Abstract
Allergenic proteins such as grass pollen and house dust mite (HDM) proteins are known to trigger hypersensitivity reactions of the immune system, leading to what is commonly known as allergy. Key allergenic proteins including sequence variants have been identified but characterization of their post-translational modifications (PTMs) is still limited.
Here, we present a detailed PTM1 characterization of a series of the main and clinically relevant allergens used in allergy tests and vaccines. We employ Orbitrap-based mass spectrometry with complementary fragmentation techniques (HCD/ETD) for site-specific PTM characterization by bottom-up analysis. In addition, top-down mass spectrometry is utilized for targeted analysis of individual proteins, revealing hitherto unknown PTMs of HDM allergens. We demonstrate the presence of lysine-linked polyhexose glycans and asparagine-linked N-acetylhexosamine glycans on HDM allergens. Moreover, we identified more complex glycan structures than previously reported on the major grass pollen group 1 and 5 allergens, implicating important roles for carbohydrates in allergen recognition and response by the immune system. The new findings are important for understanding basic disease-causing mechanisms at the cellular level, which ultimately may pave the way for instigating novel approaches for targeted desensitization strategies and improved allergy vaccines.
Allergic respiratory disease is a global health problem and current clinical guidelines recommend a combination of allergen avoidance, pharmacotherapy, and allergen specific immunotherapy for treatment (1–4). At present allergy testing and vaccines are based on isolated crude antigen preparations from natural sources (i.e. HDM, pollens, etc.), but a move toward recombinant allergen design is ongoing (5, 6). This could have important functional implications because the production host will determine the repertoire of post-translational modifications (PTMs) and in particular glycan modifications presented on allergens.
The carbohydrate structures found on allergens are in most cases not found in mammals and therefore frequently lead to the induction IgE antibodies named Cross-reactive Carbohydrate Determinants (CCD) (7–11). Moreover, glycans may directly be involved in and promote uptake and target allergens to carbohydrate lectin receptors on antigen presenting cells (APC) (12–14). Therefore, a full structural characterization of the glycans on the natural allergens is a prerequisite for understanding both antibody reactivity and lectin receptor mediated allergen recognition and modulation of the immune response (15, 16). Furthermore, a detailed characterization of PTMs of allergens is important for standardization of allergen products for diagnostic purposes as well as for vaccine use (17, 18). Although many major allergens and their etiology have been characterized in some detail, structural information on for example their immunological important PTM status is still incomplete (19–21).
Mass spectrometry-based technologies offer sensitive and accurate analyses for identification and characterization of proteins. The common proteomics workflow typically adopts the bottom-up approach, i.e. in vitro proteolytic digestion of proteins followed by nanoflow-liquid chromatography-tandem mass spectrometry (nLC-MS/MS) for protein identification and PTM characterization. Electron- or collision-driven fragmentation techniques, e.g. electron transfer dissociation (ETD) (22) or higher energy collisional dissociation (HCD) (23) have enabled accurate identification of peptides of purified proteins, e.g. allergens (21, 24), or complex biological samples (25–27) with concurrent characterization of their PTMs. One advantage of bottom-up mass spectrometry is the ability to resolve modified peptides within a narrow chromatographic time frame thereby enabling in-depth characterization of site-specific features, e.g. glycoforms, on peptides. This peptide-level information is subsequently used to generate a protein-level view on the PTM status for a given protein. Importantly, the PTM connectivity of the protein (28) is lost upon proteolytic digestion, and alternative approaches are often required for comprehensive characterization of all proteoforms (29). Top-down mass spectrometry has emerged as an alternative approach to bottom-up proteomics, offering complementary MS and MS/MS information that may be used for protein identification and characterization (30, 31). With top-down MS, intact proteins are typically analyzed by high-resolution FTMS and characterized at the MS/MS level by CID, HCD, ECD, or ETD. This technique provides instant protein-level information on analytes, e.g. sequence variants, amino acid substitutions, PTMs, etc., which can be verified at the MS/MS level by different fragmentation modes. The combination of bottom-up and top-down mass spectrometry is therefore a powerful tool for the identification and characterization of proteins. Here, we combine top-down and bottom-up mass spectrometry for comprehensive characterization of seven major allergens as a first step toward unraveling the molecular mode of action of allergens with complex PTMs. By these methods, we demonstrate hitherto unknown PTMs of HDM allergens and identify more complex glycan structures than previously reported on the major grass pollen group 1 and 5 allergens. The new findings implicate important roles for carbohydrates in allergen recognition and response by the immune system.
EXPERIMENTAL PROCEDURES
Purification of HDM Dermatophagoides Pteronyssinus (Der p) and Dermatophagoides Farina (Der f) Major Allergens Group 1 and 2
Spent house dust mite (HDM) cultures of either specie were extracted 1:10 (w/v) in 44% (v/v) acetone and 56% (v/v) 0.125 m NH4HCO3, pH 8.3, for one hour at 5 °C. After centrifugation, the supernatant was mixed with an equal volume of acetone and the precipitate was collected by centrifugation and dried in a ventilated hood over night at room temperature. The dried precipitate was dissolved in 0.125 m NH4HCO3, pH 8.3, dialyzed overnight against Milli Q water at 5 °C, and lyophilized.
The purification procedure for Der p and f allergens is almost identical. Three to four hundred mg culture extract was dissolved in 10 ml 0.05 m Na Acetate, pH 5.0, and mixed with 20 ml 2.0 m NH4SO4, and the mixture was subjected to Hydrophobic interaction chromatography (HIC) on a Butyl-S FF column (25 ml) equilibrated with 0.05 m Na Acetate and 1.5 m NH4SO4, pH 5 (Buffer A). The flow-through fractions were collected and pooled (∼100 ml). This pool contains primarily the group 2 allergen and other HDM extract proteins, whereas group 1 allergen bind to the column. The group 1 allergens were eluted from the column by a stepwise decrease of the NH4SO4 molarity, changing the buffers from A to B (0.05 m Na Acetate and 1.05 m NH4SO4, pH 5.0), to C (0.05 m Na Acetate and 0.98 m NH4SO4, pH 5.0), and finally to D (0.05 m Na Acetate, pH 5.0). The fractions eluted in buffer D (∼5 column volumes) were pooled and dialyzed toward E (0.05 m Na Phosphate and 0.5 m NaCl, pH 7.5). This sample was applied on a HiTrap Chelating Sepharose HP column (5.0 ml) charged with 0.1 m CuSO4 and equilibrated with buffer E. The proteins were eluted by a stepwise change in buffer composition and pH from E to F (0.05 m Na Phosphate and 0.5 m NH4Cl, pH 7.5), to G (0.65 m Na Acetate, pH 4.5), and finally H (0.05 m Na Phosphate, 0.5 m NaCl, and 0.1 m EDTA, pH 7.5). The HDM group 1 allergen(s) were eluted in buffer G (approx. 100 ml), dialyzed against buffer E and applied on HiTrap Chelating Sepharose HP (5 ml) charged with 0.1 m ZnSO4 and equilibrated in buffer E. The proteins were eluted from the column using the same stepwise procedure as for the Cu chelating column and Der p 1 was eluted in buffer G (∼20 ml) and dialyzed against 0.005 m NH4HCO3, pH 8.3, and lyophilized. The yield of purified Der p 1 was ∼3% (w/w) of the starting material. Der f 1 was purified by the same procedure excluding the Cu Chelating chromatography with a final yield of approx. 4% (w/w) of the starting material.
The pooled fraction containing HDM group 2 in buffer A from the above HIC (100 ml) was dialyzed against buffer E and subjected to Cu Chelate chromatography as described for the HDM group 1 allergen purification. The HDM group 2 allergens eluted in buffer G (approx. 80 ml) and were concentrated in an Amicon Ultra 15, 3K centrifugation tube to 2–3 ml. This sample was applied on a Size Exclusion column, HiLoad Superdex 75 16/60, equilibrated with 0.15 m NH4HCO3, pH 8.3. The fractions were analyzed by SDS-PAGE and the fractions containing HDM group 2 allergens were pooled (approx. 50 ml) and concentrated as above to 3 ml. The concentration step involves three dilutions with water followed by concentration in order to decrease the salt content of the sample to less than 0.01 m and the final sample is subjected to preparative flatbed Iso Electric Focusing (IEF) on a Agilent 3100 OffGel fractionator, using precast PG IEF immobilized pH gradient poly acryl amide strips, pH 3.5 to 10, in a 24 well setup. The IEF runs overnight and the fractions containing HDM group 2 allergens (Identified by SDS-PAGE and/or Fused Rocket Immuno Electrophoresis) was dialyzed against 0.025 m NaHCO3, pH 8.5, and stored in aliquots at −20 °C.
Purification of Pollen Major Allergens
Pollens from Birch trees (Betula verrucosa) and Timothy grass (Phleum pretense) were obtained from Greer, Nuway Circle, NE, and extracted 1:10 (w/v) in 0.125 m NH4HCO3, pH 8.3, overnight. After removal of particulate matter by centrifugation, the supernatant(s) were dialyzed against 0.005 m NH4HCO3 utilizing frequent changes of dialysis buffer, and lyophilized.
Five hundred thirty mg Birch pollen extract was dissolved in 30 ml 1.8 m NH4SO4 and applied on a column of two serial connected HiTrap Butyl S FF columns equilibrated in buffer A1 (0.05 m Na Phosphate and 1.8 m NH4HCO3, pH 7.0). The proteins were eluted from the column by stepwise change in buffer to A2 (0.05 m Na Phosphate, pH 7.0) and finally to A3 (0.05 m Na2CO3 and 50% (v/v) ethylene glycol, pH 10). Fractions eluted with A2 were pooled (50 ml) and concentrated in an Amicon Filter Cell using PLBC filter from Millipore. The sample (concentrated six times) was subjected to Size Exclusion Chromatography on HiLoad Superdex 75 16/60 120 ml column equilibrated in 0.125 m NH4HCO3, pH 8.3. The fractions containing the Bet v 1 allergen (identified by SDS-PAGE of the individual fractions) were pooled and dialyzed toward 0.02 m TRIS HCl, pH 8.0. The sample was subjected to ion exchange chromatography on a HiTrap Q HP column equilibrated in the same buffer. Bet v 1 was eluted with 0.02 m BIS-TRIS and 1.0 m NaCl, pH 6.5. The fractions were pooled and dialyzed against 0.05 m NA Phosphate, 0.5 m NaCl, and subjected to Cu Chelate chromatography on a HiTrap chelating HP column charged with 0.1 m CuSO4 and equilibrated with the same buffer. The column was eluted by stepwise changes in buffer and pH. Bet v 1 was primarily eluted with 0.65 m Na Acetate pH 5.0 and pH 4.8. These fractions were pooled and concentrated on an Amicon Filter Cell (see above). Finally, the concentrated sample was polished on a HiLoad Superdex 75 16/0 SEC column equilibrated in 0.150 m NH4HCO3, pH 8.3. The fraction containing Bet v 1 was dialyzed toward 0.05 m NH4HCO3, pH 8.3, and mixed with glycerol (final concentration 50% (v/v)) and stored at −20 °C. The yields were 3.7% (w/w).
The grass pollen major allergens Phl p 1 and 5 were purified from pollen extracts by the following procedures: The extract (450 mg) dissolved in 0.15 m NH4HCO3, pH 8.3 (150 ml) was precipitated by addition of solid ammonium sulfate to a concentration of 3.0 m, after centrifugation the supernatant containing Phl p 1 was diluted to 1.0 m ammonium sulfate and subjected to HIC on an Octyl FF 16/30 20 ml column equilibrated in 0.05 m Na Acetate and 1.0 m NH4SO4, pH 5.0. The flow-through fractions (containing Phl p 1) were concentrated (Amicon Ultra spin tubes) to 10 ml and further fractionate on two consecutive SEC columns HiLoad Superdex-75 16/60 equilibrated in 0.159 m NH4HCO3, pH 8.3. The fractions containing Phl p 1 (identified by SDS-PAGE) from the first SEC were pooled, concentrated (Amicon Ultra spin tubes) to 5 ml and run a second time on the same SEC column, yielding 10 mg purified Phl p 1 after lyophilization.
Phl p 5 was purified from the above 3.0 m ammonium sulfate precipitate. The precipitate was dissolved in 0.15 m NH4HCO3, pH 8.3, and subjected to SEC (see above). The fractions containing Phl p 5 (identified by SDS-PAGE) were pooled and dialyzed against 0.1 m Na Acetate, pH 5.0. After addition of 2.0 m NH4SO4 to a final concentration of 1.0 m, the solution was subjected to HIC on an Octyl FF. The column was eluted with a stepwise change in buffer and pH and the Phl p 5 eluted in 0.05 m NaCO3, pH 10, and 50% (v/v) ethylene glycol. The fractions were pooled and fractioned further on two consecutive SECs (see Phl p 1 purification), yielding ∼10 mg lyophilized Phl p 5.
SDS-PAGE and MALDI-TOF Analysis
Purified allergens (5 μg) were mixed with an equal volume of x2 Novex LDS sample buffer (Life Technologies) supplemented with 100 mm dithiothreitol and heated at 95 °C for 5 min. Two μg of each allergen was separated (200 V, 35 min) using MES-buffer and Novex Bis-Tris 4–12% gradient gels (Life Technologies, Naerum, Denmark). Bands were visualized by staining with Novex SimplyBlue SafeStain (Life Technologies). Allergens (1 μg) were analyzed on a MALDI-TOF (Autoflex Speed, Bruker Daltonics, Bremen, Germany) instrument operated in the positive mode using sinapinic acid as matrix. Mass spectra were acquired in the linear mode (mass range m/z 10,000–30,000 or m/z 10,000–40,000) using 2000 laser shots/spot.
Bottom-up Mass Spectrometry
10 μg of each allergen was reduced (5 mm dithiothreitol, 60 °C, 30 min) and alkylated (10 mm iodoacetamide, RT, 30 min) before ON treatment at 37 °C with 1 μg trypsin (Roche, Hvidovre, Denmark). The tryptic digest of each allergen was purified using in-house packed Stagetips (Empore disk-C18, 3 m) as described with minor modifications (32). The C-18 material was packed to a height of ∼2 mm in a standard 200 μl pipette tip and washed consecutively with methanol, 50% methanol in 0.1% formic acid, and with 0.1% trifluoroacetic acid. The tryptic digest was acidified by adding 2 μl trifluoroacetic acid and loaded on the Stagetip. The Stagetip was washed once again with 0.1% formic acid and tryptic peptides were eluted by 50% methanol in 0.1% formic acid. The tryptic digests were separately analyzed using a setup composed of an EASY-nLC 1000 UHPLC (Thermo Scientific) interfaced via a nanoSpray Flex ion source to an LTQ-Orbitrap Velos Pro hybrid mass spectrometer. The EASY-nLC 1000 was operated using a single analytical column setup (PicoFrit Emitters, New Objectives, 75 μm inner diameter) packed in-house with Reprosil-Pure-AQ C18 phase (Dr. Maisch, 1.9 μm particle size). Peptides were separated using a 90 min LC gradient operated at 200 nL/min. The mobile phases were composed of solvent A (H2O) and solvent B (acetonitrile); both solvents containing 0.1% formic acid (v/v). The gradient was 5–30% B for 65 min, followed by 30–80% B for 10 min, and finally 80% B for 15 min. Mass spectra were acquired essentially as previously described (27) with minor modifications. Briefly, MS1 precursor scan (m/z 350–1700) acquisition was performed in the orbitrap using a nominal resolution of 30,000 followed by HCD-MS2 fragmentation of the five most abundant multiply charged precursor ions. A minimum signal threshold of 50,000 ions was used for triggering HCD-MS2 and ETD-MS2 fragmentation. MS2 scans (m/z 100–2000) were acquired in the orbitrap mass analyzer using a resolution setting of 15,000. The tryptic digest of Bet v 1 was analyzed on an LTQ-Orbitrap XL (Thermo Scientific) instrument interfaced with an EASY-nLC II system (Thermo Scientific).The analytical column was packed in-house using the same materials as above with the exception of particle size for C18 material that was 3 μm. Peptides were separated using a 60 min gradient and characterized only by HCD fragmentation for the three most abundant multiply charged precursor ions.
Top-down Mass Spectrometry
Each allergen (1 pmol/μl in 50% methanol and 0.1% formic acid, v/v) was analyzed on an LTQ-Orbitrap Velos Pro hybrid mass spectrometer (Thermo Fisher Scientific) by direct infusion. The samples were introduced via a TriVersa NanoMate ESI-Chip interface (Advion BioSystems, Ithaca, NY); direct infusion was controlled by Chipsoft 8.1.0 (Advion Biosciences). MS1 precursor scan was acquired in the Orbitrap with 100,000 resolving power at m/z 400. MS2 fragmentation events utilizing HCD, CID, and ETD modes were performed and product ions were detected in the Orbitrap with 30,000–60,000 resolving power at m/z 400.
Quantification of Carbohydrates
The C-terminally amidated IRPTMTIPGYVEPTAV peptide standard, modified by a single O-linked mannose on Thr4, was used to generate a standard curve. 500 pmol of peptide standard, Der p 2 and Der f 2 allergens were each dissolved in 40 μl 50 mm sodium acetate, pH 4.8. To each sample, 20 mm sodium periodate was added and the reaction was allowed to proceed at RT for 60 min before being quenched by the addition of 1 μl 96% glycerol. Biotin-hydrazide was added to a final concentration of 500 μm and the mixture was left to react in darkness for 16 h at RT. Control reactions for the glycopeptide standard and allergens were as described above with the exception of omitting sodium periodate from the reaction. The products were desalted by reversed-phase chromatography (C18 Stagetip microcolumns). ELISA plates (Nunc-Immuno MaxiSorp F96 plates (Nunc, Roskilde)) were coated with a dilution series (1:2 starting at 100 ng/ml) of glycopeptide, indicated allergens and controls in carbonate-bicarbonate buffer (pH 9.6) ON at 4 °C. Plates were washed and blocked with BSA/Triton X-100 buffer (1% BSA, 1% Triton X-100, 3 mm KCl, 0.5 m NaCl, and 8 mm phosphate buffer (pH 7.4)) for 1 h at RT followed by washing and incubated with Streptavidin-HRP (Thermo Scientific) for 1h at RT. Plates were developed with TMB + one-step substrate system (Dako), reactions stopped with 0.5 m H2SO4, and read at 450 nm. Amount biotinylated protein was calculated from the peptide standard curve.
Reductive Beta Elimination
Chemical release of O-linked glycans was done as previously described (33) with modifications. Briefly, 50 μg bovine fetuin (Sigma Aldrich), Der f 2 and Der p 2 allergens were separately incubated in 100 μl 0.1 m NaOH and 1 m NaBH4. The reductive beta elimination was allowed to proceed 16 h at 50 °C and the reaction was terminated by addition of 8 μl glacial acetic acid. The glycans were separated from protein components by Sep-pak C18 columns and desalted by Dowex AG 50W X8 cation exchange resin. Finally, glycans were dried over a stream of nitrogen gas and BH4− was removed as volatile methyl borate by repeated evaporation (5x) in 500 μl methanol containing 1% acetic acid. Released glycans were permethylated according to the procedure of Ciucanu and Costello (34) and analyzed by positive mode MALDI TOF (Autoflex Speed, Bruker Daltonics, Bremen, Germany) operated in the reflector mode (mass range m/z 500–3000; 2000 laser shots/spot) using 2,5-dihydroxybenzoic acid as matrix.
Gas Chromatrography-Mass Spectrometry
For GC-MS analysis, 50 μg of dust mite allergens were spiked with myo-inositol (internal standard) and depolymerized by incubation in 0.5N methanolic HCL (Supelco, Bellefonte, PA), 80 °C, 16h. The reaction mixtures were dried over a stream of nitrogen gas and re-N-acetylated (RT, 30 min) by the addition of 500 μl methanol, 50 μl pyridine and 50 μl acetic anhydride. The reaction mixtures were dried down once again and per-O-trimethylsilylated with Tri-Sil reagent (Thermo Scientific) at 80 °C, 30 min. The samples were dried down as above and dissolved in 1 ml n-hexane (Merck) prior to injection. GC-MS analysis was performed using a TRACE™ GC Ultra gas chromatograph coupled to a PolarisQ ion trap mass spectrometer (Thermo Scientific). Samples were injected (splitless mode) at 40 °C (1 min), oven temperature was ramped to 150 °C (25 °C/min) followed by an increase to 200 °C (2 °C/min) before a final ramp to 260 °C (10 °C/min) where it was held for 5 min. Monosaccharides were identified by comparison of retention times and mass spectra to N-acteylglucosamine, N-acetylgalactosamine, mannose, or lactose standards (depolymerized, re-N-acetylated and per-O-trimethylsilylated). All retention time comparisons were relative to the myo-inositol internal standard.
LC-MS of Bariumhydroxide Mediated Peptide Bond Hydrolysates
Briefly, 180 μg Phl p 1 and Phl p 5 were subjected to BaOH2 (0.22 m final concentration) mediated peptide bond hydrolysis in a total volume of 0.5 ml, which was incubated ON at 108 °C. After cooling to RT, HCOOH was added to 5 mm and the samples carefully neutralized by small aliquots of 4 m H2SO4. The resulting precipitate was pelleted and the supernatant obtained.
LC-MS (3 μl supernatant injected) was carried out on an Agilent 1100 Series LC (www.agilent.com) coupled to a Bruker HCT Ultra ion trap mass spectrometer (www.bruker.com). The column was a Phenomenex Luna C8(2) (3 μm, 100 A, 150 × 2.0 mm; www.phenomenex.com) preceded by a Phenomenex Gemini C18 SecurityGuard (4 × 2 mm). The oven temperature was maintained at 35 °C. The mobile phases were A, water; B, acetonitrile, both with 0.1% (v/v) HCOOH, and the flow rate was 0.2 ml min−1. The gradient was: 0 to 2 min, isocratic 1% B; 2 to 8.5 min, linear gradient 1 to 3% B; 8.6 to 9.6 isocratic 99% B; and 9.7 to 17 min, isocratic 1% B. The mass spectrometer was run in positive electrospray mode. An extensin enriched fraction from roots of wild type Arabidopsis thaliana (ecotype Columbia) was obtained as described (35) and subjected to BaOH2 mediated peptide bond hydrolysis yielding the extensin derived Hyp-Araf1–4 species as earlier reported (36). This preparation served as a Hyp-Araf1–4 positive reference. Semi quantitative integration of peaks on the extracted ion chromatograms was done using the Bruckner Compass Data analysis version 1.1 program.
Data Analysis
Analysis and processing of bottom-up mass spectra was performed essentially as previously described (27). Briefly, .raw files were processed using the Sequest HT node of Proteome Discoverer 1.4 software (Thermo Fisher Scientific) and searched individually against allergen sequences (n = 95, supplemental Table S2) downloaded from the Uniprot database (April, 2014). Enzyme setting was full cleavage specificity by trypsin, one missed cleavage; peptide mass tolerance was 10 ppm; fragment ion mass tolerance was 0.05 amu; carbamidomethyl was set as fixed modification for cysteine residues; and methionine oxidation and asparagine deamidation were used as variable modifications. In addition, attachment of hexose at Ser/Thr/Tyr/Lys and N-acetylhexoseamine at Ser/Thr/Asn residues was used as a dynamic modification for dust mite allergens. Dynamic hexose modification (at Ser/Thr/Tyr/Lys), hydroxylation of proline residues, and variable pentose modification (at hydroxyproline) was used for grass pollen allergens (including Bet v 1). All spectra at the medium confidence level (p > 0.01) and below were filtered and resubmitted to a second Sequest HT node as described above but with the exception of using semi-specific trypsin cleavage for enzyme settings. The final results were filtered for only high confidence (p < 0.01) identifications. Spectra matched to peptides containing PTMs were inspected manually to verify the accuracy of the assignments and only fragment ions of 0.01 amu mass accuracy or better were considered. The Xtract tool (Xcalibur Qual Browser software, Thermo Fisher Scientific) was used to deconvolute MS and MS/MS spectra using fit factor setting of 40% and remainder setting of 25%. Masses calculated by Xtract were used to assign the correct monoisotopic mass of precursor and product ions.
RESULTS
The natural major allergens Der p 1, Der p 2, Der f 1, Der f 2, Bet v 1, Phl p 1, and Phl p 5 were purified to more than 95% purity as revealed by SDS-PAGE analysis (supplemental Fig. S1). The total molecular mass for the individual allergen proteins was obtained by MALDI-TOF analysis (supplemental Figs. S2 and S3). The characterization of PTMs of Der p 1, Der p 2, Der f 1, Der f 2, Bet v 1, Phl p 1, and Phl p 5 is summarized in Fig. 1 together with Table I, and the detailed structural analysis of the individual allergens is provided below.
Fig. 1.
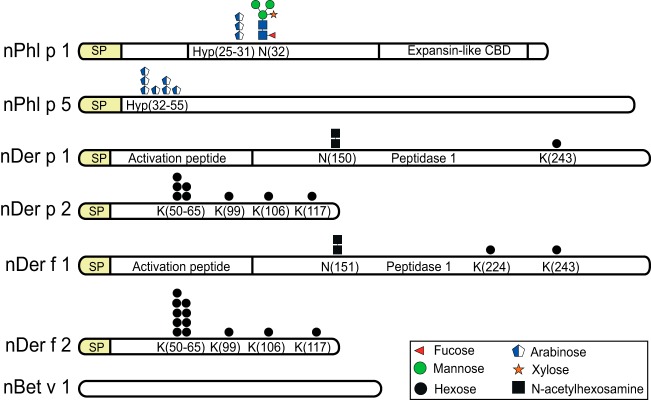
Graphic depiction of the identified glycans in seven major natural allergens studied. The most complex form of glycans identified for each allergen is shown. From top to bottom: Phl p 1, Phl p 5, Der p 1, Der p 2, Der f 1, Der f 2, and Bet v 1. The modified residues are indicated below the graphic depiction of glycans. The n prefix is short for natural; SP is short for signal peptide. Glycans are represented by symbols and colors according to recommendations from Consortium for Functional Glycomics. The cartoons shown are based upon mass measurements and assumptions of the known biosynthetic pathways.
Table I. Glycopeptides from tryptic digest of natural grass pollen and dust mite allergens identified by bottom up mass spectrometry.
| Allergen | Peptide | Modificationsa |
|---|---|---|
| Phl p 1 | V27PPGPNITATYGDK40 | Hyp1; Pen1dHex1Hex2–3HexNAc2 |
| I24PKVPPGPNITATYGDK40 | Hyp2; Pen1–4dHex1Hex 2–3HexNAc2 | |
| Phl p 5a | A26DLGYGPATPAAPAAGYTPATPAAPAEAAPAGK58 | Hyp6–7; Pen0,2 |
| Phl p 5.0103 | A26DLGYGPATPAAPAAGYTPATPAAPAGAEPAGK58 | Hyp0–7; Pen0–7 |
| Phl p 5b | A20DAGYAPATPAAAGAAAGK38 | Hyp2; Pen0–1 |
| Der p 1 | N150QSLDLAEQELVDCASQHGCHGDTIPR176 | HexNAc1,2 |
| E227ALAQTHSAIAVIIGIKDLDAFR249 | Hex | |
| Der f 1 | N151TSLDLSEQELVDCASQHGCHGDTIPR177 | HexNAc1,2 |
| R204PNSQHYGISNYCQIYPPDVKQIR227 | Hex | |
| E228ALTQTHTAIAVIIGIKDLR247 | Hex | |
| Der p 2 | G49KPFQLEAVFEANQNTKTAK68 | Hex1–4 |
| C95PLVKGQQYDIK106 | Hex | |
| G100QQYDIKYTWNVPK113 | Hex | |
| I114APKSENVVVTVK126 | Hex | |
| Der f 2 | G49KPFTLEALFDANQNTK65 | Hex1–9 |
| C95PLVKGQQYDIK106 | Hex | |
| G100QQYDIKYTWNVPK113 | Hex1,2 | |
| I114APKSENVVVTVK126 | Hex |
a Variations in number of monosaccharide units shown in bold. Characters: - represent “between” and, represent “or”. Hyp, hydroxyproline; Pen, pentose; dHex, deoxyhexose; Hex, hexose; HexNAc, N-acetylhexosamine.
Phl p 1 Allergen
The common timothy allergen Phl p 1 was observed at m/z 27,532, which is ∼1400 Da above the predicted molecular weight. The tryptic digest of the natural Phl p 1 was analyzed by bottom-up FTMS and identified with 85% sequence coverage (supplemental Data). For identification of N-glycan modifications of Phl p 1 the ion chromatogram was filtered for the HCD-MS2 m/z 204.08–204.09 ion trace (herein referred to as m/z 204), indicative for the presence of N-acetylhexosamine (HexNAc), and inspected manually. Precursor ions of full scan MS1 spectra, found at specific retention times that coincide with HCD-MS2 m/z 204 ion traces, were summed and deconvoluted into their monoisotopic [M+H]+ masses (Fig. 2A). These precursor ions were matched with good mass accuracy (<5ppm) to several N-glycoforms of the tryptic (V27PPGPNITATYGDK40) and semitryptic (I24PKVPPGPNITATYGDK40) peptides (supplemental Table S1) of Phl p 1. These glycopeptides additionally contained 1–2 hydroxyproline (Hyp) modifications together with the presence of 1–4 pentose (Pen) residues, one of which was the single xylose (Xyl) linked to the N-glycan. The glycan modifications were confirmed for all precursor ions by HCD fragmentation (supplemental Fig. S4). For example, the precursor ions at m/z 2454.098, z = 1 and m/z 2616.151, z = 1 were identified as the tryptic V27PPGPNITATYGDK40 peptide with the N-linked glycans Pen1dHex1Hex2–3HexNAc2 as earlier reported (21, 24). In addition, we identified semitryptic I24PKVPPGPNITATYGDK40 peptides with the N-linked glycans Pen1dHex1Hex2–3HexNAc2 with two Hyps and additional 1–3 Pen residues. Wicklein et al. previously identified a single Hyp linked arabinose (Ara) on Phl p 1 but no reports were made on additional Ara extensions (21). No other modifications were identified on Phl p 1.
Fig. 2.
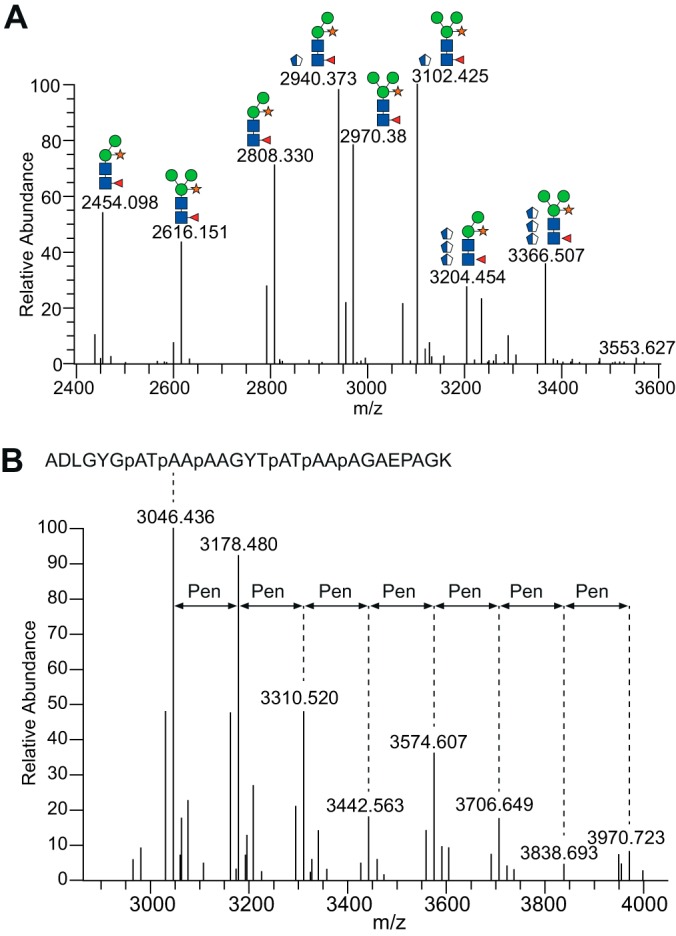
Glycan heterogeneity observed on Phl p 1 and Phl p 5 grass pollen allergens. A, Deconvoluted MS1 spectra of co-eluting glycoforms on the tryptic V27PPGPNITATYGDK40 (including one Hyp) and semitryptic I24PKVPPGPNITATYGDK50 (including two Hyp) peptides originating from Phl p 1. Glycoforms at and above m/z 2808.33 are found on the semitryptic peptide. B, Glycan heterogeneity of the Phl p 5 peptide containing six Hyp residues (indicated as p) and up to seven Pen glycans. All m/z values are represented as [M+H]+. The cartoons shown are based upon mass measurements and assumptions of the known biosynthetic pathways.
HCD-MS2 of Phl p 1 glycopeptides revealed sequential loss of individual monosaccharides with prominent B- and Y-type ions, corresponding to fragmentations at glycosidic bonds (supplemental Fig. S4). The peptide ion with a single HexNAc and Pen residue was typically observed as the most abundant fragment ion for arabinosylated glycopeptides. This suggests that the Hyp linked Pen residue is relatively stable during HCD fragmentation and not eliminated as easily as other forms of O-glycosylations (e.g. GalNAc-type). However, the Pen residue was not stable enough for glycosite assignment and was usually eliminated from b- and y-type fragments, thus precluding confident identification of Pen modified Hyp residues by HCD-MS2.
The HCD spectra were also found to contain fragment ions that were useful for mapping the location of the Hyp resides (supplemental Fig. S4B–4C). The b4 fragment ion (m/z = 438.31, z = 1), present in all I24PKVPPGPNITATYGDK40 glycopeptide spectra, showed that P25 is not modified by hydroxylation. For glycopeptides with single Hyp modifications, we observed the y10+HexNAc ion (P31NITATYGDK40) at m/z 1298.61, z = 1, which shows that the first hydroxylation occurs on P31. The y10 ion, with Hyp on P31, was also observed without HexNAc modification (m/z = 1095.53, z = 1), but typically at low abundance and not always isotopically resolved. Importantly, y10 ions, without hydroxylation on P31, were not observed. For Phl p 1 glycopeptides with two Hyp residues, we observed y10+HexNAc and y11+HexNAc ions (G30PNITATYGDK40) with hydroxylation on P31 at m/z = 1289.61, z = 1 and m/z = 1355.63, z = 1, respectively. In addition, the y12+HexNAc fragment (P29GPNITATYGDK40, m/z = 1452.69, z = 1) was also observed with one Hyp. This fragment includes two proline residues, P29 and P31, but is modified by only one hydroxylation on P31. Again, the b4 fragment was also observed at m/z = 438.31, corresponding to I24PKV27 without hydroxylation, which rules out Hyp at P25. However, the b5 fragment ion was observed at m/z = 551.35, z = 1, which corresponds to the I24PKVP28 fragment with one hydroxylation, thereby showing that the second Hyp residue is on P28. Taken together, the HCD results show that the first and second hydroxylation occurs on P31 and P28, respectively. These results are in agreement with previous reports where Hyp residues were localized through peptide sequencing using Edman degradation (21).
To investigate the number of Pen substitutions on Hyps we performed barium hydroxide mediated peptide bond hydrolysis of Phl p 1 followed by subsequent LC-ESI-MS analysis. With this strategy we identified the presence of the ions at m/z 264, 396, and 528/550 (H+/Na+ forms) corresponding to Hyp-Pen1, Hyp-Pen2, and Hyp-Pen3, with Hyp-Pen1 and Hyp-Pen3 being most abundant as evidenced by visual inspection and integration of peaks in the extracted ion chromatograms (supplemental Fig. S5). The Hyp-Pen1–3 eluted with identical retention times as barium hydroxide hydrolyzed extensin derived Hyp-Arabinofuranose1–3 (Hyp-Araf1–3) on a HPLC column. Structural confirmation of the Hyp-Pen1, Hyp-Pen2, and Hyp-Pen3 species (m/z 264, 396, and 528/550) was obtained by MS2 analysis (supplemental Fig. S5). Other Hyp substituted species, such as the extensin specific Hyp-Araf4, were not identified. Our results show that Phl p 1, besides the previously reported singly Hyp linked Ara (24), also harbors two or three Hyp linked Pen residues (Fig. 2A). Taken together with the HCD-MS2 analysis (supplemental Fig. S4), these results further suggest that only one site is occupied by the Hyp-Pen1–3 modifications.
Phl p 5 Allergen
The MALDI-TOF analysis of the natural Phl p 5 allergen revealed two peaks at m/z 26,373 and m/z 28,746 (supplemental Fig. S2), which deviate by ∼10 kDa in mass compared with the apparent molecular weight observed on the SDS-PAGE gel (supplemental Fig. S1). The two molecular species correspond to two of the main Phl p 5 isoform groups: Phl p 5.010i (MW approx. 27.7 to 18.5) and Phl p 5.020i (MW approx. 26.1), which differ in their primary amino acid sequences (www.allergen.org). Although both Phl p 5 isoforms were identified with >86% sequence coverage (supplemental data) using bottom-up MS, we did not detect a m/z 204 ion trace in any of the HCD-MS2 spectra for these pollen allergens, indicating that Phl p 5 is not modified by any N- or O-linked HexNAc residues. Data mining using Proteome Discoverer 1.4 did, however, identify 0–7 Hyp residues on Phl p 5.0103 (Uniprot accession O81341) found in the N-terminal A26DLGYGPATPAAPAAGYTPATPAAPAGAEPAGK58 peptide (supplemental Fig. S6A). By allowing variable Pen modifications, up to seven Pen residues were also found on this peptide (Fig. 2B, supplemental Fig. S7). Additional Phl p 5 peptides modified by Hyp and Pen are presented in supplemental Fig. S6C-E. No other modifications were identified on Phl p 5. ETD experiments of Phl p 5 glycopeptides were not successful, that is, ETD fragmentation did not produce fragment ions useful for localization of arabinosylated Hyp residues. However, HCD-MS2 demonstrated that all seven prolines (P32, P35, P38, P44, P47, P50, and P55) are hydroxylated (supplemental Fig. S6A). Barium hydroxide mediated peptide bond hydrolysis of Phl p 5 and subsequent ESI-MS analysis identified the presence of the ions at m/z 396 and 528, 550 corresponding to Hyp-Pen2 and Hyp-Pen3, which migrated as barium hydroxide hydrolyzed extensin derived Hyp-Arabinofuranose2 (Hyp-Araf2) and Hyp-Araf3 (supplemental Fig. S5). Structural confirmation of the m/z 396 and 528, 550 species was obtained by MS/MS analysis (supplemental Fig. S5). Other Hyp substituted species were not identified. Our results show that Phl p 5 harbors Hyp substitutions of one to three Pen in length (Fig. 1), which has not been reported previously.
Der p 2 and Der f 2 Allergens
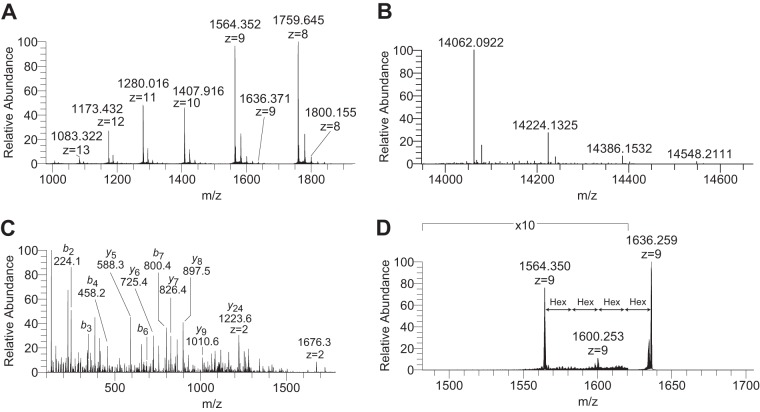
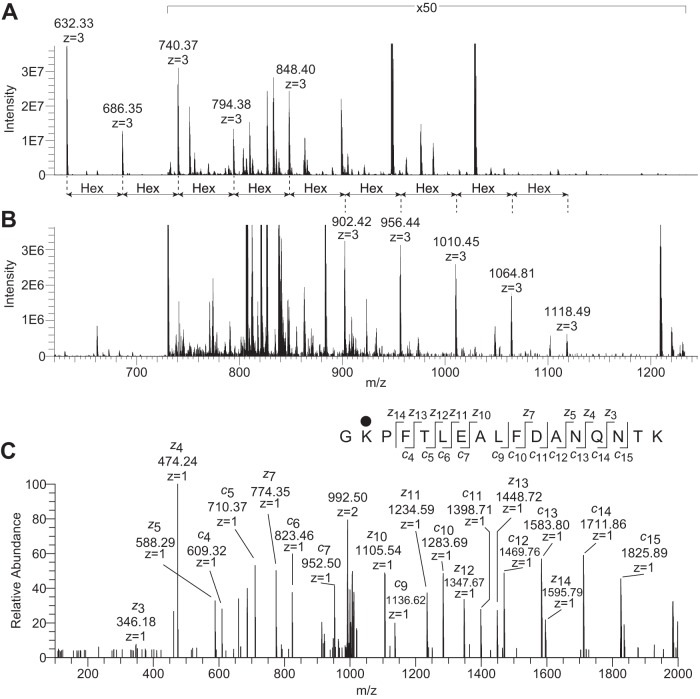
MALDI-TOF analysis of Der p 2 and Der f 2 revealed a major peak at m/z 14,116 (theoretical mass: 14,121 Da) and m/z 14,088 (theoretical mass: 14,076 Da), respectively (supplemental Fig. S2). However, both Der p 2 and Der f 2 also displayed a second peak with a 162 amu increment at m/z 14,278 and m/z 14,250 (supplemental Fig. S3). For precise mass measurements and identification, we subsequently analyzed Der p 2 and Der f 2 by top-down FTMS. A cluster of multiply charged (z = 8–14) ions were observed for Der f 2 (Fig. 3A) and deconvoluted into their monoisotopic masses (Fig. 3B). The most abundant ion (m/z 14062.092, z = 1) was found to deviate by −2.6 ppm from the theoretical mass of the natural Der f 2 protein (Uniprot accession Q00855) containing three intramolecular disulfide bonds. The precursor ion at m/z 1564.35, z = 9 (Fig. 3A) was subjected to HCD fragmentation and revealed several b- and y-type fragments (Fig. 3C) that verified the protein identity. The mass increments were now accurately measured and found to be 162.05 amu, suggesting the presence of Hex residues (Fig. 3B). The precursor ion modified by four potential Hex (m/z 1636.37, z = 9, Fig. 3A) was subsequently subjected to CID fragmentation (Fig. 3D). This revealed a prominent product ion at m/z 1564.35, z = 9 resulting from the loss of all four Hex residues. In addition, a second product ion at 1600.25, z = 9 resulting from the loss of two Hex residues was also evident (Fig. 3D). Although both allergens were identified with >85% sequence coverage using bottom up MS (supplemental data), we failed to detect m/z 204 ion traces in the HCD-MS2 spectra, indicating that these allergens are not modified by HexNAc residues. However, data mining using Proteome Discoverer 1.4 confirmed the presence of Hex modifications (supplemental Table S1) for both Der f 2, shown in Fig. 4, and Der p 2, shown in Fig. 5. Additional spectra and are presented in supplemental Figs. S8 and S9. In Fig. 4A, the Der f 2 G49KPFTLEALFDANQNTK65 peptide is shown to be modified with 0–9 Hex residues. Although the most extensive modification with nine Hex residues occurs with relatively low stoichiometry, these results show the presence of a polyhexose glycan, which could be organized as a linear or branched structure. The relatively low abundance of precursor ions representing peptides modified by more than two Hex residues precluded further MS/MS characterization of the polyhexose sequence (Fig. 4). Next, Der p 2 and Der f 2 allergens were subjected to reductive β-elimination in order to release O-linked glycans for further structural characterization. Although a typical pattern of sialyl-T, disialyl-T, and disialyl core-2 O-glycans were observed for the bovine fetuin standard (not shown), we did not detect the expected glycans from Der p 2 or Der f 2. Depolymerization of the polyhexose sequence using methanolic HCl followed by trimethylsilyl-derivatization and GC-MS did not reveal the epimeric identity of the Hex residues attached to Der p 2 and Der f 2 allergens (not shown). Finally, the attachment site of the PTMs were unambiguously identified through bottom-up ETD analysis of the tryptic digest followed by data mining allowing variable Hex modifications at lysine residues (supplemental Figs. S8 and S9). The ETD-MS2 spectrum of the Der p 2 G49KPFTLEALFDANQNTK68 peptide, with a Hex modification at K50, is shown in Fig. 4C. The z14 fragment, which includes both T53 and T67, was observed without Hex modification at m/z 1595.79. In addition, the c4 fragment, which excludes both T53 and T67 but includes K50, was observed with a 162.05 amu mass increment, corresponding to a Hex residue, at m/z 609.32. These results clearly demonstrate that K50, and not T53 or T67, is modified by Hex.
Fig. 3.
Top-down mass spectrometry of the natural Der f 2 allergen. A, Charge-state distribution of Der f 2 following direct infusion and full MS acquisition with 100,000 resolving power at m/z 400. B, Deconvoluted MS1 of multiply charged precursor ions from panel A. Monoisotopic masses of [M+H]+ forms are shown. C, HCD-MS2 of the m/z 1564.35, z = 9 precursor from panel A. The b- and y-type fragments identify the N- and C terminus of Der f 2. All annotated fragment ions are [M+H]+ unless otherwise indicated. D, CID-MS2 of the m/z 1636.37, z = 9 precursors ion from panel A showing the loss of two and ultimately four hexose residues.
Fig. 4.
Der f 2 tryptic G49KPFTLEALFDANQNTK65 peptide modified by 0–9 Hex units. A, MS1 averaged over eight full scans showing the nonmodified peptide (m/z 632.33, z = 3) as the most intense precursor ion in the m/z 600–1200 mass range. The sequential mass increments of 54 (z = 3) represent individual Hex units. B, MS1 averaged over nine full scans ∼2.5 min earlier in the LC chromatogram showing the G49KPFTLEALFDANQNTK65 modified by 5–9 Hex units. The m/z 735–1230 range is amplified ×50 in panel A and B. C, ETD-MS2 spectrum of the precursor ion at m/z 686.35, z = 3 showing the unambiguous identification of Lys50 as the modification site. Black circle represents hexose.
Fig. 5.
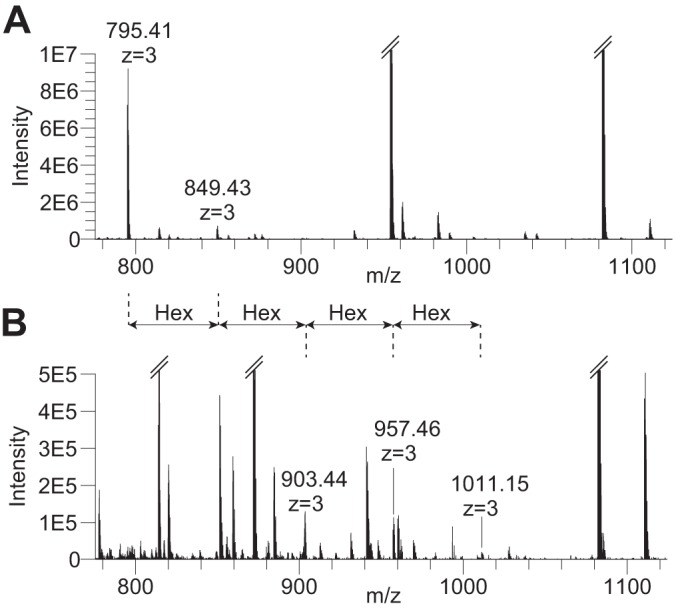
Der p 2 semitryptic G49KPFQLEAVFEANQNTKTAK68 peptide modified by 1–5 Hex units. A, MS1 averaged over seven full scans showing the Hex1 (m/z 795.41, z = 3) and Hex2 (m/z 849.43, z = 3) modified peptides. B, MS1 averaged over four full scans ∼90 s earlier in the LC chromatogram showing the same peptide modified by 2–5 Hex units. The sequential mass increments of 54 (z = 3) represent individual Hex units.
Similar results were obtained for the Der f 2 allergen. The C95PLVKGQQYDIK106 peptide was identified with a single Hex although it lacks serine or threonine residues, thus leaving only tyrosine as a potential site of O-linked glycosylation. ETD fragmentation produced multiple c- and z-fragments, which confirmed the C95PLVKGQQYDIK106 peptide identity (supplemental Fig. 9A). However, fragment ion masses in support of tyrosine O-glycosylation were not found. Instead, a continuous series of both c- and z-fragments in support of Hex modification at K99 were observed in the ETD spectrum that unambiguously identified the lysine residue as the site of modification. These results demonstrate that Der p 2 and Der f 2 are modified by mono- or polyhexoses at multiple lysine residues (Fig. 1, supplemental Table S1).
ELISA Quantification of Carbohydrate Content
The O-mannosylated peptide standard was specifically biotinylated on the hexose residue through periodate oxidation followed by aldehyde-hydrazide chemistry and subsequently used in an ELISA format to determine the carbohydrate amount of the allergens. The molar ratio of carbohydrate containing molecules was determined to be 32% for Der p 2 and 42% for Der f 2, as quantitated by ELISA (supplemental Fig. S10).
Der p 1 and Der f 1 Allergens
The masses m/z 25,554 and m/z 25,365 obtained by MALDI-TOF (supplemental Fig. S2) of the homologous dust mite allergens Der p 1 and Der f 1, respectively, are relatively close to the predicted molecular masses (Der p 1: 25,019 amu and Der f 1: 25,192 amu) of the mature and processed forms of both allergens. Although this indicated that some PTMs might be present, the MALDI-TOF measurement was unable to provide any details other than the mass increments of ∼535 amu and 173 amu for Der p 1 and Der f 1, respectively. The tryptic digests of Der p 1 and Der f 1 were analyzed by bottom up MS, resulting in >74% sequence coverage for both HDM allergens (supplemental Data). A distinct m/z 204 ion trace was observed at ∼48 min in the ion chromatogram of the Der p 1 allergen (not shown) and the HCD-MS2 spectrum of the precursor ion at m/z 814.12, z = 4, shown in Fig. 6, was matched to the tryptic N150QSLDLAEQELVDCASQHGCHGDTIPR176 peptide sequence with a single HexNAc modification (supplemental Fig. S11A). The single HexNAc moiety was mapped to Asn150 by ETD fragmentation (supplemental Fig. S11B), which is located in an N-glycosylation consensus sequence. Furthermore, a second precursor ion (m/z 865.39, z = 4) with a mass increment corresponding to an additional HexNAc residue was also observed and matched to the same peptide sequence (supplemental Table S1, supplemental Fig. S11C). Taken together, these data show that natural Der p 1 is N-glycosylated with one or two HexNAc residues at Asn150. Similar results were obtained for the natural Der f 1 allergen. A prominent m/z 204 trace was found in the tryptic digest of Der f 1 and the HCD-MS2 spectrum of the precursor ion at m/z 811.36, z = 4 (Fig. 6), was matched to the HexNAc modified N151TSLDLSEQELVDCASQHGCHGDTIPR177 peptide of Der f 1 (supplemental Fig. S12A). An additional precursor ion (m/z 862.39, z = 4), corresponding to same peptide modified by two HexNAc residues, was also characterized (supplemental Table S1, supplemental Fig. S12B). Glycans elongated beyond the HexNAc2 structure were not observed for the natural Der p 1 or Der f 1 allergens and peptides harboring these modifications were not observed in their nonglycosylated forms. Both Der p 1 and Der f 1 were also found to be modified by Hex monosaccharides at lysine residues (supplemental Table S1, supplemental Fig. S13). Compositional monosaccharide analysis by GC-MS did not reveal the identity of Hex and HexNAc residues on Der p 1 and Der f 1 allergens. In conclusion, our results show that Der p 1 and Der f 1 allergens are modified by HexNAc glycans at Asn residues and Hex glycans at Lys residues (Fig. 1, supplemental Table S1).
Fig. 6.
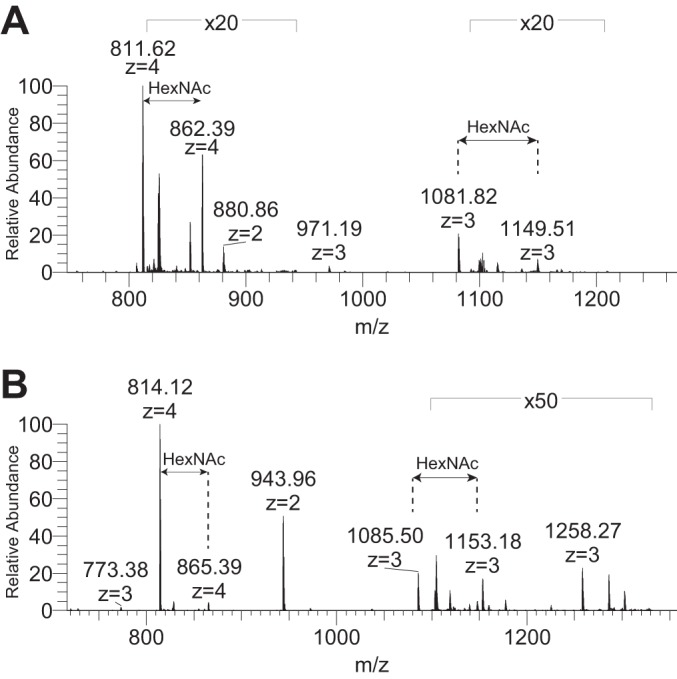
HexNAc modifications of HDM group 1 allergens. A, MS1 full scan showing the precursor ion masses for Der f 1 N151TSLDLSEQELVDCASQHGCHGDTIPR177 peptide modified by one (m/z 811.62, z = 4) and two (m/z 862.39, z = 4) HexNAc glycans at Asn151. B, MS1 full scan showing precursor ion masses for Der p 1 N150QSLDLAEQELVDCASQHGCHGDTIPR176 modified by one (m/z 814.38, z = 4) and two (m/z 865.39, z = 4) HexNAc glycans at Asn150.
Bet v 1 Allergen
The white birch allergen Bet v 1 was mass measured by MALDI (supplemental Fig. S2) and observed as a single peak at m/z 17,474 (theoretical average mass: 17,440 Da), supporting the notion that major modifications are not present on this allergen (37). Bottom up MS of the tryptic digest identified Bet v 1 with 97% sequence coverage (supplemental data), and did not reveal an m/z 204 ion trace or any other glycan modifications in accordance with lack of PTMs on the natural Bet v 1 allergen. Top down FTMS analysis (supplemental Fig. S14) revealed several multiply charged precursor ions (z = 12–22) that were deconvoluted into two prominent peaks at m/z 17429.134, z = 1 and 17429.840, z = 1. The latter mass was matched (-5.5 ppm) to the major isoform of Bet v 1 lacking the initiator methionine. Top down HCD and ETD analysis verified the Bet v 1 sequence identity (supplemental Fig. S14). The precursor ion at m/z 17429.134, z = 1 was found to deviate from the monoisotopic mass of Bet v 1 by −46 ppm. This could be attributed to a single amino acid substitution, for example, Asp to Asn, but has not been confirmed by MS2 analysis. In conclusion, our results demonstrate that Bet v 1 is not subjected to post-translational modifications (Fig. 1).
DISCUSSION
Here we demonstrate that a number of major allergens important in human allergic diseases carry carbohydrate modifications with an hitherto unknown level of complexity that may have implications for pathogenesis, clinical diagnostics, and immune based treatment strategies (19, 38–40). We have adopted a proteocentric approach for mass spectrometric characterization, which we have applied on key grass, tree pollen, and HDM allergens. The data presented provide an in depth analysis of the carbohydrate structure of Der p 1, Der p 2, Der f 1, Der f 2, Bet v 1, Phl p 1, and Phl p 5 exceeding previous investigations in the obtained level of detail (21, 24, 41). We identified novel PTMs of HDM allergens as well as more complex glycan structures than previously reported on the major grass pollen group 1 and 5 allergens. Our findings may have implications for the theoretical understanding of how specific glycans may affect antibody (IgE) reactivity to allergens, antigen uptake, and immune presentation.
By employing bottom-up analysis of tryptic digested Phl p 1 the previously published number of structural variants for Phl p 1 was increased from three (21, 24) to eight glycoforms (Table I). The N-terminal part of the mature Phl p 1 has previously been reported to contain the N-linked Pen1dHex1Hex2–3HexNAc2 core structures with the additional presence of a single Hyp-linked furanosidic arabinose in its close vicinity (21, 24). Here, we confirmed the presence of the Pen1dHex1Hex2–3HexNAc2 core structures and further identified the presence of one to three Pen (supplemental Fig. S4) on Phl p 1. Barium hydroxide treated Phl p 1 analyzed by LC-ESI-MS demonstrated the presence of Hyp substituted with one to three pentose residues (Hyp-Pen1–3) with Hyp-Pen1and Hyp-Pen3 being most abundant. The presence of Hyp substitutions with more than a single Pen on Phl p 1 has not previously been reported. Interestingly, it has earlier been demonstrated that a large proportion of patients with grass allergy have enhanced IgE reactivity to epitopes containing the carbohydrate structure and/or hydroxylation of the proline residues (42). It is therefore plausible that the two N-linked glycoforms (Pen1dHex1Hex2–3HexNAc2) together with the additional presence of the heterogeneous Hyp linked arabinosylations contribute to facilitate the reported high IgE binding and cross reactivity potential of Phl p 1.
The second grass pollen allergen that we examined, Phl p 5, does not have a N-glycosylation consensus motif and is not reported to carry any N-linked glycans. Here we report the presence of 0–7 Hyps and up to seven Pen residues on a defined N-terminal region of Phl p 5 (supplemental Table S1, supplemental Fig. S7). Barium hydroxide treated Phl p 5 analyzed by LC-ESI-MS revealed the presence of Hyps substituted with one to three Pen residues with Hyp-Pen3 being most abundant. This is a first report of Hyp formation and Hyp-substitutions with confirmed Pen1–3 glycans, which may confer significant immunogenic properties to this major allergen. In a recent study, deletion or substitution of selected proline residues in E. coli expressed Phl p 5 yielded a hypoallergenic variant that displayed a highly diminished IgE binding while maintaining T-cell reactivity suggesting involvement of prolines and probably hydroxyprolines in IgE binding to Phl p 5 (43).
N-linked heterogeneous glycosylation of the major allergens of Dermatophagoides pteronyssinus HDM 1 and 2 (Der p 1 and Der p 2) has been demonstrated to influence structure, processing, function, and immunogenicity of these allergens (44–46), although the precise glycan structures remain unresolved (41, 47). With bottom up mass spectrometry, we demonstrated that the natural Der p 1 is N-glycosylated with one or two HexNAc residues at Asn150 (supplemental Fig. S11). A similar glycosylation was obtained for the natural Der f 1 allergen (supplemental Fig. S12). Glycans elongated beyond the HexNAc2 structure were not observed for the natural Der p 1 or Der f 1 allergens. This is, to our knowledge, the first report of confirmed N-linked truncated HexNAc's in mite group 1 allergens.
In both Der p 2 and Der f 2, top down and bottom up FTMS revealed hexose modifications at multiple sites. The combination of top down and bottom up mass spectrometry was of central importance for the successful identification of the hexose modifications for HDM allergens. Initial profiling of total molecular weight using low resolution MALDI-TOF mass spectrometry hinted toward potential hexose modifications, which were verified only when high resolution FTMS was employed followed by top down characterization utilizing collisional activation at the MS2 level. The main advantage of the FTMS top down approach was thus to provide a reference point from which bottom up experiments and, perhaps more importantly, data mining and analysis could be based on. Once the hexose modifications were identified, bottom up FTMS analysis of proteolytic digest was employed not only for detailed mapping of the site-specific locations but also for characterization of glycan heterogeneities. In other words, to achieve complete characterization it was important to first understand the total molecular composition (top down) followed by detailed analysis of PTMs at the peptide level (bottom up).
Initially, the CID and HCD induced loss of hexose units, a typical feature for labile O-linked glycans, mislead us into believing that the mono- and polysaccharide glycans of Der p 2/f 2 were O-linked to Ser, Thr and, for the C95PLVKGQQYDIK106 peptide (supplemental Table S1), also to Tyr residues (supplemental Fig. S15A). Surprisingly, reductive beta elimination, a procedure that specifically releases O-linked sugars from the peptide backbone, failed to liberate the expected glycans of Der p 2 and Der f 2 allergens. The alkaline-stable nature of these PTMs thus indicated that the polyhexose glycans of these allergens are linked by other means than the classical O-linkage to Ser/Thr residues. In addition, the Hex modified G49KPFQLEALFEANQNSKTAK68 peptide of one Der p 2 isoform (Uniprot accession I2CMD6) was identified with an almost complete series of c- and z-ions, mapping the modification to S64 (supplemental Fig. S16A). However, one of the few ions missing in the series was the c16/z4 ion pair that separates S64 and K65. Notably, both the C95PLVKGQQYDIK106 and G49KPFQLEALFEANQNSKTAK68 peptide contained a missed trypsin cleavage site at lysine residues. Taken together, these results raised suspicions that lysines, and not Ser/Thr/Tyr, might be the modified residues. Attachment of sugars at lysine residues would account for the missed tryptic cleavages and the alkaline-stable nature of the modifications. We therefore re-analyzed our data by allowing variable modifications also at lysine residues. This approach resulted in the identification of all but one c- and z-ions between K99 and Y103 of the C95PLVKGQQYDIK106 sequence, thereby leading to the unambiguous identification of K99 as the modified residue (supplemental Fig. S15B). For the G49KPFQLEALFEANQNSKTAK68 peptide, the c16 ion was now also identified, which mapped the Hex modification to K65 (supplemental Fig. S16B). The attachment of Hex to lysine, which occurs through N-linkage, also explains the beta-elimination resistant properties of these modifications. In an attempt to identify the monosaccharide identities of the attached Hex residues, GC-MS was employed but did not reveal the epimeric configurations of the attached sugars. The Hex1 modification was observed on ∼30% (molar ratio) of Der p 2 but, because of the N-linkage, this lysine-linked monosaccharide is not expected to be release by acid methanolysis (48). Although significant amounts of Der p 2 are modified by this initial Hex, this allergen pool does not contribute with any free Hex residues to the final sample subjected to GC-MS analysis. The O-glycosidic linkages connecting individual monosaccharides in the polyhexose sequence are cleaved by methanolic HCl but the Hex2–9 polyhexose sequence is ≤50 times less abundant relative to the Hex1 form. In other words, only 5 ng or less Hex is expected to be released (from 50 μg Der p 2) for TMS-derivatization and GC-MS analysis. The same principle applies to the Der p 1 and Der f 1 allergens, that is, the N-linkage of HexNAc at asparagine is stable to methanolysis and ≤ 15 ng HexNAc is expected to be released for TMS-derivatization. These amounts are not sufficient for successful GC-MS analysis.
Given that the relative ion abundances are not reliable for determining the stoichiometry of these modifications, we also designed an assay for absolute quantification of carbohydrate content in Der p 2 and Der f 2 allergens. Periodate oxidation of cis-diols is a well-known procedure for introduction of reactive aldehyde functionalities specifically on glycans (49). This property may thus be exploited to tag oxidized glycans by hydrazide-containing molecules of choice, e.g. biotin-hydrazide, which could then be used in conventional ELISA format to quantify the carbohydrate content of specific glycoproteins. Using this approach, we were able to conclude that 30 and 40% (molar ratio) of Der p 2 and Der f 2, respectively, are modified by glycans, thereby confirming that a significant amount of these allergens are subjected to Hex modifications. Of note, the ELISA values are in agreement with the ion abundances observed for modified Der p 2 and Der f 2 as measured by MALDI-TOF and top down ESI-MS (Fig. 2, supplemental Fig. S3).
Although an enzymatic mechanism for the attachment of Hex to lysine residues in HDM allergens cannot be completely excluded, it is likely that the modifications may be a result of nonenzymatic glycation reactions. We do not believe that these modifications are introduced during purification procedures because all purifications steps of HDM allergens were performed at 4–12 °C using buffers that did not contain free-end mono- or polysaccharides. The results therefore strongly suggest that these glycans on the allergens originate from the mites. HDM are believed to have a repertoire of up to 30 distinct allergens (50, 51) and it is likely that the identified glycans may be present on many other allergens from mites. Glycations have previously been described on peanut allergens Ara h 1 and Ara h 3 as well as on ovalbumin (52, 53). The modification has been implicated in interactions with APCs and might have caused increased immunogenicity (52, 53). Furthermore, the receptor for advanced glycation end products (RAGE) has been shown to be an important mediator in allergic airway sensitization after HDM challenge (54), although the exact mechanisms and role of glycation in immune recognition and response is still poorly understood.
Glycosylation on allergens may play other roles in allergy than simply presenting antigen epitopes. APCs are equipped with an array of glycan receptors and targeting such receptors has been shown to induce tolerance (15, 55–62). The idea that carbohydrates are able to boost and shape the immune response has been tested previously in numerous studies, systematically reviewed in (63). In a food allergy model it has, for example, been shown that oral delivery of bovine serum albumin (BSA) carrying mannoside residues significantly reduce BSA-induced anaphylactic response in the prophylactic setting (64). Furthermore, in the study by Thunberg et al. allergen rFel d 1 bound to carbohydrate based particles promoted uptake, induced strong antibody and cytokine response, and prevented induction of allergic immune responses (65). In addition, a tolerogenic activity has been shown for the carbohydrate receptor DC-SIGN in humans (66, 67). Taken together, these and other studies support the importance of the CLR-ligand axis in the development of targeted therapies aimed at inducing immune modulation (12, 68). Thus, it will be interesting to test the potential immunogenic properties of the individual PTMs and their potential influence of the allergenicity of major allergens. Furthermore, the detailed characterization of glycans could also be important for the development of well-defined allergens for diagnostic purposes (8–11).
In conclusion, our findings demonstrate that most major allergens harbor extensive and diverse glycans, and detailed analysis of individual allergens are necessary to obtain full characterization. The presence of glycans on allergens may have profound importance for the immunogenicity of allergens as well as their presentation to the immune system. The detailed characterization presented here, now opens for direct analysis of the implications of these glycans for designing allergens for future vaccine formulations and diagnostic purposes.
Supplementary Material
Acknowledgments
We thank Ola Blixt for synthesizing and providing the O-mannose peptide standard.
Footnotes
Author contributions: A.H. and H.H.W. designed research; A.H., M.C.C., C.B.M., S.B., S.R.M., C.E.O., and S.Y.V. performed research; A.H., C.E.O., J.B., P.A.W., H.I., and B.L.P. contributed new reagents or analytic tools; A.H., M.C.C., C.B.M., S.B., S.R.M., C.E.O., S.Y.V., J.B., P.A.W., H.I., B.L.P., and H.H.W. analyzed data; A.H., B.L.P., and H.H.W. wrote the paper.
* This work was in part supported by The Danish Council for Strategic Research, The Danish Council for Independent Research (Sapere Aude program), a program of excellence 2016 (CDO2016) from the University of Copenhagen, The Danish National Research Foundation (DNRF107), The Friis Foundation, A.P. Møller og Hustru Chastine Mc-Kinney Møllers Fond til Almene Formaal, Kirsten og Freddy Johansen Fonden, The Carlsberg Foundation and The Novo Nordisk Foundation
 This article contains supplemental Figs. S1 to S16 and Tables S1 and S2.
This article contains supplemental Figs. S1 to S16 and Tables S1 and S2.
1 The abbreviations used are:
- PTM
- Post-translational modification
- APC
- Antigen presenting cells
- Ara
- Arabinose
- Bet v
- Betula verrucosa
- BSA
- Bovine serum albumin
- CCD
- Cross-reactive carbohydrate determinant
- CID
- Collision induced dissociation
- CLR
- C-type lectin receptor
- Der f
- Dermatophagoides farinae
- Der p
- Dermatophagoides pteronyssinus
- dHex
- Deoxyhexose
- ECD
- Electron capture dissociation
- ETD
- Electron transfer dissociation
- Fuc
- Fucose
- HCD
- Higher-energy collisional dissociation
- Hex
- Hexose
- HexNAc
- N-acetylhexosamine
- HIC
- Hydrophobic interaction chromatography
- HMD
- House dust mite
- Hyp
- Hydroxyproline
- IgE
- Immunoglobulin E
- Pen
- Pentose
- Phl p
- Phleum pretense
- RAGE
- Receptor for advanced glycation end products
- SEC
- Size exclusion chromatography
- Xyl
- Xylose.
REFERENCES
- 1. Bousquet J., Khaltaev N., Cruz A. A., Denburg J., Fokkens W. J., Togias A., Zuberbier T., Baena-Cagnani C. E., Canonica G. W., van Weel C., Agache I., Ait-Khaled N., Bachert C., Blaiss M. S., Bonini S., Boulet L. P., Bousquet P. J., Camargos P., Carlsen K. H., Chen Y., Custovic A., Dahl R., Demoly P., Douagui H., Durham S. R., van Wijk R. G., Kalayci O., Kaliner M. A., Kim Y. Y., Kowalski M. L., Kuna P., Le L. T., Lemiere C., Li J., Lockey R. F., Mavale-Manuel S., Meltzer E. O., Mohammad Y., Mullol J., Naclerio R., O'Hehir R. E., Ohta K., Ouedraogo S., Palkonen S., Papadopoulos N., Passalacqua G., Pawankar R., Popov T. A., Rabe K. F., Rosado-Pinto J., Scadding G. K., Simons F. E., Toskala E., Valovirta E., van Cauwenberge P., Wang D. Y., Wickman M., Yawn B. P., Yorgancioglu A., Yusuf O. M., Zar H., Annesi-Maesano I., Bateman E. D., Ben Kheder A., Boakye D. A., Bouchard J., Burney P., Busse W. W., Chan-Yeung M., Chavannes N. H., Chuchalin A., Dolen W. K., Emuzyte R., Grouse L., Humbert M., Jackson C., Johnston S. L., Keith P. K., Kemp J. P., Klossek J. M., Larenas-Linnemann D., Lipworth B., Malo J. L., Marshall G. D., Naspitz C., Nekam K., Niggemann B., Nizankowska-Mogilnicka E., Okamoto Y., Orru M. P., Potter P., Price D., Stoloff S. W., Vandenplas O., Viegi G., Williams D. (2008) Allergic Rhinitis and its Impact on Asthma (ARIA) 2008 update (in collaboration with the World Health Organization, GA(2)LEN and AllerGen). Allergy 86, 8–160 [DOI] [PubMed] [Google Scholar]
- 2. Bousquet J., Lockey R., Malling H. J. (1998) Allergen immunotherapy: therapeutic vaccines for allergic diseases. A WHO position paper. J. Allergy Clin. Immunol. 102, 558–562 [DOI] [PubMed] [Google Scholar]
- 3. Pawankar R., Canonica G. W., Holgate S. T., Lockey R. F. (2012) Allergic diseases and asthma: a major global health concern. Curr. Opin. Allergy Cl. 12, 39–41 [DOI] [PubMed] [Google Scholar]
- 4. Jacobsen L., Wahn U., Bilo M. B. (2012) Allergen-specific immunotherapy provides immediate, long-term and preventive clinical effects in children and adults: the effects of immunotherapy can be categorised by level of benefit -the centenary of allergen specific subcutaneous immunotherapy. Clin. Transl. Allergy 2, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pauli G., Malling H. J. (2010) The current state of recombinant allergens for immunotherapy. Curr. Opin. Allergy Cl. 10, 575–581 [DOI] [PubMed] [Google Scholar]
- 6. Valenta R., Campana R., Marth K., van Hage M. (2012) Allergen-specific immunotherapy: from therapeutic vaccines to prophylactic approaches. J. Intern. Med. 272, 144–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jin C., Altmann F., Strasser R., Mach L., Schahs M., Kunert R., Rademacher T., Glossl J., Steinkellner H. (2008) A plant-derived human monoclonal antibody induces an anti-carbohydrate immune response in rabbits. Glycobiology 18, 235–241 [DOI] [PubMed] [Google Scholar]
- 8. Kaulfurst-Soboll H., Rips S., Koiwa H., Kajiura H., Fujiyama K., von Schaewen A. (2011) Reduced immunogenicity of Arabidopsis hgl1 mutant N-glycans caused by altered accessibility of xylose and core fucose epitopes. J. Biol. Chem. 286, 22955–22964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaulfurst-Soboll H., Mertens M., Brehler R., von Schaewen A. (2011) Reduction of cross-reactive carbohydrate determinants in plant foodstuff: elucidation of clinical relevance and implications for allergy diagnosis. PloS One 6, e17800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muller U. R., Johansen N., Petersen A. B., Fromberg-Nielsen J., Haeberli G. (2009) Hymenoptera venom allergy: analysis of double positivity to honey bee and Vespula venom by estimation of IgE antibodies to species-specific major allergens Api m1 and Ves v5. Allergy 64, 543–548 [DOI] [PubMed] [Google Scholar]
- 11. Mari A. (2002) IgE to cross-reactive carbohydrate determinants: analysis of the distribution and appraisal of the in vivo and in vitro reactivity. Int. Arch. Allergy Immunol. 129, 286–295 [DOI] [PubMed] [Google Scholar]
- 12. Geijtenbeek T. B., Gringhuis S. I. (2009) Signalling through C-type lectin receptors: shaping immune responses. Nat. Rev. Immunol. 9, 465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Emara M., Royer P. J., Mahdavi J., Shakib F., Ghaemmaghami A. M. (2012) Retagging identifies dendritic cell-specific intercellular adhesion molecule-3 (ICAM3)-grabbing nonintegrin (DC-SIGN) protein as a novel receptor for a major allergen from house dust mite. J. Biol. Chem. 287, 5756–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Royer P. J., Emara M., Yang C., Al-Ghouleh A., Tighe P., Jones N., Sewell H. F., Shakib F., Martinez-Pomares L., Ghaemmaghami A. M. (2010) The mannose receptor mediates the uptake of diverse native allergens by dendritic cells and determines allergen-induced T cell polarization through modulation of IDO activity. J. Immunol. 185, 1522–1531 [DOI] [PubMed] [Google Scholar]
- 15. Unger W. W., van Kooyk Y. (2011) “Dressed for success” C-type lectin receptors for the delivery of glyco-vaccines to dendritic cells. Curr. Opin. Immunol. 23, 131–137 [DOI] [PubMed] [Google Scholar]
- 16. Takeuchi O., Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 [DOI] [PubMed] [Google Scholar]
- 17. Seppala U., Selby D., Monsalve R., King T. P., Ebner C., Roepstorff P., Bohle B. (2009) Structural and immunological characterization of the N-glycans from the major yellow jacket allergen Ves v 2: the N-glycan structures are needed for the human antibody recognition. Mol. Immunol. 46, 2014–2021 [DOI] [PubMed] [Google Scholar]
- 18. Leonard R., Petersen B. O., Himly M., Kaar W., Wopfner N., Kolarich D., van Ree R., Ebner C., Duus J. O., Ferreira F., Altmann F. (2005) Two novel types of O-glycans on the mugwort pollen allergen Art v 1 and their role in antibody binding. J. Biol. Chem. 280, 7932–7940 [DOI] [PubMed] [Google Scholar]
- 19. Salazar F., Sewell H. F., Shakib F., Ghaemmaghami A. M. (2013) The role of lectins in allergic sensitization and allergic disease. J. Allergy Clin. Immunol. 132, 27–36 [DOI] [PubMed] [Google Scholar]
- 20. Manduzio H., Fitchette A. C., Hrabina M., Chabre H., Batard T., Nony E., Faye L., Moingeon P., Gomord V. (2012) Glycoproteins are species-specific markers and major IgE reactants in grass pollens. Plant Biotechnol. J. 10, 184–194 [DOI] [PubMed] [Google Scholar]
- 21. Wicklein D., Lindner B., Moll H., Kolarich D., Altmann F., Becker W. M., Petersen A. (2004) Carbohydrate moieties can induce mediator release: a detailed characterization of two major timothy grass pollen allergens. Biol. Chem. 385, 397–407 [DOI] [PubMed] [Google Scholar]
- 22. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 24. Fenaille F., Nony E., Chabre H., Lautrette A., Couret M. N., Batard T., Moingeon P., Ezan E. (2009) Mass spectrometric investigation of molecular variability of grass pollen group 1 allergens. J. Proteome Res. 8, 4014–4027 [DOI] [PubMed] [Google Scholar]
- 25. Halim A., Nilsson J., Ruetschi U., Hesse C., Larson G. (2012) Human urinary glycoproteomics; attachment site specific analysis of N- and O-linked glycosylations by CID and ECD. Mol, Cell. Proteomics 11, M111 013649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Bennett E. P., Mandel U., Brunak S., Wandall H. H., Levery S. B., Clausen H. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vakhrushev S. Y., Steentoft C., Vester-Christensen M. B., Bennett E. P., Clausen H., Levery S. B. (2013) Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells. Mol. Cell. Proteomics 12, 932–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lanucara F., Eyers C. E. (2013) Top-down mass spectrometry for the analysis of combinatorial post-translational modifications. Mass Spectrom. Rev. 32, 27–42 [DOI] [PubMed] [Google Scholar]
- 29. Smith L. M., Kelleher N. L., and Consortium for Top Down, P. (2013) Proteoform: a single term describing protein complexity. Nat. Methods 10, 186–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Catherman A. D., Durbin K. R., Ahlf D. R., Early B. P., Fellers R. T., Tran J. C., Thomas P. M., Kelleher N. L. (2013) Large-scale top-down proteomics of the human proteome: membrane proteins, mitochondria, and senescence. Mol. Cell. Proteomics 12, 3465–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Catherman A. D., Skinner O. S., Kelleher N. L. (2014) Top Down proteomics: facts and perspectives. Biochem. Biophys. Res. Commun. 445, 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micropurification, enrichment, prefractionation, and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 33. Jensen P. H., Karlsson N. G., Kolarich D., Packer N. H. (2012) Structural analysis of N- and O-glycans released from glycoproteins. Nat. Protoc. 7, 1299–1310 [DOI] [PubMed] [Google Scholar]
- 34. Ciucanu I., Costello C. E. (2003) Elimination of oxidative degradation during the per-O-methylation of carbohydrates. J. Am. Chem. Soc. 125, 16213–16219 [DOI] [PubMed] [Google Scholar]
- 35. Egelund J., Obel N., Ulvskov P., Geshi N., Pauly M., Bacic A., Petersen B. L. (2007) Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1, and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol. Biol. 64, 439–451 [DOI] [PubMed] [Google Scholar]
- 36. Velasquez S. M., Ricardi M. M., Dorosz J. G., Fernandez P. V., Nadra A. D., Pol-Fachin L., Egelund J., Gille S., Harholt J., Ciancia M., Verli H., Pauly M., Bacic A., Olsen C. E., Ulvskov P., Petersen B. L., Somerville C., Iusem N. D., Estevez J. M. (2011) O-glycosylated cell wall proteins are essential in root hair growth. Science 332, 1401–1403 [DOI] [PubMed] [Google Scholar]
- 37. Swoboda I. S., Jilek A. J., Ferreira F. F., Engel E. E., Hoffmann-Sommergruber K., Scheiner O. S., Kraft D., Breiteneder H., Pittenauer E., Schmid E., Erwin Heberle-Bors E., Horst A., Michael Breitenbach M. (1995) Isoforms of Bet v 1, the major birch pollen allergen, analyzed by liquid chromatography, mass spectrometry, and cDNA cloning. J. Biol. Chem. 270, 2607–2613 [DOI] [PubMed] [Google Scholar]
- 38. Foetisch K., Westphal S., Lauer I., Retzek M., Altmann F., Kolarich D., Scheurer S., Vieths S. (2003) Biological activity of IgE specific for cross-reactive carbohydrate determinants. J. Allergy Clin. Immunol. 111, 889–896 [DOI] [PubMed] [Google Scholar]
- 39. Bublin M., Radauer C., Wilson I. B., Kraft D., Scheiner O., Breiteneder H., Hoffmann-Sommergruber K. (2003) Cross-reactive N-glycans of Api g 5, a high molecular weight glycoprotein allergen from celery, are required for immunoglobulin E binding and activation of effector cells from allergic patients. FASEB J. 17, 1697–1699 [DOI] [PubMed] [Google Scholar]
- 40. Schmid-Grendelmeier P., Holzmann D., Himly M., Weichel M., Tresch S., Ruckert B., Menz G., Ferreira F., Blaser K., Wuthrich B., Crameri R. (2003) Native Art v 1 and recombinant Art v 1 are able to induce humoral and T cell-mediated in vitro and in vivo responses in mugwort allergy. J. Allergy Clin. Immunol. 111, 1328–1336 [DOI] [PubMed] [Google Scholar]
- 41. Al-Ghouleh A., Johal R., Sharquie I. K., Emara M., Harrington H., Shakib F., Ghaemmaghami A. M. (2012) The glycosylation pattern of common allergens: the recognition and uptake of Der p 1 by epithelial and dendritic cells is carbohydrate dependent. PloS One 7, e33929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petersen A., Schramm G., Schlaak M., Becker W. M. (1998) Post-translational modifications influence IgE reactivity to the major allergen Phl p 1 of timothy grass pollen. Clin. Exp. Allergy 28, 315–321 [DOI] [PubMed] [Google Scholar]
- 43. Wald M., Kahlert H., Reese G., Krontal N., Zafred D., Keller W., Cromwell O., Fiebig H., Nandy A. (2012) Hypoallergenic mutants of the Timothy grass pollen allergen Phl p 5 generated by proline mutations. Int. Arch. Allergy Immunol. 159, 130–142 [DOI] [PubMed] [Google Scholar]
- 44. Dumez M. E., Teller N., Mercier F., Tanaka T., Vandenberghe I., Vandenbranden M., Devreese B., Luxen A., Frere J. M., Matagne A., Jacquet A., Galleni M., Chevigne A. (2008) Activation mechanism of recombinant Der p 3 allergen zymogen: contribution of cysteine protease Der p 1 and effect of propeptide glycosylation. J. Biol. Chem. 283, 30606–30617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang J., Saint-Remy J. M., Garrod D. R., Robinson C. (2009) Comparative enzymology of native and recombinant house dust mite allergen Der p 1. Allergy 64, 469–477 [DOI] [PubMed] [Google Scholar]
- 46. Takai T., Mizuuchi E., Kikuchi Y., Nagamune T., Okumura K., Ogawa H. (2006) Glycosylation of recombinant proforms of major house dust mite allergens Der p 1 and Der f 1 decelerates the speed of maturation. Int. Arch. Allergy Immunol. 139, 181–187 [DOI] [PubMed] [Google Scholar]
- 47. Meno K., Thorsted P. B., Ipsen H., Kristensen O., Larsen J. N., Spangfort M. D., Gajhede M., Lund K. (2005) The crystal structure of recombinant proDer p 1, a major house dust mite proteolytic allergen. J. Immunol. 175, 3835–3845 [DOI] [PubMed] [Google Scholar]
- 48. Merkle R. K., Poppe I. (1994) Carbohydrate composition analysis of glycoconjugates by gas-liquid chromatography/mass spectrometry. Methods Enzymol. 230, 1–15 [DOI] [PubMed] [Google Scholar]
- 49. Van Lenten L., Ashwell G. (1971) Studies on the chemical and enzymatic modification of glycoproteins. A general method for the tritiation of sialic acid-containing glycoproteins. J. Biol. Chem. 246, 1889–1894 [PubMed] [Google Scholar]
- 50. An S., Chen L., Long C., Liu X., Xu X., Lu X., Rong M., Liu Z., Lai R. (2013) Dermatophagoides farinae allergens diversity identification by proteomics. Mol. Cell. Proteomics 12, 1818–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Le Mao J., Mayer C. E., Peltre G., Desvaux F. X., David B., Weyer A., Senechal H. (1998) Mapping of Dermatophagoides farinae mite allergens by two-dimensional immunoblotting. J. Allergy Clin. Immunol. 102, 631–636 [DOI] [PubMed] [Google Scholar]
- 52. Mueller G. A., Maleki S. J., Johnson K., Hurlburt B. K., Cheng H., Ruan S., Nesbit J. B., Pomés A., Edwards L. L., Schorzman A., Deterding L. J., Park H., Tomer K. B., London R. E., Williams J. G. (2013) Identification of Maillard reaction products on peanut allergens that influence binding to the receptor for advanced glycation end products. Allergy 68, 1546–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hilmenyuk T., Bellinghausen I., Heydenreich B., Ilchmann A., Toda M., Grabbe S., Saloga J. (2010) Effects of glycation of the model food allergen ovalbumin on antigen uptake and presentation by human dendritic cells. Immunology 129, 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ullah M. A., Loh Z., Gan W. J., Zhang V., Yang H., Li J. H., Yamamoto Y., Schmidt A. M., Armour C. L., Hughes J. M., Phipps S., Sukkar M. B. (2014) Receptor for advanced glycation end products and its ligand high-mobility group box-1 mediate allergic airway sensitization and airway inflammation. J. Allergy Clin. Immunol. 134(2), 440–50 [DOI] [PubMed] [Google Scholar]
- 55. Singh S. K., Streng-Ouwehand I., Litjens M., Kalay H., Burgdorf S., Saeland E., Kurts C., Unger W. W., van Kooyk Y. (2011) Design of neo-glycoconjugates that target the mannose receptor and enhance TLR-independent cross-presentation and Th1 polarization. Eur. J. Immunol. 41, 916–925 [DOI] [PubMed] [Google Scholar]
- 56. Madsen C. B., Petersen C., Lavrsen K., Harndahl M., Buus S., Clausen H., Pedersen A. E., Wandall H. H. (2012) Cancer associated aberrant protein O-glycosylation can modify antigen processing and immune response. PloS One 7, e50139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Madsen C. B., Lavrsen K., Steentoft C., Vester-Christensen M. B., Clausen H., Wandall H. H., Pedersen A. E. (2013) Glycan elongation beyond the mucin associated Tn antigen protects tumor cells from immune-mediated killing. PloS One 8, e72413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Joshi M. D., Unger W. J., Storm G., van Kooyk Y., Mastrobattista E. (2012) Targeting tumor antigens to dendritic cells using particulate carriers. J. Control. Release 161, 25–37 [DOI] [PubMed] [Google Scholar]
- 59. Unger W. W., van Beelen A. J., Bruijns S. C., Joshi M., Fehres C. M., van Bloois L., Verstege M. I., Ambrosini M., Kalay H., Nazmi K., Bolscher J. G., Hooijberg E., de Gruijl T. D., Storm G., van Kooyk Y. (2012) Glycan-modified liposomes boost CD4+ and CD8+ T-cell responses by targeting DC-SIGN on dendritic cells. J. Control. Release 160, 88–95 [DOI] [PubMed] [Google Scholar]
- 60. Garcia-Vallejo J. J., Unger W. W., Kalay H., van Kooyk Y. (2013) Glycan-based DC-SIGN targeting to enhance antigen cross-presentation in anticancer vaccines. Oncoimmunology 2, e23040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. van Kooyk Y., Unger W. W., Fehres C. M., Kalay H., Garcia-Vallejo J. J. (2013) Glycan-based DC-SIGN targeting vaccines to enhance antigen cross-presentation. Mol. Immunol. 55, 143–145 [DOI] [PubMed] [Google Scholar]
- 62. Garcia-Vallejo J. J., Ambrosini M., Overbeek A., van Riel W. E., Bloem K., Unger W. W., Chiodo F., Bolscher J. G., Nazmi K., Kalay H., van Kooyk Y. (2013) Multivalent glycopeptide dendrimers for the targeted delivery of antigens to dendritic cells. Mol. Immunol. 53, 387–397 [DOI] [PubMed] [Google Scholar]
- 63. Johnson J. L., Jones M. B., Ryan S. O., Cobb B. A. (2013) The regulatory power of glycans and their binding partners in immunity. Trends Immunol. 34, 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhou Y., Kawasaki H., Hsu S. C., Lee R. T., Yao X., Plunkett B., Fu J., Yang K., Lee Y. C., Huang S. K. (2010) Oral tolerance to food-induced systemic anaphylaxis mediated by the C-type lectin SIGNR1. Nat. Med. 16, 1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Thunberg S., Neimert-Andersson T., Cheng Q., Wermeling F., Bergstrom U., Swedin L., Dahlen S. E., Arner E., Scheynius A., Karlsson M. C., Gafvelin G., van Hage M., Gronlund H. (2009) Prolonged antigen-exposure with carbohydrate particle based vaccination prevents allergic immune responses in sensitized mice. Allergy 64, 919–926 [DOI] [PubMed] [Google Scholar]
- 66. Geijtenbeek T. B., Van Vliet S. J., Koppel E. A., Sanchez-Hernandez M., Vandenbroucke-Grauls C. M., Appelmelk B., Van Kooyk Y. (2003) Mycobacteria target DC-SIGN to suppress dendritic cell function. J. Exp. Med. 197, 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shan M., Gentile M., Yeiser J. R., Walland A. C., Bornstein V. U., Chen K., He B., Cassis L., Bigas A., Cols M., Comerma L., Huang B., Blander J. M., Xiong H., Mayer L., Berin C., Augenlicht L. H., Velcich A., Cerutti A. (2013) Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science 342, 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gringhuis S. I., den Dunnen J., Litjens M., van der Vlist M., Geijtenbeek T. B. (2009) Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat. Immunol. 10, 1081–1088 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.