Abstract
Herein, we demonstrate the efficacy of an unbiased proteomics screening approach for studying protein expression changes in the KC-Tie2 psoriasis mouse model, identifying multiple protein expression changes in the mouse and validating these changes in human psoriasis. KC-Tie2 mouse skin samples (n = 3) were compared with littermate controls (n = 3) using gel-based fractionation followed by label-free protein expression analysis. 5482 peptides mapping to 1281 proteins were identified and quantitated: 105 proteins exhibited fold-changes ≥2.0 including: stefin A1 (average fold change of 342.4 and an average p = 0.0082; cystatin A, human ortholog); slc25a5 (average fold change of 46.2 and an average p = 0.0318); serpinb3b (average fold change of 35.6 and an average p = 0.0345; serpinB1, human ortholog); and kallikrein related peptidase 6 (average fold change of 4.7 and an average p = 0.2474; KLK6). We independently confirmed mouse gene expression-based increases of selected genes including serpinb3b (17.4-fold, p < 0.0001), KLK6 (9-fold, p = 0.002), stefin A1 (7.3-fold; p < 0.001), and slc25A5 (1.5-fold; p = 0.05) using qRT-PCR on a second cohort of animals (n = 8). Parallel LC/MS/MS analyses on these same samples verified protein-level increases of 1.3-fold (slc25a5; p < 0.05), 29,000-fold (stefinA1; p < 0.01), 322-fold (KLK6; p < 0.0001) between KC-Tie2 and control mice. To underscore the utility and translatability of our combined approach, we analyzed gene and protein expression levels in psoriasis patient skin and primary keratinocytes versus healthy controls. Increases in gene expression for slc25a5 (1.8-fold), cystatin A (3-fold), KLK6 (5.8-fold), and serpinB1 (76-fold; all p < 0.05) were observed between healthy controls and involved lesional psoriasis skin and primary psoriasis keratinocytes. Moreover, slc25a5, cystatin A, KLK6, and serpinB1 protein were all increased in lesional psoriasis skin compared with normal skin. These results highlight the usefulness of preclinical disease models using readily-available mouse skin and demonstrate the utility of proteomic approaches for identifying novel peptides/proteins that are differentially regulated in psoriasis that could serve as sources of auto-antigens or provide novel therapeutic targets for the development of new anti-psoriatic treatments.
One in three individuals in the United States is afflicted with a skin disease, with ∼2–3% of the American population suffering from psoriasis (1–3) a chronic, immune-mediated inflammatory skin disease characterized by well-demarcated areas of “involved” red, raised, and scaly skin adjacent to areas of “uninvolved” normal appearing skin. The underlying cause of psoriasis remains unknown and the specific signals that trigger disease onset have yet to be identified; however, several lines of evidence suggest the involvement of antigen-specific T cells, although the antigens involved remain elusive (4). A combination of human and animal studies have led to the understanding that in patients with a genetically susceptible background, some initiating stimulus, often a stressful event, an injury to the skin, or an infection, leads to a coordinated series of signaling events involving cytokines, resident skin cells, and skin-infiltrating immune cells, that once started, initiates a vicious pro-inflammatory hyperproliferative cycle. Once initiated, this cycle perpetuates sustained inflammatory responses. Intervention at several points in this cycle results in clinical resolution, however, durable remission and/or permanent clearance has not yet been achieved.
Current psoriasis therapies are directed toward symptomatic relief and none of them represent a cure for this chronic illness. Current treatments include topical therapies, phototherapy, and systemic administration of immune-suppressants, anti-metabolites, oral retinoids, and biologics targeting immune cells or inflammatory cytokines (5, 6). Many of the most effective therapeutics however, also have the greatest adverse reactions; moreover, psoriasis can become resistant to specific therapies over time. Therefore, an ongoing need for discovery of new biological pathways and targets for psoriasis is obvious.
We studied a mouse model of psoriasis using an unbiased proteomics screening approach to identify dysregulated peptides of interest in psoriasiform-inflamed mouse skin and ultimately compared these findings to psoriasis patient skin. We used in-gel label-free protein expression analysis to observe quantitative changes in protein expression. The peptides that were determined to be top-scoring from a statistical perspective and of biological interest were subjected to further analysis and confirmation using a targeted mass spectrometry approach along with qRT-PCR to assess gene expression in a distinct set of animal samples. Further validation of the translational importance of these novel proteins was then conducted in primary keratinocytes expanded from psoriasis skin as well as human skin taken directly from psoriasis patient lesional and nonlesional areas. The results of these experiments confirm the ability of a discovery in-gel label-free expression model to identify proteins that are in the moderate to high abundance range that are significantly different in their distribution between control and genetically modified psoriasiform mouse skin and demonstrate the usefulness of mouse models and proteomic approaches for identifying novel proteins that are differentially regulated in human psoriasis, providing future biomarkers and targets for development of translational approaches to disease improvement.
EXPERIMENTAL PROCEDURES
Mice
Adult KC-Tie2 mice (C57Bl/6) and littermate controls (n = 3 each) were euthanized and dorsal skin harvested as previously described for use in the label-free expression discovery experiments (7). A second distinct group of mice (n = 8 each) was used for validation at the RNA and protein levels. All animal protocols were approved by the Case Western Reserve University Institutional Animal Care and Use Committee and conformed to the American Association for Accreditation of Laboratory Animal Care guidelines.
Sample Preparation for Gel-based Label-free Expression Discovery
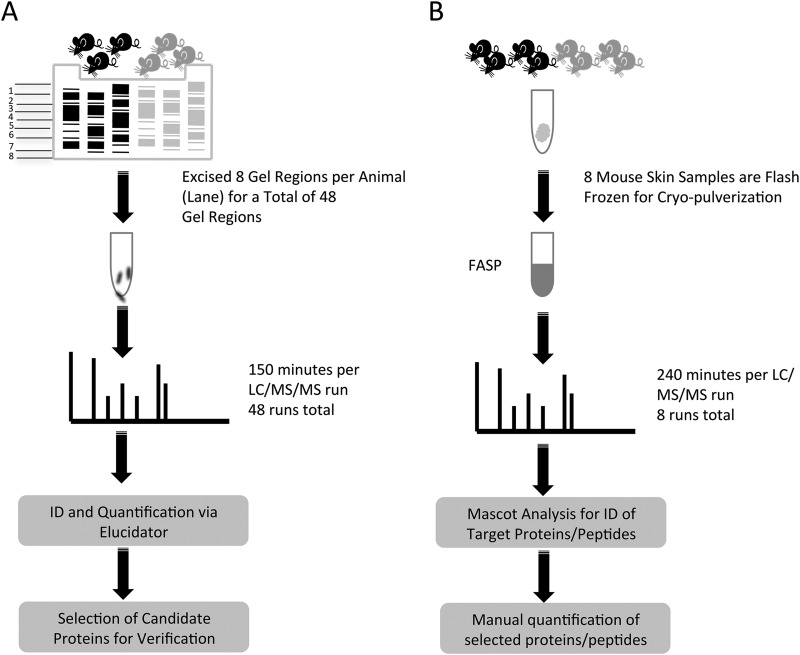
Tissue protein samples that had been isolated and prepared as previously described (7) were thawed and total protein concentrations were determined using a Bio-Rad Protein Assay as described by the manufacturer. Samples were reduced for 15 min at 80 °C using DTT with a final concentration of 10 mm. The samples were then cooled to room temperature and alkylated with iodacetamide for 30 min with a final concentration of 25 mm. Fig. 1 summarizes the gel-based workflow used. Briefly, 30 μg of each sample were loaded per well of a 1D-SDS-PAGE gel (4–20% Tris-HCl). The gel was stained using Bio-Rad Biosafe Coomassie (BioRad Laboratories, Hercules, CA) and then cut into eight distinct regions based on apparent protein molecular weight (supplemental Fig. S1).
Fig. 1.
Workflow summaries for the gel based label-free discovery and verification of target proteins approaches. Gel based label-free discovery workflow: Lysates from control and KC-Tie2 mice were separated by 1D SDS-PAGE. Eight gel regions per sample were excised resulting in 48 total regions. Each region was digested with trypsin individually and analyzed for a total of 48 LC/MS/MS runs. Proteins and peptides were identified and quantified using Rosetta Elucidator software. This analysis yielded candidate proteins that were selected for verification A. Verification of candidate proteins workflow: Tissue from four control and 4 KC-Tie2 mice was flash frozen in liquid nitrogen and then cryo-pulverized in preparation for verification of identified candidate proteins. The pulverized tissues were then subjected to FASP and analyzed individually by LC/MS/MS. Raw data files were searched using Mascot and target proteins were identified for verification. Peptides from elected candidate proteins were manually quantified and fold-changes were determined, B.
Isolated gel regions were washed using 25 mm ammonium bicarbonate (ABC; Fisher Scientific, Fairlawn, NJ) followed by 50% acetonitrile (ACN; Burdick and Jackson, Morristown, NJ) twice. Then the gel regions were crushed and proteolytic digestion was performed using 200ng of bovine trypsin (Promega, Madison, WI) for 18 h at 37 °C. Digests were then removed and stored at −80 °C until needed. The remaining gel pieces were washed with 25 mm ABC followed by 50% ACN three times to extract any remaining peptides. These supernatants were combined with the original digests and dried completely via speed vac. Resulting peptides were then stored at −80 °C for LC-MS/MS analysis and resolubilized in 11 μl of 0.1% formic acid (Acros, Geel, Belgium) for LC/MS/MS analysis.
Quantitative (q)RT-PCR Verification on Mouse Skin
RNA was isolated from the second, independent group of KC-Tie2 and control littermates (n = 8 each) and qRT-PCR performed according to the manufacturer's instructions and as described previously (13) using the following primer-probe sets Rab18 (Mm00441057_m1), Serpinb3b (Mm03032256_uH), Stefin A1 (Mm01973758_gH), KLK6 (Mm00478322_m1), and Slc25a5 ADP/ATP translocase 2 (Mm00846873_g1; Applied Biosystems, Grand Island, NY).
Sample Preparation for Solution-based Label-free Expression Verification
Skin adjacent to that used for RNA isolation and analyses described above was collected and used to perform a second solution-based label free expression assay. KC-Tie2 and control mouse skin (n = 4 each) was placed into ice-cold 25 mm Tris, pH 8.5, containing protease inhibitor mixture (Sigma Aldrich, St. Louis, MO), and thoroughly rinsed to remove any remaining blood from the tissue. Fig. 1 highlights the solution-based workflow. The tissue was transferred to a cryo bag (Covaris, Woburn, MA), capped, and frozen in liquid nitrogen prior to being pulverized. Once pulverized, each sample was again frozen in liquid nitrogen and subjected to an additional round of pulverization. From each sample, ∼10 mg was transferred to a 1.5 ml tube and 300 μl of 3% SDS was added. Pulverized tissue was then incubated on ice for 30 min. Aliquots were sonicated at 50% amplitude followed by vortexing; this cycle was repeated four times with samples sitting on ice between each round. SDS detergent removal and reduction and alkylation was performed using the Filter Assisted Sample Preparation method (FASP) (8). Total protein concentration was determined using a modified Bradford assay (Bio-Rad Laboratories, Hercules, CA), and 10 μg of protein was digested as previously described (9).
Liquid Chromatography and Mass Spectrometry for Discovery and Verification
LC/MS/MS was performed using a Waters ultra high-pressure liquid chromatography NanoAcquity (Waters Corporation, Milford, MA), an LTQ Orbitrap Velos, and an Orbitrap Elite (ThermoFisher Scientific, Waltham, MA). The order of sample injections was randomized across all samples. The instrument was mass calibrated immediately before the analysis using the instrument protocol. Mobile phase A (aqueous) contained 0.1% formic acid in 5% ACN and mobile phase B (organic) contained 0.1% formic acid in 85% ACN. Samples were trapped and desalted on-line in mobile phase A at 10 μl/min for 10 min using a Waters UPLC PST C18 nanoACQUITY 300 (75 μm × 25 cm) reversed phase column with 5% mobile phase B. Gel based discovery separation was obtained by employing a gradient of 6–28% mobile B at 0.300 μl/min over 150 min. The column was washed at 99% mobile phase B for 10 min, followed by a re-equilibration at 100% A for 15 min. Positive mode electrospray was conducted using a nanospray source and the mass spectrometer was operated at a resolution of 60,000. Quantitative and qualitative data were acquired using alternating full MS scan and MS/MS scans in normal mode. Survey data were acquired from m/z of 400 to 1600 and up to 20 precursors based on intensity were interrogated by MS/MS per switch. Two micro scans were acquired for every precursor interrogated and MS/MS was acquired as centroid data. All mass spectrometry analytical parameters for the verification samples were the same as the discovery samples with exception of the LC/MS/MS analytical run time that was extended to 240 min. For the gel based discovery study, excised gel regions for each animal were processed and analyzed as a separate LC/MS/MS for a total of 48 individual LC/MS/MS runs. One LC/MS/MS run was acquired for each animal in the verification set for a total of eight LC/MS/MS runs. The discovery set was processed via Rosetta Elucidator (Rosetta Biosoftware, Seattle, WA) (Version 3.3.01 SP4 25). The MS/MS peak lists were subsequently searched by Mascot (version 2.2.0, IPI_mouse_06_2010) (Matrix Science, London, UK). The database used was mouse International Protein Index (IPI) (56,957 sequences). Search settings were as follows: trypsin enzyme specificity; mass accuracy window for precursor ion, 10 ppm; mass accuracy window for fragment ions, 0.8Da; variable modifications including carbamidomethlylation of cysteins, one missed cleavage, and oxidation of methionine. The false discovery rate for these settings was calculated to be 1.13% for the discovery set. Automated differential quantification of peptides was performed with Rosetta Elucidator as previously described (10, 11). The processing workflow that was used for differential quantification in the discovery set is shown in supplemental Fig. S2. LC/MS/MS raw data was imported and for each MS spectrum profile of each LC/MS/MS run, chromatographic peaks and monoisotopic masses were extracted and aligned. Specifically, chromatographic peaks were first aligned by retention time and monoisotopic mass across each sample by gel region. Next, intensities of monoisotopic masses found in multiple gel regions (within a RT tolerance) of a sample were summed. Peaklists with the monoisotopic mass and corresponding MS/MS data were then generated for each sample and searched using Mascot. Resultant peptide identifications were imported into Elucidator and monoisotopic masses annotated with peptide identifications. Statistical analysis of the dataset was performed using a Student's t test and peptides from the same protein were summed together to provide fold changes for each protein identified. The verification dataset was searched using the same database and search engine parameters as described above with a false discovery rate of 3.7% for the verification set. Peak areas from peptides of targets that were identified via the Mascot database search were subsequently determined by manual extraction and integration using the Qual browser in the Xcalibur software (version 2.2) (ThermoFisher Scientific, Waltham, MA). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (12) via the PRIDE partner repository with the dataset identifier PXD001387 and 10.6019/PXD001387.
Psoriasis Patient Population
Seven individuals with chronic moderate to severe plaque psoriasis and six healthy controls were recruited (age range 18–75 years) with inclusion criteria including the presence of one or more well-demarcated, scaly, erythematous psoriatic plaques not limited to the scalp. Individual patients had no systemic anti-psoriatic treatments for 4 weeks and no topical treatments for 2 weeks prior to skin biopsy. Biopsy sites varied between psoriasis patients depending on location of active plaques, whereas biopsies of uninvolved and control skin were taken from the buttocks.
Informed consent was obtained from all subjects under protocols approved either by the Institutional Review Board of the University of Michigan or University Hospitals Case Medical Center/Case Western Reserve University. Moreover, all aspects of the study were conducted in compliance with good clinical practice and according to the Declaration of Helsinki Principles.
Verification on Human Psoriasis Skin
RNA was isolated from harvested skin and qRT-PCR performed according to the manufacturer's instructions and as described previously (13) using the following primer-probe sets KLK6 (Hs00160519_m1), Slc25a5 (Hs00854499_g1), Rab18 (Hs00222021_m1), Cystatin A (CSTA; Hs00193257_m1), and SerpinB1 (Hs00961948_m1; all Applied Biosystems, Grand Island, NY).
Immunohistochemistry was completed on control human skin and psoriasis patients' uninvolved and involved lesional skin using primary antibodies targeting human KLK6 (10 μg/ml; polyclonal, goat IgG; R&D, Minneapolis, MN), human Slc25a5 (10 μg/ml, polyclonal, rabbit IgG; Proteintech, Chicago, IL), Cystatin A (0.5 μg/ml, clone WR-23/2/3/3, mouse IgG; Abcam, Cambridge, MA), SerpinB1 (1 μg/ml, polyclonal, rabbit IgG; Sigma-Aldrich), and Rab18 (5 μg/ml, polyclonal, rabbit IgG; Novus Biologicals, Chicago, IL) following manufacturer's instructions.
Primary Human Keratinocytes
Control and psoriasis primary human keratinocyte (KC) cultures were established from adult skin (n = 5 each) and grown to confluence as previously described (13–16). RNA was isolated (RNeasy, Qiagen, Germantown, MD), reverse transcribed (High capacity RNA transcription kit, Applied Biosystems), and qRT-PCR performed (Applied Biosystems, Grand Island, NY) as described previously (13) using the same human primer-probe sets described above.
RESULTS
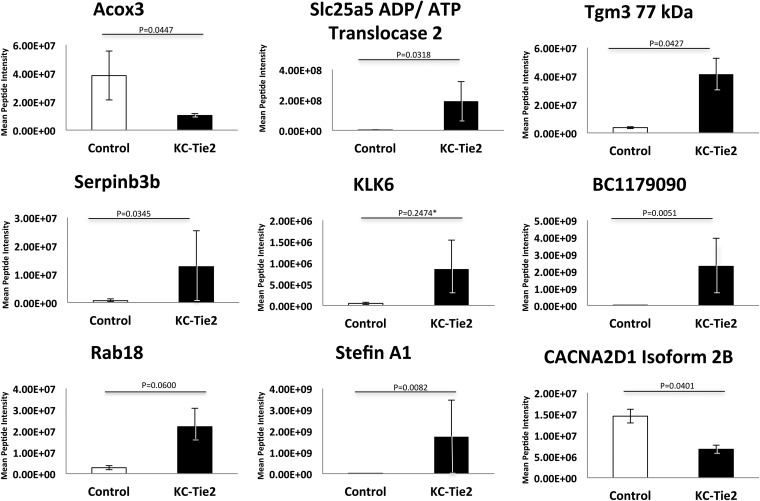
To identify differentially expressed peptides in KC-Tie2 mouse skin (n = 3) compared with littermate controls (n = 3), we used a gel-based label-free expression approach to reduce sample complexity by fractionation of the sample by molecular weight over eight gel regions (Fig. 1). The regions were selected to differentially fractionate multiple abundant skin proteins, such as albumin, keratin, and myosin. This allowed us to probe proteins of lower abundance, which would otherwise have been difficult to detect in a combined sample. This protocol resulted in high reproducibility across all samples (supplemental Fig. S1), with good separation of abundant proteins. Peptides were quantified using label-free expression analysis by determining the intensity of the precursor ion and its retention time across each sample. The isotopic pattern was integrated and a charge state was assigned to produce a unique signal for each peptide. Intensities of peptides shared by the same protein were then summed and a Student's t test was performed to identify peptides whose expression changed between control and KC-Tie2 mice. Overall, 5482 peptides mapping to 1281 proteins were identified (supplemental Table S1 and http://proteomics.case.edu/LabelFreePsoriasis/). Of these, 105 proteins had peptides that exhibited intensity changes significant at the level of p < 0.05 with overall protein fold-changes greater than 2.0 (supplemental Table S2 and http://proteomics.case.edu/LabelFreePsoriasis/). Of the 105 differentially expressed proteins, 14 had fold-change values reflecting a decrease in expression, whereas 91 were increased (supplemental Table S2). The proteins showing most significance (i.e. smallest p values) included: Stefin A1, Slc25a5 ADP/ATP translocase 2, Hsd17b10 3-hydroxyacyl-CoA dehydrogenase type-2, Serpinb3b, Tgm3 77 kDA protein, and BC117090 cDNA sequence BC1179090, while decreases in Cacna2d1 Isoform 2B of voltage-dependent calcium channel subunit alpha-2/delta-1 and acox3 peroxisomal acyl-coenzyme A oxidase 3 were observed. Although the calculated p value of KLK6 was not significant (p = 0.2474), because of biological interest and the overall fold change it was included as a protein target in our verification study (Fig. 2).
Fig. 2.
Discovery study using Rosetta Elucidator identifies highly significant differential protein expression between KC-Tie2 and control mouse skin. Proteins showing the highest levels of significance between control (n = 3) and KC-Tie2 (n = 3) skin include increases in Stefin A1, Slc25a5 ADP/ATP translocase 2, Serpinb3b, Tgm3 77 kDA protein, and BC117090 cDNA sequence BC1179090 and decreases in Cacna2d1 Isoform 2B of voltage-dependent calcium channel subunit alpha-2/delta-1, acox3 peroxisomal acyl-coenzyme A oxidase 3. * The calculated p value of KLK6 was not significant (p = 0.2474). However, because of biological interest and the overall fold change it was included as a protein target in our verification study.
Proteins were selected for verification based upon either statistical or biological significance or correlation with microarray data (17–20) and included murine Stefin A1 (average fold change 342.4; average p = 0.0082; cystatin A, human ortholog); Solute Carrier Family 25 (Mitochondrial Carrier; Adenine Nucleotide Translocator), Member 5 (average fold change 46.2; average p = 0.0318; Slc25A5); Serpinb3b (average fold change 35.6; average p = 0.0345; serpinB1, human ortholog); Ras-related protein (average fold change 4.9; average p = 0.0600; Rab18), and Kallikrein related peptidase 6 (4.7-fold change; average p = 0.2474; KLK6). In KC-Tie2 mouse skin, verification approaches included examining expression changes using qRT-PCR and performing label-free targeted mass spectrometry analysis. The latter was pursued to verify protein expression changes for two reasons. First, we had identified detectible peptides with favorable ionization efficiency in the discovery analysis and second, we were limited by commercially available antibodies suitable for Western blotting, therefore, an alternative MS-based approach was necessary. These confirmatory experiments were followed up by examining changes in protein and gene expression of the identified targets in human psoriasis samples.
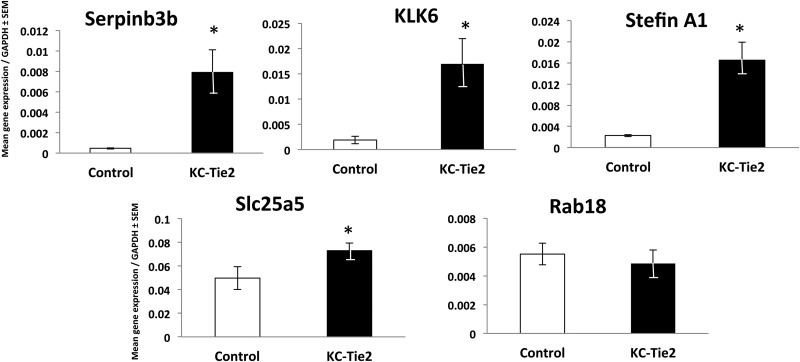
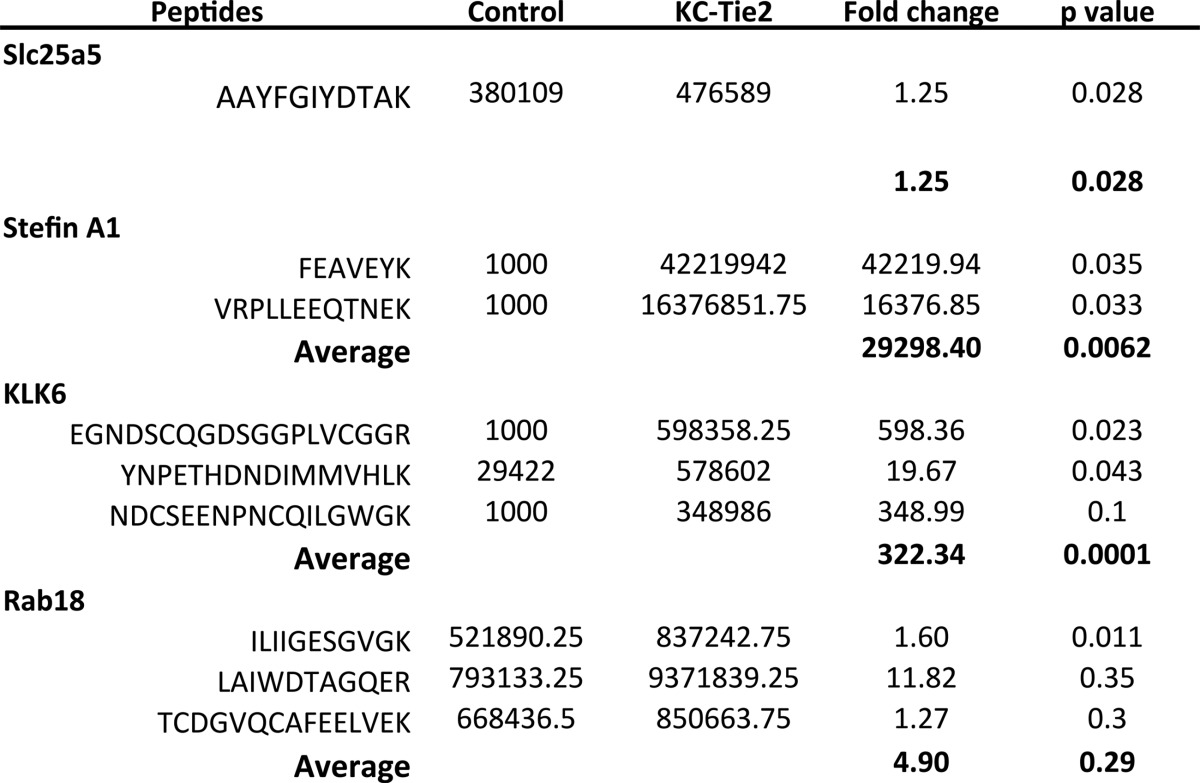
Using a distinct group of mice (n = 8 each group), we performed qRT-PCR on the identified targets of interest, and confirmed significant increases in four of the five identified targets, including Serpinb3b (17.4-fold, p < 0.0001), KLK6 (9-fold, p = 0.002), Stefin A1 (7.3-fold; p < 0.001), Slc25A5 (1.5-fold; p = 0.05), and no change in expression of Rab18 (Fig. 3). Additionally, from a subset of these mice (n = 4), newly isolated protein was subjected to LC/MS/MS analysis using the FASP protocol for detergent cleanup as opposed to the gel-fractionation method, to verify the targets identified in the initial screen and confirmed as being increased at the RNA level. Peptides were quantified in all samples for the verification analysis. Of the five proteins selected for verification, solution- based label-free analysis identified one peptide for ADP/ATP translocase 2 (Slc25a5), two peptides for Stefin A1, three peptides for KLK6 serine protease, and three peptides for Rab18 (Table I). Overall, the solution-based method identified 1544 proteins (6775 peptides). This total was larger than the number seen in the gel-based method (1281 proteins, 5482 peptides) presumably because of two factors; first, longer gradients were used in the solution-based LC/MS; and second, the Orbitrap Elite was used as the MS instrument rather than the Velos. Nonetheless, comparable numbers of peptides for the proteins selected for verification were observed with a total of nine identified in the solution-based verification and 12 identified in the gel-based discovery dataset. We observed an increase in protein coverage for KLK6 using the solution based method with three peptides identified in the verification and one identified in the initial discovery analyses. However, we were unable to detect any peptides specific to Serpinb3b using the solution-based workflow. Overall, there was a 50% overlap in peptide identifications for target proteins between the discovery and verification analyses with six of the initial 12 peptides identified in the verification. Using the peptide intensities we detected in the verification assay to infer protein changes, we observed increases in the average protein intensity of KC-Tie2 psoriasiform animals and control mice to be ∼1.3-fold for Slc25a5, ∼29,000-fold for Stefin A1, and ∼322-fold for KLK6. Rab18 showed an increase but it was not significant (4.9-fold; Table 1).
Fig. 3.
Candidate molecules identified from the gel-based label-free expression experiments are verified at the RNA level in control and KC-Tie2 mouse skin using qRT-PCR. qRT-PCR results for control (n = 8) and KC-Tie2 (n = 8) mice verified significant increases in Serpinb3b, KLK6, Stefin A1, and Slc25a5. No significant change in gene expression of Rab18 was observed (* p < 0.05).
Table I. Candidate molecules identified from the gel-based label-free expression experiments are verified at the protein level in control and KC-Tie2 mouse skin. Fold- changes are computed for peptides identified in control and KC-Tie2 mice of candidate proteins. An imputed intensity value of 1000 was used in samples for which the target peptide mass was not observed.
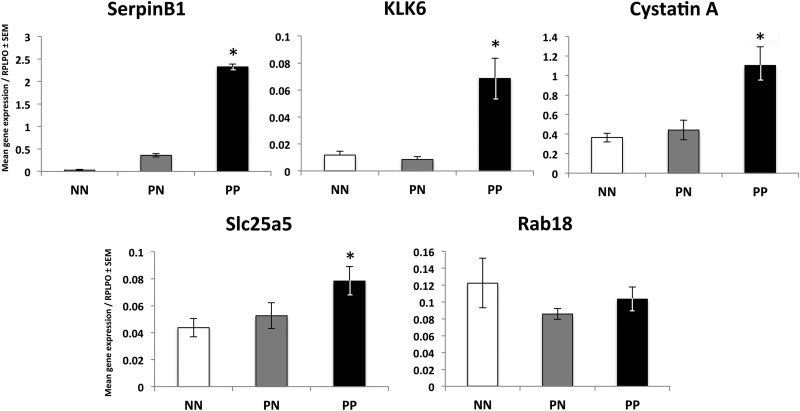
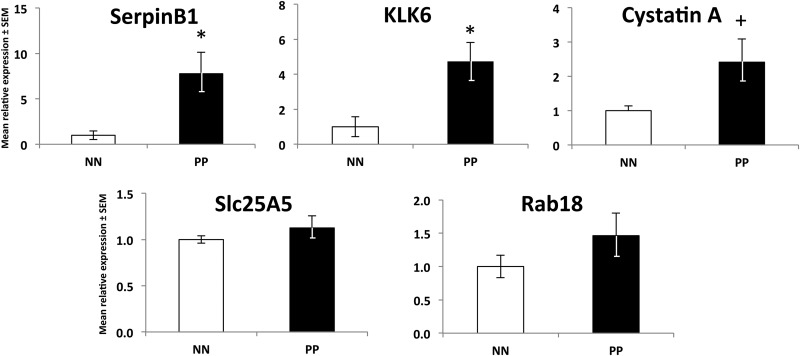
To emphasize the utility of a preclinical mouse model and our label-free proteomic screening approach in the identification of potential novel targets directly translatable to human psoriasis, we next examined the expression of our five target molecules in psoriasis patient skin and primary keratinocytes expanded from patient skin. In accord with our observations in mouse skin, we found significant increases (p values all < 0.05) in gene expression between healthy controls (NN)1 and involved lesional psoriasis skin (PP) for SerpinB1 (76-fold), KLK6 (5.8-fold), Cystatin A (3-fold), and Slc25a5 (∼1.8-fold) but no significant change in Rab18 (Fig. 4). These increases in gene expression were also observed in primary keratinocytes expanded from involved lesional psoriasis skin, where we observed a ∼7.8-, ∼4.7-, and ∼2.4-fold increase in SerpinB1 (p = 0.02), KLK6 (p = 0.02), and Cystatin A (p = 0.07), respectively (Fig. 5) compared with control keratinocytes expanded from skin biopsies obtained from individuals without skin disease. No significant change was observed for Slc25a5 or Rab18.
Fig. 4.
Translation to human psoriasis verifies the increases in candidate molecules identified using proteomic assessment of the KC-Tie2 mouse model. Using qRT-PCR we confirmed significant increases in mRNA for SerpinB1 (the human ortholog of Serpinb3b), KLK6, Cystatin A (the human ortholog of Stefin A1), and Slc25a5; and a lack of change in Rab18 between healthy control skin (NN) and involved lesional psoriasis skin (PP; *p < 0.05). No significant differences were observed between nonlesional psoriasis skin (PN) and NN skin.
Fig. 5.
Human psoriatic keratinocytes express higher levels of candidate gene transcripts compared with healthy control keratinocytes. Primary keratinocytes were isolated and grown from skin of healthy controls (NN; n = 5) and involved lesional psoriasis skin (PP; n = 5) and gene expression measured and normalized to RPLPO. Significant increases (*p < 0.05) were observed for SerpinB1 and KLK6 and modest increases in Cystatin A (+ p = 0.07) indicative of keratinocytes serving as a key cellular source of each molecule. No increases were observed for Slc25a5 and Rab18.
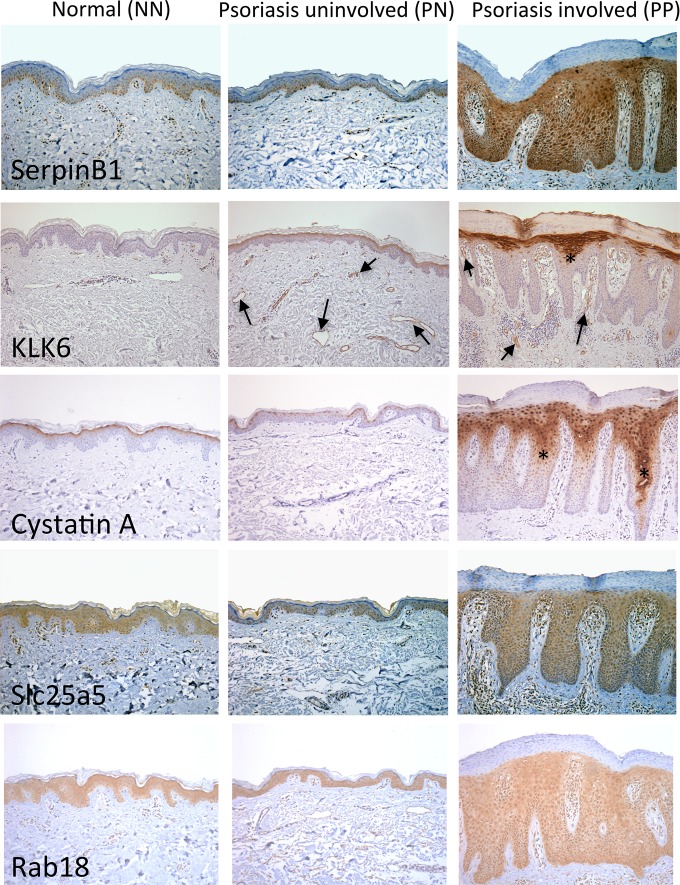
The final critical confirmation of changes in these proteins in psoriasis patient skin was completed using skin biopsies and in situ immunohistochemical (IHC) staining approaches. All five proteins were increased in lesional (PP) psoriasis skin compared with normal skin (NN); each with a unique staining pattern (Fig. 6). SerpinB1 was expressed in normal skin epidermis by basal keratinocytes and peri-vascular cells and this expression did not change between NN and nonlesional psoriasis (PN) skin. In PP skin, staining was increased in all layers of the proliferating epidermis and was no longer confined to the basal layer. KLK6 levels were very low in NN and PN skin; however, modest increases in KLK6 staining was seen in the endothelium of dermal blood vessels (arrows in Fig. 6) in PN skin; and this staining increased in the dermal vasculature of PP skin, specifically between the rete pegs (arrows in 6); KLK6 staining was strongly evident in the upper layers of keratinocytes in the epidermis. Cystatin A IHC in NN skin labeled the most apical keratinocytes in the epidermis, confined to a single layer of cells at the outer most terminally differentiated region. Similar staining was observed in PN skin. In PP skin, increased numbers of keratinocytes stained positive for Cystatin A in the apical layers of the epidermis and with greater intensity, and staining was also seen in keratinocytes deep in the rete pegs (asterisks in Fig. 6). Slc25a5 was expressed in the epidermis of healthy skin by keratinocytes and this level did not change between NN and uninvolved nonlesional psoriasis (PN) skin. In PP skin, Slc25a5 was increased in the proliferating epidermis and was also evident in infiltrating immune cells abundant in the inflamed dermis. Rab18 was present in epidermal keratinocytes and blood vessel endothelial cells in healthy skin, and levels remained unchanged in uninvolved nonlesional psoriasis (PN) skin. Staining intensity remained consistent in involved lesional skin, confined to keratinocytes and vascular cells, however, protein levels appeared to increase, perhaps reflective of the presence of more keratinocytes and blood vessels in lesional psoriasis skin (Fig. 6). Non-specific staining related to antibody host IgG was controlled by performing separate IgG control staining at matching concentrations and development times for each primary antibody (supplemental Fig. S3).
Fig. 6.
Candidate proteins identified from gel-based label-free expression analyses of KC-Tie2 mouse skin are increased in lesional psoriasis patient skin. Immunohistochemistry of SerpinB1, KLK6, Cystatin A, Slc25a5, and Rab18 in normal control skin (NN), and uninvolved (PN) and lesional involved (PP) psoriasis skin reveals increases in protein staining in PP skin. Arrows indicate blood vessel staining; asterisks' indicate staining deep in the rete pegs.
DISCUSSION
We utilized a gel-based label-free MS proteomic approach to fractionate and identify differentially expressed proteins in the skin of an established mouse model of psoriasis, the KC-Tie2 mouse (7, 21). We identified 5482 peptides that mapped to 1281 proteins, of these, 105 proteins were either increased or decreased by more than twofold in KC-Tie2 skin. We then verified increases in serpinb3b, KLK6, stefin A1, and Slc25A5 at the mRNA and protein levels using a second cohort of mice. Importantly, we confirmed these proteins were increased in lesional skin from psoriasis patients, demonstrating the ability and translational relevance of identifying novel proteins in a mouse model and then confirming these changes in human disease. This approach permitted an unbiased comparison of over 1200 skin proteins using readily available mouse skin; and demonstrates the utility of proteomic approaches for identifying proteins of interest in human disease using a bench-to-bedside approach. Further proteomics analysis of similarly prepared samples could assess protein response following anti-psoriatic treatments well as changes in potential post-translational modifications significant to disease.
Previous proteomic approaches for studying skin have used both human and mouse tissue (22–24) and have examined chronic plaque psoriasis (25–27). An early manuscript (24) utilized MALDI-TOF to study skin from Balb/c and C3H/HeN mice in order to identify proteins from different skin regions and different skin compartments. They identified 28 proteins, 25 of which were expressed in the epidermis where the majority of proteins identified included keratins (K10, K14, and K15), albumins, and myosins; abundant essential proteins of the epidermal compartment. They failed to identify any integral membrane proteins or signaling molecules and attributed this to solubility constraints and difficulties with extraction, as well as the inability to detect less abundant proteins, membrane associated proteins, and proteins with extreme pI values based on their 2D approach. More recent studies (22) sought to examine differences between human leg and breast skin. Using a combination of laser microdissection and MS, they identified differences in protein content based upon anatomical location. An abundance of collagens, keratins, and cellular proteins similar to those previously reported were identified and were typically related to the extracellular matrix. Eleven highly expressed proteins were identified in leg skin that were absent from breast tissue, including two collagen isoforms, an isoform of elastin, annexin A5, desmoglein 1, histone H2A.V, tubulin beta-2C chain, serum amyloid P-component, an isoform of fibrinogen alpha chain, and several uncharacterized proteins. Little difference in the proteomes between the reticular and papillary dermis were observed (22).
Proteomics approaches have also been used to study and identify differentially expressed proteins between healthy skin and nonlesional and/or lesional psoriasis skin. An initial study (23) examined fluid proteins collected from clinically induced suction blisters from normal and nonlesional psoriasis skin following intradermal IL-1β exposure using 2D gel electrophoresis (PAGE) proteomic profiling (MALDI-TOF). Only nine proteins were identified as different between one psoriasis patient and one healthy control, and included ceruloplasmin precursor, apolipoprotein A1, vitamin D binding protein (DBP), haptoglobin, and endoplasmic reticulum luminal protein 28. More recent work (26), also attempting to identify differential expressed proteins in psoriasis skin, modified their tissue collection approach to reduce the cellular and proteome complexity. Using a keratome approach, they focused on isolating epidermal tissues, using the logic that decreasing sampling depth (they only went to 0.5 mm depth) would reduce the cellular and proteome complexity. Albeit true, this approach also biases the proteins that will be identified, and because psoriasis is an immune-centric disease, this approach may not necessarily identify proteins critical to disease pathogenesis. Moreover, they also enriched the “secretome” by short-term culture of dissociated tissue to enhance their analysis. Stable isotope dimethyl labeling was then used to identify 128 proteins, 59 of which were ranked as possibly important based on their prior scaling system; including 22 proteins that previously had an association to psoriasis, including FABP, S100A7, CRABP2, and IL-1F9, and 36 proteins which had not been described in psoriasis skin, including gasdermis A, PA2G4, CRB1, and CRB3. One additional group has reported protein differences in psoriasis versus healthy controls using a proteomic approach; Ryu and colleagues (25) used 2D LC/MS/MS approach to examine nonlesional and lesional psoriasis skin and identified 74 and 145 differently expressed proteins compared with healthy control skin (respectively) and grouped these broadly using GO and IPA analytic tools. The peptides identified were all epidermal derived, with many redox-specific peptides reported. Of particular relevance to the results we present here, was the identification across many studies of psoriasis-associated increases in serine protease inhibitors, Serpins (25–27).
Multiple -omic based approaches for studying differences in psoriasis patient skin and serum are widely reported, and include cytokine profiling, transcript array analyses, and RNAseq; however, very few studies have examined psoriasis at the proteome level. In addition to those mentioned above, a study evaluating acute guttate versus chronic plaque psoriasis (28) as well as several original papers studying protein expression differences from epidermal keratinocytes from involved psoriasis skin (29, 30) have appeared; however, global proteomic analyses of psoriasis skin is still rather uncommon compared with the abundant gene expression based approaches (see a meta analyses of genomics approaches (20)).
Our approach utilized a well-established mouse model of psoriasis, providing readily available and abundant levels of full-thickness skin for sensitive nano-proteomic analyses. We identified more than 1200 proteins with differentially expressed peptides many with biological functions of interest in psoriasis; from these over 100 proteins were changed >twofold comparing lesional to healthy skin. Both gel- and solution-based methods (Fig. 1) were effective for probing a wide range of proteins with excellent relative quantification of peptide isoforms in a label-free workflow. An efficient detergent removal approach, coupled with long liquid chromatography gradients using ultra-high pressure liquid chromatography, was effective for probing the skin proteome. For coverage beyond 1200–1500 proteins identified in this study, a combination of the fractionation-based method and long gradients using the FASP protocol would likely be recommended. However, each method showed acceptable quantification variances suitable for both proteomics discovery and targeted validation of proteins of interest.
Increases in serpins, cystatin, and kallikreins, representing biological proteases or protease inhibitors known to contribute to inflammation and to inflammatory skin diseases, including Netherton syndrome, atopic dermatitis, rosacea, and others were identified in the discovery set. Our ability to identify increases initially in mouse skin, and then verify the increase in psoriasis lesional skin from patients, provides further proof of the translational ability of proteomics and takes better advantage of access to human skin, which can be difficult to obtain in sufficient quantity, and specific to a diseased state, such as psoriasis, for a full proteomic screen with accurate quantitation.
Recent findings have identified key functions for proteases (like KLK6, cathepsins), protease-inhibitors (serpins, cystatin) and protease targets (keratinocytes, endothelial cells, processing of fillagrin, desquamation, antimicrobial defense, activation of receptors, and cytokines) in inflammatory skin diseases (for review see (31)). Serine protease inhibitors (SERPINS) are involved in epidermal permeability barrier homeostasis; serpinB1 (serpinb3b in mouse) is a protease inhibitor and is known to be anti-apoptotic (31). Cystatin A is known to have antimicrobial properties, is an epidermal protease and a skin barrier gene and is an early differentiation marker of keratinocytes. Interestingly, cystatin A is one of the candidate genes in the PSORS5 susceptibility locus for psoriasis (32); and it has been suggested that the cystatin A risk allele confers a 100-fold increased risk for developing psoriasis (33). The identification of cystatin in lesional mouse skin using an unbiased free label approach provides an added validation of the KC-Tie2 mouse model and allows us to examine newly identified peptides to determine what their importance may be specifically in human psoriasis.
KLK6 was also increased in KC-Tie2 mouse skin and lesional psoriasis skin; and appears to increase in the endothelium of nonlesional psoriasis skin, preceding significant changes in skin anatomy characteristic of psoriasis. KLKs are secreted proteases (34), with KLK5, 7, 8, and 14 being the most well understood (35, 36), specifically for their role in other skin diseases such as Netherton syndrome (KLK5) (37) and wound healing (KLK7 and 8) (38). Less is known about KLK6, although its regulation in psoriasis at the transcript level (17–20) and its quick response to therapeutics is also well documented (39). We present data demonstrating early increases in KLK6 protein expression in the vasculature of uninvolved psoriasis skin; which further increases in both the keratinocytes of the epidermis and the immune cell infiltrate of lesional skin. A recent report (40) demonstrates KLK6 increases in response to stimulation of organotypic epidermal cultures with a cytokine mixture of IFN-γ, IL-3, and GM-CSF.
In summary, we present data demonstrating a discovery-based, in-gel, label-free expression platform suitable for identifying differentially-expressed proteins in the skin of a mouse model of psoriasis and in the lesional skin of human psoriasis. We identified and verified in both the KC-Tie2 mouse model and psoriasis patient skin significant increases in the proteins serpinB1 (serpinb3b), KLK6, cystatin A (stefin A1), and slc25a5. These results demonstrate the usefulness of mouse models and proteomic approaches for identifying novel peptides/proteins that are differentially regulated in human psoriasis.
Supplementary Material
Footnotes
Author contributions: K.C.L., A.J., T.S.M., M.R.C., and N.L.W. designed research; K.C.L., Y.F., A.J., A.M.F., J.B., J.E.G., D.S., G.G., and N.L.W. performed research; M.R.C. contributed new reagents or analytic tools; K.C.L., Y.F., A.J., A.M.F., J.B., J.E.G., D.S., G.G., M.R.C., and N.L.W. analyzed data; K.C.L., A.J., J.E.G., D.S., T.S.M., M.R.C., and N.L.W. wrote the paper.
* This work was supported by the National Psoriasis Foundation (JEG), The Lozick Discovery Grant, National Psoriasis Foundation (NLW), Babcock Foundation Endowment (AJ, JEG), National Institutes of Health (P30 AR39750, P50 AR05508, RO1 AR063437, RO1 AR062546; R21 AR063852 NLW; K01 AR064765 AJ; K08 AR060802, JEG; UL1 TR000439 MRC and NLW), Murdough Family Center for Psoriasis (NLW and TSM), A. Alfred Taubman Medical Research Institute Frances and Kenneth Eisenberg Emerging Scholar Award (JEG).
 This article contains supplemental Figs. S1 to S3 and Tables S1 and S2.
This article contains supplemental Figs. S1 to S3 and Tables S1 and S2.
1 The abbreviations used are:
- NN
- normal skin
- PN
- nonlesional psoriasis skin
- PP
- lesional psoriasis skin
- KC
- keratinocyte
- Rab18
- Ras-related protein 18
- KLK6
- kallikrein related peptidase 6
- Slc25a5
- Solute Carrier Family 25 (Mitochondrial Carrier
- Adenine Nucleotide Translocator)
- Member 5
- qRT-PCR
- quantitative reverse-transcriptase PCR
- LC/MS
- Liquid chromatography–tandem mass spectrometry
- ABC
- ammonium bicarbonate
- IPI
- International Protein Index.
REFERENCES
- 1. Helmick C. G., Sacks J. J., Gelfand J. M., Bebo B., Jr., Lee-Han H., Baird T., Bartlett C. (2013) Psoriasis and psoriatic arthritis: a public health agenda. Am. J. Prev. Med. 44, 424–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gelfand J. M., Weinstein R., Porter S. B., Neimann A. L., Berlin J. A., Margolis D. J. (2005) Prevalence and treatment of psoriasis in the United Kingdom: a population-based study. Arch. Dermatol. 141, 1537–1541 [DOI] [PubMed] [Google Scholar]
- 3. Kurd S. K., Gelfand J. M. (2009) The prevalence of previously diagnosed and undiagnosed psoriasis in US adults: results from NHANES 2003–2004. J. Am. Acad. Dermatol. 60, 218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Valdimarsson H., Thorleifsdottir R. H., Sigurdardottir S. L., Gudjonsson J. E., Johnston A. (2009) Psoriasis–as an autoimmune disease caused by molecular mimicry. Trend Immunol. 30, 494–501 [DOI] [PubMed] [Google Scholar]
- 5. Tran B., Feldman S. R. (2011) Insight into psoriasis management: commercial perspectives for the U.S. psoriasis market. J. Dermatol. Treat. 22, 18–26 [DOI] [PubMed] [Google Scholar]
- 6. Menter A., Korman N. J., Elmets C. A., Feldman S. R., Gelfand J. M., Gordon K. B., Gottlieb A., Koo J. Y., Lebwohl M., Lim H. W., Van Voorhees A. S., Beutner K. R., Bhushan R. (2010) Guidelines of care for the management of psoriasis and psoriatic arthritis: Section 5. Guidelines of care for the treatment of psoriasis with phototherapy and photochemotherapy. J. Am. Acad. Dermatol. 62, 114–135 [DOI] [PubMed] [Google Scholar]
- 7. Wolfram J. A., Diaconu D., Hatala D. A., Rastegar J., Knutsen D. A., Lowther A., Askew D., Gilliam A. C., McCormick T. S., Ward N. L. (2009) Keratinocyte but not endothelial cell specific overexpression of Tie2 leads to the development of psoriasis. Am. J. Pathol. 174, 1443–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 9. Schlatzer D. M., Sugalski J., Dazard J. E., Chance M. R., Anthony D. D. (2012) A quantitative proteomic approach for detecting protein profiles of activated human myeloid dendritic cells. J. Immunol. Methods 375, 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan E. Y., Sutton J. N., Jacobs J. M., Bondarenko A., Smith R. D., Katze M. G. (2009) Dynamic host energetics and cytoskeletal proteomes in human immunodeficiency virus type 1-infected human primary CD4 cells: analysis by multiplexed label-free mass spectrometry. J. Virol. 83, 9283–9295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neubert H., Bonnert T. P., Rumpel K., Hunt B. T., Henle E. S., James I. T. (2008) Label-free detection of differential protein expression by LC/MALDI mass spectrometry. J. Proteome Res. 7, 2270–2279 [DOI] [PubMed] [Google Scholar]
- 12. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnston A., Fritz Y., Dawes S. M., Diaconu D., Al-Attar P. M., Guzman A. M., Chen C. S., Fu W., Gudjonsson J. E., McCormick T. S., Ward N. L. (2013) Keratinocyte overexpression of IL-17C promotes psoriasiform skin inflammation. J. Immunol. 190, 2252–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elder J. T., Fisher G. J., Zhang Q. Y., Eisen D., Krust A., Kastner P., Chambon P., Voorhees J. J. (1991) Retinoic acid receptor gene expression in human skin. J. Invest. Dermatol. 96, 425–433 [DOI] [PubMed] [Google Scholar]
- 15. Johnston A., Gudjonsson J. E., Aphale A., Guzman A. M., Stoll S. W., Elder J. T. (2011) EGFR and IL-1 signaling synergistically promote keratinocyte antimicrobial defenses in a differentiation-dependent manner. J. Invest. Dermatol. 131, 329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnston A., Xing X., Guzman A. M., Riblett M., Loyd C. M., Ward N. L., Wohn C., Prens E. P., Wang F., Maier L. E., Kang S., Voorhees J. J., Elder J. T., Gudjonsson J. E. (2011) IL-1F5, -F6, -F8, and -F9: a novel IL-1 family signaling system that is active in psoriasis and promotes keratinocyte antimicrobial peptide expression. J. Immunol. 186, 2613–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yao Y., Richman L., Morehouse C., de los Reyes M., Higgs B. W., Boutrin A., White B., Coyle A., Krueger J., Kiener P. A., Jallal B. (2008) Type I interferon: potential therapeutic target for psoriasis? PloS One 3, e2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou X., Krueger J. G., Kao M. C., Lee E., Du F., Menter A., Wong W. H., Bowcock A. M. (2003) Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol. Genomics 13, 69–78 [DOI] [PubMed] [Google Scholar]
- 19. Gudjonsson J. E., Ding J., Johnston A., Tejasvi T., Guzman A. M., Nair R. P., Voorhees J. J., Abecasis G. R., Elder J. T. (2010) Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J. Invest. Dermatol. 130, 1829–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suarez-Farinas M., Lowes M. A., Zaba L. C., Krueger J. G. (2010) Evaluation of the psoriasis transcriptome across different studies by gene set enrichment analysis (GSEA). PloS One 5, e10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Swindell W. R., Johnston A., Carbajal S., Han G., Wohn C., Lu J., Xing X., Nair R. P., Voorhees J. J., Elder J. T., Wang X. J., Sano S., Prens E. P., Digiovanni J., Pittelkow M. R., Ward N. L., Gudjonsson J. E. (2011) Genome-wide expression profiling of five mouse models identifies similarities and differences with human psoriasis. PloS One 6, e18266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mikesh L. M., Aramadhaka L. R., Moskaluk C., Zigrino P., Mauch C., Fox J. W. (2013) Proteomic anatomy of human skin. J. Proteomics 84, 190–200 [DOI] [PubMed] [Google Scholar]
- 23. Macdonald N., Cumberbatch M., Singh M., Moggs J. G., Orphanides G., Dearman R. J., Griffiths C. E., Kimber I. (2006) Proteomic analysis of suction blister fluid isolated from human skin. Clin. Exp. Dermatol. 31, 445–448 [DOI] [PubMed] [Google Scholar]
- 24. Huang C. M., Foster K. W., DeSilva T., Zhang J., Shi Z., Yusuf N., Van Kampen K. R., Elmets C. A., Tang D. C. (2003) Comparative proteomic profiling of murine skin. J. Invest. Dermatol. 121, 51–64 [DOI] [PubMed] [Google Scholar]
- 25. Ryu J., Park S. G., Park B. C., Choe M., Lee K. S., Cho J. W. (2011) Proteomic analysis of psoriatic skin tissue for identification of differentially expressed proteins: up-regulation of GSTP1, SFN, and PRDX2 in psoriatic skin. Int. J. Mol. Med. 28, 785–792 [DOI] [PubMed] [Google Scholar]
- 26. Williamson J. C., Scheipers P., Schwammle V., Zibert J. R., Beck H. C., Jensen O. N. (2013) A proteomics approach to the identification of biomarkers for psoriasis utilising keratome biopsy. J. Proteomics 94, 176–185 [DOI] [PubMed] [Google Scholar]
- 27. Piruzian E., Bruskin S., Ishkin A., Abdeev R., Moshkovskii S., Melnik S., Nikolsky Y., Nikolskaya T. (2010) Integrated network analysis of transcriptomic and proteomic data in psoriasis. BMC Sys. Biol. 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carlen L. M., Sanchez F., Bergman A. C., Becker S., Hirschberg D., Franzen B., Coffey J., Jornvall H., Auer G., Alaiya A. A., Stahle M. (2005) Proteome analysis of skin distinguishes acute guttate from chronic plaque psoriasis. J. Invest. Dermatol. 124, 63–69 [DOI] [PubMed] [Google Scholar]
- 29. Celis J. E., Cruger D., Kiil J., Lauridsen J. B., Ratz G., Basse B., Celis A. (1990) Identification of a group of proteins that are strongly up-regulated in total epidermal keratinocytes from psoriatic skin. FEBS Lett. 262, 159–164 [DOI] [PubMed] [Google Scholar]
- 30. Celis J. E., Madsen P., Rasmussen H. H., Leffers H., Honore B., Gesser B., Dejgaard K., Olsen E., Magnusson N., Kiil J., Celis A., Lauridsen J. B., Basse B., Ratz G. P., Andersen A. H., Walbum E., Brandstrup B., Pedersen P. S., Brandt N. J., Puype M., Van Damme J., Vandekerckhove J. (1991) A comprehensive two-dimensional gel protein database of noncultured unfractionated normal human epidermal keratinocytes: towards an integrated approach to the study of cell proliferation, differentiation and skin diseases. Electrophoresis 12, 802–872 [DOI] [PubMed] [Google Scholar]
- 31. Meyer-Hoffert U. (2009) Reddish, scaly, and itchy: how proteases and their inhibitors contribute to inflammatory skin diseases. Arch. immunol. Ther. Ex. 57, 345–354 [DOI] [PubMed] [Google Scholar]
- 32. Samuelsson L., Stiller C., Friberg C., Nilsson C., Inerot A., Wahlstrom J. (2004) Association analysis of cystatin A and zinc finger protein 148, two genes located at the psoriasis susceptibility locus PSORS5. J. Invest. Dermatol. 122, 1399–1400 [DOI] [PubMed] [Google Scholar]
- 33. Vasilopoulos Y., Sagoo G. S., Cork M. J., Walters K., Tazi-Ahnini R. (2011) HLA-C, CSTA and DS12346 susceptibility alleles confer over 100-fold increased risk of developing psoriasis: evidence of gene interaction. J. Hum. Genet. 56, 423–427 [DOI] [PubMed] [Google Scholar]
- 34. Komatsu N., Takata M., Otsuki N., Toyama T., Ohka R., Takehara K., Saijoh K. (2003) Expression and localization of tissue kallikrein mRNAs in human epidermis and appendages. J. Invest. Dermatol. 121, 542–549 [DOI] [PubMed] [Google Scholar]
- 35. Sotiropoulou G., Pampalakis G. (2010) Kallikrein-related peptidases: bridges between immune functions and extracellular matrix degradation. Biol. Chem. 391, 321–331 [DOI] [PubMed] [Google Scholar]
- 36. Sotiropoulou G., Pampalakis G., Diamandis E. P. (2009) Functional roles of human kallikrein-related peptidases. J. Biol. Chem. 284, 32989–32994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Furio L., de Veer S., Jaillet M., Briot A., Robin A., Deraison C., Hovnanian A. (2014) Transgenic kallikrein 5 mice reproduce major cutaneous and systemic hallmarks of Netherton syndrome. J. Exp. Med. 211, 499–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kishibe M., Bando Y., Tanaka T., Ishida-Yamamoto A., Iizuka H., Yoshida S. (2012) Kallikrein-related peptidase 8-dependent skin wound healing is associated with upregulation of kallikrein-related peptidase 6 and PAR2. J. Invest. Dermatol. 132, 1717–1724 [DOI] [PubMed] [Google Scholar]
- 39. Russell C. B., Kerkof K., Bigler J., Timour M., Welcher A. A., Bautista E., Cueto I., Khatcherian A., Krueger J. G., Rand H., Martin D. A. (2011) Blockade of the IL-17R with AMG 827 leads to rapid reversal of gene expression and histopathological abnormalities in psoriatic skin, including substantial pathway-specific effects within one week. J. Invest. Dermatol. 131, S11 [Google Scholar]
- 40. Szabo K., Bata-Csorgo Z., Dallos A., Bebes A., Francziszti L., Dobozy A., Kemeny L., Szell M. (2014) Regulatory Networks Contributing to Psoriasis Susceptibility. Acta dermato-venereologica 94(4), 380–385 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.