Abstract
Advances in phosphopeptide enrichment methods enable the identification of thousands of phosphopeptides from complex samples. Current offline enrichment approaches using TiO2, Ti, and Fe immobilized metal ion affinity chromatography (IMAC) material in batch or microtip format are widely used, but they suffer from irreproducibility and compromised selectivity. To address these shortcomings, we revisited the merits of performing phosphopeptide enrichment in an HPLC column format. We found that Fe-IMAC columns enabled the selective, comprehensive, and reproducible enrichment of phosphopeptides out of complex lysates. Column enrichment did not suffer from bead-to-sample ratio issues and scaled linearly from 100 μg to 5 mg of digest. Direct measurements on an Orbitrap Velos mass spectrometer identified >7500 unique phosphopeptides with 90% selectivity and good quantitative reproducibility (median cv of 15%). The number of unique phosphopeptides could be increased to more than 14,000 when the IMAC eluate was subjected to a subsequent hydrophilic strong anion exchange separation. Fe-IMAC columns outperformed Ti-IMAC and TiO2 in batch or tip mode in terms of phosphopeptide identification and intensity. Permutation enrichments of flow-throughs showed that all materials largely bound the same phosphopeptide species, independent of physicochemical characteristics. However, binding capacity and elution efficiency did profoundly differ among the enrichment materials and formats. As a result, the often quoted orthogonality of the materials has to be called into question. Our results strongly suggest that insufficient capacity, inefficient elution, and the stochastic nature of data-dependent acquisition in mass spectrometry are the causes of the experimentally observed complementarity. The Fe-IMAC enrichment workflow using an HPLC format developed here enables rapid and comprehensive phosphoproteome analysis that can be applied to a wide range of biological systems.
Protein phosphorylation is a reversible post-translational modification with pivotal roles in cellular signaling. It is implicated in many essential biological processes, and aberrant protein phosphorylation is causally linked to numerous diseases (1). At least one-third to one-half of all human proteins are thought to be phosphorylated at some point (2, 3), and hundreds of thousands of different phosphorylation sites are believed to exist. Because of this molecular complexity and the often substoichiometric extent of phosphorylation, specific enrichment prior to analysis via liquid chromatography–tandem mass spectrometry (LC-MS/MS)1 is generally required (4, 5). Most current such strategies exploit the affinity of phosphate groups for metals immobilized on carrier resins. These approaches include Fe3+ (6), Ga3+ (7), and Zr4+ (8) immobilized metal ion affinity chromatography (IMAC); metal oxide affinity chromatography (TiO2 (9), ZrO2 (10), and others); and the recently introduced Ti-IMAC, which is a hybrid of the other two methods (11, 12). It is often stated, if not generally accepted, that these enrichment methods are capable of purifying complementary parts of the phosphoproteome with unique physicochemical characteristics (11, 13, 14). For example, Ti-IMAC is thought to be better for purifying basophilic phosphopeptides than TiO2 (11), whereas Fe-IMAC is credited with the more efficient enrichment of multiply phosphorylated peptides (15). The current consensus in the field is that no method is superior over others and none alone is sufficient for the comprehensive purification of the phosphoproteome, a view that we challenge in this study. Most phosphopeptide enrichments are conducted either in a batch format or in self-constructed microcolumns packed into pipette tips. Both formats have been found to suffer from considerable variability introduced by various manual steps in the process. Moreover, differences in loading conditions including additives (16, 17), acid concentration (17), incubation time (17, 18), and wash volume (18) can lead to different results. One prominent parameter that was found to have a considerable effect on the enrichment quality is the ratio of bead amount to protein digest quantity, and finding the optimal ratio is often a tradeoff between comprehensiveness and selectivity (13, 18–21). Consequently, careful a priori evaluation and optimization of enrichment conditions is generally required on a case-by-case basis for every experimental system. Even then, the use of such formats often comes at the expense of intra-experimental and, even more so, inter-experimental accuracy. As a means to overcome these issues, direct coupling of the chromatographic enrichment step with the LC-MS/MS system has been applied. Although these online systems increase reproducibility, sensitivity, and robustness, they suffer from limited capacity, which is why they have been primarily used for the analysis of limited sample amounts or samples of rather low complexity (9, 22–27).
In order to address this, phosphopeptide enrichment workflows often employ an upstream peptide separation step followed by phosphopeptide enrichment from each fraction. Examples of first-dimension separations include hydrophilic interaction liquid chromatography (28), electrostatic repulsion hydrophilic interaction chromatography (29), strong anion exchange (30), and strong cation exchange (11, 31). Although powerful and frequently used, enrichment after fractionation can be irreproducible and time consuming. As a consequence, reversing the order (i.e. enrichment of phosphopeptides first, followed by fractionation of the phosphopeptide pool) has recently become popular (17, 18, 32). This, however, requires that the phosphopeptide enrichment step be of exquisite selectivity, have sufficient capacity, and allow complete elution (17, 18). Phosphopeptides purified in this way can be directly analyzed via LC-MS/MS (33, 34), typically using shallow reversed phase LC gradients (35). However, it turns out that current mass spectrometers still lack the scan speed and dynamic range required to reach complete (phospho)peptide sampling in direct LC-MS/MS measurements (36, 37).
In this article, we describe a robust and flexible workflow based on offline chromatographic enrichment of phosphopeptides using a commercially available Fe-IMAC column. This approach offers selective, comprehensive, and reproducible enrichment and scales over a wide range of sample quantities. We show that this method outperformed all Ti-IMAC and TiO2 methods tested, and our data argue that the apparent orthogonality of all three methods is caused by a combination of format inadequacies, inefficient elution, and insufficient data acquisition speed, rather than the different physicochemical characteristics of phosphopeptides.
MATERIALS AND METHODS
Cell Culture and Protein Digest
Human epidermoid A431 cells were grown in Iscove's modified Dulbecco's medium supplemented with 10% (v/v) fetal bovine serum and 1% antibiotic/antimycotic solution. For all phosphopeptide enrichment optimization experiments, cells were treated with 1 mm pervanadate for 5 min prior to lysis. After harvesting, cells were washed two times with ice-cold PBS and lysed in 8 m urea, 40 mm Tris/HCl (pH 7.6), 1× EDTA-free protease inhibitor mixture (Complete Mini, Roche), and 1× phosphatase inhibitor mixture (Sigma). The lysate was centrifuged at 20,000 rpm for 1 h at 4 °C. The protein concentration was determined using the Bradford method (Coomassie (Bradford) Protein Assay Kit, Thermo Scientific). The supernatant was reduced with 10 mm DTT at 56 °C for 1 h and alkylated with 25 mm iodoacetamide for 45 min at room temperature in the dark. The protein mixture was diluted with 40 mm Tris/HCl to a final urea concentration of 1.6 m. We induced digestion by adding sequencing-grade trypsin (Promega, Mannheim, Germany; 1:100 enzyme:substrate ratio) and allowing samples to incubate at 37 °C for 4 h. Subsequently another 1:100 quantity of trypsin was added for overnight digestion at 37 °C. Samples were acidified with TFA to pH 2 to stop the trypsin activity. SepPack columns (C18 cartridges, Sep-Pak Vac, 1 cc (50 mg), Waters Corp., Eschborn, Germany; solvent A, 0.07% TFA; solvent B, 0.07% TFA, 50% ACN) were used for peptide desalting according to the manufacturer's instructions, and eluates were dried down and stored at −80 °C. Smaller sample amounts of up to 100 μg were desalted as described elsewhere (38).
Fe-IMAC Batch Enrichment
Fe-IMAC batch enrichments were essentially performed as described in Ref. 31. Briefly, 100 μl of Fe-IMAC beads (PhosSelect iron affinity gel, Sigma) were washed four times with 1 ml of binding solvent (25 mm FA, 40% ACN), and a 50% slurry in binding solvent was prepared. Dried-down peptides were resuspended in binding solvent at a concentration of 1 μg/μl. After the beads and dissolved peptides had been combined, samples were incubated for 1 h at room temperature with vigorous shaking. Subsequently, Fe-IMAC beads were transferred on top of previously equilibrated C18 StageTips and washed by being sequentially passed through 50 μl of Fe-IMAC binding solvent (twice) and 40 μl of 1% FA. Elution was achieved via the application of 70 μl of 500 mm K2HPO4, pH 7 (twice). Peptides that were retained on the C18 material were washed with 40 μl of 1% FA and eluted directly into an MS plate using 40 μl of 60% ACN, 0.1% FA. Eluates were dried down and stored at −80 °C.
Fe-IMAC Column Phosphopeptide Enrichment
The desalted digest was reconstituted in 0.5 ml of Fe-IMAC solvent A (30% ACN, 0.07% (v/v) TFA) and loaded onto an analytical Fe-IMAC column (4 × 50 mm or 9 × 50 mm ProPac IMAC-10, Thermo Fisher Scientific) connected to an HPLC system (ÄKTA Explorer FPLC system, Amersham Biosciences Pharmacia) with a 1-ml sample loop. The column was charged with iron according to the manufacturer's instructions. Briefly, the column was rinsed with 3 column volumes of 20 mm formic acid and charged using 3 column volumes of 25 mm FeCl3, 100 mm acetic acid. To wash out unbound iron ions, we flushed the column with 20 column volumes of 20 mm formic acid and subsequently removed it from the HPLC system. The HPLC lines were flushed first with 20 ml of double-deionized H2O and then with 20 ml of 50 mm EDTA and 10 ml of double-deionized H2O to remove remaining iron ions. After column reconnection and baseline equilibration, the gradient was started. Sample was loaded (0.1 ml/min over 10 min), and unbound peptides were washed out with Fe-IMAC solvent A (0.3 ml/min over 16 min). Subsequent phosphopeptide elution was achieved with a linear gradient from 0% to 45% Fe-IMAC solvent B (0.5% (v/v) NH4OH) (0.2 ml/min over 60 min). After an increase to 100% solvent B and a 5-min holding step, the column was re-equilibrated with Fe-IMAC solvent A (0.5 ml/min over 30 min). Flow-through and a phosphopeptide fraction were collected according to the UV signal (280 nm), dried down, and stored at −80 °C. The column was recharged after a maximum of three enrichments.
Ti-IMAC Synthesis and Phosphopeptide Enrichment
Ti-IMAC beads were synthesized as previously described by Zhou et al. (39), with some modifications and additional quality control steps. A detailed description can be found in the supplemental “Materials and Methods” section. Enrichment of up to 250 μg of sample was performed as described in Ref. 39; the phosphopeptide enrichment protocol was modified for sample amounts exceeding 250 μg. Sep-Pak cartridges (C18 cartridges, Sep-Pak Vac, 1 cc (50 mg), Waters Corp.) were attached to a vacuum manifold and flushed with 1 ml of Ti-IMAC loading solvent (80% ACN, 6% TFA). Subsequently, 50 mg of Ti-IMAC beads were loaded on top of the C18 material and equilibrated with 5 × 1 ml loading solvent. Dried-down digests were dissolved in 1 ml of binding solvent and slowly passed through the cartridges. After reapplication of the flow-through, columns were washed with 10 ml of washing solution 1 (50% (v/v) ACN, 0.5% (v/v) TFA, 200 mm NaCl) and 10 ml of washing solution 2 (50% (v/v) ACN, 0.1% (v/v) TFA). Bound peptides were eluted sequentially with 0.8 ml of 10% (v/v) NH4OH and 0.2 ml of 80% (v/v) ACN, 2% (v/v) FA. Flow-through and elution fractions were dried down and stored at −80 °C.
TiO2 Batch Enrichment
TiO2 batch enrichment was performed as described by Kettenbach and Gerber (17), with some modifications. TiO2 beads (5 μm, GL Sciences Inc., Mainz, Germany) were washed twice with 1 ml of washing solvent (50% ACN, 0.1% TFA) and four times with 1 ml of binding solvent (2 m lactic acid, 50% ACN, 0.1% TFA). In between, beads were spun down and the supernatant was discarded. Peptides were dissolved in 0.5 ml of binding solvent, and after the addition of 0.25 ml of equilibrated bead slurry, the mixture was incubated for 1 h at room temperature with vigorous shaking. Subsequently, the beads were washed four times with 0.2 ml of binding solvent and five times with 1 ml of washing solution. Bound peptides were eluted in two 10-min incubation steps with 200 μl of elution solvent (50 mm KH2PO4, 0.5% (v/v) NH4OH, pH 11.3). The supernatant was quenched by the addition of 30 μl of 100% FA, dried down, and stored at −80 °C.
Phosphopeptide Enrichment with TiO2 and Ti-IMAC Columns
For chromatographic separation of phosphorylated peptides, TiO2 beads (5 μm; GL Sciences Inc.) were packed into a column (10 × 4 mm; Dr. Maisch, Ammerbuch, Germany; Amersham Biosciences) and connected to an HPLC system (ÄKTA Explorer FPLC, Amersham Biosciences). Dried samples were reconstituted in 0.5 ml of TiO2 solvent A (80% ACN, 6% TFA) and injected. The system was operated at a flow rate of 0.2 ml/min with a linear gradient from 0% to 85% solvent B (1.5% (v/v) NH4OH, pH 11.8) over 30 min followed by an increase to 95% over 15 min. For elution of bound peptides, the concentration of solvent B was kept at 95% for 50 min. In total, 13 fractions were collected. In-house-synthesized Ti-IMAC beads were also packed into a column (50 × 4 mm; Dr. Maisch, Amersham Biosciences). The gradient corresponded to the one used for Fe-IMAC column enrichments, except for some modifications; because of the strong metal ion–peptide interaction, 10% (v/v) NH4OH was used for elution.
Hydrophilic Strong Anion Exchange Separation
A Dionex Ultimate 3000 HPLC system (Dionex Corp., Idstein, Germany) equipped with an IonPac AG24 guard column (2 × 50 mm, Thermo Fisher Scientific) and a IonPac AS24 strong anion exchange column (2 × 250 mm, Thermo Fisher Scientific) was used for hydrophilic strong anion exchange (hSAX) separation of both enriched phosphopeptides and digested full proteome samples. The system was operated at 30 °C with a flow rate of 0.25 ml/min and an initial 3-min equilibration step with 100% hSAX solvent A (5 mm Tris/HCl, pH 8.5) followed by elution with a linear 17-min gradient up to 40% hSAX solvent B (5 mm Tris/HCl, pH 8.5, 1 m NaCl). Solvent B was increased to 100% in 10 min and held constant for another 10 min. A subsequent switch to 100% solvent A in 3 min was followed by column re-equilibration with 100% solvent A for 10 min. 24 fractions were collected in 1-min intervals (starting 2 min into the gradient), dried down, and measured after on-trap desalting.
LC-MS/MS Analysis
Nanoflow LC-MS/MS was performed by coupling an Eksigent nanoLC-Ultra 1D+ (Eksigent, Dublin, CA) to an Oribtrap Velos (Thermo Scientific, Bremen, Germany). Peptides were delivered to a trap column (100-μm inner diameter × 2 cm, packed with 5-μm C18 resin (Reprosil PUR AQ, Dr. Maisch)) at a flow rate of 2 μl/min for 12 min and 5 μl/min for 13 min in 100% solvent A (0.1% FA in HPLC-grade water). After 25 min of loading and washing, peptides were transferred to an analytical column (75 μm × 40 cm C18 column; Reprosil PUR AQ (3 μm), Dr. Maisch) and separated using a linear gradient from 2% to 27% (4% to 32% for samples not previously enriched) solvent B (0.1% FA, 5% dimethyl sulfoxide in ACN) at a flow rate of 300 nL/min. For one direct measurement, a 210-min gradient (220-min turnaround) was applied, whereas hSAX factions were measured for 110 min per fraction (135-min turnaround including an on-trap desalting step). TiO2 column fractions were measured for 45 min each (60-min turnaround including an on-trap desalting step). To prevent potential Fe3+ and Ti4+ ion adsorption to the reversed phase stationary phase, 5 μl of 1 μm deferoxamine dissolved in double-deionized H2O was injected between phosphopeptide direct measurements (40).
Peptides were ionized using a 2.2-kV spray voltage and a capillary temperature of 275 °C. The mass spectrometer was operated in data-dependent acquisition mode, automatically switching between MS and MS2. Full-scan MS spectra (m/z 360–1300) were acquired in the Orbitrap at 30,000 (m/z 400) resolution and with an automatic gain control target value of 1e6. For internal calibration, the signal at m/z 401.922718 was used as a lock mass (41). High-resolution higher energy collision-induced dissociation MS2 spectra were generated for up to 10 precursors with a normalized collision energy of 30% (for non-enriched samples) or 35% (for phosphopeptide samples). The precursor ion count for triggering an MS2 event was set at 500 with a dynamic exclusion of 20 s. Fragment ions were read out in the Orbitrap mass analyzer at a resolution of 7500 (isolation window: 2 Th). For phosphopeptide samples, the MS2 automatic gain control target value was set at 4e4 with a maximum ion injection time of 250 ms, whereas samples that had not been previously enriched were measured with values of 3e4 and 200 ms.
Peptide and Protein Identification and Quantification
For peptide and protein identification, peak lists were extracted from raw files using Mascot Distiller v2.2.1 (Matrix Science, London, UK) and subsequently searched against the Human IPI database (v3.68; 87,061 entries) using Mascot (v2.3.0) with carbamidomethyl cysteine as a fixed modification. Phosphorylation of serine, threonine, and tyrosine; oxidation of methionine; and N-terminal protein acetylation were allowed as variable modifications. The precursor tolerance was set to 10 ppm, and the fragment ion tolerance to 0.05 Da. Trypsin/P was specified as the proteolytic enzyme, with up to two missed cleavage sites allowed. Identified phosphopeptides were filtered using two criteria: (i) peptide–spectrum matches were filtered for 1% FDR using Rockerbox (42) in combination with Percolator (v1.0) (43), and (ii) remaining peptides corresponding to accepted peptide–spectrum matches were filtered according to a 5% global peptide FDR cutoff based on the formula published by Marx et al. (44) (higher energy collision-induced dissociation, global FDR). This translated to Mascot ion scores of >13.96 for phosphopeptides and >27.65 for non-phosphorylated peptides. The peptide table created after Rockerbox processing was used for further analysis. We established three different categories for phosphopeptide identifications: (i) unique phosphopeptides were based on non-redundant phosphosequences including phosphoisomers, (ii) unique phosphosequences were based on the sequence combined with the number of phosphomodifications (phosphoisomers were not considered), and (iii) confidently localized phosphopeptides were unique phosphopeptides that were additionally filtered with a 5% false localization cutoff based on the Mascot delta score, which was calculated according to the formula introduced by Marx et al. (44) (higher energy collision-induced dissociation). In order to calculate phosphopeptide selectivity, we applied the global 5% FDR cutoff separately to identified phospho- and non-phosphopeptides.
For label-free experiments, Progenesis (v4.1, Nonlinear Dynamics, Newcastle, UK) was used essentially as described previously (45). Briefly, after one sample had been selected as a reference, the retention times of all eluting precursor m/z values within the experiment were aligned. Precursors between 2 and 10 charges were included, and those with two isotopes or less were excluded. After filtering, precursor abundances were normalized, and replicate samples were grouped together. The corresponding MS2 spectra were exported and searched against the Human IPI database (v3.68) using Mascot (v2.3.0). The parameters corresponded to those used for previously mentioned peptide identification. The search results were exported in a .dat file format and filtered for 1% peptide–spectrum match FDR and Mascot ion scores > 13.96 (or > 27.65 for non-phosphopeptides) using Rockerbox in combination with percolator (v1.0). Filtered peptide–spectrum matches were re-imported into Progenesis and matched to the respective features. Raw data were normalized by VSN using the R environment.
All data associated with protein and peptide quantification and identification have been deposited as supplementary tables in ProteomeXchange with the identifier PXD001060 (supplemental Table S1, supplemental Figs. S1–S4, supplemental Fig. S6).
RESULTS AND DISCUSSION
Chromatographic Phosphopeptide Enrichment Using an Fe-IMAC Column
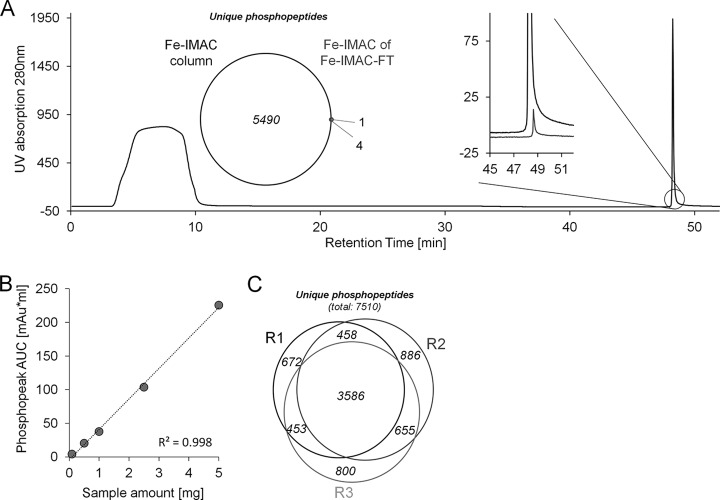
As discussed above, batch and tip-based enrichment protocols have been found to suffer from irreproducibility due to a number of factors including significant manual intervention. In addition, enrichment selectivity and efficiency in such formats are also strongly dependent on the ratio of enrichment material to protein digest used, which is hard to predict for any specific experimental context (13, 18–21). We hypothesized that much of the quoted complementarity between different enrichment materials originates in practical shortcomings rather than differences in the molecular properties of phosphopeptides. We conducted a series of experiments (outlined below) that supported this hypothesis, and the data collectively show that these practical issues can be substantially reduced by the use of a conventional Fe-IMAC HPLC column (Fig. 1). First, batch and tip-based enrichments using TiO2, Ti-IMAC, and Fe-IMAC beads with different bead-to-sample ratios were re-evaluated. Our results confirm previous observations showing that comprehensiveness and selectivity are directly related to the bead-to-sample ratio (supplemental Fig. S1A). Next, we revisited the use of a column format, employing a commercially available IMAC HPLC column charged with Fe3+ ions. The UV trace shown in Fig. 2A (1 mg of a trypsin-digested A431 cell lysate) showed a clear separation of two different peptide pools. As expected, LC-MS/MS analysis only detected non-phosphorylated peptides in the column breakthrough. In contrast, 5494 non-redundant phosphopeptides were identified in the retained peptide pool with >90% selectivity in a single 4-h LC-MS/MS run on an Orbitrap Velos (higher energy collision-induced dissociation fragmentation). No peptides were detected in any of the other fractions of the chromatogram. The performance of the Fe-IMAC column was further characterized with respect to (i) comprehensiveness, (ii) scalability, and (iii) reproducibility. To address comprehensiveness, we dried down the flow-through of the initial enrichment and subjected it to a second round of chromatographic enrichment on the same Fe-IMAC column. Both the weak UV trace and the very low number of identified phosphopeptides (five, of which four were also identified in the first enrichment) indicate virtually complete phosphopeptide depletion in a single enrichment step (Fig. 2A). Next, the scalability of the Fe-IMAC column (4 × 50 mm) was evaluated. Fig. 2B shows that the chromatographic peak area scaled linearly between 100 μg and 5 mg of digest on column, indicating sufficient capacity within that range. Importantly, phosphopeptide selectivity exceeded 90% in all cases (supplemental Fig. S1B), demonstrating that the use of an Fe-IMAC column solved the issue of capacity without compromising selectivity. As a result, we did not observe any overrepresentation of multiply phosphorylated peptides relative to other methods (8% in this cell line; Table I), which would be expected to occur if the capacity of the enrichment material were exceeded (15, 46).
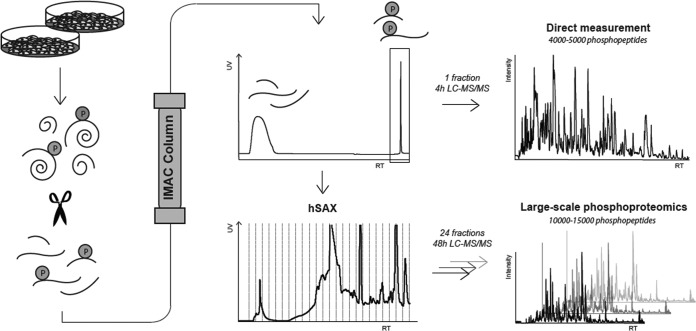
Fig. 1.
Workflow utilizing an Fe-IMAC HPLC column setup for the purification of phosphopeptides from complex digests. Fe-IMAC eluates can be either directly measured via LC-MS/MS, providing considerable throughput, or further fractionated via hSAX to provide increased analytical depth.
Fig. 2.
Fe-IMAC column characterization. A, Fe-IMAC UV chromatogram of 1 mg of A431 cell digest. The phosphopeptide fraction (reaction time: ∼49 min) is well separated from the column breakthrough that contains the non-phosphorylated peptides. The inset on the left shows the number of phosphopeptides identified from the IMAC eluate and the number of phosphopeptides identified from a second IMAC enrichment of the column flow-through (FT) of the first Fe-IMAC enrichment. The inset on the right shows the respective UV traces. Both indicate that the A431 digest was essentially depleted of phosphopeptides. B, UV signal-based quantification of Fe-IMAC enriched phosphopeptides as a function of the amount of cellular digest applied. It is apparent that the column captured phosphopeptides over a wide linear range. C, Venn diagram of the number of unique phosphopeptide identifications across three technical replicates of Fe-IMAC purification of phosphopeptides from 1 mg of A431 lysate digest.
Table I. Summary of phosphopeptide identifications purified via Fe-IMAC column, Ti-IMAC tip, or TiO2 batch format.
| Fe-IMAC column |
Fe-IMAC column |
Fe-IMAC column |
Ti-IMAC tip |
Ti-IMAC tip |
Ti-IMAC tip |
TiO2 batch |
TiO2 batch |
TiO2 batch |
|
|---|---|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R1 | R2 | R3 | R1 | R2 | R3 | |
| Unique phosphopeptides | 5494 | 5585 | 5169 | 5397 | 4727 | 3706 | 4187 | 4173 | 3709 |
| Unique phosphosequences | 4467 | 4597 | 4206 | 4427 | 3879 | 3055 | 3617 | 3572 | 3135 |
| Confidently localized phosphopeptides | 3987 | 4127 | 3799 | 3956 | 3511 | 2694 | 3192 | 3168 | 2765 |
| Non-p | 561 | 681 | 856 | 26 | 77 | 377 | 267 | 217 | 156 |
| Selectivity (%) | 91 | 89 | 86 | 100 | 98 | 91 | 94 | 95 | 96 |
| 1p (%) | 92 | 93 | 94 | 90 | 91 | 92 | 96 | 95 | 94 |
| >2p (%) | 8 | 7 | 6 | 10 | 9 | 8 | 4 | 5 | 6 |
| Unique phosphopeptides across triplicates | 7510 | 6599 | 5763 | ||||||
| Unique phosphosequences across triplicates | 5552 | 4906 | 4518 | ||||||
| Confidently localized phosphopeptides across triplicates | 5490 | 4822 | 4315 | ||||||
The reproducibility of the Fe-IMAC column format was evaluated through analysis and quantification of phosphopeptides from three independent Fe-IMAC enrichments from the same A431 cell lysate digest. Collectively, 7510 unique phosphopeptides were identified (direct LC-MS/MS of IMAC eluates), of which 3586 were identified in all three replicates (Table I, Fig. 2C). To account for the fact that many phosphopeptides are not picked for fragmentation because of the stochastic nature of data-dependent acquisition (see below), the retention times of all peptide precursors were aligned using Progenesis LCMS. This boosted the number of unique phosphopeptides quantified across all replicates to 5009, still indicating undersampling by the mass spectrometer (supplemental Table S1). To analyze quantitative reproducibility, we binned the quantifiable phosphopeptides by their coefficient of variation (cv). The median cv of phosphopeptide quantification was 15% even for raw feature abundances (supplemental Fig. S1C), and, as expected, variation was a function of feature abundance itself (supplemental Fig. S1D). The excellent reproducibility between the replicates was further underscored by an R2 value of 0.96 when we plotted the raw phosphopeptide feature abundances of the respective replicates (supplemental Fig. S1E).
The above results indicate that our Fe-IMAC HPLC column enrichments enabled the comprehensive, reproducible, and selective isolation of phosphopeptides over a wide range of input amounts. The workflow does not the require special additives often used in, for example, TiO2 methods to enhance selectivity, and it benefits from sample loading and elution conditions that are downstream compatible (e.g. for full proteome measurements or enrichment of different post-translational modifications from the same digest (47)) without time-consuming and sample-losing desalting steps.
Apparent Complementarity of Methods Is Primarily Caused by Inefficient Elution and Insufficient Mass Spectrometric Capacity
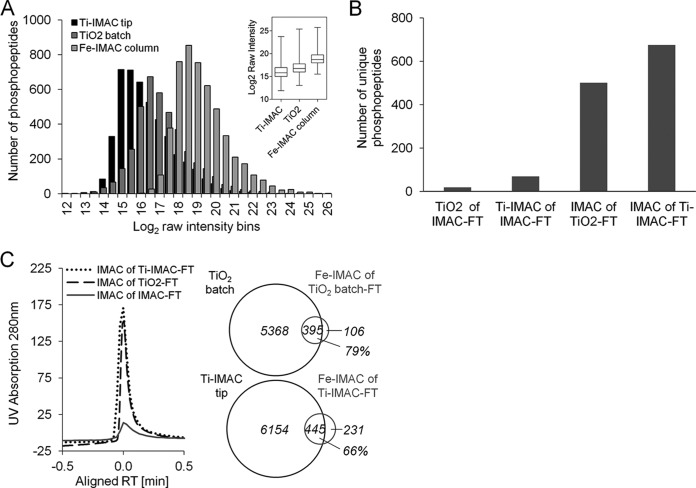
The Fe-IMAC column enrichment characterized above was benchmarked against Ti-IMAC in tip and TiO2 batch enrichment. Ti-IMAC beads were synthesized according to published procedures (48) (see also supplemental “Materials and Methods” section). 1 mg of A431 digest and optimized bead-to-sample ratios were used to perform triplicate enrichments for each method (supplemental Fig. S1A). Competitive numbers were identified in the two methods: 6599 phosphopeptides (cumulative) for Ti-IMAC tip, and 5763 (cumulative) for TiO2 batch (Table I). The overlap in replicates, phosphopeptide selectivity, and the percentage of multiply phosphorylated peptides also did not differ profoundly from that in Fe-IMAC column triplicates (supplemental Fig. S2A and Table I). We next extracted phosphopeptide abundance (intensity) information using Progenesis and found that the intensity of triplicate Fe-IMAC column enrichments was seven and four times higher than that of the respective Ti-IMAC tip and TiO2 batch triplicates (Fig. 3A, supplemental Fig. S2B). This striking finding suggests not only that the Fe-IMAC column depleted phosphopeptides from complex digests (see above), but also that it was possible to elute the peptides very efficiently (see below). We note that the Ti-IMAC beads were synthesized in house, and thus their quality might not have been entirely comparable to that of beads used in other studies (39, 48). Still, this observation prompted the question of whether phosphopeptide binding to or elution from Ti-IMAC or TiO2 material is as efficient as that with Fe-IMAC. A further observation was that the overlap between phosphopeptides identified by the different materials did not exceed 20% (supplemental Fig. S2C). This strong apparent complementarity led us to perform sequential enrichments from the same sample but using different materials. Surprisingly, neither Ti-IMAC tip nor TiO2 batch purification recovered a considerable amount of phosphopeptides from Fe-IMAC column flow-throughs (a total of 19 for Ti-IMAC tip and 69 for TiO2 batch; Fig. 3B). We then reversed the enrichment order such that desalted TiO2 batch and Ti-IMAC tip enrichment flow-throughs were subjected to Fe-IMAC column enrichment. Although several hundred phosphopeptides were recovered in this way (676 for Ti-IMAC and 501 for TiO2; triplicate experiments (Fig. 3C)), the respective Fe-IMAC UV traces indicated that the enrichment efficiency of the Ti-IMAC tip and TiO2 batch methods was likely still at least 90%. Interestingly, when we compared the recovered phosphopeptides from the Fe-IMAC flow-through enrichments to those identified in the respective original triplicate analyses, a considerable overlap of 79% for Ti-IMAC tip and 66% for TiO2 batch was found (Fig. 3C), which is much higher than the 20% figure quoted above, indicating that there is much less complementarity in the methods than previously thought.
Fig. 3.
Comparison of Fe3+-IMAC, Ti-IMAC tip, and TiO2 batch enrichments. A, MS signal intensity distribution of unique phosphopeptides for each method showing the superiority of Fe-IMAC for phosphopeptide recovery from complex digests. The box plot (inset) summarizes the same data for a triplicate measurement. B, summary of the number of unique phosphopeptide identifications (in triplicate) from experiments aiming at recovering phosphopeptides from the unbound fractions of one enrichment method using an alternative enrichment method. The data show that Fe-IMAC recovered a noticeable number of phosphopeptides from the unbound fraction of TiO2 batch and Ti-IMAC tip enrichments. TiO2 batch and Ti-IMAC tip purifications from the unbound Fe-IMAC fraction did so to a much lesser extent. C, UV chromatogram of Fe-IMAC enrichments from the unbound fractions of TiO2 batch and Ti-IMAC tip experiments. The Venn diagrams (triplicate experiments) show that the phosphopeptides recovered by Fe-IMAC largely overlapped with those identified in the original TiO2 batch and Ti-IMAC tip enrichments, indicating insufficient capacity of the amount of TiO2 and Ti-IMAC used.
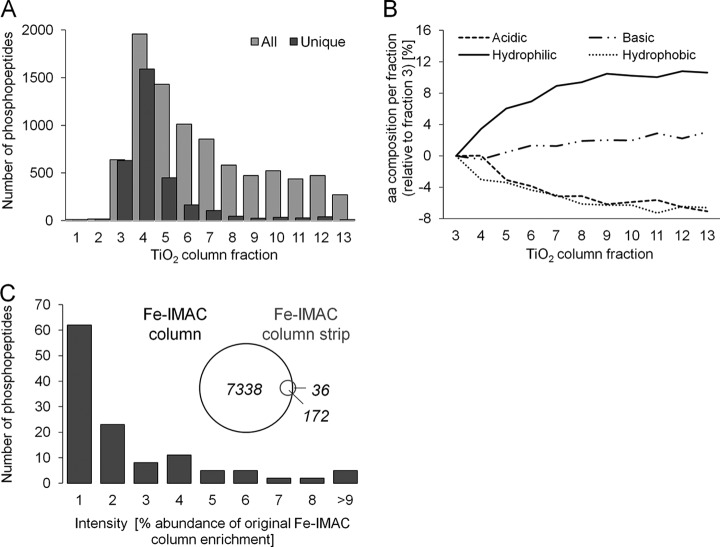
Next, in order to distinguish material from format, we had a Ti-IMAC column packed by a professional service and purchased a TiO2 HPLC column containing the very same materials as used in tip or batch format. For the Ti-IMAC column, the same loading conditions as for Fe-IMAC columns were used. Phosphopeptides were eluted in a single peak but over a wider retention time window than for Fe-IMAC columns (supplemental Fig. S3A). The flow-through of the Ti-IMAC column was then subjected to Fe-IMAC column enrichment, and the low UV signal and the identification of just 18 phosphopeptides in this analysis indicate more efficient enrichment of phosphopeptides by Ti-IMAC in column than in tip format (supplemental Fig. S3B). For the TiO2 column, very acidic (6% TFA) loading conditions were needed in order to separate phosphopeptides from non-phosphorylated peptides. In addition, 10-fold higher ammonia concentrations had to be used in order to elute any phosphopeptides from both TiO2 and Ti-IMAC HPLC columns. Surprisingly, phosphopeptide eluted from the TiO2 column over a range of nearly 50 min (or 10 ml of solvent; supplemental Fig. S3C), indicating rather inefficient elution, presumably owing to very strong binding to the TiO2 stationary phase. Fraction 4 (Fig. 4A; supplemental Fig. S3C) contained most of the unique phosphopeptides, and in subsequent fractions the extent of redundant phosphopeptide identifications increased—another sign of inefficient elution. Further evidence of this was seen in an analysis of the amino acid composition of phosphopeptides identified in the different TiO2 fractions (Fig. 4B). Between the first and last fraction, the absolute percentage of acidic and hydrophobic amino acids within each fraction decreased by roughly 7%. In contrast, the frequency of hydrophilic amino acids (mainly serine residues; supplemental Fig. S3D) increased by 11%. In other words, whereas acidic and hydrophobic phosphopeptides eluted relatively easily off the column, hydrophilic peptides were more strongly retained. Some peptides may in fact never elute when ammonia is used as a solvent, and as a consequence the observed bias in identified phosphopeptides between Fe-IMAC and TiO2 can, at least in part, be attributed to inefficient elution from the TiO2 material. In contrast, for Fe-IMAC columns, both the sharp elution peak (supplemental Fig. S3A) and the absence of phosphopeptide identifications in collected fractions before and after the main peak indicate efficient elution. We surmise that the elution bias for Ti-IMAC was less pronounced than for TiO2 because the amino acid compositions of phosphopeptides eluting from Ti-IMAC and Fe-IMAC were largely the same. To determine whether complete elution of phosphopeptides could be achieved from Fe-IMAC columns with ammonia, we performed two consecutive phosphopeptide enrichments using 1 mg of A431 digest each, after which we stripped the iron off the column using EDTA and analyzed the phosphopeptide content of the EDTA fraction. In total, 208 unique phosphopeptides were identified, of which 172 were high-abundance peptides present in the prior ammonia elution (Fig. 4C). The vast majority of these were 100 times less abundant in the EDTA fraction, demonstrating that phosphopeptide elution with ammonia from Fe-IMAC columns is virtually complete.
Fig. 4.
Elution efficiency. A, number of unique and redundant phosphopeptides identified across fractions from a TiO2 HPLC column enrichment of 1 mg of A431 cell digest. The broad elution profile and the high degree of redundant identifications indicate inefficient elution. B, analysis of the absolute frequency of basic, acidic, hydrophilic, and hydrophobic amino acids of phosphopeptides in the different TiO2 column fractions (relative to the earliest fraction containing phosphopeptides, fraction 3). It is apparent that hydrophilic phosphopeptides were more difficult to elute from TiO2 columns than other phosphopeptides. C, when we stripped the iron from Fe-IMAC columns, only very few phosphopeptides were identified in the stripped fraction. The majority of these were identified with merely 1% of the intensity with which they were identified in a regular Fe-IMAC eluate, indicating that the elution of Fe-IMAC columns was virtually complete.
A separate aspect that somewhat complicates the dissection of the various factors determining the phosphopeptide enrichment efficiency of the different methods tested is the fact that in complex samples, the mass spectrometer cannot keep up with the number of peptides entering the instrument at any one time. For data-dependent acquisition, peptide abundance is of course a major factor in determining which precursor is picked for fragmentation. Once the precursor ion complexity exceeds the fragmentation capacity of the mass spectrometer, the process becomes rather stochastic (36). In our experiments, only 31% of the eluting peptide-like features (across triplicates) were actually identified (26% for Ti-IMAC and 22% for TiO2), and those that were predominantly represented highly abundant precursors (supplemental Fig. S3E). Thus, the inherently biased nature of data-dependent acquisition is a major factor in the observed apparent complementarity between the different enrichment methods (13).
Large-scale Phosphoproteomics by Coupling Fe-IMAC Column Enrichment to hSAX Separation
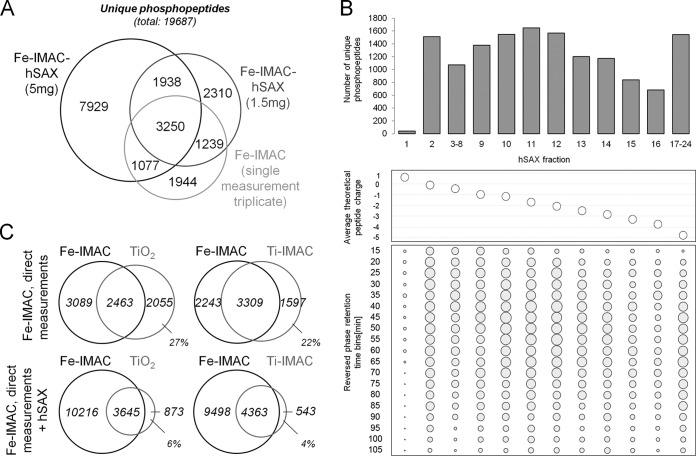
As shown above, Fe-IMAC columns enabled unbiased and comprehensive phosphopeptide enrichment and generally outperformed the other two methods/formats. Still, the restricted analytical capacity of the mass spectrometer used prevented very deep phosphoproteome analysis in direct LC-MS/MS measurements (supplemental Fig. S3E). To increase analytical depth, fractionation of an isolated phosphopeptide pool can be performed prior to LC-MS/MS analysis. We also hypothesized that the differences in the three phosphopeptide enrichment methods investigated in this study would decrease in such a setting, as the aforementioned limitations of data-dependent acquisition would become less pronounced (49). The Trost laboratory has recently shown good orthogonality of hydrophilic strong anion exchange and reversed phase chromatography for ordinary peptides (50). We therefore attempted to determine whether hSAX would show the same desirable characteristics for phosphopeptides (50). In a first step, we reproduced hSAX performance for the fractionation of full proteome samples by separating 300 μg of A431 digest into 24 fractions and measuring each of these via LC-MS/MS using a 110-min reversed phase gradient. The identification of 48,250 unique peptides and 7343 proteins in 2 days of instrument time corroborated the published results (supplemental Figs. S4A and S4B). Next, 1.5 mg of A431 digest was subjected to Fe-IMAC column enrichment and separated into 24 hSAX fractions. This increased the number of identifications to 6321 unique phosphosequences (+13% relative to direct measurements of 1-mg triplicates), but at the expense of a 3-fold increase in measurement time (Fig. 5A). To test the merits of the combination of Fe-IMAC and hSAX further, we scaled up the experiment to 5 mg of digest, leading to the identification of 10,978 unique phosphosequences (∼3700 proteins, of which 70% were also identified in the full proteome (supplemental Fig. S4B)). As expected, hSAX separation of phosphopeptides closely followed the computed theoretical charge states (calculated for pH 8.5), and good orthogonality and separation efficiency relative to reversed phase separation were observed (Fig. 5B; supplemental Fig. S4C). Also in line with expectations, our comparison of hSAX retention time differences between phosphopeptides and their respective non-phosphorylated counterparts (identified in the full proteome dataset) showed that the addition of a phosphate group considerably increased binding to the hSAX material (supplemental Fig. S4E), rendering the application of hSAX particularly well suited for phosphopeptide fractionation. With a working three-dimensional phosphopeptide separation scheme in hand, we revisited the apparent complementarity of phosphopeptide enrichment materials yet again. We compared Fe-IMAC data with and without hSAX separation to the data obtained from triplicate direct LC-MS/MS measurement of TiO2 and Ti-IMAC enriched samples. Fig. 5C shows that the overlap of phosphopeptide identifications increased substantially when the analytical depth increased. Specifically, the fraction of exclusively identified phosphosequences for TiO2 enrichment decreased from 27% to 6% and from 22% to 4% for Ti-IMAC enrichment, showing that these were actually purified by Fe-IMAC and were therefore not specific to purification on TiO2 or Ti-IMAC resins. The remaining small discrepancy can likely be attributed to the insufficient sampling speed of the mass spectrometer (supplemental Fig. S4F).
Fig. 5.
Hydrophilic strong anion exchange separation of Fe-IMAC eluates increases depth of phosphoproteome coverage. A, comparison of phosphosequences identified in Fe-IMAC column-based workflows with or without hSAX separation of Fe-IMAC eluates. B, top: number of phosphopeptide identifications across an hSAX separation of Fe-IMAC eluates. Middle: as one would expect, the average theoretical peptide charge decreased along the hSAX separation. Bottom: identification of phosphopeptides from hSAX fractions by means of reversed phase LC-MS/MS. The size of the dot indicates the number of identified phosphopeptides. The data clearly demonstrate the strong orthogonality of the hSAX and reversed phase separations. C, comparison of phosphosequences identified from Fe-IMAC column (direct measurement, triplicate) to TiO2 batch (direct measurement, triplicate) and Ti-IMAC tip (direct measurement, triplicate) enrichments with or without Fe-IMAC hSAX separation. It is apparent that the number of phosphopeptides exclusively detected in TiO2 batch or Ti-IMAC tip experiments decreased strongly when the analytical depth of Fe-IMAC column enrichment was increased by hSAX fractionation. This shows that the often quoted orthogonality of the enrichment materials is much smaller than anticipated.
CONCLUSION
In this paper we have shown that the often quoted complementarity of different materials for phosphopeptide enrichment can actually be attributed to limited binding capacity, biased or incomplete elution, shortcomings in the physical formats (batch, tip), and limited analytical capacity of the mass spectrometer. All these can be largely overcome by using an Fe-IMAC HPLC column for bulk phosphopeptide enrichment from complex digests and an optional hSAX separation step prior to LC-MS/MS analysis. These two chromatographic elements can be integrated into a flexible workflow (Fig. 1) that allows the identification of 4000 to 5000 phosphopeptides within 4 h of measurement time or 10,000 to 15,000 phosphopeptides within 48 h of measurement time on a modestly performing mass spectrometer. Although our data show that the Fe-IMAC column methodology scaled over a wide range of digest quantities, we note that down-scaling samples would also require down-scaling of the column. The Ti-IMAC material in tip format also gave good overall results at sample quantities less than 100 μg of digest. As the Fe-IMAC column used here is commercially available, the method should be readily implemented. It can also be anticipated that the overall simplicity of the approach will lead to improved reproducibility of results within one laboratory, as well as better comparability of data acquired in different laboratories.
Supplementary Material
Acknowledgments
We thank Svenja Petzoldt for excellent technical assistance.
The mass spectrometry proteomics data, including search result files containing annotated spectra (Scaffold files), have been deposited to the ProteomeXchange Consortium (www.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD001060.
Footnotes
Author contributions: B.R., B.K., and S.L. designed research; B.R., H.K., and M.M. performed research; G.M. and M.M. contributed new reagents or analytic tools; B.R. and H.K. analyzed data; B.R., B.K., and S.L. wrote the paper.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- LC-MS/MS
- liquid chromatography–tandem mass spectrometry
- FDR
- false discovery rate
- IMAC
- immobilized metal ion affinity chromatography
- ACN
- acetonitrile
- FA
- formic acid
- hSAX
- hydrophilic strong anion exchange
- HPLC
- high-performance liquid chromatography
- TiO2
- titanium dioxide.
REFERENCES
- 1. Blume-Jensen P., Hunter T. (2001) Oncogenic kinase signalling. Nature 411, 355–365 [DOI] [PubMed] [Google Scholar]
- 2. Cohen P. (2000) The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem. Sci. 25, 596–601 [DOI] [PubMed] [Google Scholar]
- 3. Wilhelm M., Schlegl J., Hahne H., Moghaddas Gholami A., Lieberenz M., Savitski M. M., Ziegler E., Butzmann L., Gessulat S., Marx H., Mathieson T., Lemeer S., Schnatbaum K., Reimer U., Wenschuh H., Mollenhauer M., Slotta-Huspenina J., Boese J. H., Bantscheff M., Gerstmair A., Faerber F., Kuster B. (2014) Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587 [DOI] [PubMed] [Google Scholar]
- 4. Lemeer S., Heck A. J. (2009) The phosphoproteomics data explosion. Curr. Opin. Chem. Biol. 13, 414–420 [DOI] [PubMed] [Google Scholar]
- 5. Ruprecht B., Lemeer S. (2014) Proteomic analysis of phosphorylation in cancer. Expert Rev. Proteomics 11, 259–267 [DOI] [PubMed] [Google Scholar]
- 6. Andersson L., Porath J. (1986) Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal. Biochem. 154, 250–254 [DOI] [PubMed] [Google Scholar]
- 7. Posewitz M. C., Tempst P. (1999) Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal. Chem. 71, 2883–2892 [DOI] [PubMed] [Google Scholar]
- 8. Zhou H., Xu S., Ye M., Feng S., Pan C., Jiang X., Li X., Han G., Fu Y., Zou H. (2006) Zirconium phosphonate-modified porous silicon for highly specific capture of phosphopeptides and MALDI-TOF MS analysis. J. Proteome Res. 5, 2431–2437 [DOI] [PubMed] [Google Scholar]
- 9. Pinkse M. W., Uitto P. M., Hilhorst M. J., Ooms B., Heck A. J. (2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-nanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 10. Kweon H. K., Håkansson K. (2006) Selective zirconium dioxide-based enrichment of phosphorylated peptides for mass spectrometric analysis. Anal. Chem. 78, 1743–1749 [DOI] [PubMed] [Google Scholar]
- 11. Zhou H., Low T. Y., Hennrich M. L., Toorn H. v. d., Schwend T., Zou H., Mohammed S., Heck A. J. R. (2011) Enhancing the identification of phosphopeptides from putative basophilic kinase substrates using Ti (IV) based IMAC enrichment. Mol. Cell. Proteomics 10, M110.006452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lai A. C.-Y., Tsai C.-F., Hsu C.-C., Sun Y.-N., Chen Y.-J. (2012) Complementary Fe3+- and Ti4+-immobilized metal ion affinity chromatography for purification of acidic and basic phosphopeptides. Rapid Commun. Mass Spectrom. 26, 2186–2194 [DOI] [PubMed] [Google Scholar]
- 13. Bodenmiller B., Mueller L. N., Mueller M., Domon B., Aebersold R. (2007) Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat. Methods 4, 231–237 [DOI] [PubMed] [Google Scholar]
- 14. Tsai C.-F., Hsu C.-C., Hung J.-N., Wang Y.-T., Choong W.-K., Zeng M.-Y., Lin P.-Y., Hong R.-W., Sung T.-Y., Chen Y.-J. (2014) Sequential phosphoproteomic enrichment through complementary metal-directed immobilized metal ion affinity chromatography. Anal. Chem. 86, 685–693 [DOI] [PubMed] [Google Scholar]
- 15. Thingholm T. E., Jensen O. N., Robinson P. J., Larsen M. R. (2008) SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol. Cell. Proteomics 7, 661–671 [DOI] [PubMed] [Google Scholar]
- 16. Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., Jorgensen T. J. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 17. Kettenbach A. N., Gerber S. A. (2011) Rapid and reproducible single-stage phosphopeptide enrichment of complex peptide mixtures: application to general and phosphotyrosine-specific phosphoproteomics experiments. Anal. Chem. 83, 7635–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Montoya A., Beltran L., Casado P., Rodríguez-Prados J.-C., Cutillas P. R. (2011) Characterization of a TiO2 enrichment method for label-free quantitative phosphoproteomics. Methods 54, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Q. R., Ning Z. B., Tang J. S., Nie S., Zeng R. (2009) Effect of peptide-to-TiO2 beads ratio on phosphopeptide enrichment selectivity. J. Proteome Res. 8, 5375–5381 [DOI] [PubMed] [Google Scholar]
- 20. Yue X.-S., Hummon A. B. (2013) Combination of multistep IMAC enrichment with high-pH reverse phase separation for in-depth phosphoproteomic profiling. J. Proteome Res. 12, 4176–4186 [DOI] [PubMed] [Google Scholar]
- 21. Zhou H., Di Palma S., Preisinger C., Peng M., Polat A. N., Heck A. J. R., Mohammed S. (2013) Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 12, 260–271 [DOI] [PubMed] [Google Scholar]
- 22. Ficarro S. B., Salomon A. R., Brill L. M., Mason D. E., Stettler-Gill M., Brock A., Peters E. C. (2005) Automated immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry platform for profiling protein phosphorylation sites. Rapid Commun. Mass Spectrom. 19, 57–71 [DOI] [PubMed] [Google Scholar]
- 23. Wang J., Zhang Y., Jiang H., Cai Y., Qian X. (2006) Phosphopeptide detection using automated online IMAC-capillary LC-ESI-MS/MS. Proteomics 6, 404–411 [DOI] [PubMed] [Google Scholar]
- 24. Pinkse M. W. H., Mohammed S., Gouw J. W., van Breukelen B., Vos H. R., Heck A. J. R. (2008) Highly robust, automated, and sensitive online TiO2-based phosphoproteomics applied to study endogenous phosphorylation in Drosophila melanogaster. J. Proteome Res. 7, 687–697 [DOI] [PubMed] [Google Scholar]
- 25. Ndassa Y. M., Orsi C., Marto J. A., Chen S., Ross M. M. (2006) Improved immobilized metal affinity chromatography for large-scale phosphoproteomics applications. J. Proteome Res. 5, 2789–2799 [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y., Wolf-Yadlin A., Ross P. L., Pappin D. J., Rush J., Lauffenburger D. A., White F. M. (2005) Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics 4, 1240–1250 [DOI] [PubMed] [Google Scholar]
- 27. Zarling A. L., Ficarro S. B., White F. M., Shabanowitz J., Hunt D. F., Engelhard V. H. (2000) Phosphorylated peptides are naturally processed and presented by Major Histocompatibility Complex Class I molecules in vivo. J. Exp. Med. 192, 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zarei M., Sprenger A., Gretzmeier C., Dengjel J. (2012) Combinatorial use of electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) and strong cation exchange (SCX) chromatography for in-depth phosphoproteome analysis. J. Proteome Res. 11, 4269–4276 [DOI] [PubMed] [Google Scholar]
- 29. Zarei M., Sprenger A., Metzger F., Gretzmeier C., Dengjel J. (2011) Comparison of ERLIC–TiO2, HILIC–TiO2, and SCX–TiO2 for global phosphoproteomics approaches. J. Proteome Res. 10, 3474–3483 [DOI] [PubMed] [Google Scholar]
- 30. Dai J., Wang L.-S., Wu Y.-B., Sheng Q.-H., Wu J.-R., Shieh C.-H., Zeng R. (2009) Fully automatic separation and identification of phosphopeptides by continuous pH-gradient anion exchange online coupled with reversed-phase liquid chromatography mass spectrometry. J. Proteome Res. 8, 133–141 [DOI] [PubMed] [Google Scholar]
- 31. Villén J., Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Engholm-Keller K., Birck P., Størling J., Pociot F., Mandrup-Poulsen T., Larsen M. R. (2012) TiSH—a robust and sensitive global phosphoproteomics strategy employing a combination of TiO2, SIMAC, and HILIC. J. Proteomics 75, 5749–5761 [DOI] [PubMed] [Google Scholar]
- 33. Thakur S. S., Geiger T., Chatterjee B., Bandilla P., Fröhlich F., Cox J., Mann M. (2011) Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol. Cell. Proteomics 10, M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pirmoradian M., Budamgunta H., Chingin K., Zhang B., Astorga-Wells J., Zubarev R. A. (2013) Rapid and deep human proteome analysis by single-dimension shotgun proteomics. Mol. Cell. Proteomics 12, 3330–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Köcher T., Pichler P., Swart R., Mechtler K. (2012) Analysis of protein mixtures from whole-cell extracts by single-run nanoLC-MS/MS using ultralong gradients. Nat. Protoc. 7, 882–890 [DOI] [PubMed] [Google Scholar]
- 36. Michalski A., Cox J., Mann M. (2011) More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J. Proteome Res. 10, 1785–1793 [DOI] [PubMed] [Google Scholar]
- 37. Hebert A. S., Richards A. L., Bailey D. J., Ulbrich A., Coughlin E. E., Westphall M. S., Coon J. J. (2014) The one hour yeast proteome. Mol. Cell. Proteomics 13, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 39. Zhou H., Ye M., Dong J., Corradini E., Cristobal A., Heck A. J., Zou H., Mohammed S. (2013) Robust phosphoproteome enrichment using monodisperse microsphere-based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc. 8, 461–480 [DOI] [PubMed] [Google Scholar]
- 40. Seidler J., Zinn N., Haaf E., Boehm M. E., Winter D., Schlosser A., Lehmann W. D. (2011) Metal ion-mobilizing additives for comprehensive detection of femtomole amounts of phosphopeptides by reversed phase LC-MS. Amino Acids 41, 311–320 [DOI] [PubMed] [Google Scholar]
- 41. Hahne H., Pachl F., Ruprecht B., Maier S. K., Klaeger S., Helm D., Médard G., Wilm M., Lemeer S., Kuster B. (2013) DMSO enhances electrospray response, boosting sensitivity of proteomic experiments. Nat. Methods 10, 989–991 [DOI] [PubMed] [Google Scholar]
- 42. van den Toorn H. W. P., Muñoz J., Mohammed S., Raijmakers R., Heck A. J. R., van Breukelen B. (2011) RockerBox: analysis and filtering of massive proteomics search results. J. Proteome Res. 10, 1420–1424 [DOI] [PubMed] [Google Scholar]
- 43. Käll L., Canterbury J. D., Weston J., Noble W. S., MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 44. Marx H., Lemeer S., Schliep J. E., Matheron L., Mohammed S., Cox J., Mann M., Heck A. J. R., Kuster B. (2013) A large synthetic peptide and phosphopeptide reference library for mass spectrometry-based proteomics. Nat. Biotechnol. 31, 557–564 [DOI] [PubMed] [Google Scholar]
- 45. Wu Z. X., Doondeea J. B., Gholami A. M., Janning M. C., Lemeer S., Kramer K., Eccles S. A., Gollin S. M., Grenman R., Walch A., Feller S. M., Kuster B. (2011) Quantitative chemical proteomics reveals new potential drug targets in head and neck cancer. Mol. Cell. Proteomics 10, M111.011635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ye J., Zhang X., Young C., Zhao X., Hao Q., Cheng L., Jensen O. N. (2010) Optimized IMAC-IMAC protocol for phosphopeptide recovery from complex biological samples. J. Proteome Res. 9, 3561–3573 [DOI] [PubMed] [Google Scholar]
- 47. Mertins P., Qiao J. W., Patel J., Udeshi N. D., Clauser K. R., Mani D. R., Burgess M. W., Gillette M. A., Jaffe J. D., Carr S. A. (2013) Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods 10, 634–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou H., Ye M., Dong J., Corradini E., Cristobal A., Heck A. J. R., Zou H., Mohammed S. (2013) Robust phosphoproteome enrichment using monodisperse microsphere–based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc. 8, 461–480 [DOI] [PubMed] [Google Scholar]
- 49. Matheron L., van den Toorn H., Heck A. J., Mohammed S. (2014) Characterization of biases in phosphopeptide enrichment by Ti(4+)-immobilized metal affinity chromatography and TiO2 using a massive synthetic library and human cell digests. Anal. Chem. 86, 8312–8320 [DOI] [PubMed] [Google Scholar]
- 50. Ritorto M. S., Cook K., Tyagi K., Pedrioli P. G. A., Trost M. (2013) Hydrophilic strong anion exchange (hSAX) chromatography for highly orthogonal peptide separation of complex proteomes. J. Proteome Res. 12, 2449–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.