Abstract
Intracellular pathogens need to establish a growth-stimulating host niche for survival and replication. A unique feature of the gastrointestinal pathogen Salmonella enterica serovar Typhimurium is the creation of extensive membrane networks within its host. An understanding of the origin and function of these membranes is crucial for the development of new treatment strategies. However, the characterization of this compartment is very challenging, and only fragmentary knowledge of its composition and biogenesis exists. Here, we describe a new proteome-based approach to enrich and characterize Salmonella-modified membranes. Using a Salmonella mutant strain that does not form this unique membrane network as a reference, we identified a high-confidence set of host proteins associated with Salmonella-modified membranes. This comprehensive analysis allowed us to reconstruct the interactions of Salmonella with host membranes. For example, we noted that Salmonella redirects endoplasmic reticulum (ER) membrane trafficking to its intracellular niche, a finding that has not been described for Salmonella previously. Our system-wide approach therefore has the potential to rapidly close gaps in our knowledge of the infection process of intracellular pathogens and demonstrates a hitherto unrecognized complexity in the formation of Salmonella host niches.
Bacterial pathogens have evolved sophisticated mechanisms enabling them to invade, reside in, and proliferate in a large range of eukaryotic hosts. This often involves hijacking the host phagosomal system, interfering with the host cell signaling and trafficking machinery, and establishing a replication niche to avoid clearance (1). Whereas some pathogens escape phagosomes and replicate in the host cytoplasm, most of the described pathogens replicate in membrane-bound, vacuole-like compartments (2). Such intracellular niches of various pathogens are diverse, and biogenesis often depends on the delivery of bacterial effector proteins into the host cell cytoplasm.
Salmonella enterica, the causative agent of localized gastroenteritis and the life-threatening systemic infection known as typhoid fever, forms so-called Salmonella-containing vacuoles (SCVs)1 inside host cells (3). SCVs mature through continuous interactions with endocytic and recycling pathways, accompanied by a spatial shift from the side of internalization to the juxtanuclear position close to the microtubule-organizing center (4, 5). Whereas the initial maturation steps are similar to the canonical phagosome biogenesis, the formation of an extensive tubular membrane network extending from the mature SCV is unique to Salmonella-infected host cells. This network contains various tubular structures such as Salmonella-induced filaments (SIFs), sorting nexin tubules, Salmonella-induced secretory carrier membrane protein 3 tubules, and lysosome-associated membrane protein 1-negative tubules (5–7), distinguishable by individual organelle marker proteins. For instance, tubules decorated with lysosome-associated membrane protein 1 (LAMP1) are known as SIFs (8, 9). In this paper we refer to all host membranes modified by intracellular Salmonella as Salmonella-modified membranes (SMMs).
In general, the appearance of SMMs coincides with the onset of bacterial replication, and both phenomena are dependent on the translocation of effector proteins of the Salmonella Pathogenicity Island 2 (SPI2)-encoded type III secretion system (T3SS) (10, 11). These effector proteins manipulate a large number of host cell processes and force the host cell to create a suitable microenvironment for Salmonella (7, 12, 13). Although many Salmonella effector proteins have been described (14), much less is known about the host proteins that are manipulated to foster bacterial growth.
A systematic proteome-wide analysis would be an important step toward understanding the mechanisms used by Salmonella to reorganize the host cell endosomal system during intracellular proliferation. However, one major challenge is the need to distinguish host proteins directed toward the Salmonella-induced compartments from those that are present independent of an infection.
In this report we describe a novel method for the enrichment of SMMs and utilize a comparative strategy to identify proliferation-relevant host proteins. This systematic characterization of the SMM proteome provides new insights into the cellular origin and biogenesis of SMMs and identifies host cell proteins modified by the activity of intracellular Salmonella.
EXPERIMENTAL PROCEDURES
Chemicals
All chemicals used in this study were obtained from Sigma Aldrich, unless otherwise indicated.
Cell Lines, Bacterial Strains, and Their Cultivation
Human epithelial cell line HeLa (ATCC No. CCL-2) and stable lentiviral-transfected HeLa LAMP1-GFP cells (15) were maintained in DMEM containing 4.5 g/l glucose, 4 mm l-glutamine, and sodium pyruvate (Biochrom, Berlin, Germany) supplemented with 10% inactivated FCS in an atmosphere of 5% CO2 and 90% humidity at 37 °C. Cells from the murine macrophage-like cell line RAW264.7 (ATCC No. TIB-71) were cultured in DMEM containing 4.5 g/l glucose and 4 mm stable glutamine (Biochrom) supplemented with 6% FCS at 37 °C in an atmosphere containing 5% CO2 and 90% humidity. S. enterica serovar Typhimurium strains NCTC12023 (wild type (WT)) or HH107 and P2D6 harboring p3711 for the synthesis of SseF-2TEV-2M45 were used. Strain P2D6 is defective in the SPI2-encoded T3SS due to ssaV mutation. For live cell imaging, strains harboring plasmids p3589 or pFPV-mCherry/2 were used for constitutive expression of mCherry. Strain characteristics are summarized in supplemental Table S1A. Salmonella strains were routinely cultured in Luria–Bertani broth containing 50 μg/ml carbenicillin (Roth, Karlsruhe, Germany) or 12.5 μg/ml chloramphenicol if required for the selection of plasmids.
Construction of Recombinant DNA Molecules
For construction of the plasmid p3711 encoding the bait protein SseF, the following sequence was synthesized by GeneArt (Invitrogen): CCCGGGGGATCCGCCATGGAGAATCTTTATTTTCAGGGCGGCGACGTCGAAAACCTTTATTTCCAAGGAGGGTCCGGCGATCGGAGTAGGGATCGCCTACCTCCTTTTGAGACAGAGACGCGCATCCTCGGCTCGGGCAGCAGAGACCGTCTGCCGCCGTTCGAAACCGAGACGCGCATCCTCTAGAGCGGCCGC. The sequence encodes two recognition sites for the tobacco etch mosaic virus protease (bold) and two sites encoding the M45 epitope for antibody binding (italic). The synthetic DNA molecule was inserted into pSK via SmaI/NotI to generate p3673. Next, a fragment containing ProsseA sscB sseF1–258 was released from p2810 (16) via digestion with KpnI/EcoRV and inserted into p3673. Subsequently, the insert was released via KpnI/NotI digestion and subcloned into pWSK29 to generate p3711.
To generate a plasmid expressing LAMP1-mCherry, we amplified mcherry from pFPV-mCherry using mCherry-For-BamHI and mCherry-Rev-XbaI-NotI and cloned in pEGFP-N1 (Clontech, Mountain View, CA) to replace eGFP (supplemental Table S1B). Next, human lamp1 was amplified from cDNA clone IRAU p969C0275D (ImaGenes, Berlin, Germany) using hLAMP1-For-EcoRI and hLAMP1-Rev-BamHI (supplemental Table S1B), EcoRI/BamHI restricted, and inserted into the pmCherry-N1 transfection vector to generate p3451.
Infection of HeLa Cells
Host cell infections were performed as described previously (17). In short, HeLa cells were infected with 3.5 h subcultures of Salmonella with a multiplicity of infection (m.o.i.) of 50 or 75. The bacteria were centrifuged onto the cells at 500 × g for 5 min, and the cells were then incubated for 25 min at 37 °C in an atmosphere of 5% CO2 before extracellular bacteria were removed by three washes with pre-warmed PBS. Subsequently, host cells were maintained in cell culture media containing 100 μg/ml gentamicin (AppliChem, Darmstadt, Germany) for 1 h. Afterward, cells were cultivated in media with a decreased gentamicin concentration of 10 μg/ml for the rest of the experiment.
Intracellular Replication Assays
Gentamicin protection assays were performed according to Ref. 18. Briefly, Salmonella strains were grown to stationary phase. The A600 values of the cultures were adjusted with PBS to 0.2, and cultures were added with an m.o.i. of 1 to the seeded RAW264.7 cells. After centrifugation for 5 min at 500 × g and incubation for 25 min at 37 °C in an atmosphere of 5% CO2, macrophages were washed three times with pre-warmed PBS before being incubated in cell culture medium containing 100 μg/ml gentamicin for 1 h. The medium was replaced with medium containing 10 μg/ml gentamicin, and the macrophages were kept in this medium for the remaining time of the experiment. To determine the amount of intracellular bacteria, we washed macrophages three times with PBS and lysed them with 0.1% Triton X-100 for 10 min at room temperature at 2 h and 16 h post-infection (p.i.). Serial dilutions of the lysates were plated on Müller-Hinton agar plates. Statistical analyses were performed using one-way analysis of variance with SigmaPlot 11.0 (Sysstat Software, San Jose, CA).
Confocal Laser-scanning Microscopy
Fluorescence imaging was partially performed using a Leica SP5 confocal laser-scanning microscope with live cell periphery equipped with an HCX PL APO CS ×100 (numerical aperture 0.7–1.4) oil immersion objective (Leica, Wetzlar, Germany). Images were acquired using the LAS AF (Leica Application Suite Advanced Fluorescence) software and the following filter combinations: GFP/Alexa Fluor 488 and mCherry/Alexa Fluor 568 with polychroic mirror DD 488/543 or the combination of GFP/Alexa Fluor 488, mCherry/Alexa Fluor 568, and Cy5 with the polychroic mirror TD 488/543/633. All images obtained were processed by Leica LAS AF. Scale bars were added with ImageJ (National Institutes of Health), and figures were arranged in Photoshop CS6 (Adobe, San Jose, CA).
Live cell imaging was performed as described elsewhere (15). SIF formation was monitored from 4 h to 16 h p.i. For SMM validation, HeLa cells were co-transfected using FuGENE® HD Transfection Reagent (Promega, Madison, WI) with plasmids encoding GFP-fusion proteins Rab2a, Rab5c, Rab7a, Rab10a, Rab11a, Rab14, and UtrCH and pLAMP1-mCherry (supplemental Table S1A) and then subsequently infected with Salmonella WT harboring pFPV-mCherry/2 at an m.o.i. of 75. Confocal laser-scanning microscopy (CLSM) images were taken at 8 h p.i.
Immunostaining was performed as described elsewhere (19). Briefly, infected HeLa LAMP1-GFP cells (m.o.i. 50) were fixed with 3% paraformaldehyde at 8 h p.i., washed, and incubated for 30 min in blocking solution (2% goat serum, 2% BSA, and 0.1% saponin in PBS) before being stained with primary antibodies — anti-M45 (1:500), Salmonella O antiserum (1:1000), anti-human EEA1 (1:500), or anti-human TfR (1:100) for 1 h at room temperature and anti-human CopA (1:17), anti-human CopG1 (1:12.5), anti-human Sec23a (1:17), anti-human Sar1a (1:12.5), or anti-human Mitofilin (1:250) overnight at 4 °C (supplemental Table S1A). Secondary antibodies were selected accordingly (supplemental Table S1A), and samples were incubated for 1 h at room temperature.
Spinning Disk Microscopy
Fluorescence imaging was partially performed using a Zeiss Cell Observer Spinning Disk microscope with Yokogawa Spinning Disc Unit CSU-X1a 5000, an Evolve EMCCD camera from Photometrics (Tucson, AZ), and live cell periphery, equipped with an Alpha Plan-Apochromat ×63 (numerical aperture 1.46) oil immersion objective (Zeiss, Jena, Germany). Images were acquired using the ZEN (Zeiss) software and the following filter combinations: GFP with BP 525/50, mCherry with LP 580, and mTurquoise2 with BP 485/30. All images obtained were processed by ZEN2012 software. Micrographs and live cell images were prepared as described before. HeLa cells were co-transfected using FuGENE® HD Transfection Reagent (Promega) with plasmids encoding GFP-fusion proteins Rab2a, Rab7a, Rab10a, and UtrCH, as well as pmTurquoise2-Golgi, pmTurquoise2-ER, pLifeAct-mTurquoise2, and pLAMP1-mCherry.
Quantitation via Flow Cytometry Analysis
HeLa cells were infected with either HH107 [p3711] or P2D6 [p3711] at an m.o.i. of 50 for 25 min. At 4, 8, 12, and 16 h p.i. cells were fixed, permeabilized, and stained with primary anti-M45 (1:1000) and secondary anti-mouse IgG Alexa Fluor 488 (1:1000) for subsequent flow cytometry analysis using FACSCalibur (BD Biosciences). Experiments were performed in triplicate at least three times. Data were analyzed with FACS Express 4 (De Novo Software, Los Angeles, CA). Statistical analyses were performed using Student's t test with SigmaPlot 11 (Sysstat Software).
Enrichment of GEMN Fraction
Roughly 7 × 107 HeLa LAMP1-GFP cells were used per immunoprecipitation (IP) and biological replicate. Before cell homogenization, the infected host cells were rinsed thrice with PBS. Scraped cells were resuspended in osmostabilizing homogenization buffer (250 mm sucrose, 20 mm HEPES, 0.5 mm EGTA, pH 7.4), centrifuged at 1000 × g for 10 min, and resuspended in 1 ml of 4 °C pre-cooled homogenization buffer with 1× protease inhibitor mixture (Serva, Heidelberg, Germany). Host cells were mechanically disrupted with 0.5-mm glass beads (Scientific Industries, New York, NY) using a Vortex-2 Genie with Turbomix (Scientific Industries; three 1-min strokes) with intermediate cooling. The lysate was centrifuged at 100 × g for 10 min at 4 °C, and the resulting GEMN pellet was washed twice with pre-cooled homogenization buffer with protease inhibitor mixture. The final GEMN pellet was resuspended in 500 μl of homogenization buffer supplemented with 1.5 mm MgCl2 and treated with DNaseI (50 μg/ml) for 30 min at 37 °C. The protein concentration was determined via Bradford assay (Bio-Rad).
Immunoprecipitation
For IP, 25 μl of Protein G magnetic beads (GE Healthcare) were coated with 40 μg of purified anti-M45 antibody on a rotary shaker at 4 °C overnight. The beads were washed twice with PBS, cross-linked according to the manufacturer's instructions, and blocked for 30 min with 1% BSA in PBS at 4 °C. A total of 500 μg of GEMN proteins were adjusted to a final volume of 200 μl in resuspension mix (1.5 mm MgCl2, 10 mm KCl, 0.1% Nonidet P-40) and then incubated with 25 μl of cross-linked anti-M45 antibody-labeled Protein G magnetic beads on a rotary shaker at 4 °C overnight. To remove unbound proteins, we washed the sample five times with 0.1% Nonidet P-40 in PBS. Finally, bound proteins were eluted in 25 μl of 1× SDS sample buffer (12.5% glycerol, 4% SDS, 2% mercaptoethanol, 50 mm Tris, pH 6.8).
SDS-PAGE and Western Blotting
Proteins were separated on NuPAGE Novex 4–12% gradient gels. For Western blot analysis, 2 μl of the protein sample were used per lane. Proteins were transferred to 0.2-μm nitrocellulose membranes (Protran, Whatman, Dassel, Germany), blocked with 5% w/v BSA, 0.1% v/v TWEEN in TBS, and then incubated in TBS containing 1% w/v BSA with primary and secondary antibodies as follows: anti-M45 (1:5000) and peroxidase-conjugated anti-mouse IgG (1:20000) (supplemental Table S1A). Proteins were detected by chemiluminescence with ECL detection reagent (Pierce, ThermoScientific, Rockford, IL) and blue-light-sensitive film (Agfa Healthcare NV, Morstel, Belgium).
Protein Digest, RP-LC Separation, MS, and Data Analysis
In total we performed four IP proteome experiments. For each profiling, the precipitated proteins were one-dimensionally separated via SDS-PAGE and immediately Coomassie Blue–stained (20). Gel lanes of each biological replicate were sliced into 36 gel pieces. Each gel slice was subjected to standard in-gel de-staining and trypsinolysis procedures (21). Afterward the digest was transferred into vials, resulting in a total of 144 digested samples.
The LC-MS/MS analysis was performed using an UltiMate 3000 NCS-3500 nano-HPLC system (Dionex, Sunnyvale, CA) controlled by Chromeleon chromatography software coupled to an amaZon speed ETD ion trap mass spectrometer with a CaptiveSpray source (Bruker Daltonics, Bremen, Germany). The UltiMate 3000 NCS-3500 nano-HPLC system (Dionex) was configured with a 2-cm PepMap 75-μm inner diameter C18 sample trapping pre-column (Thermo Fisher Scientific) and a 15-cm PepMap 75-μm inner diameter C18 microcapillary column (Thermo Fisher Scientific). Samples of 7 μl each were applied to the columns and separated by a 60-min linear gradient from 5% to 50% solvent B (80% acetonitrile, 0.1% v/v formic acid) with a flow rate of 300 nl/min. For each MS scan, up to eight abundant multiply charged species in the m/z 400–1600 range were automatically selected for MS/MS but excluded for 30 s after having been selected twice. The HPLC system was controlled using Compass 1.5 (Bruker).
Acquired MS/MS data were processed by the ProteinScape 3.1 software (Bruker) and searched against the UniProt human database (October 2013; 20272 entries) using ProteinExtraktor (Bruker). Spectral data are available in PeptideAtlas (ftp://PASS00480:SU9795nb, ftp.peptideatlas.org). Data analyses were conducted according the published guidelines (22). Mass tolerance values for MS and MS/MS were set at 200 ppm and 0.5 Da. Fixed search parameters were semi-tryptic digestion and up to 1 missed cleavage. Variable search parameters used for the search were deamidation (NQ) and oxidation (M). Proteins were considered as identified with a ProteinScape score of >40 and two unique peptides with >95% confidence. Peptide Decoy (Mascot) and false discovery rates were adjusted to 1% at the protein and peptide levels for all experiments.
All identified proteins were searched against the UniProt-GO Annotation database (23). Only proteins identified in two biological replicates were considered as candidates in the SMM proteome (supplemental Table S2A). Protein abundance was estimated using the exponentially modified protein abundance index approach (24). Identified proteins were converted using UniProt IP mapping (25) into STRING numbers and searched against the STRING database (version 8.3; minimal confidence score of 0.4 (26, 27)) to identify potential protein–protein interactions.
To compare the compositions of pathogen-modulated host compartments from different pathogens and hosts, we used the MGI vertebrate homology database to extract human–mouse homologues and the inparanoid tool (28) to determine human–Dictyostelium homologues.
RESULTS
Enrichment of SMMs Using a Salmonella Translocated Membrane-integral Effector Protein
Enrichment of pathogen-containing compartments is conventionally based on subcellular fractionation after disruption of infected cells. However, a lack of specificity, dilution effects, and fragmentation of the complex and extensive membrane structures minimize the success achievable in determining the intrinsic SMM protein composition. Therefore, we developed an alternative approach for enriching SMMs (Fig. 1). This approach utilizes SPI2-T3SS effector proteins that are embedded in SMMs as bait (Fig. 1A). IP of a tagged version of a membrane-integral SPI2-T3SS effector protein allows the enrichment of SMMs and additional associated proteins (Fig. 1B). A similar approach was recently used by the Hilbi group (29, 30) to analyze the composition of Legionella-containing vacuoles (LCVs). We selected effector protein SseF as bait for the IP. After translocation by the SPI2-T3SS, SseF has characteristics of an integral membrane protein in SCVs (19). As indicated by previous work, SseF is translocated in high amounts into host cells, is present within SIFs, and shows a long half-life (18). Furthermore, SseF is amenable as a fusion partner for various heterologous antigens and tags (31), rendering SseF as attractive bait for SMM enrichment.
Fig. 1.
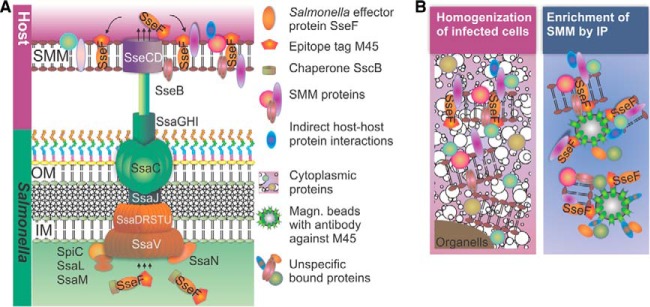
Schematic representation of Salmonella SPI2-T3SS injectosome (A) and approach for enrichment of Salmonella-modified membranes (SMMs) (B). Salmonella translocates effector proteins into the host cell cytosol via the SPI2-encoded T3SS. A subset of SPI2-T3SS effectors are associated with or integral to SMMs. Modified effector proteins, such as M45-tagged SseF, are efficiently translocated by intracellular Salmonella into the host cell and integrate into membranes. After lysis of infected cells, immunoprecipitation of M45-tagged SseF using magnetic beads coated with anti-M45 antibody enriches the SMM decorated with SseF-2TEV-2M45.
We generated a low-copy-number vector for co-expression of sseF and its cognate chaperone sscB under control of the promoter ProsseA (supplemental Fig. S1A). Two TEV cleavage sites and a tandem M45 tag were fused to the C terminus of SseF, thus allowing immunoprecipitation with anti-M45 monoclonal antibodies. The resulting plasmid for synthesis of SseF-2TEV-2M45 was designated as p3711. We first tested the translocation and function of SseF-2TEV-2M45 in the sseF-deficient mutant strain HH107. The strain HH107 is attenuated in intracellular proliferation and induces an altered SIF network relative to the Salmonella WT (18, 32). Complementation with SseF-2TEV-2M45 restored intracellular proliferation of HH107 to WT levels (supplemental Fig. S2).
To exclude potential influences of the SseF-2TEV-2M45 construct on SIF network formation, we infected lentiviral-transfected HeLa LAMP1-GFP cells with HH107 translocating SseF-2TEV-2M45 and analyzed SIF formation via CLSM. LAMP1 is a prominent membrane-integral component of SIFs, and the fusion with GFP enabled us to continuously monitor their development. LAMP1-GFP transfection of HeLa cells had no influence on the infection process of Salmonella as previous experiments demonstrated (15, 33). As expected, the tagged effector protein SseF was present in SIFs and co-localized with LAMP1-GFP (Fig. 2). Furthermore, HH107 expressing SseF-2TEV-2M45 showed no impairment in SIF development (Fig. 3A). SIF formation, dynamics, and positioning were comparable to what was observed in HeLa cells infected with WT Salmonella (supplemental Fig. S1B).
Fig. 2.
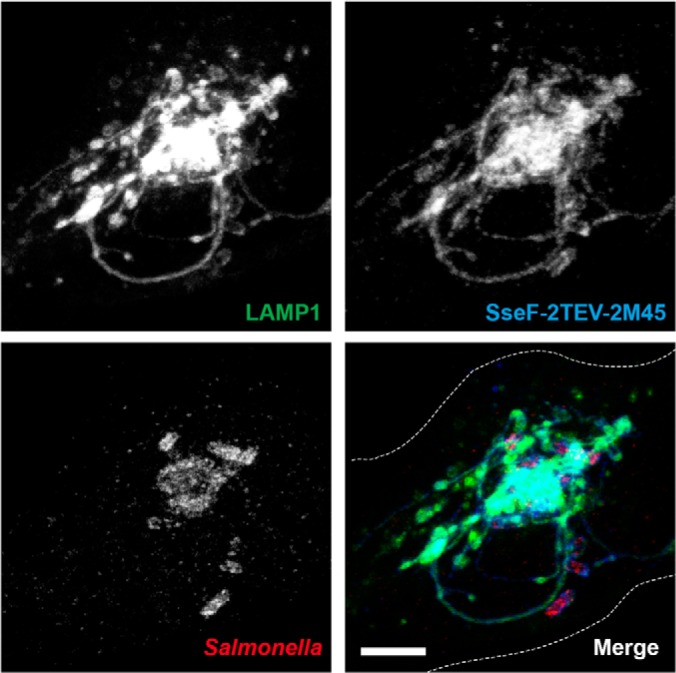
Epitope-tagged Salmonella effector protein SseF is an integral component of SMMs, indicated by its co-localization with LAMP1. HeLa cells constitutively expressing LAMP1-GFP (green) were infected with Salmonella HH107 expressing SseF-2TEV-2M45 fixed at 8 h p.i., immunostained against Salmonella O antiserum (red) and the M45 tag (blue), and imaged using CLSM. Images are shown as maximum-intensity projections. Scale bar: 10 μm.
Fig. 3.
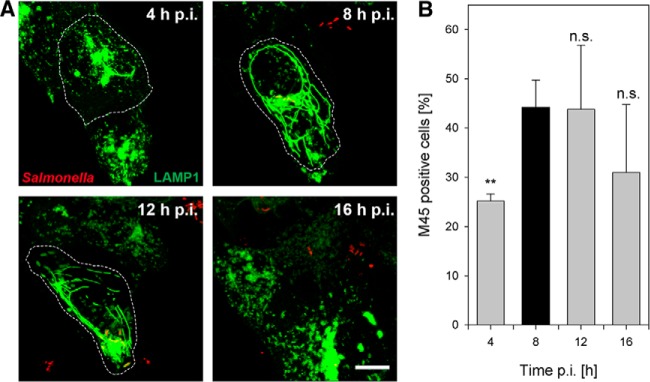
Extension of the SMM network (A) and the number of SseF-positive host cells (B) are maximal at 8 h p.i. A, HeLa cells expressing LAMP1-GFP (green) were infected with Salmonella HH107 expressing SseF-2TEV-2M45 and mCherry (red). Continuous live cell imaging was performed and representative images of Salmonella-infected cells at various time points p.i. show the extent and variation in SIF morphology. Images are shown as maximum intensity projections. Scale bar: 10 μm. B, infected cells were fixed at the indicated time points p.i., immunostained for SseF-2TEV-2M45, and analyzed via flow cytometry. The percentage of M45-positive cells at various time points p.i. is indicated. Shown are the mean values (+ S.D.) of three independent experiments. Student's t-test relative to 8 h p.i. values: **p < 0.01. n.s., not significant.
To improve the capture yield of SMMs for a proteomics survey, we next screened for infection conditions in which the extension of the SMM network and the bait concentration were optimal. CLSM revealed an extensively widespread network of the LAMP1-GFP-positive SMM in Salmonella HH107 SseF-2TEV-2M45-infected HeLa cells around 8 h p.i. (Fig. 3A), which is consistent with previous findings from HeLa cells infected by WT Salmonella (34). Furthermore, we quantified the number of SseF-2TEV-2M45-positive Salmonella-infected HeLa cells at 4, 8, 12, and 16 h p.i. using flow cytometry. As indicated in Fig. 3B, the cell number positive for translocated SseF-2TEV-2M45 was maximal at 8 h p.i., which is in good agreement with the CLSM analyses of the SMM network.
In summary, the data suggest that at 8 h p.i. the SMM network is fully developed and decorated with SseF. Therefore, this time point is optimal for the extraction of the SMM proteome for subsequent analyses.
Proteome Profiling of SMM
To determine the SMM proteome, we used the illustrated procedure integrating subcellular fractionation and IP with LC-MS/MS (Fig. 4). Because the SMM network of infected HeLa cells (8 h p.i.) is strongly intertwined with the host cells' GEMN complex, we decided to increase the amount of SMM-located proteins by enriching the GEMN fraction. To this end we first isolated the GEMN fraction using a combination of gentle mechanical lysis and centrifugation (supplemental Fig. S3B/C). Subsequently, the GEMN pellet was solubilized using mild detergents. The resulting mixture was then used for IP experiments. To elucidate Salmonella proteins specific to SMM and necessary for intracellular survival, we conducted comparative proteome analysis using the same methods with HeLa cells infected by Salmonella strain P2D6 harboring p3711. This strain is able to infect HeLa cells, but it is deficient in the translocation of SPI2-T3SS effector proteins and unable to induce SIFs, and it shows attenuated proliferation in host cells (supplemental Fig. S1C). We first used Western blots to confirm that the immune-precipitated fraction contained the bait protein SseF (supplemental Fig. S4B). Afterward the IP proteomes were profiled via LC-MS/MS. In total we performed 144 LC-MS/MS runs and identified 583 host cell proteins. Of these hits, 336 proteins were also detected in the immune-precipitated fraction of cells infected with control strain P2D6 [p3711]. Thus, 247 host cell proteins were found to be unique in the SMM proteome (supplemental Table S2A). The 20 most abundant proteins are listed in supplemental Table S3.
Fig. 4.
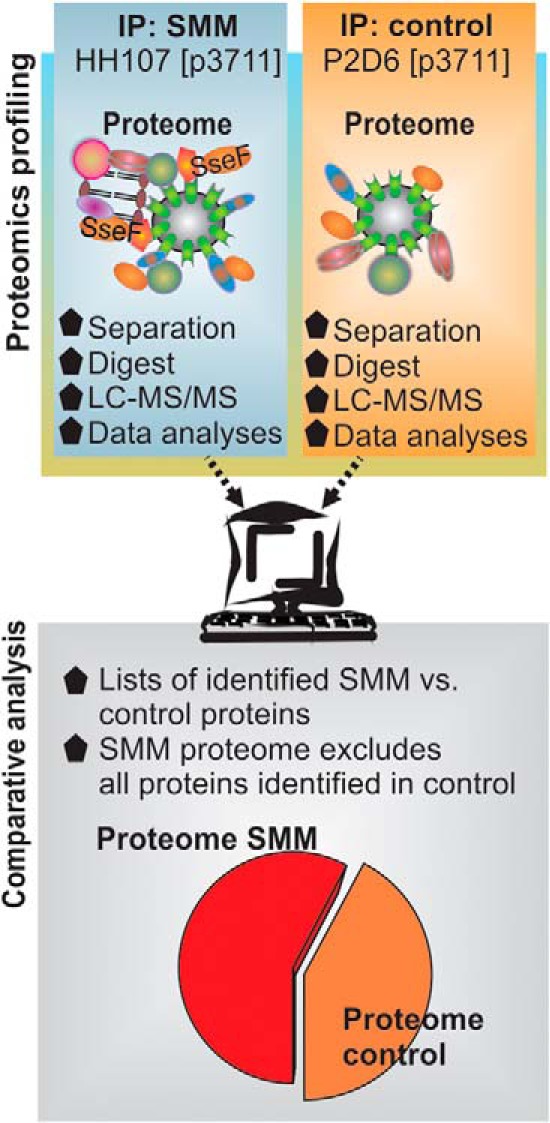
Schematic overview of experimental approach and data analysis.
Classification by subcellular location of the 247 identified host cell proteins using the UniProt database (23) revealed that a significant proportion of our identified SMM proteins are predicted to be of cytoplasmic origin (Fig. 5A). This might seem surprising considering that the enrichment primarily targeted the SMM. However, proteins that form tight interactions with SMM proteins are also likely to be co-isolated (Fig. 1A). This notion was supported using the STRING database (35), which revealed a highly intertwined protein–protein interaction network of the identified SMM proteins (supplemental Fig. S5). In total, 86% of all identified proteins showed potential physical and functional protein–protein interactions.
Fig. 5.
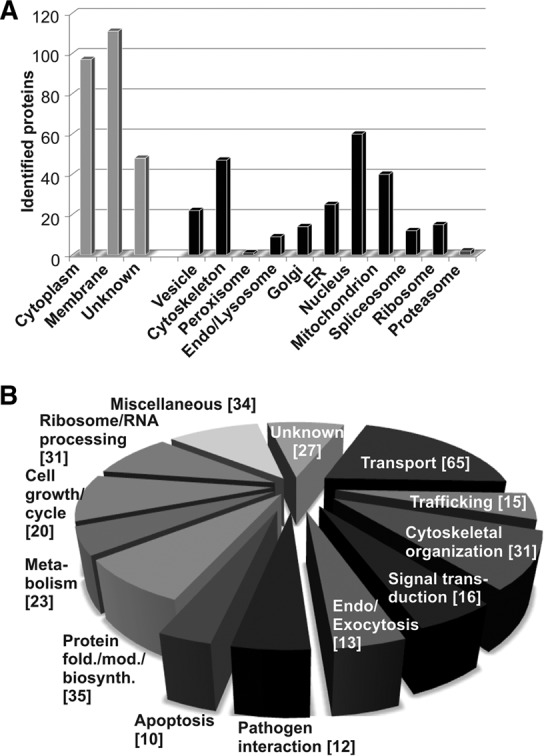
Classification of the identified SMM proteome according to Gene Ontology subcellular localization (A) and biological processes (B), based on the annotations in the UniProt database. The term “cytoskeleton” includes proteins involved in stress fibers, intermediate filaments, microtubules, focal adhesion, and tight and cell junctions. Some proteins have more than one functional annotation. Numbers of identified proteins are indicated in brackets.
Origin and Function of the Identified SMM Proteome
It is still not clear how precisely SMMs are formed within the host cell. However, several compartments and cellular structures have been implicated in their generation. Salmonella actively recruits host membranes for SIF formation from endosomes, lysosomes, and the trans-Golgi network (9, 36, 37). In addition, the cytoskeleton stabilizes SIFs and guides their elongation (38). In line with these observations, a number of cytoskeleton proteins (47 proteins), endosomal and lysosomal proteins (9), Golgi proteins (14), and vesicle-transport-related proteins (24) have been identified (Fig. 5A). Interestingly, proteins were also found to originate from other compartments such as the ER, nucleus, and mitochondria. This might hint at interplay between these compartments during the formation of SMMs.
The identified SMM proteins were further grouped according to their experimentally determined or predicted biological function (UniProt (23)) (Fig. 5B). A considerable number of SMM proteins were implicated in transport (65 proteins), vesicle trafficking (15 proteins), endo-/exocytosis (13 proteins), cytoskeletal organization processes (31 proteins), and signal transduction processes (16 proteins). Selected candidates are summarized in Table I. A subset of host cell proteins identified in our SMM proteome survey have previously been reported as associated with or involved in the biogenesis of SCVs or SIFs. These include GTPases Rab7a, Rab5c, Rab11a, and Rab14 (39, 40); filamin (41); myosin II (42); dynein (36); desmoplakin (43); and actin (44).
Table I. MS-identified SMM proteins involved in trafficking, transport, or cytoskeleton.
| Name (*_human) | Protein description | Localization |
|---|---|---|
| Transport (amino acids, sugar calcium, phosphate, and electrons) | ||
| 4F2 | 4F2 cell-surface antigen, amino acid transport | Plasma membrane |
| CTP | Tricarboxylate transport protein | Membrane, mitochondrion |
| CMC2 | Calcium-binding mitochondrial carrier protein Aralar2 | Membrane, mitochondrion |
| MPCP | Phosphate carrier protein | Membrane, mitochondrion |
| MTCH2 | Mitochondrial carrier homolog 2 | Membrane, mitochondrion |
| VCP | Transitional endoplasmic reticulum ATPase | ER |
| ATP5C/D/O | F0F1-ATPase complex | Membrane, mitochondrion |
| GTPases | ||
| RAB2A | Rab GTPase Rab2a | ER, Golgi |
| RAB5C | Rab GTPase Rab5c | Vesicle, endosomes |
| RAB7A | Rab GTPase Rab7a | Vesicle, endosome, lysosome |
| RAB10 | Rab GTPase Rab10 | Vesicle, endosome, ER |
| RB11A | Rab GTPase Rab11a | Vesicle, endosome, lysosome |
| RAB14 | Rab GTPase Rab14 | Vesicle, endosome, lysosome |
| RALA | Multifunctional GTPase RalA | Vesicle, membrane |
| RAP1B | Ras GTPase Rap1b | Cell junctions, membrane |
| OPA1 | Dynamin-like 120-kDa protein | Membrane, mitochondrion |
| SAR1A | COPII-associated GTPase Sar1A | ER |
| Clathrin, coatomer I and II–mediated transport | ||
| AP2A1/B1 | AP-2 complex subunits α and β | Coated pits, membrane |
| TFR | Transferrin receptor protein 1 | Coated pits, vesicle, membrane, endosome |
| COPA/G1 | Coatomer subunits α and γ | Vesicle, Golgi |
| BAP31 | B-cell receptor–associated protein 31 | Membrane, ER |
| TMED9/10 | Transmembrane emp24 domain-containing proteins 9 and 10 | Vesicle, Golgi, ER |
| SC23A | Protein transport protein Sec23a | Vesicle, Golgi, ER |
| VAPA/B | Vesicle-associated membrane protein-associated proteins A and B | Vesicle, membrane, microtubule, ER |
| ARF4 | ADP-ribosylation factor 4 | Membrane, Golgi |
| Cytoskeleton and membrane linker | ||
| MYH9/10 | Myosin II | Membrane, cytoskeleton |
| MYO1B/C | Myosin I | Cytoskeleton |
| ACTS | Actin | Cytoskeleton |
| DYHC1/7 | Dynein | Cytoskeleton, microtubule |
| ARPC4 | Arp2/3 complex | Cytoskeleton |
| DYST | Dystonin | Cytoskeleton, microtubule, ER |
| FLNA | Filamin-A | Cytoskeleton, endosome |
| CAPZB | F-Actin capping protein | Cytoskeleton |
| SYN1/2 | Nesprin 1 and 2 | Cytoskeleton, Golgi |
| PLST | Plastin-3 | Cytoskeleton |
| ANAXA1 | Annexin A1 | Membrane, nucleus, cell projection |
| TCPE/G/Z | T-complex protein 1 subunit ϵ/γ/ζ | Cytoskeleton, microtubule |
| UTRO | Utrophin | Cell junction, cytoskeleton |
| COR1C | Coronin | Membrane, cytoskeleton |
| TPM2/3 | Tropomyosin α and β | Cytoskeleton |
The majority of identified proteins were not reported to be located within SIFs, although some are known to be involved in the Salmonella infection process. These include, for instance, annexin A1 (45), α-actinin (46), actin-related protein 2/3 complex (47), BAG chaperone regulator 2 (43), cullin family proteins (48), coronin (49), tropomyosin (46), serine/threonine-protein phosphatase 2A (50), catenin α-1 (51), dynamitin (52), endoplasmin (50), coatomer I protein (48), elongation factors (50), T-plastin (53), small GTPase Ral-A (54), protein disulfide-isomerases (50), ezrin (55), spectrin (56), and ubiquitin-like modifier-activating proteins (50).
However, to the best of our knowledge, most of the identified proteins have not been mentioned before in relation to the intracellular lifestyle of Salmonella. Interesting candidates include trafficking-related components (coatomer II–associated small GTPase Sar1a, GTPase Rab2a, ADF-ribosylation factor 4, signal recognition particle subunits Srp72 and SrpR, Ras GTPase-activating protein-binding protein 2, B-cell receptor–associated protein 31, transmembrane emp24 domain–containing proteins 9 and 10, vesicle-associated membrane protein–associated proteins A and B, protein transport protein Sec23a, vacuolar protein sorting-associated protein 26B), nutrient transporter parts (tricarboxylate transport protein, phosphate transport protein, aspartate glutamate carrier 2, 4F2 cell-surface antigen), signaling components (Ras-related protein Rap1b, serine/threonine-protein phosphatase PGAM5), and cytoskeleton-associated elements (utrophin, protein spire homolog 2 Arf-GAP with Rho-GAP domain-ANK repeat and PH domain-containing protein 2, caldesmon). These host proteins, or organelles containing these proteins, are likely hijacked by Salmonella to provide the SCVs with nutrients, to avoid antimicrobial activities, to modify the normal endosomal maturation, and to support further intracellular proliferation. However, further validations are required to assess their impact on the survival of Salmonella inside host cells.
Intracellular Salmonella Redirects Host Traffic to SMM
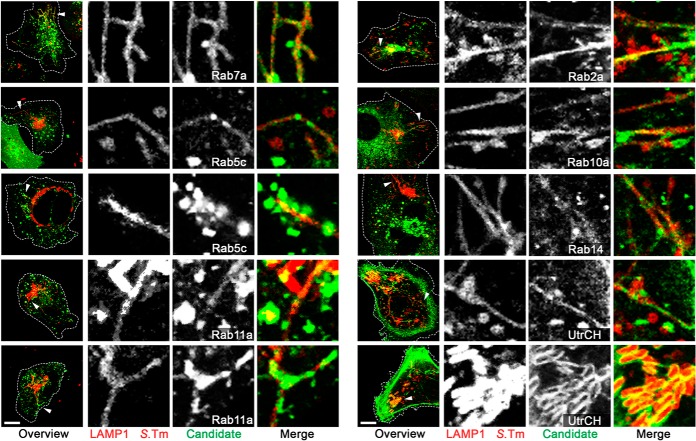
Small Rab GTPases perform a fundamental role in membrane dynamics and are known key targets of intracellular bacterial pathogens (1, 57). In the presented SMM proteome analysis we identified six small Rab GTPases (Rab2a, Rab5c, Rab7a, Rab10a, Rab11a, and Rab14), of which only Rab7a was previously linked to SIF formation (1). To determine whether the other proteins are also involved in SIF formation and associated with the SMM network, we transiently co-transfected HeLa cells for synthesis of pLAMP1-mCherry and fusion proteins of GFP to Rab2a, Rab5c, Rab7a, Rab10a, Rab11a, or Rab14. Subsequently, cells were infected with WT Salmonella expressing mCherry and imaged via CLSM at 8 h p.i. All identified and analyzed Rab GTPase GFP-fusion proteins were localized at SCV and SIF membranes (Fig. 6), as indicated by co-localization with the marker protein LAMP1. Similarly, we analyzed the localization of the F-actin binding protein utrophin (UtrCH). Here, we observed co-localization of UtrCH along SIFs, as well as an accumulation of UtrCH surrounding individual Salmonella assumed to be part of the vacuole-associated actin polymerization (Fig. 6). Controls verifying that the protein localizations in uninfected cells were not affected by overexpression of GFP fusion proteins are presented in supplemental Fig. S6.
Fig. 6.
Co-localization of identified Rab GTPases and the F-actin binding protein utrophin with SMMs. HeLa cells were co-transfected with p3451 for expression of LAMP1-mCherry (red) and various plasmids for the synthesis of fusion proteins of Rab2a, Rab5c, Rab7a, Rab10a, Rab11a, Rab14, or UtrCh to GFP (green) (supplemental Table S1A). Afterward, cells were infected with Salmonella WT (red) and imaged via CLSM at 8 h p.i. The lysosomal glycoprotein LAMP1 served as a marker for SIFs and SCVs. Controls verifying that the protein localizations were not affected by overexpression of GFP fusion proteins are presented in supplemental Fig. S6. Overview images are shown as maximum intensity projections. White arrowheads indicate the structures of interest shown as magnifications for each channel. Scale bar: 10 μm.
Components of COPI- and COPII-mediated transport processes including CopA, CopG1 (both COPI), Sec23A, and Sar1A (both COPII) are also part of the SMM proteome. Although these proteins have been implicated in the replication niche formation of other pathogens (58), they have not been shown to be involved in Salmonella. We therefore immunostained these marker proteins in Salmonella WT mCherry-infected HeLa LAMP1-GFP and revealed localizations of COP vesicles along SIF membranes (Fig. 7). Similarly, we also observed the association of a representative mitochondrial marker protein, mitofilin, along SIFs. The association of proteins of the SMM proteome with SIFs further validates the usefulness of the SMM proteome approach.
Fig. 7.
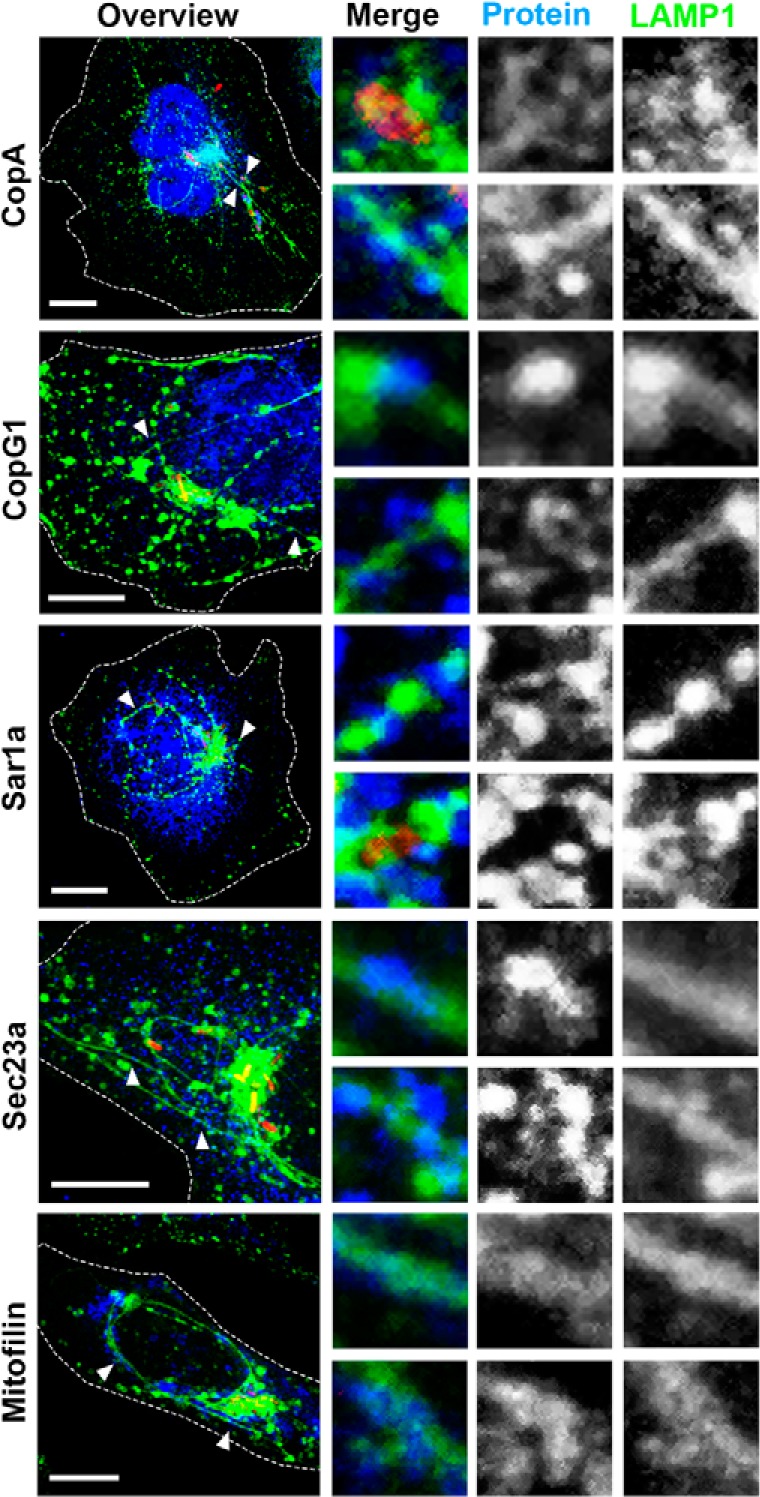
Localization of COPI, COPII, and mitochondria in Salmonella-infected cells. HeLa LAMP1-GFP cells were infected with Salmonella WT mCherry (red) and fixed at 8 h p.i. Immunostaining was performed for COPI (CopA, CopG1), COPII (Sar1a, Sec23a), or mitochondria (Mitofilin) (blue), after which cells were imaged using a Leica SP5 CLSM. Images are shown as maximum intensity projections. White arrowheads indicate the structures of interest shown as magnifications for each channel. Scale bar: 10 μm.
In summary, these findings confirm that the presented SMM proteome enrichment and analysis strategy identified proteins located to the Salmonella compartment. In addition, the results clearly indicate the redirection of proteins of various host trafficking pathways (endosomal, recycling, Golgi–ER, and ER–ER), some of which were not known to be connected to SIFs, the formation of the replication niche inside the host, and Salmonella infection in general. Our findings of host transport processes redirected by Salmonella are summarized in Fig. 8. Overall, the identified SMM proteins provide compelling insights into host processes that are usurped by Salmonella to ensure its survival.
Fig. 8.
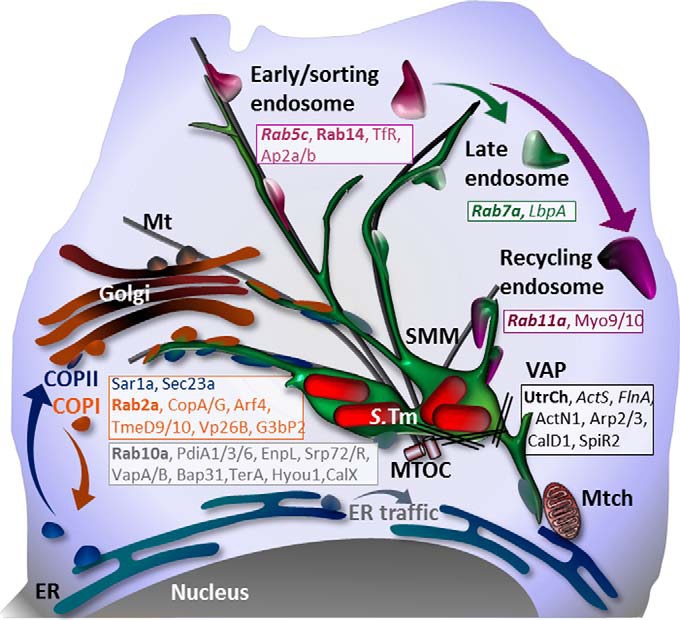
Model for the origin of SMMs. SMM networks contain relocated host proteins from endocytic and intracellular host trafficking routes, as well as components of mitochondrial, ribosomal, ER, and nuclear origin identified in this study. Proteins known to be involved in SIF or SCV formation are italicized or bolded, respectively. Mt, microtubule; Mtch, mitochondrium; VAP, vacuole-associated actin polymerization; S.Tm, Salmonella Typhimurium.
DISCUSSION
The success of an intracellular pathogen depends on its ability to establish a niche for proliferation within its host. Although the infection process of Salmonella has been intensively studied (7, 50, 59–63), less is known about the host niche of this bacterium.
Salmonella-infected cells are characterized by a complex network of SCVs linked to various types of tubular membrane compartments, collectively termed the SMM. The extensive and highly dynamic nature of SMMs prevented classical subcellular fractionation, and we developed a new procedure for the enrichment of SMMs. In this paper we describe immunoproteomic analyses that allowed, for the first time, a survey of the proteome of the SMM.
It is assumed that the host niche of Salmonella is formed during its continuous interactions with the host endosomal and recycling pathway (4, 5). We identified several components of these host pathways such as Rab7a, Rab5c, Rab11a, and Rab14 that are known to be involved in the formation of SCVs or SIFs (36, 39–44). These findings demonstrate that our approach is well suited to identify infection-relevant host proteins from the SMM.
In addition we observed proteins indicative of other intracellular host membrane trafficking pathways. For instance, we identified proteins associated with the anterograde and retrograde transport system, responsible for trafficking between the ER and Golgi (58). This includes components of the COPI and COPII (COPI: coatomer subunits α and γ, transmembrane emp24 domain–containing proteins 9 and 10, vacuolar protein sorting–associated protein 26B, ADF-ribosylation factor 4, small GTPase Rab2a, and Rab2a effector glyceraldehyde-3-phosphate dehydrogenase (64); COPII: small coatomer II–associated GTPase SAR1, protein transport protein Sec23). Salcedo and Holden (65) stated in an earlier report that only Salmonella cells closely associated with the Golgi network are able to multiply. However, no vesicle trafficking or direct association or fusion with either the Golgi or the ER system was observed. Our study thus provides the first evidence of direct interaction of SMMs with the ER system.
In addition to COPI and COPII components, we further identified the small GTPase Rab10a involved in ER dynamics (66), several ER chaperones (protein disulfide-isomerase PDIA1, PDIA3, PDIA6, endoplasmin, hypoxia up-regulated protein 1), SNARE proteins (VAMP-associated proteins A and B), vesicle recognition particles (subunit Srp72 and receptor SrpR), B-cell receptor–associated protein 31, and diverse ER membrane proteins (transitional ER ATPase, calnexin, estradiol 17-β dehydrogenase, dolichyl-diphosphooligosaccharide-glycosyltransferase, transmembrane protein 43).
In summary, the data indicate that Salmonella at the stage of efficient intracellular proliferation specifically intercepts intracellular membrane trafficking pathways and recruits membranes and proteins from endosomes (Fig. 8). However, this is a common strategy among other intracellular pathogens (58). For example, Legionella pneumophilia tethers and subsequently fuses transitional ER-derived vesicles to its vacuolar host compartment (LCV) (1). Similarly, Brucella spp. intercept COPI and COPII vesicle trafficking and target the ER to create a replicating niche termed the Brucella-containing vacuole (67). Interaction of the Brucella-containing vacuole with the ER leads, for instance, to the presence of several ER marker proteins such as calnexin, Sec61, Rab2a, and the Rab2a effector glyceraldehyde-3-phosphate dehydrogenase in the Brucella-containing vacuole. Interestingly, we observed many of these proteins in the SMM.
This raises the interesting question of whether there might be more commonalities between the host niche composition and mechanisms of intracellular survival of these bacteria. Recently, the proteome of LCVs has been profiled (29, 30). Unfortunately, these studies did not distinguish between proteins that are relevant to bacterial persistence and nonspecific host proteins. Nonetheless, this offers the opportunity to compare these host niches of different intracellular pathogens and identify common host proteins. One interesting finding of the comparative analyses was that 45% (109 of 247 proteins) of the SMM proteome isolated from HeLa was also found in LCVs isolated from Dictyostelium cells or mouse macrophages (supplemental Table S4). This is quite remarkable considering that the studies were based on different pathogens and host systems. Thus it appears that although Salmonella and Legionella utilize distinct mechanisms to manipulate their host cells, the pathogen-containing compartments share a remarkable degree of similarity.
The 109 proteins common to the SMM proteome and LCVs are of diverse origin, including ER, mitochondria, nucleus, ribosomes, and endocytic pathways (supplemental Table S4). Again, we found that a large number of the proteins identified in both studies were involved in transport processes (43 proteins), which indicates that these transport processes might be essential for survival for a wide range of intracellular pathogens. Further notable groups of proteins were involved in vesicle trafficking (12 proteins), endo-/exocytosis (10 proteins), protein folding/modification and biosynthesis (17 proteins), and metabolism (16 proteins). In contrast, only relatively few (eight) proteins with cytoskeletal functions were found in LCVs and SMMs. This includes actin; two motor proteins (myosin 9 and myosin 10); and actin-binding proteins coronin-1C, α-actinin, F-actin capping protein, Arp2/3 complex, and desmoplakin. Actin is involved in the formation of a stabilizing meshwork around pathogen-containing compartments such as SCVs (41, 68). Motor proteins are important mediators of vesicular transport, and interference with host cell motor protein function enables intracellular pathogens to segregate their own membrane-bound compartments from the default endosomal maturation pathway (59, 69). For example, SCVs maintain a juxtanuclear position and are restricted in motility (4, 44, 70). However, dynein, a motor protein identified here, was found only on SMMs. This is consistent with the finding that the SMM network extension is dynein mediated (52, 71). Interestingly, tubular networks are not known for Legionella-infected host cells. Thus, despite the fact that similar host proteins are recruited, the bacteria appear to elicit different intracellular phenotypes. This might hint at a novel mechanism that needs further exploration. Together, these findings suggest the interesting possibility that despite the differences in the structures of the niches occupied by the pathogens, they might commandeer similar host proteins to establish their intracellular niches.
A general limitation of proteome studies is that typically the separation of proteins or peptides prior to MS is insufficient to fully resolve complex protein mixtures. As a consequence, low-abundance proteins in particular might be absent from an individual run. However, the use of immunoprecipitation appears to reduce the complexity to a manageable degree, as we found from our combined dataset. Nonetheless, the main purpose of this analysis was to provide priority targets to be investigated in further detail using targeted methods. Although validating all targets would be beyond the scope of this study, we found that at least 11 proteins identified via the proteome approach were indeed associated with SMMs, including a number of new proteins previously not related to the intracellular lifestyle of Salmonella.
In summary, our proteomic study provides global insights into the molecular machinery involved in the formation and maintenance of SMMs. The data indicate that more membrane trafficking systems than hitherto realized are involved, and we have provided numerous targets for further investigation. It is also interesting to note that despite the unique structure of SMMs, the host-hijacking mechanisms are markedly similar to those of other intracellular pathogens.
Supplementary Material
Acknowledgments
We thank Dr. Christian Ungermann, Dr. Francis Barr, and Dr. Martin Aepfelbacher for providing pEGFP-C2-Rab5c, pEGFP-C2-Rab2a, pEGFP-C2-Rab10a, pEGFP-C2-Rab14, and pGL-Rab7a constructs. We also thank Dr. Karin Busch for the anti-Mitofilin antibody. We are grateful to Dr. Stefan Walter, Masha Namakchian, Monika Nietschke, Ursula Krehe, und Daniela Jäckel for technical support and to Laura Spelmink for preparatory experiments.
Footnotes
Author contributions: M.H. and N.H. designed research; S.V. and V.K. performed research; J.D. contributed new reagents or analytic tools; S.V. and N.H. analyzed data; M.H. and N.H. wrote the paper.
* This work was supported by Grant No. HE1964/18-1 within Priority Program SPP1580 of the Deutsche Forschungsgemeinschaft and by the Niedersächsisches Ministerium für Wissenschaft und Kultur.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- SCV
- Salmonella-containing vacuole
- CLSM
- confocal laser-scanning microscopy
- ER
- endoplasmic reticulum
- GEMN
- Golgi–endoplasmic reticulum–microtubule organizing center–nucleus
- IP
- immunoprecipitation
- LAMP
- lysosome-associated membrane protein
- LC
- liquid chromatography
- MS/MS
- tandem mass spectrometry
- SIF
- Salmonella-induced filament
- SMM
- Salmonella-modified membrane
- SPI
- Salmonella Pathogenicity Island
- T3SS
- type III secretion system
- WT
- wild type
- m.o.i.
- multiplicity of infection
- p.i.
- post-infection
- LCV
- Legionella-containing vacuole
- COP
- coat protein complex.
REFERENCES
- 1. Brumell J. H., Scidmore M. A. (2007) Manipulation of rab GTPase function by intracellular bacterial pathogens. Microbiol. Mol. Biol. Rev. 71, 636–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Casadevall A. (2008) Evolution of intracellular pathogens. Ann. Rev. Microbiol. 62, 19–33 [DOI] [PubMed] [Google Scholar]
- 3. Galan J. E. (2001) Salmonella interactions with host cells: type III secretion at work. Ann. Rev. Cell Dev. Biol. 17, 53–86 [DOI] [PubMed] [Google Scholar]
- 4. Abrahams G. L., Müller P., Hensel M. (2006) Functional dissection of SseF, a type III effector protein involved in positioning the Salmonella-containing vacuole. Traffic 7, 950–965 [DOI] [PubMed] [Google Scholar]
- 5. Malik-Kale P., Jolly C. E., Lathrop S., Winfree S., Luterbach C., Steele-Mortimer O. (2011) Salmonella—at home in the host cell. Front. Microbiol. 2, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bakowski M. A., Braun V., Brumell J. H. (2008) Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic 9, 2022–2031 [DOI] [PubMed] [Google Scholar]
- 7. Schroeder N., Mota L. J., Meresse S. (2011) Salmonella-induced tubular networks. Trends Microbiol. 19, 268–277 [DOI] [PubMed] [Google Scholar]
- 8. Knodler L. A., Vallance B. A., Hensel M., Jackel D., Finlay B. B., Steele-Mortimer O. (2003) Salmonella type III effectors PipB and PipB2 are targeted to detergent-resistant microdomains on internal host cell membranes. Mol. Microbiol. 49, 685–704 [DOI] [PubMed] [Google Scholar]
- 9. Brumell J. H., Tang P., Mills S. D., Finlay B. B. (2001) Characterization of Salmonella-induced filaments (Sifs) reveals a delayed interaction between Salmonella-containing vacuoles and late endocytic compartments. Traffic 2, 643–653 [DOI] [PubMed] [Google Scholar]
- 10. Garcia-del Portillo F., Zwick M. B., Leung K. Y., Finlay B. B. (1993) Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 90, 10544–10548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McGhie E. J., Brawn L. C., Hume P. J., Humphreys D., Koronakis V. (2009) Salmonella takes control: effector-driven manipulation of the host. Curr. Opin. Microbiol. 12, 117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Waterman S. R., Holden D. W. (2003) Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 5, 501–511 [DOI] [PubMed] [Google Scholar]
- 13. Boucrot E., Beuzon C. R., Holden D. W., Gorvel J. P., Meresse S. (2003) Salmonella typhimurium SifA effector protein requires its membrane-anchoring C-terminal hexapeptide for its biological function. J. Biol. Chem. 278, 14196–14202 [DOI] [PubMed] [Google Scholar]
- 14. Haraga A., Ohlson M. B., Miller S. I. (2008) Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6, 53–66 [DOI] [PubMed] [Google Scholar]
- 15. Zhang Y., Hensel M. (2013) Evaluation of nanoparticles as endocytic tracers in cellular microbiology. Nanoscale 5, 9296–9309 [DOI] [PubMed] [Google Scholar]
- 16. Husseiny M. I., Wartha F., Hensel M. (2007) Recombinant vaccines based on translocated effector proteins of Salmonella Pathogenicity Island 2. Vaccine 25, 185–193 [DOI] [PubMed] [Google Scholar]
- 17. Rajashekar R., Liebl D., Seitz A., Hensel M. (2008) Dynamic remodeling of the endosomal system during formation of Salmonella-induced filaments by intracellular Salmonella enterica. Traffic 9, 2100–2116 [DOI] [PubMed] [Google Scholar]
- 18. Kuhle V., Hensel M. (2002) SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell. Microbiol. 4, 813–824 [DOI] [PubMed] [Google Scholar]
- 19. Müller P., Chikkaballi D., Hensel M. (2012) Functional dissection of SseF, a membrane-integral effector protein of intracellular Salmonella enterica. PLoS One 7, e35004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hansmeier N., Albersmeier A., Tauch A., Damberg T., Ros R., Anselmetti D., Puhler A., Kalinowski J. (2006) The surface (S)-layer gene cspB of Corynebacterium glutamicum is transcriptionally activated by a LuxR-type regulator and located on a 6 kb genomic island absent from the type strain ATCC 13032. Microbiology 152, 923–935 [DOI] [PubMed] [Google Scholar]
- 21. Chao T. C., Kalinowski J., Nyalwidhe J., Hansmeier N. (2010) Comprehensive proteome profiling of the Fe(III)-reducing myxobacterium Anaeromyxobacter dehalogenans 2CP-C during growth with fumarate and ferric citrate. Proteomics 10, 1673–1684 [DOI] [PubMed] [Google Scholar]
- 22. Taylor G. K., Goodlett D. R. (2005) Rules governing protein identification by mass spectrometry. Rapid Commun. Mass Spectrom. 19, 3420. [DOI] [PubMed] [Google Scholar]
- 23. Dimmer E. C., Huntley R. P., Alam-Faruque Y., Sawford T., O'Donovan C., Martin M. J., Bely B., Browne P., Mun Chan W., Eberhardt R., Gardner M., Laiho K., Legge D., Magrane M., Pichler K., Poggioli D., Sehra H., Auchincloss A., Axelsen K., Blatter M. C., Boutet E., Braconi-Quintaje S., Breuza L., Bridge A., Coudert E., Estreicher A., Famiglietti L., Ferro-Rojas S., Feuermann M., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., James J., Jimenez S., Jungo F., Keller G., Lemercier P., Lieberherr D., Masson P., Moinat M., Pedruzzi I., Poux S., Rivoire C., Roechert B., Schneider M., Stutz A., Sundaram S., Tognolli M., Bougueleret L., Argoud-Puy G., Cusin I., Duek-Roggli P., Xenarios I., Apweiler R. (2012) The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 40, D565–D570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 25. David F. P., Yip Y. L. (2008) SSMap: a new UniProt-PDB mapping resource for the curation of structural-related information in the UniProt/Swiss-Prot Knowledgebase. BMC Bioinformatics 9, 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L. J., von Mering C. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. von Mering C., Huynen M., Jaeggi D., Schmidt S., Bork P., Snel B. (2003) STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 31, 258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Remm M., Storm C. E., Sonnhammer E. L. (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314, 1041–1052 [DOI] [PubMed] [Google Scholar]
- 29. Hoffmann C., Finsel I., Otto A., Pfaffinger G., Rothmeier E., Hecker M., Becher D., Hilbi H. (2013) Functional analysis of novel Rab GTPases identified in the proteome of purified Legionella-containing vacuoles from macrophages. Cell Microbiol. 16, 1034–1052 [DOI] [PubMed] [Google Scholar]
- 30. Urwyler S., Nyfeler Y., Ragaz C., Lee H., Müller L. N., Aebersold R., Hilbi H. (2009) Proteome analysis of Legionella vacuoles purified by magnetic immunoseparation reveals secretory and endosomal GTPases. Traffic 10, 76–87 [DOI] [PubMed] [Google Scholar]
- 31. Xiong G., Husseiny M. I., Song L., Erdreich-Epstein A., Shackleford G. M., Seeger R. C., Jackel D., Hensel M., Metelitsa L. S. (2010) Novel cancer vaccine based on genes of Salmonella pathogenicity island 2. Int. J. Cancer 126, 2622–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hensel M., Shea J. E., Waterman S. R., Mundy R., Nikolaus T., Banks G., Vazquez-Torres A., Gleeson C., Fang F. C., Holden D. W. (1998) Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30, 163–174 [DOI] [PubMed] [Google Scholar]
- 33. Krieger V., Liebl D., Zhang Y., Rajashekar R., Chlanda P., Giesker K., Chikkaballi D., Hensel M. (2014) Reorganization of the endosomal system in Salmonella-infected cells: the ultrastructure of Salmonella-induced tubular compartments. PLoS Pathogens 10, e1004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Drecktrah D., Levine-Wilkinson S., Dam T., Winfree S., Knodler L. A., Schroer T. A., Steele-Mortimer O. (2008) Dynamic behavior of Salmonella-induced membrane tubules in epithelial cells. Traffic 9, 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. (2009) STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ramsden A. E., Holden D. W., Mota L. J. (2007) Membrane dynamics and spatial distribution of Salmonella-containing vacuoles. Trends Microbiol. 15, 516–524 [DOI] [PubMed] [Google Scholar]
- 37. Mota L. J., Ramsden A. E., Liu M., Castle J. D., Holden D. W. (2009) SCAMP3 is a component of the Salmonella-induced tubular network and reveals an interaction between bacterial effectors and post-Golgi trafficking. Cell. Microbiol. 11, 1236–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haglund C. M., Welch M. D. (2011) Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. J. Cell Biol. 195, 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith A. C., Heo W. D., Braun V., Jiang X., Macrae C., Casanova J. E., Scidmore M. A., Grinstein S., Meyer T., Brumell J. H. (2007) A network of Rab GTPases controls phagosome maturation and is modulated by Salmonella enterica serovar Typhimurium. J. Cell Biol. 176, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kuijl C., Pilli M., Alahari S. K., Janssen H., Khoo P. S., Ervin K. E., Calero M., Jonnalagadda S., Scheller R. H., Neefjes J., Junutula J. R. (2013) Rac and Rab GTPases dual effector Nischarin regulates vesicle maturation to facilitate survival of intracellular bacteria. EMBO J. 32, 713–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miao E. A., Brittnacher M., Haraga A., Jeng R. L., Welch M. D., Miller S. I. (2003) Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol. Microbiol. 48, 401–415 [DOI] [PubMed] [Google Scholar]
- 42. Wasylnka J. A., Bakowski M. A., Szeto J., Ohlson M. B., Trimble W. S., Miller S. I., Brumell J. H. (2008) Role for myosin II in regulating positioning of Salmonella-containing vacuoles and intracellular replication. Infect. Immun. 76, 2722–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Auweter S. D., Bhavsar A. P., de Hoog C. L., Li Y., Chan Y. A., van der Heijden J., Lowden M. J., Coombes B. K., Rogers L. D., Stoynov N., Foster L. J., Finlay B. B. (2011) Quantitative mass spectrometry catalogues Salmonella pathogenicity island-2 effectors and identifies their cognate host binding partners. J. Biol. Chem. 286, 24023–24035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Odendall C., Rolhion N., Forster A., Poh J., Lamont D. J., Liu M., Freemont P. S., Catling A. D., Holden D. W. (2012) The Salmonella kinase SteC targets the MAP kinase MEK to regulate the host actin cytoskeleton. Cell Host Microbe 12, 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smyth T., Totemeyer S., Haugland S., Willers C., Peters S., Maskell D., Bryant C. (2008) Dexamethasone modulates Salmonella enterica serovar Typhimurium infection in vivo independently of the glucocorticoid-inducible protein annexin-A1. FEMS Immunol. Med. Microbiol. 54, 339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Finlay B. B., Ruschkowski S., Dedhar S. (1991) Cytoskeletal rearrangements accompanying Salmonella entry into epithelial cells. J. Cell Sci. 99, 283–296 [DOI] [PubMed] [Google Scholar]
- 47. Criss A. K., Casanova J. E. (2003) Coordinate regulation of Salmonella enterica serovar Typhimurium invasion of epithelial cells by the Arp2/3 complex and Rho GTPases. Infect. Immun. 71, 2885–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Misselwitz B., Dilling S., Vonaesch P., Sacher R., Snijder B., Schlumberger M., Rout S., Stark M., von Mering C., Pelkmans L., Hardt W. D. (2011) RNAi screen of Salmonella invasion shows role of COPI in membrane targeting of cholesterol and Cdc42. Mol. Syst. Biol. 7, 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Galkin V. E., Orlova A., Brieher W., Kueh H. Y., Mitchison T. J., Egelman E. H. (2008) Coronin-1A stabilizes F-actin by bridging adjacent actin protomers and stapling opposite strands of the actin filament. J. Mol. Biol. 376, 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vogels M. W., van Balkom B. W., Heck A. J., de Haan C. A., Rottier P. J., Batenburg J. J., Kaloyanova D. V., Helms J. B. (2011) Quantitative proteomic identification of host factors involved in the Salmonella typhimurium infection cycle. Proteomics 11, 4477–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lu R., Liu X., Wu S., Xia Y., Zhang Y. G., Petrof E. O., Claud E. C., Sun J. (2012) Consistent activation of the beta-catenin pathway by Salmonella type-three secretion effector protein AvrA in chronically infected intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G1113–G1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marsman M., Jordens I., Kuijl C., Janssen L., Neefjes J. (2004) Dynein-mediated vesicle transport controls intracellular Salmonella replication. Mol. Biol. Cell 15, 2954–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou D., Mooseker M. S., Galan J. E. (1999) An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proc. Natl. Acad. Sci. U.S.A. 96, 10176–10181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nichols C. D., Casanova J. E. (2010) Salmonella-directed recruitment of new membrane to invasion foci via the host exocyst complex. Curr. Biol. 20, 1316–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Agbor T. A., Demma Z. C., Mumy K. L., Bien J. D., McCormick B. A. (2011) The ERM protein, ezrin, regulates neutrophil transmigration by modulating the apical localization of MRP2 in response to the SipA effector protein during Salmonella typhimurium infection. Cell Microbiol. 13, 2007–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruetz T. J., Lin A. E., Guttman J. A. (2012) Enterohaemorrhagic Escherichia coli requires the spectrin cytoskeleton for efficient attachment and pedestal formation on host cells. Microb. Pathog. 52, 149–156 [DOI] [PubMed] [Google Scholar]
- 57. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 58. Roy C. R., Salcedo S. P., Gorvel J. P. (2006) Pathogen-endoplasmic-reticulum interactions: in through the out door. Nat. Rev. Immunol. 6, 136–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Steele-Mortimer O. (2008) The Salmonella-containing vacuole: moving with the times. Curr. Opin. Microbiol. 11, 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brumell J. H., Grinstein S. (2004) Salmonella redirects phagosomal maturation. Curr. Opin. Microbiol. 7, 78–84 [DOI] [PubMed] [Google Scholar]
- 61. Honer zu Bentrup K., Ramamurthy R., Ott C. M., Emami K., Nelman-Gonzalez M., Wilson J. W., Richter E. G., Goodwin T. J., Alexander J. S., Pierson D. L., Pellis N., Buchanan K. L., Nickerson C. A. (2006) Three-dimensional organotypic models of human colonic epithelium to study the early stages of enteric Salmonellosis. Microbe Infect. 8, 1813–1825 [DOI] [PubMed] [Google Scholar]
- 62. Becker D., Selbach M., Rollenhagen C., Ballmaier M., Meyer T. F., Mann M., Bumann D. (2006) Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440, 303–307 [DOI] [PubMed] [Google Scholar]
- 63. Imami K., Bhavsar A. P., Yu H., Brown N. F., Rogers L. D., Finlay B. B., Foster L. J. (2013) Global impact of Salmonella pathogenicity island 2-secreted effectors on the host phosphoproteome. Mol. Cell. Proteomics 12, 1632–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tisdale E. J., Azizi F., Artalejo C. R. (2009) Rab2 utilizes glyceraldehyde-3-phosphate dehydrogenase and protein kinase Ci to associate with microtubules and to recruit dynein. J. Biol. Chem. 284, 5876–5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Salcedo S. P., Holden D. W. (2003) SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 22, 5003–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. English A. R., Voeltz G. K. (2013) Rab10 GTPase regulates ER dynamics and morphology. Nat. Cell Biol. 15, 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. de Bolle X., Letesson J. J., Gorvel J. P. (2012) Small GTPases and Brucella entry into the endoplasmic reticulum. Biochem. Soc. Trans. 40, 1348–1352 [DOI] [PubMed] [Google Scholar]
- 68. Poh J., Odendall C., Spanos A., Boyle C., Liu M., Freemont P., Holden D. W. (2008) SteC is a Salmonella kinase required for SPI-2-dependent F-actin remodelling. Cell Microbiol. 10, 20–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Alix E., Mukherjee S., Roy C. R. (2011) Subversion of membrane transport pathways by vacuolar pathogens. J. Cell Biol. 195, 943–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ramsden A. E., Mota L. J., Munter S., Shorte S. L., Holden D. W. (2007) The SPI-2 type III secretion system restricts motility of Salmonella-containing vacuoles. Cell Microbiol. 9, 2517–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Guignot J., Caron E., Beuzon C., Bucci C., Kagan J., Roy C., Holden D. W. (2004) Microtubule motors control membrane dynamics of Salmonella-containing vacuoles. J. Cell Sci. 117, 1033–1045 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.