Abstract
Background
Glioblastoma multiforme (GBM) is the most aggressive and invasive brain tumor, for which novel prognostic markers and predictors of therapeutic response are urgently needed. We reported previously that levels of peptide-O-linked mannose β-1,2-N-acetylglucosaminyltransferase 1 (PomGnT1) in glioma specimens correlated with tumor grade. However, the prognostic significance of PomGnT1 in glioma patients and its function in GBM progression remain unknown.
Methods
Clinical relevance of PomGnT1 in GBM patients' prognosis was analyzed both in a clinically annotated expression dataset of 446 GBM tumor specimens and in 82 GBM tumor samples collected at our institution. The function of PomGnT1 in glioma growth and invasion, and the underlying mechanisms of PomGnT1 regulation were explored in vitro and in vivo.
Results
PomGnT1 expression in GBM tissues was closely associated with poor prognosis in GBM patients. Forced overexpression of PomGnT1 in glioblastoma cells impaired cell adhesion and increased their proliferation and invasion in vitro. Subsequent in vivo experiments showed that overexpression of PomGnT1 promoted tumor growth and shortened the survival time of tumor-bearing mice in an orthotopic model. Conversely, stable short hairpin RNA–mediated knockdown of PomGnT1 expression produced opposite effects both in vitro and in vivo. Mechanistic studies revealed that activation of epidermal growth factor receptor (EGFR) resulted in EGFR/extracellular signal-regulated kinase–dependent upregulation of PomGnT1, downregulation of receptor-type protein tyrosine phosphatase β, and activation of β-catenin pathway signaling.
Conclusion
These findings suggest that PomGnT1 promotes GBM progression via activation of β-catenin and may serve as a prognostic factor for glioma patient survival as well as a novel molecular target for anticancer therapy in malignant glioma.
Keywords: glioblastoma, PomGnT1, prognosis, progression
Gliomas are the most frequent primary tumors in the brain and are graded on a scale of I–IV based on histopathologic criteria.1 Among the gliomas, glioblastomas have the worst prognosis, with survival time of 12–15 months despite multimodal therapy.2 Since the extremely aggressive and invasive behavior of GBM accounts for the major clinical challenge in the treatment of this tumor, there is a clear need to identify biomarkers of invasive phenotype in GBM. Such biomarkers may be useful in guiding therapy. In the past decade, intense efforts have been made to understand the molecular pathogenesis of GBM, leading to discovery and identification of candidate molecular markers for the evaluation and management of GBM patients.3 The broad spectrum of those potential markers include proteolytic enzymes, extracellular matrix proteins, cell adhesion molecules, neurodevelopmental factors, cell signaling and transcription factors, angiogenic effectors, metabolic proteins, membrane channels, and cytokines and chemokines.4 However, selection of anti-invasive therapies based on known markers in GBM has had mixed success. For instance, while an integrin inactivator cilengitide increased 2-year overall survival (OS)5 in patients with newly diagnosed glioblastoma, a matrix metalloproteinase inhibitor, marimastat, did not improve survival in patients with glioblastoma or gliosarcoma following surgery and radiotherapy.6 Nevertheless, identification of new and previously unrecognized molecular targets altered in GBM and elucidation of the mechanism involved in the development and progression of GBM are considered crucial requirements for better prediction, prognostication, and treatment of this tumor.
O-mannosyl-linked glycans comprise one third of all brain O-linked glycoproteins, but very little is understood about their functions.7 Several congenital muscular dystrophies with neuronal abnormalities such as muscle-eye-brain disease, Walker Warburg syndrome, and Fukuyama congenital muscular dystrophy are caused by genetic disruptions in glycosyltransferases that function in the O-mannosyl glycan processing pathway.8–10 Of particular interest are the defects in neuronal migration and nerve-muscle adhesion associated with muscle-eye-brain disease due to inactivating mutations in peptide-O-mannose β-1,2-N-acetylglucosaminyltransferase 1 (PomGnT1), the glycosyltransferase that catalyzes the transfer of N-acetylglucosamine (GlcNAc) to O-mannose of glycoproteins in the brain, nerve, and skeletal muscle cells of mammals.8,11 The pathology of this disorder implicates a functional role for PomGnT1 in control of cell adhesion and migration. A previous study by Abbott et al8 showed that simultaneous knockdown of both PomGnT1 and GnT-Vb, another glycosyltransferase involved in the O-mannosyl–linked glycosylation pathway, in human neuroblastoma SH-SY5Y cells significantly increased adhesion and impaired neuronal cell migration. However, to the best of our knowledge there are no published reports evaluating the biological role of PomGnT1 in human glioma cells except our most recent work showing that PomGnT1 regulates pseudopodia formation and c-myc expression in malignant glioma U87 cells.12 More importantly, we also demonstrated that PomGnT1 expression levels measured in 133 cases of grade I to IV gliomas were correlated with the histopathologic grade and could be used as a marker to distinguish between low- and high-grade gliomas.12
In the present study, we analyzed the prognostic significance of PomGnT1 mRNA expression in data collected by The Cancer Genome Atlas (TCGA) on GBM patients and the prognostic significance of PomGnT1 protein expression in a set of GBM patients from our own institution. In addition, the functional role of PomGnT1 in GBM malignant progression was characterized by engineering GBM cell lines to alter the expression of PomGnT1. Moreover, we explored the underlying mechanisms of PomGnT1 regulation, which appear to involve epidermal growth factor receptor (EGFR) and extracellular signal-regulated kinase (ERK) as upstream components of the signaling pathway that upregulate PomGnT1 expression and receptor-type protein tyrosine phosphatase β (RPTPβ)/β-catenin signaling as the PomGnT1 downstream target by which PomGnT1 exerts its effects on cell proliferation and invasion. Overall, our investigations indicate that PomGnT1 plays an essential role in GBM progression and warrants consideration as a novel prognostic factor and potential therapeutic target for GBM patients.
Materials and Methods
Reagents and Antibodies
The EGFR inhibitor AG1478 was from Cell Signaling Technology. The ERK inhibitor U0126 was obtained from the Beyotime Institute of Biotechnology. The proteasome inhibitor MG132 was purchased from Sigma. EGF was obtained from Proteintech Group. The immunizing peptide used to produce anti-PomGnT1 antibody was purchased from Abnova. Antibodies to phospho-EGFR (Tyr1068), EGFR, phospho-ERK1/2 (Thr202/Tyr204), ERK1/2, cyclin D1, and β-catenin were from Cell Signaling Technology. Anti–phospho-β-catenin (Tyr654) and anti–β-actin antibodies were from Santa Cruz Biotechnology. Anti–phospho-β-catenin (Tyr333) antibody was from Signalway Antibody. Anti-RPTPβ and anti-PomGnT1 antibodies were from BD Biosciences and Abnova, respectively.
Patients and Tumor Samples
Public TCGA (http://cancergenome.nih.gov/) data repositories were used as our primary source of samples. To analyze the data generated by TCGA, we directly accessed the input data (gene expression of the Genechip Human Genome HT-HG-U133A of TCGA and Agilent Human Genome Comparative Genomic Hybridization [CGH] Microarray 244K). PomGnT1 expression was treated as a binary variable divided into low or high PomGnT1 expression by a criterion of whether the value was greater than the median. In total, 446 GBM tumors having clinical data (date of download: April 10, 2011) were profiled for class discovery and survival analysis. Survival was defined as the time interval from surgery until the date of death. Gene-level copy number variation was estimated using the circular binary segmentation algorithm from the “snapCGH” package using an R code, as described.13
For a retrospective study, we enrolled a total of 82 patients with GBM who underwent surgical removal of tumor between June 2005 and June 2009 in the Department of Neurosurgery, Renji Hospital, Shanghai Jiao Tong University, China. All specimens were collected under an institutional review board–approved protocol and de-identified for patient confidentiality. Clinical information was available for all the patients. Patients' age ranged from 27 to 83 years (median, 58 y), with 35 females and 47 males. The median follow-up time for OS for all patients was 8 months (range, 1–80 mo). Additionally, 5 nonneoplastic brain tissues from epileptogenic patients with clinical information were collected at our department.
Tissue Microarray and Immunohistochemistry
A tissue microarray was constructed using 2 cores 1 mm in diameter per tumor with sampling of different neoplastic regions when possible using tumor samples from our own institution. Immunohistochemistry (IHC) studies were performed on formalin-fixed paraffin-embedded tissue microarrays using human anti-PomGnT1 antibody at a dilution of 1:100, followed by incubation with horseradish peroxidase (HRP)–conjugated goat anti-human IgG secondary antibody. The slides were then stained with the Dako Cytomation EnVision + System HRP (diaminobenzidine) detection kit and counterstained with hematoxylin. To evaluate expression of PomGnT1, 5 high-power fields (400×) within each tumor tissue showing nuclear staining were randomly selected and 100 cells in each field were counted. A 4-tiered semiquantitative scale was used to assess the degree of staining based on the average percentage of positive staining cells: 0 = negative/rare positive cells, 1 = 1%–25%, 2 = 25%–50%, 3 = 50%–100%. In order to analyze the prognosis between groups, staining of either <50% or ≥50% of the tumor cells was used to assign patients to a low-score or high-score group, respectively, as these cutoff values had been used in past studies.14–16
Cell Culture
The human GBM cell lines U87 and U251, purchased from the Cell Bank of the Shanghai Branch of the Chinese Academy of Sciences, were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and maintained in a humidified atmosphere with 5% CO2 at 37°C.
Cell Proliferation Assay
Proliferation of cells stably transfected with PomGnT1-overexpressing, PomGnT1-specific short hairpin (sh)RNA, or their respective controls was measured using the Cell Counting Kit 8 (CCK-8; Dojindo) according to the manufacturer's protocol.
Cell Adhesion and Invasion Assay
For cell adhesion assays, a single-cell suspension was plated in quadruplicate in a 96-well plate and observed every 5 min. Once adhesion/spreading was observed, the plate was shaken for 30 s on an automatic plate shaker to remove unattached cells. The medium was removed and the cells were washed once with phosphate buffered saline (PBS), then fixed for 15 min with 3.5% formalin followed by staining with 0.5% crystal violet. After rinsing and drying, 1% sodium dodecyl sulfate was added to wells and the plate was agitated on an orbital shaker until color was uniform with no areas of dense coloration at the bottom of the wells. Absorbance values were measured at 570 nm using a microplate reader and were directly proportional to the number of viable cells in culture. Cell invasive capability was determined using a modified Boyden chamber invasion assay as previously described.17 Cells were cultured to about 80% confluence and serum starved overnight. Transwell inserts (Corning) of 8-μm pore size were coated with Matrigel (BD Biosciences), and 40 000 cells after overnight starvation were plated onto the top of each of the coated filters in serum-free medium. The same medium containing 20% fetal bovine serum was placed in the lower chamber to act as a chemoattractant. After 20 h of culture, cells that did not migrate or invade through the pores of the Transwell inserts were manually removed with cotton swabs and the inserts were fixed in 0.5% formaldehyde for 5 min and then stained with 0.5% crystal violet. The invading cells of the filter were counted from 5 randomly selected fields (total magnification, ×200).
Gene Silencing and Overexpression
For transient knockdown of RPTPβ, U87 or U251 cells were transfected with small interfering (si)RNA oligonucleotides using Lipofectamine 2000 (Sigma) according to the manufacturer's instructions. The sequences of siRNA for RPTPβ and for nontargeting scrambled control were 5′-CCCUUAUGCACCAACUAGA-3′ and 5′-ATTGCGACCTAACCACACT-3′, respectively. These siRNA transiently transfected cells were designated here as U87-siRPTPβ and U87-siControl, and U251-siRPTPβ and U251-siControl. The PomGnT1-specific shRNA construct (shRNA sequence targeting PomGnT1: CATGGTGCTATTCCTCAACAT) was generated using pGIPZ lentiviral vector (Thermo Scientific). Lentivirus for the stable knockdown of PomGnT1 was produced from 293T cells cotransfected with the pGIPZ-PomGnT1 shRNA and the 2 packaging plasmids psPAX2 and pMD2G. U87 and U251 cells were transduced with the PomGnT1-specific shRNA lentiviral particles at a ratio of 5 particles to 1 cell for 24 h in the presence of hexadimethrine bromide to improve transduction efficiency. Afterward, the medium containing viral particles was removed and replaced with fresh medium containing 1.0 μg/mL puromycin. Before lentiviral transduction, a puromycin titration was performed that identified 1.0 μg/mL as the minimum puromycin concentration that caused complete death of U87 and U251 cells after a 5-day incubation. One “nontarget” construct containing an shRNA sequence that did not target any known human gene was transduced separately into U87 or U251 cells to serve as a scrambled negative control. The 2 paired shRNA interference (shRNAi)–transduced cell lines were named U87-shRNAi PomGnT1 and U87-shRNAi Control, and U251-shRNAi PomGnT1 and U251-shRNAi Control.
An oligonucleotide coding for PomGnT1 (NM_017739) was cloned into the plasmid murine stem cell virus (pMSCV)–puro retroviral vector (Clontech) with which 293T cells were transfected along with packaging plasmids pMD.env and pMD.gag.pol by calcium phosphate precipitation to produce the retroviruses. The produced retroviruses were used to transduce U87 or U251 cells, which were further selected in the presence of 1.0 μg/mL puromycin to establish a cell line with stable PomGnT1 overexpression (U87-PomGnT1 or U251-PomGnT1). Cells infected with empty vector (EV) pMSCV-puro derived retroviruses were used as control (U87-EV or U251-EV).
Orthotopic Glioblastoma Xenograft Model
US National Institutes of Health and institutional guidelines for animal welfare and experimental conduct were followed. This animal study was approved by the Institutional Animal Care and Use Committee of Renji Hospital, Shanghai Jiao Tong University. The orthotopic glioblastoma mouse model was prepared as we previously reported.18 In brief, PomGnT1-knockdown, -overexpressing, or control U87 cells were implanted into the corpus striatum of anesthetized athymic nude mice at 5–6 weeks of age (athymic Ncr-nu/nu, Shanghai Experimental Animal Center) using a small animal stereotactic frame (David Kopf Instruments). A sagittal incision was made through the skin to expose the cranium, and a burr hole was created in the skull at 0.2 mm anterior and 1.8 mm lateral from the bregma using a small dental drill. At a depth of 3 mm from the brain surface, 5 μL of cell suspension containing 1 × 106 cells in PBS was injected into the brain. The needle was left in place for 5 min before retracting. Bone wax was used to seal the skull cavity, and the wound was sutured immediately. One hundred percent model success rate was achieved, as reflected by imaging of the intracranial tumors with MRI (3-Tesla scanner, General Electric). A contrast agent of gadolinium diethylenetriamine pentaacetic acid (Gd/DTPA; 100 μL/20 g; Bayer Healthcare Pharmaceuticals) was injected intraperitoneally 10 min prior to scanning. T1-weighted image-enhanced scans were used, and the scanning parameters were as previously defined.18 Pro-plus software (General Electric) was used to analyze the images, and tumor volume was calculated as: (length × width2)/2 using Function Analysis software (General Electric). Mice were continuously observed until death was recorded for all animals.
Western Blot Analysis
Tissue and cell samples were lysed in a lysis buffer (Sigma) and the protein concentration in the lysates was quantified using the Bradford protein assay (Abcam). Thirty micrograms of protein were loaded for immunoblotting using antibodies against PomGnT1, phospho-EGFR (Tyr1068), EGFR, phospho-ERK1/2 (Thr202/Tyr204), ERK1/2, RPTPβ, phospho–β-catenin Tyr654, phospho–β-catenin Tyr333, β-catenin, cyclin D1, and β-actin. The blots were developed using enhanced chemiluminescence (Pierce) and the films scanned using a densitometer. The protein bands were quantified using Quantity One software.
Statistical Analysis
All statistical analyses were performed using JMP v10 software (SAS Institute). Overall survival curves were plotted according to the Kaplan–Meier method, with the Mantel–Haenszel log-rank test applied for comparison. Multiple comparisons were performed by ANOVA with Tukey correction. The t-test was used to determine differences in each 2-group comparison. All data were presented as means ± SEM. A 2-sided P-value of <.05 was considered significant.
Results
PomGnT1 Is Upregulated in GBM Tumors and Correlated With Prognosis
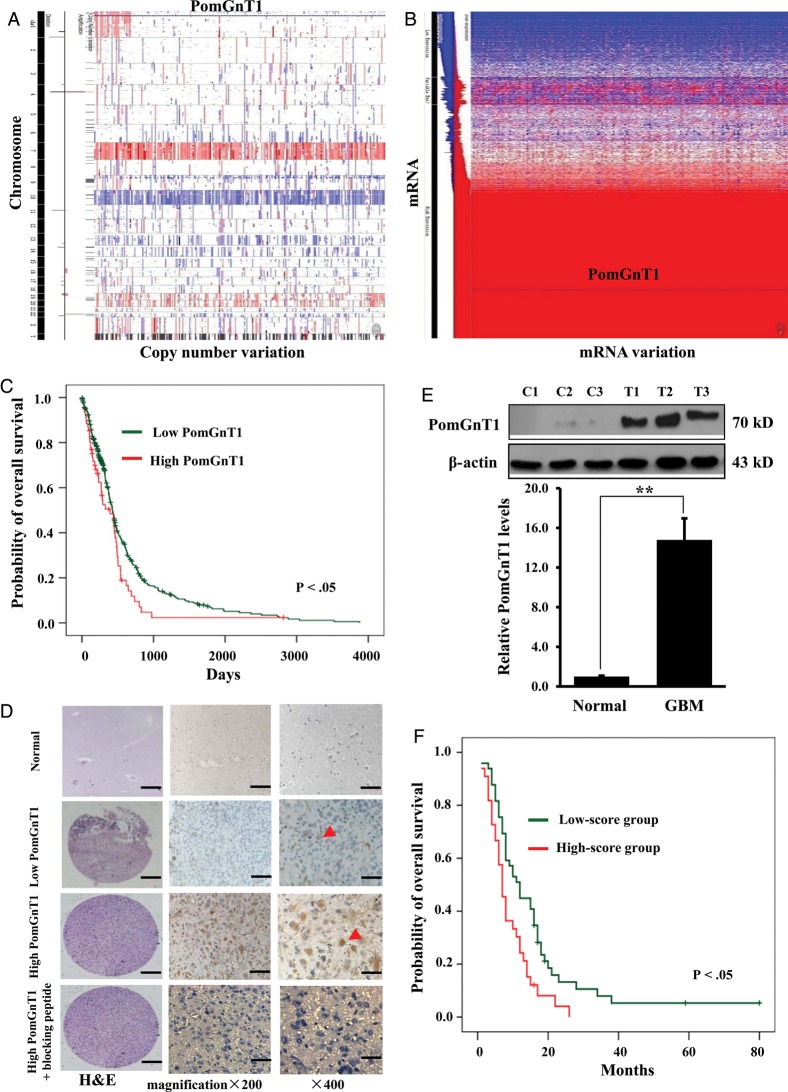
The expression data from 446 GBM samples from TCGA (http://cancergenome.nih.gov/) were used as a genomic discovery set. Genechip Human Genome HT-HG-U133A and Agilent Human Genome CGH Microarray 244K gene expression datasets and clinical data were retrieved from the Open-Access and Controlled-Access Data Tiers Portal (https://tcga-data.nci.nih.gov/tcga/) of TCGA and preprocessed for downstream analyses. As shown in Fig. 1A, no significant change in the PomGnT1 DNA copy number was observed in the heatmap of the gene expression values. The same was true for copy number variation. However, PomGnT1 mRNA was located in the highly expressed region in the mRNA data heatmap from TCGA (Fig. 1B). Notably, PomGnT1 mRNA expression was upregulated in 76.0% of GBM samples compared with normal tissues in the microarray chip. The correlation between PomGnT1 mRNA expression and OS was evaluated using Kaplan–Meier survival curve analysis with a log-rank comparison. As shown in Fig. 1C, patients whose GBM expressed higher than median levels of PomGnT1 mRNA had decreased survival relative to those whose PomGnT1 levels were lower than the median (P < .05) in TCGA data.
Fig. 1.
PomGnT1 was overexpressed in GBM and negatively correlated with patient survival. (A) DNA copy number alterations of PomGnT1 by TCGA data analysis. (B) A heatmap of PomGnT1 mRNA expression profile in TCGA dataset. (C) Kaplan–Meier survival analysis of patients with GBM (n = 446) binned into high (greater or equal to median) and low (less than median) expression of PomGnT1. (D) Representative images of IHC staining of human GBM tissues with low or high expression of PomGnT1 protein. Arrowheads, localization of PomGnT1 in the cytoplasm. Scale bar, 20 µm. H&E, hematoxylin & eosin. (E) Immunoblot analysis of PomGnT1 expression in GBM and controlled brain tissues (n = 3 for each). C: nonneoplastic control brain tissues. T: GBM tumor tissues. The histogram indicates the average levels of the PomGnT1 protein determined from 3 randomly selected GBM tissues expressed as the fold change relative to those in the control brain tissues after normalization to β-actin. **P < .01. (F) Correlation analysis of PomGnT1 protein expression with patient OS (n = 82). The low- and high-score groups represent the patients whose staining extent in GBM tissues assessed by IHC was <50% and ≥50%, respectively, as described in Materials and Methods.
To discern the prognostic relevance of PomGnT1 expression at the protein level, IHC was performed in a tissue microarray we developed that contains a collection of 82 GBM tumor samples and 5 control brain tissues. As shown in the representative Fig. 1D, there was no specific PomGnT1 staining in the control brain tissues, and high staining in the GBM tissues can be blocked with an excess of the immunizing peptide, indicating the specificity of the anti-PomGnT1 antibody. Based on the extent of staining in GBM tissues, we divided the samples into a low-score group (<50% staining) and a high-score group (≥50% staining). PomGnT1 was localized in the cytoplasm of GBM tumor cells. We next performed immunoblot analysis to more quantitatively confirm the expression level of PomGnT1 using GBM tissue from 3 randomly selected GBM patients and 3 samples of normal brain. Figure 1E shows that the level of PomGnT1 in these tumor tissues was substantially higher (14.8 ± 1.3-fold, P < .05) than that in the control brain tissues. Given the observation that PomGnT1 protein expression was increased in GBM, Kaplan–Meier analysis was used to investigate the relationship of PomGnT1 protein expression to patient outcome across all the tumor samples, as assessed by IHC. Patients in the high-score group had significantly shorter survival than patients in the low-score group (P < .05, Fig. 1F). These findings clearly suggest that higher PomGnT1 expression in tumors is associated with poor prognosis in patients with GBM.
PomGnT1 Promotes Glioma Growth in an Orthotopic Glioma Model
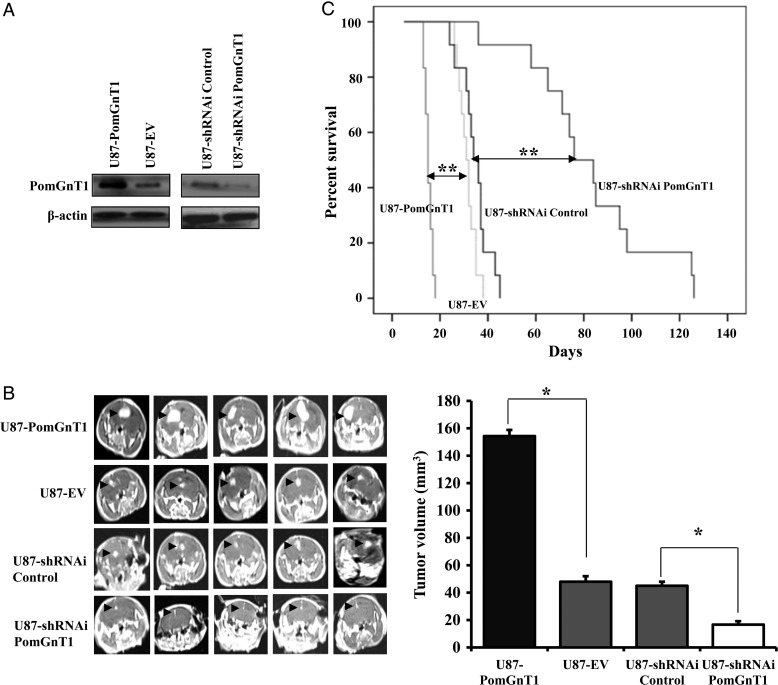
Given the evidence that PomGnT1 expression is of prognostic significance in GBM, we examined the functional role of PomGnT1 in malignant glioma progression in an orthotopic glioma model. We used both gene silencing and overexpression strategies to specifically knock down or overexpress PomGnT1 in GBM cell line U87. Stable overexpression or knockdown of PomGnT1 in U87 cells was confirmed by western blot analysis (Fig. 2A). A subline of U87-PomGnT1, U87-EV, U87-siRNA PomGnT1, or U87-siRNA Control was implanted into the corpus striatum of athymic nude mice. After 14 days, at which point a few animals started to show signs of morbidity, mice in each experimental group were assessed by MRI to confirm intracranial tumor formation and to measure tumor size (Fig. 2B). We found that in vivo tumor growth in the PomGnT1-overexpressing group was much faster than in the empty vector control group, who received cells transduced with nontargeting shRNA (tumor volume 34.9 ± 2.0 mm3 vs 13.3 ± 1.3 mm3, P < .05). In contrast, knockdown of PomGnT1 resulted in significantly reduced tumor volume compared with the control group (tumor volume 3.3 ± 1.1 mm3 vs 11.9 ± 1.1 mm3, P < .05). Consistent with the tumor growth data, mice implanted with PomGnT1-overexpressing cells died within 20 days, whereas 100% of the control mice survived for that duration with a median survival of 31 days. Strikingly, knockdown of PomGnT1 dramatically prolonged survival of the mice compared with the nontarget control group (median survival 83 days vs 35 days, P < .01). These data provide compelling evidence for an important role for PomGnT1 in GBM tumor growth in vivo.
Fig. 2.
PomGnT1 controls the growth of GBM in vivo and the survival time of the tumor-bearing mice. (A) Western blot analysis to confirm stable overexpression or knockdown of PomGnT1 in U87 cells. (B) Representative MR images of the GBM tumors orthotopically inoculated with U87-PomGnT1, U87-EV, U87-siRNA PomGnT1, or U87-siRNA Control cells on day 14 postimplantation. Arrows indicate the tumor areas. The tumor volumes were calculated by the formula: (length × width2)/2 and presented as means ± SEM from 2 independent experiments with each group of 12 mice. *P < .05. (C) Comparative survival of mice bearing U87-PomGnT1, U87-EV, U87-shRNAi Control, or U87-shRNAi PomGnT1 tumors. Time of death was recorded as days after the GBM cell implantation. **P < .01.
PomGnT1 Enhances GBM Cell Proliferation and Invasion and Reduces Cell Adhesion
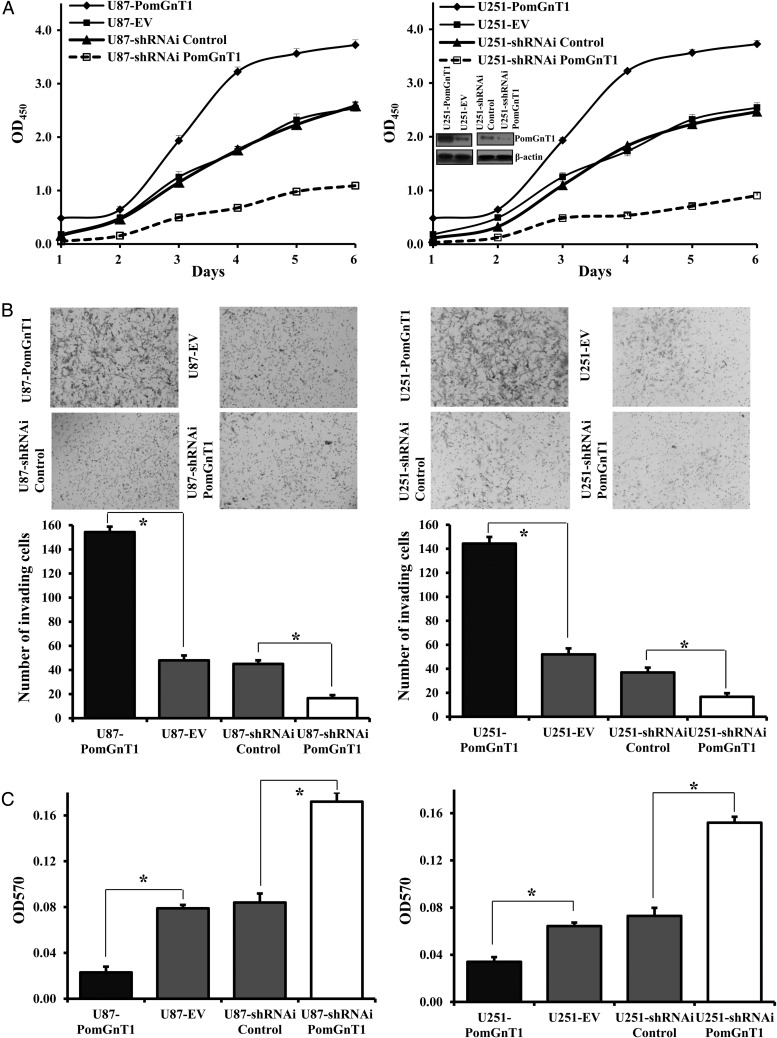
We next sought to evaluate the effect of PomGnT1 on the growth, invasion, and adhesion of the tumor cells in vitro. The large effect of altering PomGnT1 expression on cell proliferation in vivo was further confirmed using the same U87 sublines when cultured in vitro. We observed a marked increase in the proliferation rate of the PomGnT1-overexpresing U87 cells but a significant decrease in the rate of proliferation in the PomGnT1-knockdown U87 cells (Fig. 3A). To validate this finding, an additional GBM cell line, U251, was engineered to overexpress or knock down PomGnT1 expression (Fig. 3A, right panel inset), and the sublines were tested for their proliferation in vitro. As observed in the U87 cells, PomGnT1 overexpression or suppression progressively enhanced or reduced U251 cell proliferation.
Fig. 3.
PomGnT1 regulates GBM cell proliferation, invasion and adhesion in vitro. (A) Effect of PomGnT1 on GBM cell proliferation. Cells were cultured for the indicated periods and relative cell growth was determined by CCK-8 assay. Left panel: growth curve for the U87 sublines. Right panel: growth curve for the U251 sublines (the inset image showing stable overexpression or knockdown of PomGnT1 in U251 cells determined by western blot analysis). Each data point represents the mean ± SEM obtained from 3e independent experiments. OD, optical density. (B) Effect of PomGnT1 on the invasion of GBM cells. Top panels are representative microscopic images of stained cells that had invaded the matrix and migrated to the bottom side of the filter 20 h after seeding. Bottom histograms indicate the number of invading cells counted from 5 randomly selected fields. Vertical bars, ±SEM, n = 3. *P < .05. (C) Effect of PomGnT1 on cell adhesion. Relative adhesion quantified at 6 h after seeding on uncoated tissue culture dishes. Vertical bars, ±SEM, n = 3. *P < .05.
To gain further insights into a functional role of PomGnT1 in the malignant behavior of these GBM cells, we performed invasion and adhesion assays on the U87 and U251 cells with PomGnT1 overexpressed or knocked down. The effect of PomGnT1 on invasive potential was examined using a modified Boyden chamber invasion assay where the cells that invaded through a layer of Matrigel were counted at 20 h after plating the cells on Matrigel-coated Transwell inserts. As shown in Fig. 3B, the invasive ability was increased in U87 and U251 cells that overexpressed PomGnT1 (U87-PomGnT1 and U251-PomGnT1) compared with the corresponding U87-EV and U251-EV control cells (3.2 ± 1.2-fold, P < .05, and 2.8 ± 0.9-fold, P < .05, respectively), whereas knockdown of PomGnT1 in U87 and U251 cells caused a significant inhibition of their invasion by factors of 2.7 ± 1.3-fold (P < .05, U87-siRNA PomGnT1 vs U87-siRNA Control) and 2.2 ± 0.6-fold (P < .05, U251-siRNA PomGnT1 vs U251-siRNA Control), respectively. Because tumor cell motility and invasive potential in large part depend on cell attachment, the ability of the cells to attach firmly to tissue culture plates was evaluated. Figure 3C shows that overexpression of PomGnT1 in U87 and U251 cells led to a reduction in cell adhesion relative to their respective control cells (mean ± SEM, 3.4 ± 1.2-fold, P < .05, and 1.9 ± 0.8-fold, P < .05, respectively). In contrast, suppression of PomGnT1 in U87 and U251 cells induced an increase in cell adhesion compared with their scrambled negative controls (mean ± SEM, 2.0 ± 0.6-fold, P < .05, and 2.1 ± 0.7-fold, P < .05, respectively). Together, the results obtained from these functional assays support an essential role of PomGnT1 in the proliferation, invasion, and adhesion of the GBM cells in vitro.
Regulation of PomGnT1 Is Dependent on EGFR/ERK Activity
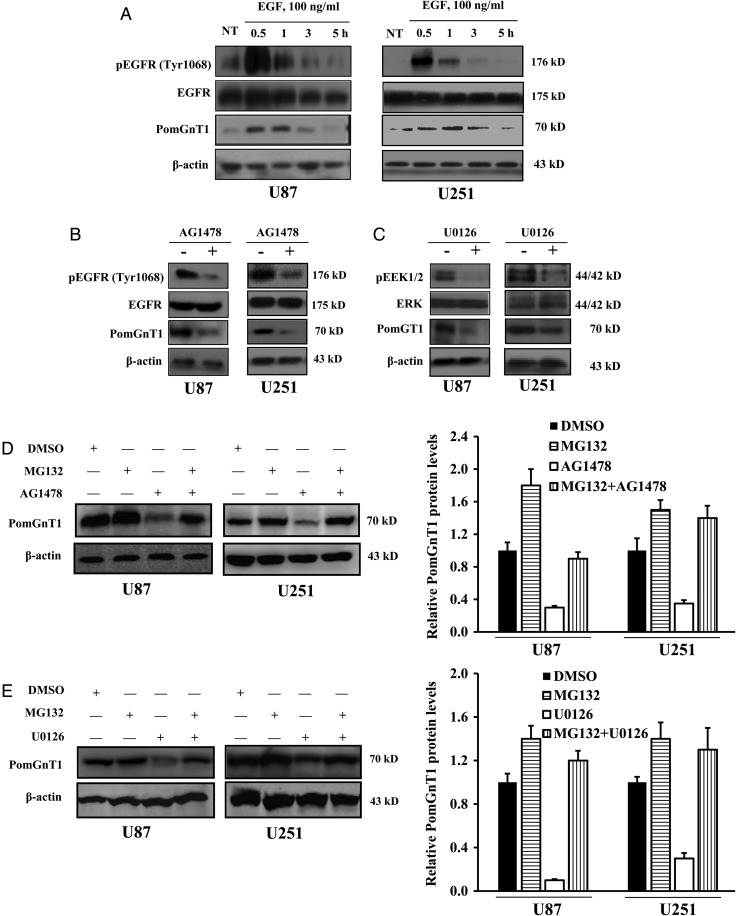
Epidermal growth factor receptor plays a prominent part in initiation and progression of many tumors, including GBM, in which it is overexpressed in >50% of cases.19 To determine whether signaling via this pathway could affect PomGnT1 expression, levels of phospho-EGFR and PomGnT1 were measured in parallel over a 5-h time course following treatment of serum-starved U87 and U251 cells with 100 ng/mL EGF. As shown in Fig. 4A, peak phosphorylation of EGFR on Tyr1068 was observed in both U87 and U251 cells at 30 min after the start of stimulation, followed by dephosphorylation of the protein back to baseline within 3 h. In concert with maximal phosphorylation of EGFR, a significant upregulation of PomGnT1 was detectable at 30 min and peaked at 60 min with relatively slow recovery to the steady state by 5 h. To further explore the role of EGFR signaling in the regulation of PomGnT1, U87 and U251 cells were pretreated with the EGFR inhibitor AG1478 for 30 min and then stimulated with 100 ng/mL EGF for 30 min. Figure 4B shows that AG1478 diminished EGFR phosphorylation and that this was accompanied by a reduction in PomGnT1 levels. Similar changes in the patterns of EGFR phosphorylation and PomGnT1 levels were observed with the inhibition of ERK, a direct EGFR downstream effector, by the ERK inhibitor U0126 (Fig. 4C). Because proteasome inhibition is well known to activate the EGFR/ERK signaling pathway,20–22 we extended our studies to determine whether PomGnT1 was induced by proteasome inhibition, and whether proteasome inhibition could counteract the downregulation of PomGnT1 induced by the EGFR or ERK inhibitor. We found that the proteasome inhibitor MG132 enhanced PomGnT1 expression in U87 and U251 cells in the absence of AG1478 and U0126 and that pretreatment with MG132 effectively prevented the reduction of PomGnT1 induced by EGFR or ERK inhibition (Fig. 4D and E). Furthermore, enhanced GBM cell proliferation driven by EGF stimulation could be effectively blocked by PomGnT1 suppression (Supplementary Fig. S1), indicating that PomGnT1 is one of the components downstream of the EGFR prosurvival signaling pathway. These observations identify EGFR/ERK pathway signaling as being capable of regulating PomGnT1 expression.
Fig. 4.
EGFR/ERK signaling mediates PomGnT1 expression. (A) EGFR phosphorylation and PomGnT1 expression in response to EGF stimulation. U87 and U251 cells were treated with 100 ng/mL EGF for 30 min and assessed for PomGnT1 levels and EGFR phosphorylation by western blot analysis at the indicated time points. NT, nontreated cells. (B and C) Western blot documenting a decrease in EGFR or ERK1/2 phosphorylation accompanied by a reduced PomGnT1 expression when cells were pretreated with either EGFR inhibitor AG1478 at 100 nM or ERK1/2 inhibitor U0126 at 100 nM for 30 min and then with 100 ng/mL EGF for 30 min. (D and E) Representative western blot showing that proteasome inhibitor MG132 counteracts the inhibitory effects of AG1478 or U0126 to increase PomGnT1 expression. The histogram shows the mean level of the PomGnT1 protein determined from 3 independent experiments expressed as the fold change relative to that in the vehicle control after normalization to β-actin. U87 and U251 cells were pretreated with 20 µM MG132 for 2 h, followed by 100 nM AG1478 or U0126 for 30 min. DMSO, dimethyl sulfoxide.
EGF Stimulation Suppresses RPTPβ Expression and Increases β-Catenin Activation
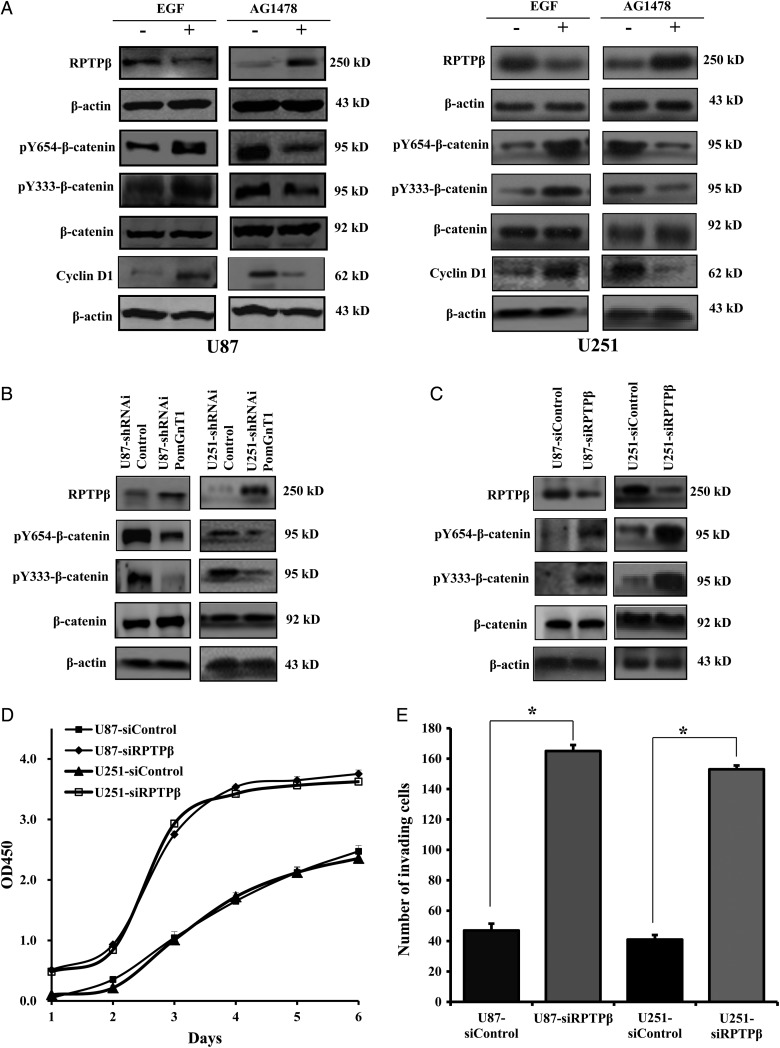
While O-mannosyl–linked glycosylation prevails in the central nervous system, very few glycoproteins with this glycan modification have been identified to date. A seminal report by Abbott and colleagues23 demonstrated that GnT-Vb mediates glycosylation of RPTPβ in neuroblastoma cells and inhibits RPTPβ intrinsic phosphatase activity, which resulted in high levels of phosphorylated β-catenin.23 We reasoned that PomGnT1 might inhibit RPTPβ expression and subsequently increase phosphorylated β-catenin in GBM cells as well. To test this hypothesis, U87 and U251 cells were treated with EGF or sequentially with the EGFR inhibitor AG1478, which had been shown capable of enhancing or reducing PomGnT1 expression, and then levels of RPTPβ, β-catenin phosphorylation, and cyclin D1 were assessed by western blot analysis. As shown in Fig. 5A, EGFR stimulation suppressed RPTPβ expression with a concomitant increase of phosphorylated β-catenin in both U87 and U251 cells. Conversely, inhibition of EGFR by AG1478 substantially elevated RPTPβ levels but reduced β-catenin phosphorylation at both of the tyrosine residues examined. Tyrosine phosphorylation of β-catenin has been implicated in its release from E-cadherin and correlates with enhanced transcriptional activity.24,25 In good agreement with this concept, we found that change in the expression of the known β-catenin–regulated target, cyclin D1, was accompanied by a similar change in the β-catenin phosphorylation level. To provide further evidence that PomGnT1 was involved in the EGF-induced changes in RPTPβ expression and β-catenin phosphorylation, we stably knocked down PomGnT1 and determined how this affected the ability of EGF to alter the levels of RPTPβ and the phosphorylated form of β-catenin. As shown in Fig. 5B, there was a clear induction in RPTPβ level but a decrease in the level of phosphorylated β-catenin in EGF-treated U87-shRNAi PomGnT1 and U251-shRNAi PomGnT1 cells. It is noteworthy that the increased RPTPβ protein level was accompanied by a proportional increase in the phosphatase activity of RPTPβ (Supplementary Fig. S2). To further document the negative regulatory effect of RPTPβ on β-catenin phosphorylation, endogenous RPTPβ was knocked down by an siRNA targeting to RPTPβ. Clearly, siRNA-mediated inhibition of RPTPβ in U87 and U251 cells resulted in much higher levels of phosphorylated β-catenin compared with their scrambled siRNA transfected controls (Fig. 5C), an effect likely contributing to the enhanced cell proliferative and invasive capability (Fig. 5D and E). Therefore, the findings from this series of experiments extend observations made in other cell lines and support the hypothesis that PomGnT1 acts as a regulator of RPTPβ that influences β-catenin signaling and cellular function.
Fig. 5.
PomGnT1 inhibits RPTPβ expression and induces β-catenin activation resulting in enhanced cell growth and invasion. (A) EGF stimulation suppressed RPTPβ expression but increased β-catenin phosphorylation at residues of Y654 and Y333 and enhanced cyclin D1 expression. (B) Knockdown of PomGnT1 induced RPTPβ expression but reduced the tyrosine phosphorylation of β-catenin. (C) Knockdown of RPTPβ increased the tyrosine phosphorylation of β-catenin. (D) Effect of RPTPβ knockdown on GBM cell proliferation. Cells were cultured for the indicated periods, and relative cell growth was determined by CCK-8 assay. OD, optical density. (E) Effect of RPTPβ knockdown on the invasion of GBM cells. The histogram indicates the number of invading cells counted from 5 randomly selected fields. Vertical bars, ±SEM, n = 3. *P < .05.
Discussion
The results of the studies reported here provide multiple lines of evidence that the expression of PomGnT1 influences the behavior of GBM. Our data mining indicates that high PomGnT1 expression in GBM tumors is associated with poor survival of the patients. The tissue microarray studies confirmed that GBM patients with high PomGnT1 expression in tumor samples had a poorer prognosis for OS than those with low PomGnT1 expression. In preclinical models, specific knockdown of PomGnT1 in GBM cell lines leads to strong inhibition of tumor growth and progression both in vitro and in vivo.
GBM is pathologically characterized by unclear boundaries within normal brain tissues, invasions into the surrounding normal tissue, and high rates of cell proliferation.26 Therefore, characterization of the invasive and proliferative properties of GBM cells is of fundamental importance to understanding the malignant development of GBM.27 A role of PomGnT1 in the control of cell adhesion and migration was initially highlighted in a transgenic mouse model where genetic disruption of PomGnT1 in mice led to failure of granule cells within the cerebellum from migration during development.28 Studies by Abbott et al7 confirmed that PomGnT1 played an essential role in neural cell migration, since suppression of PomGnT1 in human neuroblastoma cells significantly impaired cell migration. In keeping with this, we found that knockdown of PomGnT1 in U87 and U251 GBM cells diminished cell invasion and motility, whereas forced expression of PomGnT1 produced the opposite effects. It is noteworthy that an important addition from this study to current knowledge about the biological function of PomGnT1 is its previously unrecognized role in tumor cell proliferation. We observed that downregulation of PomGnT1 inhibited GBM cell proliferation both in vitro and in vivo, whereas PomGnT1 overexpression produced the opposite results. Thus, it is conceivable that PomGnT1 provides an invasive and proliferative advantage to GBM cells, and the development of an effective intervention targeting this molecule should improve the mortality rate and surgical opportunity of GBM patients. Taken together, the data from these PomGnT1 functional significance studies support our conclusions that PomGnT1 overexpression is correlated with GBM malignant progression and poor prognosis and might serve as a potential target to be exploited therapeutically in patients with GBM.
Activation of the EGFR pathway leads to phosphorylation of its downstream molecules, including the pathways of mitogen-activated protein kinase, Akt, and c-Jun NH(2)-terminal kinase, which are frequently implicated in cancer cell migration, protease induction, regulation of apoptosis, and angiogenesis.29,30 EGFR is a common molecular hallmark of GBM and promotes a pro-proliferative signal upon stimulation.31 Certain glycosyltransferases have been associated with EGFR activity and functioning probably through their ability to branch or modify glycans on EGFR. For instance, N-acetylgalactosaminyltransferase 2, GALNT2, the enzyme that mediates the initial step of mucin type-O glycosylation, is a critical mediator of malignant character in hepatocellular carcinoma that acts by modifying the activity of EGFR.32 Blocking N-glycosylation of EGFR in the absence of β1,4-galactosyltransferases inhibits EGFR cellular accumulation and EGFR-dependent signaling.33 N-acetylglucosaminyltransferase V has been shown to upregulate β-1-6-GlcNAc branched N-glycosylation of EGFR, leading to activation of EGFR as a potential novel upstream molecular event for p21-activating kinase 1–induced anoikis resistance in hepatoma cells.34 However, the molecular mechanisms remain poorly defined and no studies have examined an association of PomGnT1 with EGFR. In this study, we speculate that PomGnT1 may be one of the effector molecules downstream of EGFR signaling in the regulation of malignant behavior of GBM cells. We demonstrated that in response to EGF stimulation, induction and resolution of PomGnT1 expression was temporally followed with the levels of EGFR phosphorylation, and inhibition of EGFR signaling using either EGFR or ERK inhibitor reduced PomGnT1 expression. This observation suggests that expression of PomGnT1 was, at least in part, regulated by EGFR/ERK signaling. As a hallmark of tumor phenotype, altered glycosylation patterns are frequently detected during tumorigenesis and progression, and these tumor-associated glycans most often arise from changes in the expression levels of glycosyltransferases.35 However, it is unknown which signaling pathways are activated by PomGnT1 expression in GBM cells or how PomGnT1 links to these downstream pathways. A previous report suggests that RPTPβ is the predominant substrate of glycosyltransferase GnT-Vb in SH-SY5Y neuroblastoma cells, and its glycosylation by GnT-Vb inhibits RPTPβ phosphatase activity with the functional consequence of increased tyrosine phosphorylation of β-catenin resulting in decreased cell-cell adhesion and increased cell motility.23 In this study, we corroborate and extend these findings by demonstrating that EGF-induced PomGnT1 expression in GBM cells downregulates RPTPβ and increases tyrosine phosphorylation of β-catenin that drives the increased expression of cyclin D1. However, it should be noted that although a detailed mechanism at work is likely specific to given cell types, we could not exclude the possibility that GnT-Vb may also participate in the EGF-engaged signaling toward activation of β-catenin. GnT-Vb has been reported to activate EGFR signaling and promote cell migration via decreasing the protein level and oligosaccharide modification of RPTPκ.36 While the exact molecular mechanisms responsible for the PomGnT1-mediated malignant progression of GBM remains to be clarified and explored, it is tempting to suggest a model in which PomGnT1 functions as a key effector of EGFR-generated proliferative and invasive signals by increasing levels of the proteins related to cell proliferation, survival, and invasion through inhibition of RPTPβ and consequent activation of β-catenin. Therefore, a genetic approach or pharmacological intervention targeting PomGnT1 enzymatic activity or its interaction with downstream targets could potentially serve as an effective strategy to inhibit its oncogenic activity.
Supplementary Material
Funding
This work was supported by grants from the Shanghai Science and Technology Research Program (no. 13XD1402600) and the National Natural Science Foundation of China (no. 81372705) as well as from the State Key Laboratory of Oncogenes and Related Genes (no. 90-14-01).
Conflict of interest statement. The authors have no conflicts of interest to report.
Supplementary Material
References
- 1.Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205(12):613–621. doi: 10.1016/j.cancergen.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 3.Sawyers CL. The cancer biomarker problem. Nature. 2008;452(7187):548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 4.Sayegh ET, Kaur G, Bloch O, et al. Systematic review of protein biomarkers of invasive behavior in glioblastoma. Molec Neurobiol. 2013 doi: 10.1007/s12035-013-8593-5. 49(3):1212–1244. [DOI] [PubMed] [Google Scholar]
- 5.Stupp R, Hegi ME, Neyns B, et al. Phase I/IIa study of cilengitide and temozolomide with concomitant radiotherapy followed by cilengitide and temozolomide maintenance therapy in patients with newly diagnosed glioblastoma. J Clin Oncol. 2010;28(16):2712–2718. doi: 10.1200/JCO.2009.26.6650. [DOI] [PubMed] [Google Scholar]
- 6.Levin VA, Phuphanich S, Yung WK, et al. Randomized, double-blind, placebo-controlled trial of marimastat in glioblastoma multiforme patients following surgery and irradiation. J Neurooncol. 2006;78(3):295–302. doi: 10.1007/s11060-005-9098-5. [DOI] [PubMed] [Google Scholar]
- 7.Abbott KL, Troupe K, Lee I, et al. Integrin-dependent neuroblastoma cell adhesion and migration on laminin is regulated by expression levels of two enzymes in the O-mannosyl-linked glycosylation pathway, PomGnT1 and GnT-Vb. Exp Cell Res. 2006;312(15):2837–2850. doi: 10.1016/j.yexcr.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida A, Kobayashi K, Manya H, et al. Muscular dystrophy and neuronal migration disorder caused by mutations in a glycosyltransferase, POMGnT1. Dev Cell. 2001;1(5):717–724. doi: 10.1016/s1534-5807(01)00070-3. [DOI] [PubMed] [Google Scholar]
- 9.Beltran-Valero de Bernabe D, Currier S, Steinbrecher A, et al. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker-Warburg syndrome. Am J Hum Genet. 2002;71(5):1033–1043. doi: 10.1086/342975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi K, Nakahori Y, Miyake M, et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;394(6691):388–392. doi: 10.1038/28653. [DOI] [PubMed] [Google Scholar]
- 11.Endo T. O-mannosyl glycans in mammals. Biochim Biophys Acta. 1999;1473(1):237–246. doi: 10.1016/s0304-4165(99)00182-8. [DOI] [PubMed] [Google Scholar]
- 12.Lan J, Guo P, Chen M, et al. O-linked mannose beta-1,2-N-acetylglucosaminyltransferase 1 correlated with the malignancy in glioma. J Craniofac Surg. 2013;24(4):1441–1446. doi: 10.1097/SCS.0b013e318295378b. [DOI] [PubMed] [Google Scholar]
- 13.Bredel M, Scholtens DM, Yadav AK, et al. NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364(7):627–637. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catzavelos C, Bhattacharya N, Ung YC, et al. Decreased levels of the cell-cycle inhibitor p27Kip1 protein: prognostic implications in primary breast cancer. Nat Med. 1997;3(2):227–230. doi: 10.1038/nm0297-227. [DOI] [PubMed] [Google Scholar]
- 15.van Diest PJ, van Dam P, Henzen-Logmans SC, et al. A scoring system for immunohistochemical staining: consensus report of the task force for basic research of the EORTC-GCCG. European Organisation for Research and Treatment of Cancer–Gynaecological Cancer Cooperative Group. J Clin Pathol. 1997;50(10):801–804. doi: 10.1136/jcp.50.10.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu ZB, Cai L, Lin SJ, et al. High-mobility group box 2 is associated with prognosis of glioblastoma by promoting cell viability, invasion, and chemotherapeutic resistance. Neuro Oncol. 2013;15(9):1264–1275. doi: 10.1093/neuonc/not078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang X, Lin X, Alvarez E, et al. Tight junction proteins claudin-3 and claudin-4 control tumor growth and metastases. Neoplasia. 2012;14(10):974–985. doi: 10.1593/neo.12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen Y, Wang Y, Sheng K, et al. Serine/threonine protein phosphatase 6 modulates the radiation sensitivity of glioblastoma. Cell Death Dis. 2011;2:e241. doi: 10.1038/cddis.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan QW, Cheng CK, Gustafson WC, et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell. 2013;24(4):438–449. doi: 10.1016/j.ccr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ying WZ, Zhang HG, Sanders PW. EGF receptor activity modulates apoptosis induced by inhibition of the proteasome of vascular smooth muscle cells. J Am Soc Nephrol. 2007;18(1):131–142. doi: 10.1681/ASN.2006040333. [DOI] [PubMed] [Google Scholar]
- 21.Sloss CM, Wang F, Liu R, et al. Proteasome inhibition activates epidermal growth factor receptor (EGFR) and EGFR-independent mitogenic kinase signaling pathways in pancreatic cancer cells. Clin Cancer Res. 2008;14(16):5116–5123. doi: 10.1158/1078-0432.CCR-07-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sloss CM, Wang F, Palladino MA, et al. Activation of EGFR by proteasome inhibition requires HB-EGF in pancreatic cancer cells. Oncogene. 2010;29(21):3146–3152. doi: 10.1038/onc.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbott KL, Matthews RT, Pierce M. Receptor tyrosine phosphatase beta (RPTPbeta) activity and signaling are attenuated by glycosylation and subsequent cell surface galectin-1 binding. J Biol Chem. 2008;283(48):33026–33035. doi: 10.1074/jbc.M803646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simoneau M, Coulombe G, Vandal G, et al. SHP-1 inhibits beta-catenin function by inducing its degradation and interfering with its association with TATA-binding protein. Cell Signal. 2011;23(1):269–279. doi: 10.1016/j.cellsig.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Yang W, Xia Y, Ji H, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480(7375):118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homma T, Fukushima T, Vaccarella S, et al. Correlation among pathology, genotype, and patient outcomes in glioblastoma. J Neuropathol Exp Neurol. 2006;65(9):846–854. doi: 10.1097/01.jnen.0000235118.75182.94. [DOI] [PubMed] [Google Scholar]
- 27.Bigner SH, Wong AJ, Mark J, et al. Relationship between gene amplification and chromosomal deviations in malignant human gliomas. Cancer Genet Cytogenet. 1987;29(1):165–170. doi: 10.1016/0165-4608(87)90045-8. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Ball SL, Yang Y, et al. A genetic model for muscle-eye-brain disease in mice lacking protein O-mannose 1,2-N-acetylglucosaminyltransferase (POMGnT1) Mech Dev. 2006;123(3):228–240. doi: 10.1016/j.mod.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Reddy KB, Nabha SM, Atanaskova N. Role of MAP kinase in tumor progression and invasion. Cancer Metastasis Rev. 2003;22(4):395–403. doi: 10.1023/a:1023781114568. [DOI] [PubMed] [Google Scholar]
- 30.She QB, Solit DB, Ye Q, et al. The BAD protein integrates survival signaling by EGFR/MAPK and PI3 K/Akt kinase pathways in PTEN-deficient tumor cells. Cancer Cell. 2005;8(4):287–297. doi: 10.1016/j.ccr.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNamara MG, Sahebjam S, Mason WP. Emerging biomarkers in glioblastoma. Cancers. 2013;5(3):1103–1119. doi: 10.3390/cancers5031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu YM, Liu CH, Hu RH, et al. Mucin glycosylating enzyme GALNT2 regulates the malignant character of hepatocellular carcinoma by modifying the EGF receptor. Cancer Res. 2011;71(23):7270–7279. doi: 10.1158/0008-5472.CAN-11-1161. [DOI] [PubMed] [Google Scholar]
- 33.Gabius HJ, van de Wouwer M, Andre S, et al. Down-regulation of the epidermal growth factor receptor by altering N-glycosylation: emerging role of beta1,4-galactosyltransferases. Anticancer Res. 2012;32(5):1565–1572. [PubMed] [Google Scholar]
- 34.Liu J, Liu H, Zhang W, et al. N-acetylglucosaminyltransferase V confers hepatoma cells with resistance to anoikis through EGFR/PAK1 activation. Glycobiol. 2013;23(9):1097–1109. doi: 10.1093/glycob/cwt049. [DOI] [PubMed] [Google Scholar]
- 35.Dube DH, Bertozzi CR. Glycans in cancer and inflammation—potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4(6):477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 36.Wang C, Yang Y, Yang Z, et al. EGF-mediated migration signaling activated by N-acetylglucosaminyltransferase-V via receptor protein tyrosine phosphatase kappa. Arch Biochem Biophys. 2009;486(1):64–72. doi: 10.1016/j.abb.2009.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.