Abstract
Vertebrates produce various chondroitin sulfate proteoglycans (CSPGs) that are important structural components of cartilage and other connective tissues. CSPGs also contribute to the regulation of more specialized processes such as neurogenesis and angiogenesis. Although many aspects of CSPGs have been studied extensively, little is known of where the CS chains are attached on the core proteins and so far, only a limited number of CSPGs have been identified. Obtaining global information on glycan structures and attachment sites would contribute to our understanding of the complex proteoglycan structures and may also assist in assigning CSPG specific functions. In the present work, we have developed a glycoproteomics approach that characterizes CS linkage regions, attachment sites, and identities of core proteins. CSPGs were enriched from human urine and cerebrospinal fluid samples by strong-anion-exchange chromatography, digested with chondroitinase ABC, a specific CS-lyase used to reduce the CS chain lengths and subsequently analyzed by nLC-MS/MS with a novel glycopeptide search algorithm. The protocol enabled the identification of 13 novel CSPGs, in addition to 13 previously established CSPGs, demonstrating that this approach can be routinely used to characterize CSPGs in complex human samples. Surprisingly, five of the identified CSPGs are traditionally defined as prohormones (cholecystokinin, chromogranin A, neuropeptide W, secretogranin-1, and secretogranin-3), typically stored and secreted from granules of endocrine cells. We hypothesized that the CS side chain may influence the assembly and structural organization of secretory granules and applied surface plasmon resonance spectroscopy to show that CS actually promotes the assembly of chromogranin A core proteins in vitro. This activity required mild acidic pH and suggests that the CS-side chains may also influence the self-assembly of chromogranin A in vivo giving a possible explanation to previous observations that chromogranin A has an inherent property to assemble in the acidic milieu of secretory granules.
Chondroitin sulfates (CS)1 are complex polysaccharides present at cell surfaces and in extracellular matrices. The polysaccharides belong to a subclass of glycosaminoglycans (GAGs) and are covalently linked to various core proteins to form CS-proteoglycans (CSPGs), each with differences in the protein structures and/or numbers of CS side chains. Apart from their structural role in cartilage, CSPGs contribute to the regulation of a diverse set of biological processes such as neurogenesis, growth factor signaling, angiogenesis, and morphogenesis (1–5). Although the molecular basis of CSPGs functions remains elusive, accumulating evidence suggests that the underlying activities relate to selective ligand binding to discrete structural variants of the polysaccharides. Thus, the current strategy for understanding the biological role of CSPGs aims to identify selective CS polysaccharide–ligand interactions. However, information on the number of CS-chains and their specific attachment site(s) on any given core protein is often scarce which limits our functional understanding of CSPGs.
The biosynthesis of GAGs occurs in the endoplasmic reticulum and Golgi compartments and is initiated by the enzymatic addition of a beta-linked xylose (Xyl) to a Ser residue of the core protein. The sequential addition of two galactose residues (Gal) and a glucuronic acid (GlcA) onto the growing saccharide chain completes the formation of a tetrasaccharide linkage region (GlcAβ3Galβ3Galβ4XylβSer). This part of the biosynthesis is the same for CS and heparan sulfate (HS). However, for CS the biosynthesis continues with the addition of an N-acetylgalactosamine (GalNAcβ3), whereas HS biosynthesis continues with the addition of an N-acetylglucosamine (GlcNAcα4) (6). The CS-chains are thereafter elongated through the addition of repeating units of GlcA and GalNAc and are further modified by the addition of specifically positioned sulfate groups (7). Certain features of the core protein seem to influence if a certain Ser residue is selected for GAG attachment and whether CS or HS will be synthesized, but the selection mechanism is largely unknown. Sequence analysis of previously known GAG-substituted core proteins reveals that the glycosylated serine residues are usually flanked by a glycine residue (-SG-), and are associated with a cluster of acidic residues in close proximity (8). This motif may assist in the prediction of potential GAG-sites of core proteins; however, the use of such strategy is ambiguous because proteoglycans may also contain unoccupied motifs or motifs that are occasionally occupied (9).
Glycoproteomics strategies have recently appeared that provide site-specific information of N- and O-glycans. Such strategies are typically based on a specific enrichment of glycopeptides and a subsequent analysis with nano-liquid chromatography-tandem mass spectrometry (nLC-MS/MS) (10). By further developing this concept for proteoglycans (11), we have now analyzed CSPG linkage region glycopeptides of human samples, which enabled us to identify 13 novel human CSPGs in addition to 13 already established CSPGs. Urine and cerebrospinal fluid (CSF) samples were trypsinized and CS glycopeptides were enriched using strong anion exchange (SAX) chromatography. The CS chains were depolymerized with chondroitinase ABC, generating free disaccharides and a residual hexameric structure composed of the linkage region and a GlcA-GalNAc disaccharide dehydrated on the terminal GlcA residue (12). MS/MS analysis provided the combined sequencing of the residual hexasaccharide and of the core peptide.
EXPERIMENTAL PROCEDURES
Bikunin CS-glycopeptide Preparation
One hundred micrograms of bikunin (provided by Mochida Pharmaceutical, Japan) was incubated for 3 h at 37 °C with 10 μl digestion buffer (55 mm NaAc, pH 8.0) and 1 mU of chondroitinase ABC (C3667, Sigma-Aldrich). The sample was lyophilized and thereafter trypsinized using an in-solution digestion protocol. Briefly, the sample was incubated for 10 min with Protease Max surfactant trypsin enhancer (0.02% final concentration) (Promega, Madison, WI) in 50 mm NH4HCO3. The sample was thereafter reduced with DTT (5 mm) and alkylated with iodoacetamide (15 mm). Additional Protease Max surfactant was then added (0.03% final concentration) and the sample was trypsinized over night (37 °C) with 20 μg trypsin (Promega). The actions of the two enzymes were monitored with SDS-PAGE. Three micrograms of bikunin was saved before and after the enzymatic steps. The samples (three in total) were mixed with 5 × SDS sample buffer and loaded onto a 4–20% Novex Tris-Glycine gel (Invitrogen, Carlsbad, CA). After electrophoresis separation at 100 V for 1 h, the gel was stained with Coomassie blue and scanned.
Enrichment of CS-glycopeptides from Human Urine and CSF
Eight ml of morning urine was collected from a healthy male individual and the sample was separated from cell-debris by centrifugation (3000 × g for 10 min). The urine was mixed with SDS to a final concentration of 0.1% and run through a PD-10 column (GE Healthcare) equilibrated in 0.1% SDS to remove urochrome pigments. The eluted sample was thereafter run through a second PD-10 column and equilibrated in dH2O to remove the SDS. The sample was collected and lyophilized to a volume of ∼10 μl. The CSF sample was donated from a patient undergoing neurosurgery because of a benign condition and was kept in 2 ml aliquots at −80 °C until use. The CSF sample (2 ml) was directly lyophilized to a volume of ∼10 μl, without any prior purification. The protein amount of the two samples was ∼1 mg, as determined with Nanodrop 1000 spectrophotometer (Thermo Scientific). The samples were trypsinized using an in-solution digestion protocol, as described above. The GAG-substituted peptides were enriched using SAX-chromatography (Vivapure, Q Mini H). The trypsin-digested samples (1 mg) were diluted in 10 ml coupling buffer (50 mm NaAc, 200 mm NaCl, pH 4.0) and 400 μl of the diluted sample were applied onto the column and spun at 1000 × g for 2 min. The procedure was repeated until all sample volume had been applied onto the column. The column was thereafter washed with 400 μl of a low-salt wash solution (50 mm Tris-HCl, 200 mm NaCl, pH 8.0) to remove loosely bound material. The GAG-peptides were then eluted stepwise with three buffers of increasing NaCl-concentrations and pH; 1) 50 mm NaAc, 400 mm NaCl, pH 4.0, 2) 50 mm Tris-HCl, 800 mm NaCl, pH 8.0, and 3) 50 mm Tris-HCl, 1600 mm NaCl, pH 8.0. For each wash and elution step, the column was spun at 1000 × g for 2 min. The collected fractions were desalted using a PD10-column and individually subjected to chondroitinase-lyase degradation, as described above. Prior to MS-analysis, the samples were desalted again using a C18 spin column (8 mg resin) according to the manufacturer's protocol (Thermo Scientific, Inc., Waltham, MA). The samples were thereafter dried and stored at −18 °C until MS-analysis.
LC-MS/MS Analysis
The samples were analyzed on a Q Exactive mass spectrometer coupled to an Easy-nLC 1000 (Thermo Fisher Scientific, Inc., Waltham, MA). Glycopeptides (10 μl injection volume) were separated using an in-house constructed precolumn and analytical column set up (45 × 0.075 mm I.D. and 200 × 0.050 mm I.D., respectively) packed with 3 μm Reprosil-Pur C18-AQ particles (Dr. Maisch GmbH, Ammerbuch, Germany). The following gradient was run at 150 nL/min; 7–37% B-solvent (acetonitrile in 0.2% formic acid) over 15 or 60 min, 37–80% B over 5 min, with a final hold at 80% B for 10 min. The 15 min nLC-gradient was used for pharmaceutical grade bikunin sample, whereas the 60 min gradient was used for the complex CSF and urine samples.
Ions were generated and injected into the Q Exactive mass spectrometer under a spray voltage of 1.6 kV in positive ion mode. MS scans were performed at 70,000 resolution (at m/z 200), an Automatic Gain Control (AGC)-target value of 3E6 with a mass range of m/z 600–2000. MS/MS analysis was performed in a data-dependent mode, with the top six most abundant doubly or multiply charged precursor ions in each MS scan selected for fragmentation (MS2) by stepped higher energy collision dissociation (stepped HCD) of ± 25% around a normalized collision energy (NCE)-value of 20, 25, and 30. For MS2 scans the following parameters were employed; one microscan performed at 35,000 resolution (at m/z 200), AGC 1E6 with a maximal injection time of 128 ms, a fixed first mass of m/z 100, an isolation window of 2 Da, intensity threshold of 1.2E5 and a dynamic exclusion of 30 s.
Mascot Search for CS-glycopeptides
The HCD.raw spectra were converted to Mascot .mgf format using Mascot distiller (version 2.3.2.0, Matrix Science, London, UK). The ions were presented as singly protonated in the output Mascot file. Searches were performed using an in-house Mascot server (version 2.3.02) with the enzyme specificity set to Trypsin, and then to Semitrypsin, allowing for one or two missed cleavages, in subsequent searches on human sequences of the UniprotKB (87,613, sequences, 13/3/2013). Additional searches were made on human sequences of the NCBInr (22,826,248, sequences, 03/18/2013). The instrument parameter was set to consider the MH+ form of b- and y-ions and the corresponding ion series resulting from losses of H2O (b0 and y0 series) or NH3 (b* and y* series). The peptide tolerance was set to 10 parts per million (ppm) and fragment tolerance was set to 0.01 Da. The searches were allowed to include variable modifications at serine residues of the residual hexasaccharide structure [GlcA(-H2O)GalNAcGlcAGalGalXyl-O-] with 0 (C37H55NO30, 993.2809 Da), 1 (C37H55NO33S, 1073.2377 Da), or 2 (C37H55NO36S2, 1153.1945 Da) sulfate groups attached. Moreover, “neutral losses” of the same masses were implemented to the search constraints, as the glycan modification is absent in the obtained peptide fragments because of the efficiency of the HCD-fragmentation. The distinction between a sulfate group (79.9663 Da), as opposed to a phosphate groups (79.9568 Da), was made by manually evaluating the MS2-spectra for obtained hits. Further, the HexNAc-derived peaks were added as “Ignore Masses” to improve the Mascot-score of potential hits: C6H8NO2 (126.06 Da); C7H8NO2 (138.06 Da); C6H10NO3 (144.07 Da); C8H10NO3 (168.08 Da); C8H12NO4 (186.08 Da); C8H14NO5 (204.09 Da); C9H12NO5 (214.09 Da); and C14H20NO10 (362.11 Da).
Manual Data Evaluation
We manually evaluated all hits according to the following criteria: (1) The presence of HexNAc generated oxonium ions, most specifically m/z 362.11 and m/z 214.09 that are specific for the CS linkage region hexasaccharide structures and not found for core 1 Neu5AcHexHexNAc and (Neu5Ac)2HexHexNAc structures. (2) Stepwise glycosidic fragmentation of the linkage region must be visible and/or the peak(s) corresponding to the deglycosylated peptide ion must be present. (3) Ions originating from peptide backbone fragmentation at the N-terminal side of Pro must be prominent, with the simultaneous presence of both the b- and y-ion. Scores below the p < 0.05 Mascot score threshold were also considered to be true if all above criteria were met.
Chromogranin A and CS Interaction
Binding of Chromogranin A (Abcam, Ab85486, Abcam, MA) to CS purified from bovine trachea (Sigma-Aldrich, C9819) was evaluated using the Biosensor system (Biacore 2000, GE Healthcare). The protein was diluted into a 50 mm NaAc (pH 4.5) coupling buffer and immobilized onto a CM 5 sensor chip (2000 RU) using an amine coupling kit (GE Healthcare). All experiments and procedures used a flow rate of 20 μl/min. CS was injected over the protein ligand at a concentration of 5 μm under mild acidic conditions (pH 5.0) or under neutral conditions (pH 7.4). As a negative control, BSA was immobilized to a sensor chip to the same level as for Chromogranin A and CS was injected over surface as described above. Sequential injections of CS (5 μm) and CgA (15 nm) were made over immobilized CgA (1000 RU) at pH 5.0 to assess whether CS may promote CgA complex formation. All sensorgrams obtained were double-reference subtracted in Biacore evaluation software 2.0, using reference flow cell and buffer blank sample.
RESULTS
Structural Analysis of Chondroitin Sulfate Linkage Region Glycopeptides
Bikunin, also known as Protein AMBP (Inter-alpha-trypsin inhibitor light chain; Uniprot P02760), was chosen as model CSPG because it has been the focus of previous structural studies and is therefore perceived as relatively well characterized (13–16). Bikunin holds only one CS chain that typically contains 27–39 monosaccharides with 3–6 sulfate groups attached to GalNAc residues (13). The chain attaches to the core protein at Ser10 through the tetrasaccharide linkage region and the linkage region may also contain modifications such as a phosphate group at the xylose residue or a sulfate group at one of the two galactose residues (17). Fig. 1A shows a schematic illustration of bikunin. To obtain a defined CS-glycopeptide for structural analysis, pharmaceutical grade bikunin was degraded by chondroitinase ABC, generating free disaccharides and a residual hexasaccharide structure still attached to the core protein. Previous studies have demonstrated that the resulting hexasaccharide is composed of the linkage region and a GlcA-GalNAc disaccharide, which is dehydrated on C4-C5 on the terminal GlcA (12).
Fig. 1.
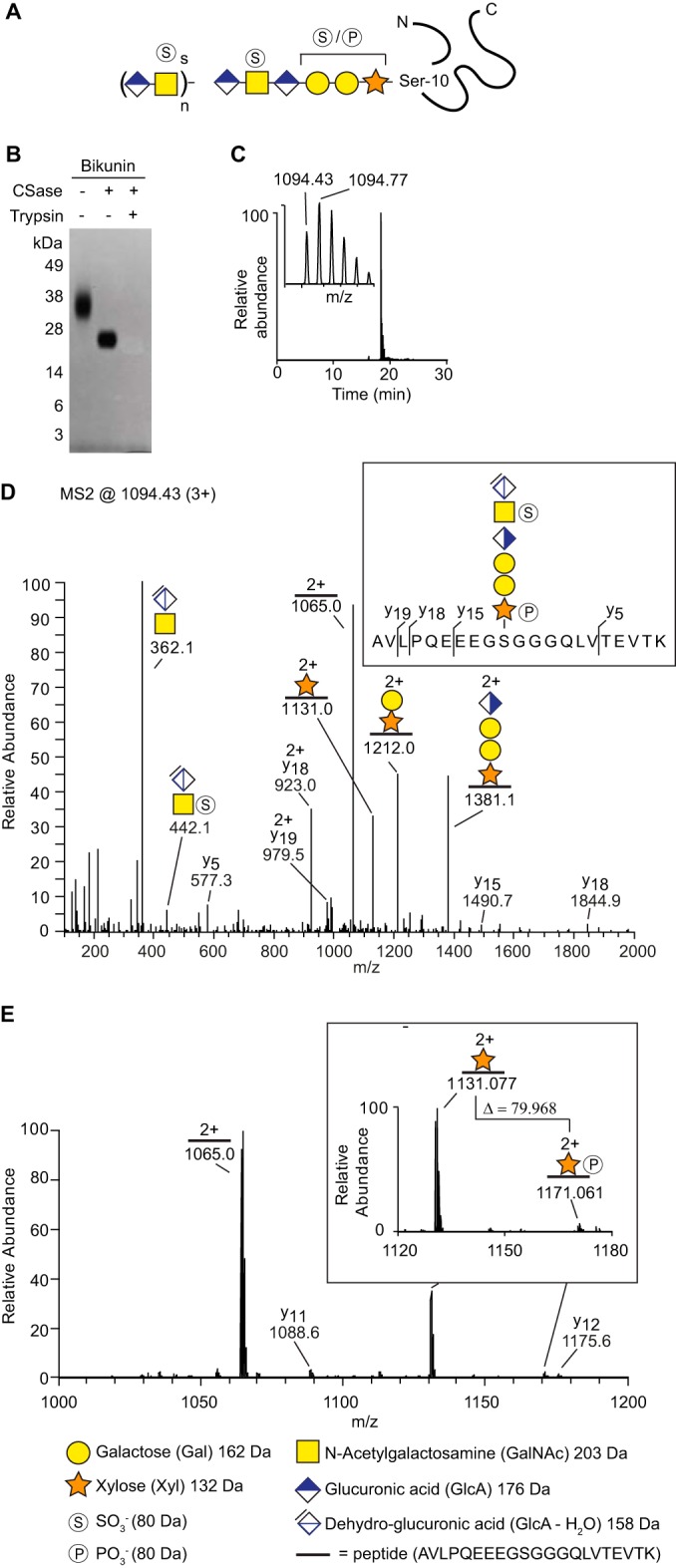
Site-specific linkage region analysis of bikunin. A, Schematic illustration of bikunin. The number of disaccharides (n) and the degree of sulfation (s) vary according to a predefined pattern. The linkage region may contain modifications such as a phosphate group at the xylose residue or a sulfate group at one of the galactose residues. B, Bikunin incubated either with chondroitinase ABC, or with chondroitinase ABC and trypsin, was analyzed with SDS-PAGE and stained with Coomassie blue. C, Reversed phase liquid chromatography-ESI-Orbitrap extracted-ion current of the bikunin sample revealed a CS-glycopeptide (m/z 1094.43; 3+), eluting as a single distinct peak at ∼18.5 min and (C, insert) Orbitrap-MS1 of the molecular ion. D, MS2-fragmentation of the CS-glycopeptide provided y-ions as well as glycosidic fragmentation. E, A mass shift of 79.968 Da was identified between m/z = 1131.077; 2+ and 1171.061; 2+ (E, enlarged view), demonstrating the presence of a phosphate group (79.966 Da) on the xylose-residue. All ions are [M + zH]+ and their charges are shown as superscript when z > 1.
Analysis of the chondroitinase-treated sample by SDS-PAGE showed a distinct band that fitted well with that of the combined core protein-hexasaccharide molecular weight (∼17 kDa) (Fig. 1B). Additional trypsin treatment generated a CS-glycopeptide that migrated out of the gel because of its low molecular weight. The preparation was thereafter analyzed with nLC-MS/MS in a 30 min program (see experimental procedures) and an extracted-ion chromatogram demonstrated the presence of a precursor ion (MS1 m/z 1094.43; 3+), which eluted as a distinct chromatographic peak at 18.5 min (Fig. 1C). The identified precursor ion equated to the molecular mass of the expected CS-glycopeptide with two phosphate/sulfate modifications (3280.2641 Da). Fragmentation of the ion with higher energy collision dissociation (HCD) at normalized collision energy (NCE) of 20% enabled the identification of several specific glycosidic and peptide fragments (Fig. 1D). A prominent diagnostic oxonium ion at m/z 362.1 was observed corresponding to the disaccharide [GlcAGalNAc-H2O+H] +. (A comparable oxonium ion at m/z 366.1 is typically found for the GalGalNAc disaccharide of mucin core 1 structures (11)). Furthermore, the glycan was modified with one sulfate group (SO3−) at the subterminal GalNAc residue ([GlcAGalNAc-H2O+SO3+H]+ oxonium ion, m/z 442.1) and one phosphate group (PO3−) at the Xyl residue (peptide+Xyl+PO3, m/z 1171.061; 2+). The identified CS-glycopeptide structure is shown in Fig. 1D (insert). The distinction between phosphate- and sulfation modification was feasible by examining the spectrum in a more narrow mass range (Fig. 1E). A mass shift of 79.968 Da between m/z = 1131.077; 2+ and 1171.061; 2+ was observed in the range of m/z 1000–1200 (Fig. 1E, enlarged view). This demonstrates the presence of a phosphate group (79.966 Da), as opposed to a sulfate group (79.957 Da), on the xylose-residue.
An additional bikunin CS-glycopeptide precursor ion (MS1 m/z 1094.43; 3+) was identified that co-eluted with the aforementioned structure at 18.5 min (supplemental Fig. S1A). Similar to the first precursor ion, the MS2 fragment spectrum of this precursor ion displayed a mass shift of 79.958 Da between m/z = 362.110; 1+ and 442.068; 1+, demonstrating the presence of a sulfate group on the GalNAc-residue (supplemental Fig. S1A, i). However, the exact structure of this structural isomer could not be determined as no additional mass shift could be identified that indicated the presence of either a phosphate- or sulfate modification.
Structural Analysis of the Bikunin Linkage Region in Human Urine
We then tested whether the strategy could be used for direct detection and sequencing of CSPG linkage region glycopeptides in a complex biological sample. Human urine was chosen as sample matrix because it contains high concentrations of bikunin and urine has also previously proven useful for glycoproteomics analyses (16, 18). Trypsinized urine sample was passed over a SAX-column that had been equilibrated with a low-salt buffer (0.2 m NaCl). The positively charged matrix retains anionic polysaccharides and their attached peptides, whereas nonsubstituted peptides flow through (19). After a subsequent wash step, the bound structures were eluted stepwise with three buffers of increasing sodium chloride concentration (0.4 m NaCl, 0.8 m NaCl and 1.6 m NaCl). These three fractions were collected, desalted and individually treated with chondroitinase ABC to depolymerize the CS-chains. LC-MS/MS analysis of the 0.8 m fraction revealed a bikunin-derived glycopeptide (m/z 1094.43; 3+) that displayed a fragmentation pattern similar to that of the pharmaceutical grade preparation (supplemental Fig. S2A–S2B). (To obtain better separation of molecules with similar polarity, a 70 min nLC-program with a slow increase in organic solvent was used for analysis of the complex urine sample). Furthermore, a NCE level of 30% was used to generate abundant peptide fragmentation that enabled the identification of several diagnostic b- and y-ions (m/z 400–1000) (supplemental Fig. S2C). This analysis also enabled the identification of several diagnostic GalNAc-derived oxonium ions. Such ions were the result of H2O losses (m/z 168.1 and m/z 186.1) and saccharide decompositions (m/z 126.1, m/z 138.1, m/z 144.1, and m/z 214.1) (supplemental Fig. S2D). Further evaluation also revealed the presence of additional bikunin CS-glycopeptide structures, including one without secondary modifications (m/z 1041.13; 3+) and one with only one phosphate group (m/z 1067.78; 3+) (supplemental Fig. S3).
Identification of Novel CSPGs in Human Urine and CSF
An automated Mascot search strategy was developed to identify all CS substituted peptides from the urine sample. The general workflow for glycopeptide enrichment, MS-analysis and subsequent data interpretation is illustrated in Fig. 2. In order to identify CS substituted peptides, standard proteomic database search settings were allowed to include also variable modifications of the residual hexasaccharide structure (993.2808 Da) for [GlcA(-H2O)GalNAcGlcAGalGalXyl-O-] with 0, 1, or 2 sulfates or phosphates attached. All generated hits were manually validated and interpreted with regard to peptide sequence, glycan structure, and sulfate- and phosphate modifications. We thereafter used the same procedure to analyze CSPGs in a CSF sample.
Fig. 2.
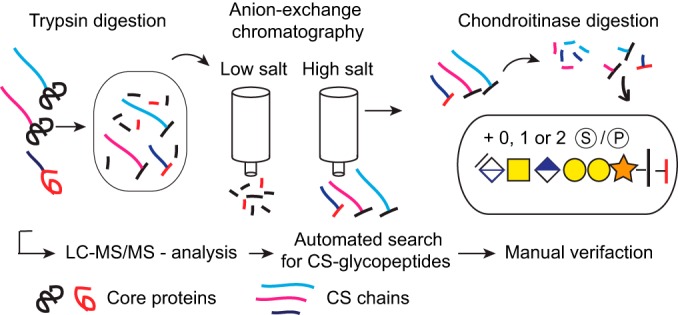
Work flow for enrichment, analysis, and identification of CSPGs. General work flow from sample preparation to LC-MS/MS-analysis and interpretation of spectra.
In total, Mascot-assisted analysis of urine and CSF samples revealed 30 different CS-glycopeptides. supplemental Table S1 shows a complete list of all the CSPGs identified in the present study, organized based on the CSPGs' cellular distribution. These groups include “cell surface,” “extracellular matrix,” “intracellular granules,” and “miscellaneous.” Moreover, the identified peptide sequences are shown together with their corresponding linkage region glycan isoforms. We identified fifteen glycopeptides on 13 novel core proteins, where two of the novel CSPGs were found with two CS-glycopeptides (osteopontin and secretogranin-1). Additionally, 14 glycopeptides were identified from 13 previously known CSPGs, where two of the glycopeptides were related to neurocan. Annotated spectra of all CS-glycopeptides are shown in supplemental Fig. S4. Moreover, supplemental Table S2 shows a complete list of all previously known human CSPGs and the novel CSPGs identified in this study.
Prohormones as a Novel Class of Proteoglycans
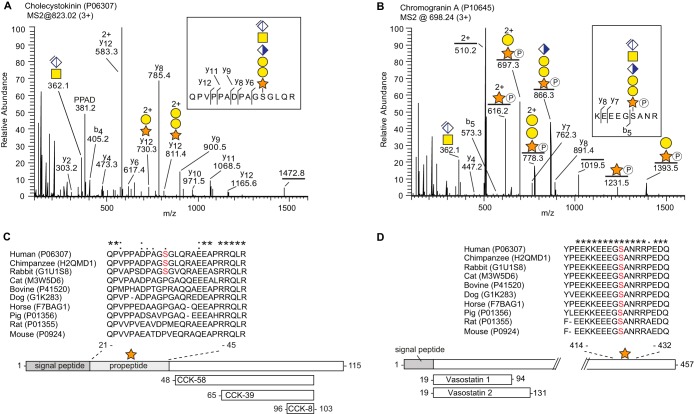
Proteoglycans are often classified as cell surface proteoglycans or extracellular matrix components (20). However, several of the novel CSPGs identified in the present study may better be described as granular or secretory proteoglycans, similar to serglycin present in mast cells and other cells of hematopoietic origin. The identified CSPGs that have a granular distribution are shown in Table I. Notably, many of the core proteins have previously been defined as prohormones, including chromogranin A (CgA), cholecystokinin, neuropeptide W, secretogranin-1, and secretogranin-3. Two examples of the CS-glycopeptides identified are shown in Fig. 3. Cholecystokinin (Fig. 3A) has not previously been reported to carry CS, whereas CgA (Fig. 3B) has been identified as a CSPG in one earlier study (21). Both proteins are prohormones as they serve as precursors for several bioactive peptides, as illustrated in Fig. 3C–3D. Alignment of mammalian cholecystokinins shows a relatively low degree of sequence homology for the CS-locus. For instance, the mouse sequence contains a proline instead of a serine at the CS-attachment site (S31), thereby excluding the possibility of a CS-modification. Because the CS-modification may influence how cholecystokinin is processed, this modification is likely relevant when studying cholecystokinin in animal models with sequences different from humans (22). Unlike cholecystokinin, CgA is highly conserved in mammals with almost complete homology for the aligned CS-site.
Table I. CSPGs identified in the present study with an intracellular distribution.
| Protein namea | Peptide sequenceb | Glycan modificationc | Sample type |
|---|---|---|---|
| Intracellular granules CSPGs | |||
| Bone marrow proteoglycan | 53ELEEEEEWGSGSEDASK69 |  |
Urine |
| aCholecystokinin | 21QPVPPADPAGSGLQR35 |  |
CSF and Urine |
| Chromogranin-A | 419KEEEGSANR427 | 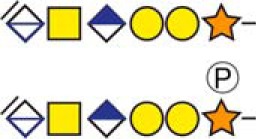 |
CSF and Urine |
| aCollagen and calcium-binding EGF domain-containing protein 1 | 382DLGSGDDHPR391 | 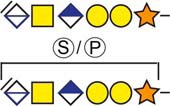 |
Urine |
| aDermcidin | 20YDPEAASAPGSGNPCHEASAAQK42 | 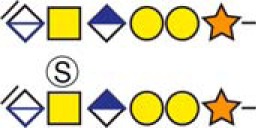 |
Urine |
| aNeuropeptide W | 120APEPALEPESLDFSGAGQR138 | 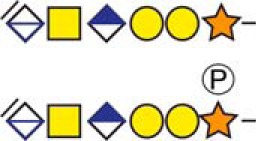 |
CSF and Urine |
| aSecretogranin-1 | 88DPADASEAHESSSR101 |  |
CSF and Urine |
| aSecretogranin-1 | 235SSQESGEETGSQENHPQESK254 |  |
CSF |
| aSecretogranin-3 | 35ELSAERPLNEQIAEAEEDK53 |  |
Urine |
a Indicate novel CSPGs identified in this study.
b Bold and underlined serine residues depict established attachment sites while bold depict probable attachment sites.
c The CS-hexasaccharide were identified either without or with sulfate (SO3−) and/or phosphate (PO3−) modifications and their positions on the hexasaccharide structure are indicated. The positioning and distinction of sulfate- (79.9663 Da) and phosphate (79.9568 Da) modifications were made by manually evaluating the MS2-spectra. When the expected modifications could not be identified a bracket including the whole hexasaccharide structure is presented as the expected modification could be, in theory, anywhere on the glycan.
Fig. 3.
Prohormones as a novel class of proteoglycans. A, MS2 fragment spectrum of cholecystokinin (P06307) (m/z 1242.5; 3+). The fragment ion of the peptide backbone (QPVPPADPAGSGLQR, m/z 1472.8; 1+) was identified with an N-terminal glutamine deamination (-NH3). B, MS2 fragment spectrum of CgA (P10645) (m/z 698.2; 3+). C–D, Alignment of the CS-sites in cholecystokinin and CgA of 10 commonly studied mammalian species. The serine residue to which the CS-chain is attached is indicated in red. The comparison shows low similarity for the CS-site in cholecystokinin across species, whereas CgA shows much greater similarity. An asterisk (*) indicates amino acids that are conserved in all species. Homologous amino acid substitutions are indicated with a colon (:) and nonhomologous substitutions with a period (.). Post-translational processing of the prohormones to a few of their known bioactive peptides are shown at the bottom.
Chromogranin A and CS form Complex Under Mild Acidic Conditions
During the formation of secretory granules, CgA is known to self-assemble into so-called “dense core aggregates” in a process that requires mild acidic conditions (23). Because CS-chains are known to promote the binding and assembly of various proteins (24, 25), we hypothesized that the CS-side chain of CgA may promote self-assembly of CgA in a similar mode. To investigate a potential core protein to CS interaction, we thus applied surface plasmon resonance (SPR) spectroscopy. CgA expressed in a bacterial system (and therefore lacking CS) was immobilized onto a Biacore sensor chip and free CS was allowed to interact with the ligand at neutral (pH 7.4) and mild acidic conditions (pH 5.0). CS binding to CgA was easily observed at pH 5.0 whereas very little binding was observed at neutral conditions, demonstrating a pH-dependence for this interaction (Fig 4A). Such drastic effect was not observed for CS binding to BSA, a protein with similar isoelectric point to that of CgA (Fig. 4B). The pH-dependence suggests that the CgA-CS interaction is specifically mediated by electrostatic interactions between histidine residues that become positively charged in an acidic milieu, and the negatively charged CS-chain of CgA. Inspection of the CgA amino acid sequence indeed reveals a histidine-motif (KERAHQQKKH) that may account for the observed effect (Fig. 4C). Sequential injection of CS and CgA at pH 5.0 demonstrated the formation of a CgA/CS/CgA complex (Fig. 4D). In contrast, CgA injected directly over the protein ligand without prior CS injection did not display any clear binding either at neutral or mild acidic conditions (Fig. 4E). These results suggest that CS may influence the assembly of CgA core proteins under mild acidic conditions, which may be of functional importance for the function of CgA in secretory granules.
Fig. 4.
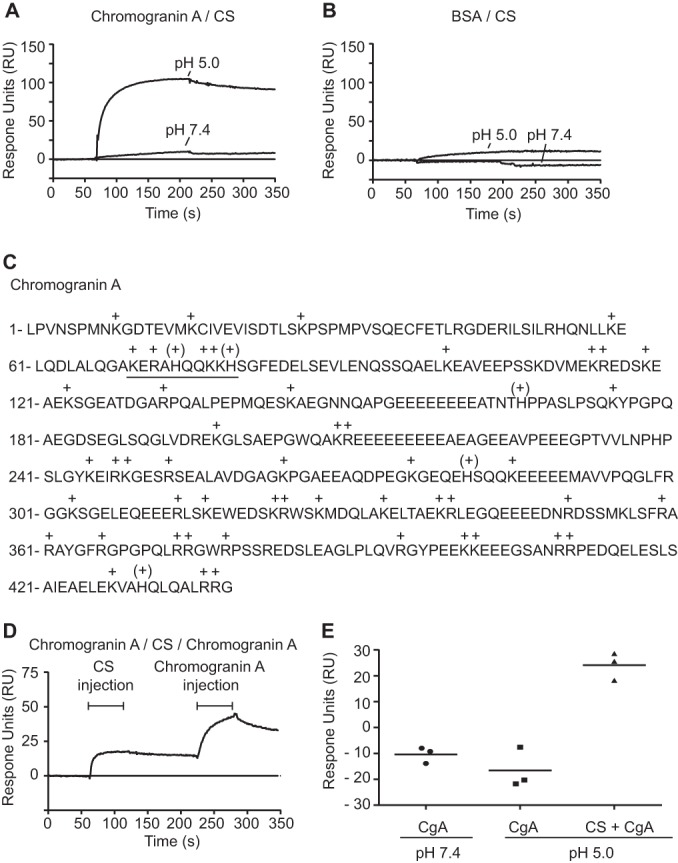
Complex formation of Chromogranin A and CS under mild acidic conditions. A–B, CgA core protein A, and BSA B, were immobilized onto a Biacore CM5 sensor chip (2000 RU) and CS (5 μm) was allowed to interact with the surfaces at neutral (pH 7.4) and under mild acidic conditions (pH 5.0). C, CgA amino acid sequence where histidine “(+)” and other basic amino acids “+” are marked. A basic peptide sequence likely to interact with negatively charged CS-chains is underlined. D, Sequential injection of CS (5 μm) and CgA (15 nm) over immobilized CgA (1000 RU) at pH 5.0 indicates CgA-CS-CgA complex formation. E, In contrast, CgA injected directly over immobilized CgA without any prior CS injection did not display any binding, neither at pH 5.0 nor at pH 7.4. The increment in CgA-binding of the three different experiments is shown. The lines represent the mean value of three independent experiments. Notably, a slight decrease in signal was observed for CgA and CgA interaction at both pH-conditions. This effect was related to a stronger binding of the analyte to the reference channel than that of the ligand channel.
DISCUSSION
Proteoglycans affect many aspects of normal cellular physiology and are essential for embryonic development. In addition, proteoglycans may also contribute to pathological conditions, such as cancer and amyloid diseases including Alzheimer's disease (26, 27). Many of the functions depend on specific interactions between the GAG side chains and the target proteins. However, in most cases, attachment sites as well as core protein identities are unknown. Such information would be of great value to further delineate proteoglycan-mediated functions. To this aim, we enriched and analyzed by LC-MS/MS CS linkage region glycopeptides in human samples, which resulted in the identification of 13 novel CSPGs. Moreover, the method allowed integrated glycopeptide structural characterization in which the positions of sulfate- and/or phosphate modifications could be determined.
Enrichment of GAG-chains using SAX-chromatography is commonly used because of the abundance of sulfate groups and GlcA on the polysaccharides. Similar to previous studies, GAG-chains were purified from biological samples using SAX-chromatography with the exception that the samples were enriched for GAG-substituted glycopeptides, rather than for intact proteoglycans or released GAGs (28). Chondroitinase ABC digestion was thereafter employed to specifically reduce the length and the structural variability of CS-chains. Enzymatic treatment generated a residual hexasacharide structure, composed of the linkage region and an additional unsaturated GlcA-GalNAc disaccharide (12). This concept of reducing glycan heterogeneity prior to analysis is similarly used for other glycoproteomics studies (10).
Earlier strategies for identifying GAG-attachment sites typically involve site-directed mutagenesis of potential “GAG-motifs” (8). If amino acid substitution by molecular engineering (e.g. into alanine) results in the reduction of the apparent molecular weight of the potential proteoglycan, this result serves a proof for the presence of a GAG-site. This strategy is useful for mapping potential sites in a given protein, but does not allow for the discrimination of the type of GAG (HS or CS) or for effective mining of the GAG-proteome. Only one unbiased proteomic approach for identifying GAG-sites has previously been described (19). Enriched GAG-peptides were then treated with sodium hydroxide, causing β-elimination of the sugar chains resulting in a reactive serine residue that was subsequently tagged with DTT. The tagged serine-residue allowed site-specific characterization with MS/MS. However, this strategy does not discriminate between the different types of GAG-chains, neither provides site-specific glycan structural information. Determining the type of GAG is essential for the biological understanding because HS- and CS-chains typically display different roles in cell functions. For instance, HS was recently shown to promote axonal guidance through clustering of receptor protein tyrosine phosphatase sigma, whereas CS had an inhibitory effect (29).
Surprisingly, several of the core proteins identified in this study belong to a family known as prohormones, including CgA, secretogranin-1, secretogranin-3, cholecystokinin, and neuropeptide W. One previous study identified CgA to be modified with CS (21), which was confirmed in the present study with the identification of a CS-substitution in the C-terminal end (S424). Prohormones are known to undergo extensive post-translational modifications, such as phosphorylation, tyrosine sulfation, and glycosylations (30). These modifications may influence the action of proprotein convertases and thereby contribute to the functional processing of the prohormones. Intriguingly, the ratio of T3 and T4, which is derived from human thyroglobulin, was affected by a CS-chain linked onto a subset of the protein. This effect was likely related to the blocking of specific proteolytic cleavage sites (30). Whether CS-modifications influence the processing of the above identified prohormones in a similar way remains to be determined.
CgA, secretogranin-1 and secretogranin-3 are acidic proteins that all belong to the granin family. In addition to being precursors of several bioactive peptides, they share a propensity to self-assemble into aggregates in the acidic milieu of secretory granules (31). Our binding studies using SPR-technique revealed that free CS and CgA-core protein interacted under mild acidic conditions but not at neutral pH, demonstrating a pH-dependence for this interaction. Inspection of the CgA amino acid sequence revealed a histidine-motif that likely becomes protonated in mild acidic milieu (Fig. 4C) and may thereby bind to the negatively charged CS through electrostatic interactions. Further, the SPR data demonstrated that sequential injection of CS and CgA over a CgA-surface at pH 5.0 resulted in the formation of a CgA/CS/CgA complex. This may reflect the in vivo aggregation of CgA in secretory granules, where the CS-side chains promote the assembly of CgA core proteins in an acidified environment. It is possible that the CS-side chains of secretogranin-1 and 3 may influence the assembly of their respective core proteins in a similar mode.
Interestingly, it was recently discovered that protein hormones in secretory granules of the endocrine system are stored in an amyloid-like cross-β-rich conformation (32). Whereas incubation of certain recombinant peptide hormones resulted in their spontaneous aggregation under conditions mimicking that of secretory granules, other hormones aggregated only after the addition of low molecular weight heparin or free CS (32). With our finding that several prohormones are actually CSPGs, one may speculate that self-induced CS-aggregation is a common mechanism for hormone storage in secretory granules.
The concept of specificity of GAG-protein interactions has been intensely discussed over the years (25). Although the clinically explored anticoagulant activity of heparin clearly needs a distinct sulfation pattern, other interactions display lower degree of specificity and rely on a less stringent sulfation distribution (27). There is no correlation between physiological importance and degree of specificity. Most likely, evolution has only developed a higher degree of specificity when it is needed. Furthermore, recent studies suggest that several ligands share binding sites in both HS and CS chains (4). With this in mind it is reasonable to believe that HS may also bind CgA core protein in a similar fashion as CS, given that the HS is of sufficient negative charge. Nevertheless, it will be interesting to determine how specific the binding between CgA and its CS-chain is. Such attempts would likely require site-specific sequencing of CS to discern whether any given core protein or peptide is associated with unique CS structures or not. We propose that the methods presented here open new possibilities for such an endeavor.
In summary, the vast structural heterogeneity of CSPGs has previously hindered the development of integrated glycopeptide characterizations. As the CS-chain(s) and core protein are usually separated prior to analysis, the possibility of site-specific glycan information is excluded. Such information is likely to be valuable for improved understanding of CSPG-dependent processes and may open new avenues in cell biology, diagnostics, and therapeutic intervention. Our novel strategy allows for the combined enrichment of CSPGs, sequencing and identification of the peptide backbone, and detailed structural characterization of the innermost six residues of the CS-polysaccharide. Analysis of human urine and CSF samples unraveled that several established prohormones carry CS-chains and should thus fit into the definition of granular CSPGs. The quantitative and functional aspects of such glycan modifications of peptide prohormones will be the objects of future studies.
Supplementary Material
Footnotes
Author contributions: The project was conceived by F.N., J.N. and G.L. Sample preparation was conducted by F.N. and A.G.T. whereas MS-analysis was carried out by C.S. and J.L. Glycopeptide analysis was performed by F.N., A.G.T. and J.N. General data interpretation was performed by F.N., A.G.T., E.F., L.K., J.N. and G.L. The manuscript was written by F.N., A.G.T., C.S., L.K., J.N. and G.L.
* This work was supported by grants from the Swedish Medical Research Council (8266), Governmental grants (ALF) to the Sahlgrenska University Hospital. The glycoproteomic LC-MS analyses were all performed at the Proteomics Core Facility, Sahlgrenska Academy at the University of Gothenburg.
 This article contains supplemental Tables S1 and S2, and Figs. S1 to S4.
This article contains supplemental Tables S1 and S2, and Figs. S1 to S4.
1 The abbreviations used are:
- CS
- chondroitin sulfate
- CgA
- chromogranin A
- CSPGs
- chondroitin sulfate proteoglycans
- GAGs
- glycosaminoglycans
- Gal
- galactose
- GalNAc
- N-acetylgalactosamine
- GlcA
- glucuronic acid
- GlcNAc
- N-acetylglucosamine
- HCD
- higher-energy collisional dissociation
- HS
- heparan sulfate
- NCE
- normalized collision energy
- nLC-MS/MS
- nano-liquid chromatography-tandem mass spectrometry
- ppm
- parts per million
- RT
- retention time
- SAX
- strong anion exchange
- SPR
- surface plasmon resonance
- Xyl
- xylose.
REFERENCES
- 1. Lau L. W., Cua R., Keough M. B., Haylock-Jacobs S., Yong V. W. (2013) Pathophysiology of the brain extracellular matrix: a new target for remyelination. Nat. Rev. Neurosci. 14, 722–729 [DOI] [PubMed] [Google Scholar]
- 2. Dityatev A., Schachner M., Sonderegger P. (2010) The dual role of the extracellular matrix in synaptic plasticity and homeostasis. Nat. Rev. Neurosci. 11, 735–746 [DOI] [PubMed] [Google Scholar]
- 3. Seidler D. G. (2012) The galactosaminoglycan-containing decorin and its impact on diseases. Curr. Opin. Struct. Biol. 22, 578–582 [DOI] [PubMed] [Google Scholar]
- 4. Le Jan S., Hayashi M., Kasza Z., Eriksson I., Bishop J. R., Weibrecht I., Heldin J., Holmborn K., Jakobsson L., Soderberg O., Spillmann D., Esko J. D., Claesson-Welsh L., Kjellen L., Kreuger J. (2012) Functional overlap between chondroitin and heparan sulfate proteoglycans during VEGF-induced sprouting angiogenesis. Arterioscler. Thromb. Vasc. Biol. 32, 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Holmborn K., Habicher J., Kasza Z., Eriksson A. S., Filipek-Gorniok B., Gopal S., Couchman J. R., Ahlberg P. E., Wiweger M., Spillmann D., Kreuger J., Ledin J. (2012) On the roles and regulation of chondroitin sulfate and heparan sulfate in zebrafish pharyngeal cartilage morphogenesis. J. Biol. Chem. 287, 33905–33916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mizumoto S., Ikegawa S., Sugahara K. (2013) Human genetic disorders caused by mutations in genes encoding biosynthetic enzymes for sulfated glycosaminoglycans. J. Biol. Chem. 288, 10953–10961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thelin M. A., Bartolini B., Axelsson J., Gustafsson R., Tykesson E., Pera E., Oldberg A., Maccarana M., Malmstrom A. (2013) Biological functions of iduronic acid in chondroitin/dermatan sulfate. FEBS J. 280, 2431–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Esko J. D., Zhang L. (1996) Influence of core protein sequence on glycosaminoglycan assembly. Curr. Opin. Struct. Biol. 6, 663–670 [DOI] [PubMed] [Google Scholar]
- 9. Oohira A., Shuo T., Tokita Y., Nakanishi K., Aono S. (2004) Neuroglycan C, a brain-specific part-time proteoglycan, with a particular multidomain structure. Glycoconj. J. 21, 53–57 [DOI] [PubMed] [Google Scholar]
- 10. Thaysen-Andersen M., Packer N. H. (2014) Advances in LC-MS/MS-based glycoproteomics: getting closer to system-wide site-specific mapping of the N- and O-glycoproteome. Biochim. Biophys. Acta 1844, 1437–1452 [DOI] [PubMed] [Google Scholar]
- 11. Nilsson J., Ruetschi U., Halim A., Hesse C., Carlsohn E., Brinkmalm G., Larson G. (2009) Enrichment of glycopeptides for glycan structure and attachment site identification. Nat. Methods 6, 809–811 [DOI] [PubMed] [Google Scholar]
- 12. Lawrence R., Brown J. R., Al-Mafraji K., Lamanna W. C., Beitel J. R., Boons G. J., Esko J. D., Crawford B. E. (2012) Disease-specific nonreducing end carbohydrate biomarkers for mucopolysaccharidoses. Nat. Chem. Biol. 8, 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ly M., Leach F. E., 3rd, Laremore T. N., Toida T., Amster I. J., Linhardt R. J. (2011) The proteoglycan bikunin has a defined sequence. Nat. Chem. Biol. 7, 827–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lord M. S., Day A. J., Youssef P., Zhuo L., Watanabe H., Caterson B., Whitelock J. M. (2013) Sulfation of the bikunin chondroitin sulfate chain determines heavy chain.hyaluronan complex formation. J. Biol. Chem. 288, 22930–22941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chi L., Wolff J. J., Laremore T. N., Restaino O. F., Xie J., Schiraldi C., Toida T., Amster I. J., Linhardt R. J. (2008) Structural analysis of bikunin glycosaminoglycan. J. Am. Chem. Soc. 130, 2617–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fries E., Blom A. M. (2000) Bikunin–not just a plasma proteinase inhibitor. Int. J. Biochem. Cell Biol. 32, 125–137 [DOI] [PubMed] [Google Scholar]
- 17. Mikami T., Kitagawa H. (2013) Biosynthesis and function of chondroitin sulfate. Biochim. Biophys. Acta 1830, 4719–4733 [DOI] [PubMed] [Google Scholar]
- 18. Halim A., Nilsson J., Ruetschi U., Hesse C., Larson G. (2012) Human urinary glycoproteomics; attachment site specific analysis of N- and O-linked glycosylations by CID and ECD. Mol. Cell Proteomics 11, M111 013649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olson S. K., Bishop J. R., Yates J. R., Oegema K., Esko J. D. (2006) Identification of novel chondroitin proteoglycans in Caenorhabditis elegans: embryonic cell division depends on CPG-1 and CPG-2. J. Cell Biol. 173, 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bishop J. R., Schuksz M., Esko J. D. (2007) Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 21. Gowda D. C., Hogue-Angeletti R., Margolis R. K., Margolis R. U. (1990) Chromaffin granule and PC12 cell chondroitin sulfate proteoglycans and their relation to chromogranin A. Arch. Biochem. Biophys. 281, 219–224 [DOI] [PubMed] [Google Scholar]
- 22. Rehfeld J. F., Lindberg I., Friis-Hansen L. (2002) Increased synthesis but decreased processing of neuronal proCCK in prohormone convertase 2 and 7B2 knockout animals. J. Neurochem. 83, 1329–1337 [DOI] [PubMed] [Google Scholar]
- 23. Kim T., Tao-Cheng J. H., Eiden L. E., Loh Y. P. (2001) Chromogranin A, an “on/off” switch controlling dense-core secretory granule biogenesis. Cell 106, 499–509 [DOI] [PubMed] [Google Scholar]
- 24. Kanato Y., Ono S., Kitajima K., Sato C. (2009) Complex formation of a brain-derived neurotrophic factor and glycosaminoglycans. Biosci. Biotechnol. Biochem. 73, 2735–2741 [DOI] [PubMed] [Google Scholar]
- 25. Kreuger J., Spillmann D., Li J. P., Lindahl U. (2006) Interactions between heparan sulfate and proteins: the concept of specificity. J. Cell Biol. 174, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Silver D. J., Silver J. (2014) Contributions of chondroitin sulfate proteoglycans to neurodevelopment, injury, and cancer. Curr. Opin. Neurobiol 27C, 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lindahl U., Kjellen L. (2013) Pathophysiology of heparan sulphate: many diseases, few drugs. J. Intern. Med. 273, 555–571 [DOI] [PubMed] [Google Scholar]
- 28. Escobar Galvis M. L., Jia J., Zhang X., Jastrebova N., Spillmann D., Gottfridsson E., van Kuppevelt T. H., Zcharia E., Vlodavsky I., Lindahl U., Li J. P. (2007) Transgenic or tumor-induced expression of heparanase upregulates sulfation of heparan sulfate. Nat. Chem. Biol. 3, 773–778 [DOI] [PubMed] [Google Scholar]
- 29. Coles C. H., Shen Y., Tenney A. P., Siebold C., Sutton G. C., Lu W., Gallagher J. T., Jones E. Y., Flanagan J. G., Aricescu A. R. (2011) Proteoglycan-specific molecular switch for RPTPsigma clustering and neuronal extension. Science 332, 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Conte M., Arcaro A., D'Angelo D., Gnata A., Mamone G., Ferranti P., Formisano S., Gentile F. (2006) A single chondroitin 6-sulfate oligosaccharide unit at Ser-2730 of human thyroglobulin enhances hormone formation and limits proteolytic accessibility at the carboxyl terminus. Potential insights into thyroid homeostasis and autoimmunity. J. Biol. Chem. 281, 22200–22211 [DOI] [PubMed] [Google Scholar]
- 31. Bartolomucci A., Possenti R., Mahata S. K., Fischer-Colbrie R., Loh Y. P., Salton S. R. (2011) The extended granin family: structure, function, and biomedical implications. Endocr. Rev. 32, 755–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maji S. K., Perrin M. H., Sawaya M. R., Jessberger S., Vadodaria K., Rissman R. A., Singru P. S., Nilsson K. P., Simon R., Schubert D., Eisenberg D., Rivier J., Sawchenko P., Vale W., Riek R. (2009) Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.