Δcas3::cat cells harboring the indicated single-spacer or multi-spacer plasmid were grown on each sugar as the sole carbon source and turbidity was measured after 24 h of growth. Values represent the geometric mean of the measured ABS600 values from independent experiments starting with three separate colonies.
To assess silencing of promoter activity, we cloned the promoter of each operon upstream of gfp in the pUA66 plasmid (Supplementary Table S5) and measured the ability of each spacer to repress its target promoter by flow cytometry analysis (Figure 4B and Supplementary Figure S3). In comparison to the spacer-free plasmid (pCRISPR.con; Supplementary Figure S2), each targeting plasmid greatly reduced fluorescence (∼80-fold to 900-fold). As expected, combining the spacers into one array strongly reduced fluorescence for each promoter (Figure 4B), although the degree of silencing was generally less than that observed for the individual spacers.
To evaluate silencing of the endogenous genes, we measured mRNA levels of each operon for cells with each single-spacer plasmid. In comparison to the no-spacer plasmid, the single-spacer plasmids greatly reduced mRNA levels (∼11-fold to 2200-fold) of the target operons (Figure 4C), paralleling that observed for the GFP reporters (Figure 4B). This wide range in repression matches the variability in gene silencing observed with dCas9 (13,17)
Finally, we explored whether targeting endogenous genes could generate defined phenotypes. Because each operon is required for the catabolism of its cognate sugar, we measured growth on each sugar as well as on two non-targeted sugars D-glucose and N-acetyl-D-glucosamine (GlcNAc). We cultured MG1655 Δcas3::cat expressing a single-spacer or four-spacer array with the different sugars as sole carbon sources and measured the turbidity of the culture after 24 h of growth (Figure 4D and Supplementary Figure S5A). We found that targeting each operon limited growth on the cognate sugar, whether using a single-spacer array or a four-spacer array. The four-spacer array silenced all target operons in individual cells, as this array limited growth in medium containing all four targeted sugars (Supplementary Figure S5A). Critically, growth was unhampered for all non-targeted sugars, supporting the specificity of targeting. The final turbidity was generally lower for all cultures grown in L-rhamnose (Figure 4D and Supplementary Figure S5A), which we attribute to L-rhamnose being a poor carbon source (Supplementary Figure S5B). We thus conclude that the Type I-E system in E. coli can be programmed to silence multiple endogenous genes and generate complex phenotypes.
DISCUSSION
We found that the Type I-E CRISPR-Cas system in E. coli can be repurposed for programmable gene repression through the deletion of cas3 and constitutive expression of the Cascade operon. An ensuing question is the extent to which this phenomenon applies to the other five Type I subtypes. Structural and phylogenetic data suggest that this same phenomenon would apply to Type I-B, I-C and I-F systems based on the stability of Cascade in the absence of Cas3 and the ability of this complex to process transcribed CRISPR arrays (3,22,27,29). Type I-A and I-D systems appear to be exceptions, as two distinct Cas3 proteins (Cas3′ and Cas3″) are required for stabilization of the Type I-A Cascade and the uncharacterized Type I-D Cascade is most closely related to that of Type I-A systems (1,45). However, these cas3 genes could be catalytically inactivated (24) as performed with Cas9 (12), albeit via point mutations that are harder to introduce with rudimentary genetic tools.
With this demonstration, another question is whether Type I systems or Type II systems should be employed for transcriptional regulation. Type II systems in the form of dCas9 are highly attractive because they offer a compact heterologous system that can be imported into diverse organisms. However, exploiting endogenous Type I systems does offer some potential advantages. For instance, once cas3 is deleted, only the CRISPR array totaling at most a few hundred bases must be introduced. Another potential advantage is that a native Type I system would be well suited for thermophilic and hyperthermophilic microorganisms that thrive in environmental conditions that would prevent proper folding of common Cas9 proteins. Type I systems also offer PAMs that are distinct from those associated with known Type II systems, including a different orientation and a bias toward T/C-rich sequences (1,23,46). Finally, Type I systems are naturally found in diverse industrially and medically relevant strains, including E. coli, Streptococcus thermophilus, Clostridium autoethanogenum and Acinetobacter baumannii (47). A drawback to this strategy is that the strains would lose immunity against some invading pathogens. Overexpression of Cascade in the absence of Cas3 may also inadvertently impact the transcriptional landscape, although this remains to be explored even for dCas9.
One interesting parallel observed for transcriptional regulation with Type I and Type II systems is the strand bias when targeting transcribed regions (Figure 2B) (13,17). Previous work with dCas9 demonstrated that targeting the non-template strand but not the template strand strongly interfered with RNA polymerase extension. We observed the same trend with the Type I-E Cascade (Figure 2B) despite structural differences and opposing PAM locations in comparison to dCas9 (21,23,48–49). Based on this parallel, we speculate that RNA polymerase extension is more sensitive to protein binding on the non-template strand rather than the particular orientation of the interfering protein or titration by the encoded mRNA. However, further investigation of the mechanisms of transcriptional repression is warranted.
An emerging concern with CRISPR technologies is the degree of off-target effects (50–53). These concerns stem from the Cas proteins accommodating mismatches between the crRNA spacer and the DNA target (5,35,48,54), potentially recognizing similar sites elsewhere in the genome. While recent genome-wide screens in human embryonic stem cells failed to detect any unintended editing events with Cas9 (55,56), off-targets would be expected to vary with the selected spacer sequence. Fortunately, using CRISPR-Cas systems for transcriptional regulation in prokaryotes would be far less likely to produce off-target effects: prokaryotes possess much smaller genomes than eukaryotes, limiting the probability of similar sequences appearing at other sites; and transcriptional repression can only occur within defined regions and strands of the genome (13,17). Accordingly, only one off-target has been reported to date for dCas9 in multiple studies in bacteria and in mammalian cells (13–15), wherein the off-target contained a recognized PAM and strong homology to the target sequence (17).
The regulatory capacity of the Type I-E system in the absence of cas3 hints at the possibility of Type I systems naturally controlling gene expression. A previous bioinformatics search for genome-targeting spacers—a potential indicator of gene regulation—identified numerous instances in natural arrays (57). The authors concluded that accidental self-targeting forced deactivation of the endogenous CRISPR-Cas system because many of the cas genes were missing. However, accidental self-targeting could also drive the loss or disruption of cas3, thereby converting the system into a gene regulator. The identification of such systems would complement the single CRISPR-Cas system known to regulate cellular processes (58,59).
In summary, our findings offer a novel strategy for exploiting an organism's native Type I CRISPR-Cas system for transcriptional regulation. In the future, we intend to explore additional Type I subtypes, broadening the scope of this method. Utilizing native Type I CRISPR-Cas systems would require elucidating the PAM, deleting cas3 and validating the functionality of Cascade (48). However, once achieved, these systems would further augment the genetic toolbox available for programmable gene regulation and offer novel approaches for genome-wide screens and strain engineering. Moreover, our findings provide a framework to identify natural Type I systems that naturally regulate gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank A. May and R. Barrangou for helpful discussions and critical reading of the manuscript. We thank S. Carrell and A. Jermusyk for technical assistance with qRT-PCR analysis and M. Shah for assistance with cloning and recombineering.
Authors' contributions: M.L.L. and C.L.B. designed the study and wrote the manuscript. M.L.L., A.S.M. and R.T.L. performed the experiments.
Disclaimer: The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
FUNDING
Kenan Institute of Engineering, Science & Technology and the National Science Foundation [CBET-1403135 to C.L.B.]; National Institutes of Health (NIH) [5T32GM008776-15 to R.T.L.]. Funding for open access charge: National Science Foundation [CBET-1403135].
Conflict of interest statement. A provisional patent has been submitted in part entailing the reported approach.
REFERENCES
- 1.Sorek R., Lawrence C.M., Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 2.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 3.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J.J., Charpentier E., Horvath P., Moineau S., Mojica F.J.M., Wolf Y.I., Yakunin A.F., et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouns S.J.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J.H., Snijders A.P.L., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Semenova E., Jore M.M., Datsenko K.A., Semenova A., Westra E.R., Wanner B., van der Oost J., Brouns S.J.J., Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl Acad. Sci. U.S.A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garneau J.E., Dupuis M.-È., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A.H., Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 8.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fineran P.C., Dy R.L. Gene regulation by engineered CRISPR-Cas systems. Curr. Opin. Microbiol. 2014;18:83–89. doi: 10.1016/j.mib.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Bikard D., Marraffini L.A. Control of gene expression by CRISPR-Cas systems. F1000prime Rep. 2013;5:47. doi: 10.12703/P5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A., et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Pinera P., Kocak D.D., Vockley C.M., Adler A.F., Kabadi A.M., Polstein L.R., Thakore P.I., Glass K.A., Ousterout D.G., Leong K.W., et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zebec Z., Manica A., Zhang J., White M.F., Schleper C. CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2014;42:5280–5288. doi: 10.1093/nar/gku161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hale C.R., Majumdar S., Elmore J., Pfister N., Compton M., Olson S., Resch A.M., Glover C.V.C., III, Graveley B.R., Terns R.M., et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jore M.M., Lundgren M., van Duijn E., Bultema J.B., Westra E.R., Waghmare S.P., Wiedenheft B., Pul U., Wurm R., Wagner R., et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 22.Nam K.H., Haitjema C., Liu X., Ding F., Wang H., DeLisa M.P., Ke A. Cas5d protein processes pre-crRNA and assembles into a Cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Struct. Lond. Engl. 1993. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westra E.R., van Erp P.B.G., Künne T., Wong S.P., Staals R.H.J., Seegers C.L.C., Bollen S., Jore M.M., Semenova E., Severinov K., et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinkunas T., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochstrasser M.L., Taylor D.W., Bhat P., Guegler C.K., Sternberg S.H., Nogales E., Doudna J.A. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc. Natl Acad. Sci. U.S.A. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson R.N., Lavin M., Carter J., Wiedenheft B. Fitting CRISPR-associated Cas3 into the helicase family tree. Curr. Opin. Struct. Biol. 2014;24:106–114. doi: 10.1016/j.sbi.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brendel J., Stoll B., Lange S.J., Sharma K., Lenz C., Stachler A.-E., Maier L.-K., Richter H., Nickel L., Schmitz R.A., et al. A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (CRISPR)-derived RNAs (crRNAs) in Haloferax volcanii. J. Biol. Chem. 2014;289:7164–7177. doi: 10.1074/jbc.M113.508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richter C., Gristwood T., Clulow J.S., Fineran P.C. In vivo protein interactions and complex formation in the Pectobacterium atrosepticum subtype I-F CRISPR/Cas system. PloS One. 2012;7:e49549. doi: 10.1371/journal.pone.0049549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiedenheft B., van Duijn E., Bultema J.B., Bultema J., Waghmare S.P., Waghmare S., Zhou K., Barendregt A., Westphal W., Heck A.J.R., et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl Acad. Sci. U.S.A. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Court D.L., Swaminathan S., Yu D., Wilson H., Baker T., Bubunenko M., Sawitzke J., Sharan S.K. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene. 2003;315:63–69. doi: 10.1016/s0378-1119(03)00728-5. [DOI] [PubMed] [Google Scholar]
- 32.Cherepanov P.P., Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 33.Zaslaver A., Bren A., Ronen M., Itzkovitz S., Kikoin I., Shavit S., Liebermeister W., Surette M.G., Alon U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 34.Afroz T., Biliouris K., Kaznessis Y., Beisel C.L. Bacterial sugar utilization gives rise to distinct single-cell behaviours. Mol. Microbiol. 2014;93:1093–1103. doi: 10.1111/mmi.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomaa A.A., Klumpe H.E., Luo M.L., Selle K., Barrangou R., Beisel C.L. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio. 2014;5:e00928–e00913. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stead M.B., Agrawal A., Bowden K.E., Nasir R., Mohanty B.K., Meagher R.B., Kushner S.R. RNAsnapTM: a rapid, quantitative and inexpensive, method for isolating total RNA from bacteria. Nucleic Acids Res. 2012;40:e156. doi: 10.1093/nar/gks680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pul U., Wurm R., Arslan Z., Geissen R., Hofmann N., Wagner R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 38.Westra E.R., Pul U., Heidrich N., Jore M.M., Lundgren M., Stratmann T., Wurm R., Raine A., Mescher M., Van Heereveld L., et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 2010;77:1380–1393. doi: 10.1111/j.1365-2958.2010.07315.x. [DOI] [PubMed] [Google Scholar]
- 39.Kovacic R.T. The 0°C closed complexes between Escherichia coli RNA polymerase and two promoters, T7-A3 and lacUV5. J. Biol. Chem. 1987;262:13654–13661. [PubMed] [Google Scholar]
- 40.Hussein R., Lim H.N. Disruption of small RNA signaling caused by competition for Hfq. Proc. Natl Acad. Sci. U.S.A. 2011;108:1110–1115. doi: 10.1073/pnas.1010082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gross J., Englesberg E. Determination of the order of mutational sites governing L-arabinose utilization in Escherichia coli B/r bv transduction with phage Plbt. Virology. 1959;9:314–331. doi: 10.1016/0042-6822(59)90125-4. [DOI] [PubMed] [Google Scholar]
- 42.Power J. The L-rhamnose genetic system in Escherichia coli K-12. Genetics. 1967;55:557–568. doi: 10.1093/genetics/55.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawlis V.B., Dennis M.S., Chen E.Y., Smith D.H., Henner D.J. Cloning and sequencing of the xylose isomerase and xylulose kinase genes of Escherichia coli. Appl. Environ. Microbiol. 1984;47:15–21. doi: 10.1128/aem.47.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beckwith J.R. Regulation of the lac operon. Recent studies on the regulation of lactose metabolism in Escherichia coli support the operon model. Science. 1967;156:597–604. doi: 10.1126/science.156.3775.597. [DOI] [PubMed] [Google Scholar]
- 45.Plagens A., Tripp V., Daume M., Sharma K., Klingl A., Hrle A., Conti E., Urlaub H., Randau L. In vitro assembly and activity of an archaeal CRISPR-Cas type I-A Cascade interference complex. Nucleic Acids Res. 2014;42:5125–5138. doi: 10.1093/nar/gku120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esvelt K.M., Mali P., Braff J.L., Moosburner M., Yaung S.J., Church G.M. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grissa I., Vergnaud G., Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuscu C., Arslan S., Singh R., Thorpe J., Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 54.Fineran P.C., Gerritzen M.J.H., Suárez-Diez M., Künne T., Boekhorst J., van Hijum S.A.F.T., Staals R.H.J., Brouns S.J.J. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc. Natl Acad. Sci. U.S.A. 2014;111:E1629–E1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith C., Gore A., Yan W., Abalde-Atristain L., Li Z., He C., Wang Y., Brodsky R.A., Zhang K., Cheng L., et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veres A., Gosis B.S., Ding Q., Collins R., Ragavendran A., Brand H., Erdin S., Talkowski M.E., Musunuru K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stern A., Keren L., Wurtzel O., Amitai G., Sorek R. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 2010;26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sampson T.R., Napier B.A., Schroeder M.R., Louwen R., Zhao J., Chin C.-Y., Ratner H.K., Llewellyn A.C., Jones C.L., Laroui H., et al. A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proc. Natl Acad. Sci. U.S.A. 2014;111:11163–11168. doi: 10.1073/pnas.1323025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sampson T.R., Saroj S.D., Llewellyn A.C., Tzeng Y.-L., Weiss D.S. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


