Abstract
Background
The current therapy for glioblastoma multiforme (GBM), the most aggressive and common primary brain tumor of adults, involves surgery and a combined radiochemotherapy that controls tumor progression only for a limited time window. Therefore, the identification of new molecular targets is highly necessary. Inhibition of kinases has become a standard of clinical oncology, and thus the oncogenic kinase Pim1 might represent a promising target for improvement of GBM therapy.
Methods
Expression of Pim1 and associated signaling molecules was analyzed in human GBM samples, and the potential role of this kinase in patients' prognosis was evaluated. Furthermore, we analyzed the in vivo role of Pim1 in GBM cell growth in an orthotopic mouse model and examined the consequences of Pim1 inhibition in vitro to clarify underlying pathways.
Results
In comparison with normal brain, a strong upregulation of Pim1 was demonstrated in human GBM samples. Notably, patients with short overall survival showed a significantly higher Pim1 expression compared with GBM patients who lived longer than the median. In vitro experiments with GBM cells and analysis of patients' GBM samples suggest that Pim1 regulation is dependent on epidermal growth factor receptor. Furthermore, inhibition of Pim1 resulted in reduced cell viability accompanied by decreased cell numbers and increased apoptotic cells, as seen by elevated subG1 cell contents and caspase-3 and -9 activation, as well as modulation of several cell cycle or apoptosis regulatory proteins.
Conclusions
Altogether, Pim1 could be a novel therapeutic target, which should be further analyzed to improve the outcome of patients with aggressive GBM.
Keywords: epidermal growth factor receptor, glioblastoma multiforme, pim1 kinase
Glioblastoma multiforme (GBM) represents the most common primary tumor of the central nervous system in adults. This highly aggressive tumor (World Health Organization [WHO] grade IV) is characterized by a poor prognosis, which is caused by exponential growth and diffuse invasiveness into the adjacent brain parenchyma. A large number of genetic alterations, including phosphatase and tensin homolog (PTEN) and epidermal growth factor receptor (EGFR), have been detected in GBM, and their impact as targets for new therapeutic strategies have been thoroughly investigated. No groundbreaking success or significant prolongation of median survival to more than 12–15 months has so far been achieved.1,2 However, inhibition of protein kinases has become a standard of modern clinical oncology, and this targeted therapy could possibly improve the survival of glioblastoma patients.3 During the last years, the serine/threonine kinase Pim1 has emerged as a relevant target in the field of cancer research. Primarily, Pim1 was discovered in murine lymphoma samples and was shown to be activated through integration of the murine leukemia virus into the 3′-untranslated region of the Pim1 gene, resulting in increased Pim1 expression.4 Since overexpression of Pim1 induced lymphomas in transgenic mice, Pim1 is considered to function as an oncogene.5 In contrast to other kinases, such as Akt1 and extracellular signal-regulated kinase (ERK)1/2, Pim1 is constitutively active after translation without required phosphorylation by upstream kinases and is both transcriptionally and translationally regulated.6–8 Two Pim1 isoforms were described, a short 34-kDa isoform (Pim1S) and a long 44-kDa isoform (Pim1L), which differ in their translational starting points and cellular localizations.9 An important function of Pim1 is the induction of cell cycle progression by phosphorylating numerous targets such as the cell cycle activating phosphatases cell division cycle Cdc25A and C, which are activated by Pim1, or the cell cycle inhibitor p21, which is inhibited by Pim1 phosphorylation.10–12 Additionally, Pim1 inhibits apoptosis by phosphorylating the pro-apoptotic Bcl-2 associated death promoter (Bad) and the pro-apoptotic transcription factor FoxO3a.13,14
Besides a role of Pim1 in the pathogenesis of lymphoma, an increased expression of Pim1 was observed in some other cancer entities, but the prognostic consequence seems to be inconsistent. In prostate cancer, elevated Pim1 expression is accompanied by a good prognosis.15,16 A similar association between Pim1 expression and patients' prognosis was seen in pancreatic and non–small cell lung cancers.17,18 In contrast, increased Pim1 expression in stomach, esophagus, bladder, and head and neck cancers results in a poor prognosis and a reduced response to antitumor therapy.16,19–21
To date, no systematic studies of expression and functional characterization of Pim1 in GBM have been conducted, but it was recently shown that Pim1 expression is under the control of EGFR,22 which is often overexpressed in GBM.1 Therefore, we analyzed the expression of Pim1 and associated signaling molecules (c-myc, Akt1, EGFR) in GBM cell lines and samples of GBM patients in comparison with nonmalignant brain tissue and evaluated the potential role of Pim1 in GBM patients' prognosis. Furthermore, the impact of pharmacological Pim1 inhibition on GBM cell survival was studied in vitro and in vivo to determine underlying signaling pathways and to assess Pim1 as a therapeutic target.
Materials and Methods
Patients’ Samples
Following an institutional review board–approved protocol (in accordance with the ethical standards of the Helsinki Declaration of the World Medical Association), fresh human GBM tissues were obtained from 75 patients who underwent surgical removal of GBM within their therapeutic regime. The 75 glioblastomas consisted of 72 primary and 3 secondary GBM. Further, 39 samples of tumor relapses were investigated, and the recurrence of the tumor was specified by radiological appearance and particularly by histological classification by a neuropathologist. Additionally, we analyzed 14 low-grade astrocytomas (10 astrocytomas WHO grade II or III, 1 pilocytic astrocytoma grade I, and 3 astrocytomas with oligodendroglial components). Vital status was available for 72 of 75 analyzed GBM patients. Among them were 47 males (65.3%) and 25 females (34.7%). At the end of the study period (October 15, 2007 to February 11, 2013, the last update of the vital status), 62 patients were deceased (86.1%) and 10 were alive (13.9%). Gender was not associated with significant differences in the patients' outcome. The histological analyses are based on the WHO criteria.23 Eight nonneoplastic brain tissues (frontal/temporal lobes) of donor specimens were kindly provided by the Institute of Neuropathology of the University Greifswald. Brain tissues of these control cases were obtained by routine autopsy. After the brain was removed, tissue samples were cut and frozen at −80°C immediately. The autopsy cases died of pneumonia, heart failure, sepsis, or carcinoma of the pancreas. There were no neurological disorders. Further, protein as well as RNA of 2 nonmalignant specimens (1 frontal and 1 temporal lobe) were obtained from BioChain Institute. For assessment of overall survival (OS), we made attempts to obtain information for all patients with glioblastoma.
Cell Culture
Cell lines used for this study were A172, GaMG, HF66, LN18, U87MG, U251MG, U373 (human glioblastoma cell lines), GL261 and green fluorescent protein–transfected GL261 (murine glioblastoma cell lines), HepG2 (human hepatocellular carcinoma cell line), Caco2 (human colon carcinoma cell line), the human embryonal kidney (HEK)293 cell line, and HeLa (human cervix carcinoma cell line), obtained from either the American Type Culture Collection or the German Collection of Cell Cultures, except the murine GL261 cells (kindly provided by Dr M. Synowitz) and A172, GaMG, HF66, U251MG, and U373 (kindly provided by Dr H. Miletic). A172, GaMG, HF66, LN18, GL261, Caco2, HEK293, U251MG, and U373 cells were cultured in Dulbecco's modified Eagle's medium, U87MG cells in Minimum Essential Medium, and HeLa cells in Roswell Park Memorial Institute 1640 medium; all media were supplemented with 10% fetal calf serum (FCS) and 2 mM glutamine. Cells were maintained at 37°C under 5% CO2. All cell culture materials were from PAA Laboratories. Reagents for cell culture experiments were as follows: LY294002, quercetagetin (QT), allylgeldanamycin (AGA) (all Calbiochem), TCS Pim1-1 (Tocris Biosciences), temozolomide (Sigma Aldrich), EGF (Invitrogen), AG1478 (Calbiochem ), and PD98059 (Calbiochem). For some experiments, cells were serum starved (0.05% FCS) for 24 h followed by incubation with various concentrations of LY294002, QT, AGA, or TCS Pim1-1.
The tumor material from patients P3, P6, P8, P17, and P22 was derived from passaged xenografts as described previously.24,25 P28 and 421k cells were derived from a GBM patient biopsy, and cells were cultured in neural basal medium with B27 supplement, Glutamax (Life Technologies), and fibroblast growth factor 2 (20 ng/mL).
Quantitative Real-time PCR Analysis
For mRNA expression analysis, total RNA was isolated using PeqGold RNAPure (PeqLab) and reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturers' protocols. The expression of the specific genes was analyzed by the following gene expression assays on demand from Applied Biosystems and normalized to 18S rRNA as the internal reference: Pim1, Hs01065498_m1; Akt1, Hs00178289_m1; c-myc, Hs00153408_m1; EGFR, Hs01076078_m1; and eukaryotic 18S rRNA endogenous control, 4310893E. Quantitative real-time PCR was performed in a 7900 HT Fast Real-Time PCR system from Applied Biosystems. Each mRNA level was normalized to 18S rRNA and was expressed relative to the mean value of all control samples (nonmalignant brain tissue) or relative to solvent control, which both were given a value of 1.
Western Blot Analysis
For preparation of cell lysates, the following lysis buffer was used: 50 mM Tris-HCl pH 7.4, 100 mM NaCl, 0.1% Triton X-100, 5 mM EDTA containing protease/phosphatase inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 mM leupeptin, 1 mM aprotinin, and 250 μg/mL sodium vanadate). Briefly, cells were centrifuged at 6000 rpm, and the resulting pellet was dissolved in the lysis buffer and incubated on ice for 45 min. Protein extracts of patients' glioblastoma samples were prepared using the Qiagen TissueLyser II. The nitrogen-cooled tumor sample (∼20–30 mg) was shredded for 90 s at a frequency of 30 Hz. The resulting tissue powder was dissolved immediately in precooled lysis buffer and incubated on ice for 45 min followed by a centrifugation step at 6000 rpm to detach cell debris. Protein concentrations were analyzed using the BCA Protein Assay Kit (Thermo Fisher Scientific). Subsequently, after denaturation in Laemmli buffer at 95°C for 5 min, 40 µg of each sample was separated on sodium dodecyl sulfate polyacrylamide gels of different concentrations (7.5%–12.5%) according to the molecular weights of the respective proteins. Immunoblotting to Whatman nitrocellulose membrane was performed with the tank blot system (Bio-Rad). The membrane was blocked in 5% FCS or skim milk in Tris-buffered saline containing 0.05% Tween 20 (TBST) and 1% bovine serum albumin for 1 h at room temperature. The following primary antibodies were diluted in TBST and 0.05% sodium azide and incubated either for 2 h at room temperature or overnight at 4°C: monoclonal rabbit anti-Pim1 (for Pim1S; Epitomics), monoclonal rabbit anti-Pim1 (for Pim1L; Bioworld Technology), monoclonal mouse anti-phosphorylated Akt1 (Ser473; Millipore), monoclonal mouse anti-phosphorylated EGFR (Tyr1173; Millipore), monoclonal rabbit anti-PTEN, monoclonal rabbit anti-phosphorylated Bad (Ser112), monoclonal rabbit anti-p21, monoclonal rabbit anti-p27, monoclonal rabbit anti–B-cell lymphoma extra large (Bcl-xL; all Cell Signaling Technology), polyclonal goat anti-p53, polyclonal rabbit anti-phosphorylated Myt1 (Ser83), polyclonal rabbit anti-phosphorylated cyclin-dependent kinase 2 (Cdc2; Thr14, Tyr15), polyclonal goat anti-actin, polyclonal rabbit anti-Bak (all Santa Cruz Biotechnology), polyclonal rabbit anti-phosphorylated Wee1 (Ser53; Abnova), monoclonal mouse anti–X-linked inhibitor of apoptosis protein (XIAP) (BD Biosciences), and monoclonal mouse anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Biodesign). The secondary horseradish peroxidase–conjugated goat anti-rabbit, goat anti-mouse, or horse anti-goat IgG antibodies (Vector Laboratories) were used at a 1:2000 dilution for 1.5 h at room temperature. Chemiluminescence signals were detected with the ChemiDoc XRS Imaging System (Bio-Rad) using ECL Plus Western Blotting Substrate (Thermo Scientific) followed by densitometric analysis (Quantity One, Bio-Rad). The relative optical densities of the specific bands were calculated and normalized to GAPDH or β-actin as loading controls.
PCR for Detection of EGFR Wild-type or Deletion Mutant Variant III
To analyze the expression of EGFR wild type (EGFRwt) and the presence of the deletion variant III (EGFRvIII), 2 µL of patients' samples cDNA were amplified using the following primers in a conventional PCR26: forward primer 5′-GGGCTCTGGAGGAAAAGAAA-3′ and reverse primer 5′-AGGCCCTTCGCACTTCTTAC-3′, which span exon 1 to exon 8, so that both EGFRwt and EGFRvIII could be detected. The amplification conditions were as follows: 95°C for 2 min, 32 cycles including 95°C for 30 s, 55°C for 45 s, 72°C for 30 s, and 10 min at 72°C for final elongation. The amplification products were separated by agarose gel electrophoresis.
Immunofluorescence Microscopy
For detection of the cellular localization of Pim1 in GBM cell lines and patients’ samples, we performed immunofluorescence microscopy using the Zeiss LSM780. For detection of the cellular localization of Pim1 in GBM cell lines, we cultured LN18 cells on coverslips at a density of 50 000 cells/well in a 12-well multiplate in 1 mL culture medium for 24 h. After aspiration of the media and 3 washing steps with phosphate buffered saline (PBS), cells were fixed in 4% paraformaldehyde for 10 min, rinsed with PBS 3 times, and then permeabilized with 1% Triton X-100 for 10 min. Afterward, cells were blocked in 5% FCS for 1 h, and the primary antibodies were incubated at 4°C overnight in blocking solution in the following concentrations: monoclonal rabbit anti-Pim1, 1:150 (for Pim1S); monoclonal rabbit anti-Pim1, 1:50 (for Pim1L); monoclonal mouse anti-phosphorylated Akt1, 1:50 (Ser473); and monoclonal mouse anti-phosphorylated EGFR, 1:50 (Tyr1173). The next day cells were washed 3 times with PBS, and the secondary antibodies goat anti-mouse-568 and chicken anti-rabbit-488 (both Alexa Fluor, Invitrogen) were incubated for 2 h at room temperature in a 1:200 dilution. Afterward cells were washed with PBS 3 times followed by staining of the nuclei with 4′,6′-diamidino-2-phenylindole (DAPI) for 10 min. Cells were embedded in Dako Fluorescence Mounting Medium and dried flat overnight before being examined at the Zeiss LSM780.
The human patients' samples were embedded in Jung Tissue Freezing Medium (Leica Microsystems), and ultrathin sections (5–7 µm) were sliced with a Feather microtome blade (Type A35) on a Leica CM1900 cryostat. The sections were transferred to microscope slides and stored at −80°C until use. Apart from fixing in 70% ethanol, the sections were stained as described above.
Small Interfering RNA Mediated Silencing of Pim1
LN18 cells were transfected using the Lipofectamine 2000 reagent protocol (Invitrogen) according to the manufacturer's instructions in either a 24-well plate for RNA analysis or a 96-well plate for cell viability analysis. Pim1 small interfering (si)RNA and control siRNA (each from Santa Cruz Biotechnology or OriGene Technologies) were used at a final concentration of 5 pmol. To optimize the knockdown effectivity, the whole procedure was repeated 24 h after the first application of siRNA. RNA knockdown was tested 48 h posttransfection using quantitative real-time PCR. Cell viability analysis was performed 48 and 72 h after transfection using resazurin (PromoCell) as described below.
Cell Viability Analysis
Cells were seeded at 5000 cells/well in 96-well multiplates in 150 µL culture medium. The growth medium was changed 1 day later and cells were incubated for different time points (48, 72, and 96 h) with fresh medium containing the indicated inhibitors. After the respective incubation periods, medium was removed and replaced by fresh medium containing 10% resazurin, and cells were incubated for 2 further hours at 37°C. The plate was read on a fluorescence plate reader (excitation, 530 nm; emission, 590 nm) (Tecan Infinite M200). Data were calculated as percentage of cell viability of dimethyl sulfoxide (DMSO)–treated control cells.
Crystal Violet Staining
Cells were seeded at 5000 cells/well in 96-well multiplates. The growth medium was changed 1 day later and cells were incubated for different time points (48, 72, and 96 h) with fresh medium containing the indicated inhibitors. After the respective incubation periods, medium was removed and cells were washed once with PBS followed by a 15-min fixation step in 4% paraformaldehyde. Afterward, fixed cells were washed 3 times with PBS, and crystal violet solution was added. Ten minutes later, crystal violet was removed and cells were washed several times with aqua dest. Finally, crystal violet–stained cells were redissolved in 1% sodium dodecyl sulfate solution and colorimetrically measured at 560 nm.
Cell Cycle Analysis
Determination of cell cycle status was performed using propidium iodide staining of DNA in fixed cells. Briefly, treated cells were harvested and pooled with floating cells. After centrifugation at 3000 rpm for 5 min, cells were washed once with PBS and fixed in ethanol overnight at 4°C. Afterward, fixed cells were centrifuged for 5 min at 2000 rpm and washed 2 times with PBS, including resuspension of cells followed by a centrifugation step at 2000 rpm for 5 min. Pellets were resuspended in 500 µL staining buffer containing 1% glucose, 2 µg/mL RNase A, and 50 µg/mL propidium iodide and incubated for 30 min in the dark. Measurement of propidium iodide–incorporated DNA was analyzed with a FACSCalibur flow cytometer (BD Biosciences) at the FL-2 channel, and the cell fractions in the respective cell cycle phases were expressed as percentages of the total cell population.
Assays for Caspase-3 and -9
Caspase-3 and -9 activities were measured by the use of commercially available colorimetric assay kits from R&D Systems according to the manufacturer's instructions. Data were normalized to the respective protein contents and expressed as percentage of DMSO-treated control cells.
Analysis of Cell Migration
To analyze the effect of Pim1 inhibition on glioma cell migration, we used 2 different techniques: the scratch wound healing assay and the transwell migration assay with a Boyden chamber. Briefly, for the wound healing assay, 1.5*105 LN18 cells were seeded per cavity of a 48-well plate until confluence was reached. Using a yellow pipette tip, a wound was set in the middle of the confluent cell layer. After washing with PBS to remove detached cells and application of 5 mM hydroxy urea as proliferation inhibitor, the wound was imaged by the PALM RoboSoftware of an AxioVision HXP120C microscope (Zeiss), and the exact position of the image was saved to analyze the same region after the respective incubation times. After a pre-incubation time of 1 h with 5 mM hydroxy urea, cells were treated with the different inhibitors for 16 h followed by the analysis of the wound width.
Additionally, we performed a transwell migration assay using a Boyden chamber (NeuroProbe). Shortly after reaching near confluence, cells were cultured in FCS-free media for 24 h followed by trypsinization and seeding of 5*104 cells/50 µL FCS-free media into the upper well of the Boyden chamber, whereas the bottom chamber was filled with media containing 10% FCS as stimulus for migration. A polycarbonate membrane with a pore size of 8 µm (Whatman) was located between the upper and the bottom chamber. After treatment of cells with the different kinase inhibitors for 3 h, cells were fixed on the membrane with 4% paraformaldehyde, stained with crystal violet solution for 30 min, and counted using the cell counting tool of ImageJ.
Orthotopic In vivo Mouse Model
For the analysis of Pim1 as a suitable therapeutic target, we used the murine glioblastoma cell line GL261, which was stereotactically implanted into the brain of C57BL/6N mice. Animal experiments were carried out in compliance with the German laws on animal welfare, and the animal protocols were approved by the Landesveterinär- und Lebensmitteluntersuchungsamt Mecklenburg-Vorpommern. Female wild-type C57BL/6 mice (Charles River Breeding Laboratories) were kept with a 12-h light/dark cycle and received food and water ad libitum. At the age of 12 weeks, anesthetized mice were immobilized and placed into a stereotactic head holder (David Kopf Instruments) in the flatskull position. Approximately 1 mm anterior and 2 mm lateral to bregma, a 2-μL 30-gauge gastight syringe (Hamilton) was inserted to a depth of 3.5 mm and retracted to a depth of 3 mm from the dural surface, and 1 μL (2.5 × 104 cells/μL) of GL261 cells in PBS was injected. After 3 min the syringe was retracted to a depth of 2 mm, and again 1 µL (2.5 × 104 cells/μL) of GL261 cells was injected. After 3 min the syringe was removed, and the wound was sewn with surgical fiber and fixed with liquid patch. Twelve days postinjection, tumor development was assessed in isoflurane-anesthetized mice by magnetic resonance tomography (MRT; 7 Tesla MR Bruker ClinScan imaging scanner) to start the treatment at a tumor volume >1.5 mm3. Standard anatomic MRI sequences consisted of T2-weighted turbo spin echo images (coronar) and gadobutrol (Gadovist, 1 mmol/mL; Bayer HealthCare Pharmaceuticals)-enhanced T1-weighted images (coronar and axial). Tumor volume was calculated with OsiriX software in coronar and axial gadolinium-enhanced T1 sections. The mice were treated with either 5% dextrose (control animals, n = 6) or 75 mg/kg TCS (n = 6) as a Pim1 inhibitor every second day by oral gavage until the end of the study (12 days of treatment). Weight and physical condition were controlled every day, and tumor size was measured using MRT 12 days after starting the treatment.
Statistical Analysis
Statistical analyses were performed with GraphPad Prism 5.0. Data of in vitro analyses represent 3 or 4 independent experiments (as indicated in the figure legends and shown as mean ± SD). Box plots of data of patients' samples are shown as the median and the 5th and 95th percentiles. Pairwise comparisons were performed using Student’s t-test or a 2-tailed Mann–Whitney U test. For comparison of frequency data, Fisher's exact test was used. More than 2 groups were compared by Wilcoxon rank sum test or ANOVA and corrected for multiple testing. Additionally, nonlinear regression analysis and the Wilcoxon signed-rank test were used for determination of half-maximal inhibitory concentration values and comparison between 2 groups, respectively. Correlations between expressions of the investigated genes were analyzed by Spearman's nonparametric correlation. The duration of a patient's OS was defined as the time from the first tumor detection until death. Information on vital status and date of death were obtained from official population registry. Based on gene expression levels, Kaplan–Meier survival functions were calculated and compared with a log-rank test using Intercooled Stata/SE 10.1 software. Glioblastoma cases were divided into the lower half versus the upper half of gene expression level as determined by real-time PCR. Statistical significances were defined as P < .05, P < .01, and P < .001.
Results
Clinicopathological Features of the Analyzed Patients
Clinicopathological features of all analyzed patients with GBM are summarized in Table 1. Vital status was available for 72 of 75 analyzed GBM patients. At the end of the study period (see above), 62 patients were deceased (86.1%) and 10 were alive (13.9%). Gender was not associated with significant differences in the patients' outcomes. Median OS of the GBM cohort was 289 days (range, 33–1116 d). The patients who lived longer than the median OS were significantly younger (median, 57 y) at the date of diagnosis compared with the subgroup with a survival time below the median OS (median, 70 y). Resection grade was significantly associated with the outcome of the GBM patients, that is, in the group with total resection more patients lived longer than the median OS (62.9%) compared with patients with a subtotal resection (30.8%). Concerning the therapy, we divided the GBM cohort into patients receiving temozolomide (68.1%) and patients without temozolomide therapy (25.0%). No therapy data were available from 5 GBM patients (6.9%). In the subgroup of GBM patients with temozolomide therapy, the proportion of patients who lived longer than the median OS (73.7%) was significantly higher compared with only 1 patient with a survival time above the median OS without temozolomide therapy (5.6%).
Table 1.
Clinicopathological features of the analyzed patients
| All Analyzed Patients | Patients by OS |
P | ||
|---|---|---|---|---|
| ≤Median (n = 31) | >Median (n = 30) | |||
| Age, y | ||||
| Median (q1;q3) | 65.5 (54;71) | 70 (63;75) | 57 (53;68) | .002 |
| Gender, n (%) | ||||
| Men | 47 (65.3) | 21(50.0) | 21 (50.0) | |
| Women | 25 (34.7) | 10 (52.6) | 9 (47.4) | 1 |
| Resection grade, n (%) | ||||
| Total | 41 (56.9) | 13 (37.1) | 22 (62.9) | |
| Nontotal | 31(43.1) | 18 (69.2) | 8 (30.8) | .02 |
| Therapy, n (%) | ||||
| With temozolomide | 49 (68.1) | 10 (26.3) | 28 (73.7) | |
| Without temozolomide | 18 (25.0) | 17 (94.4) | 1 (5.6) | .02 |
| Unknown | 5 (6.9) | 4 (80.0) | 1 (20.0) | |
| Vital status, n (%) | ||||
| Deceased | 62 (86.1) | |||
| Alive | 10 (13.9) | |||
*For 1 patient date of death unknown.
Expression of Pim1 in Glioma Cell Lines, Patient-Derived Lines, and Xenografts
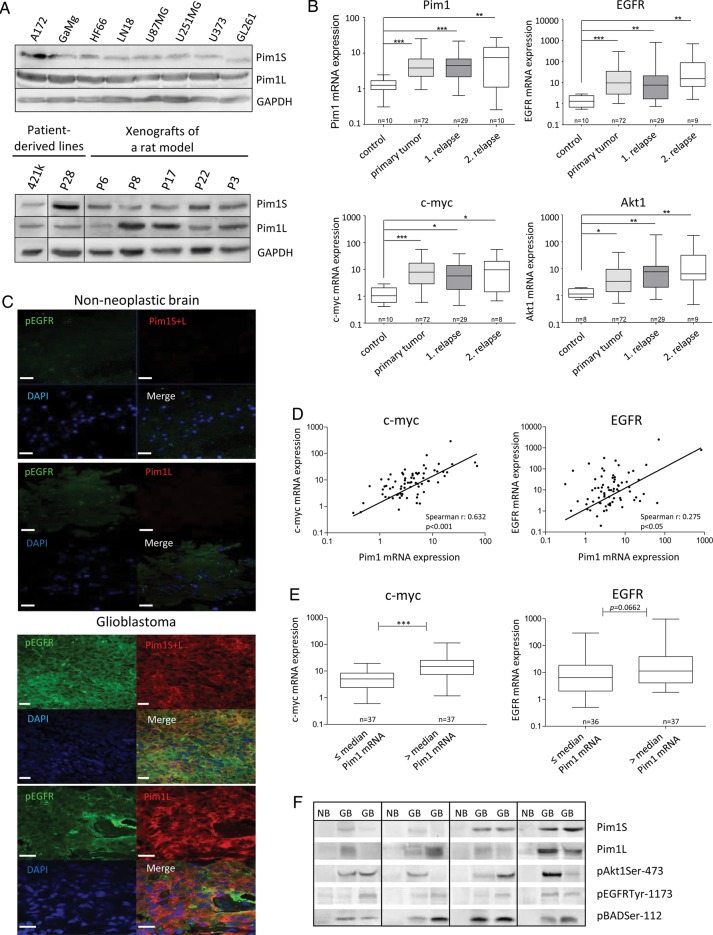
With regard to testing pharmacological inhibitors in vitro, we first analyzed the expression of Pim1 at the protein level in different glioma cell lines (Fig. 1A). The short Pim1 isoform (Pim1S) was identified at 34 kDa and the long isoform (Pim1L) at 44 kDa. Both Pim1 isoforms as well as the expression of the kinase Akt1 were detectable in all analyzed glioma cell lines. Also, phosphorylated Bad (pBad), as one of the substrates of Pim1, was detected in all glioma cell lines. The murine glioma cell line GL261 showed the highest phosphorylation of EGFR followed by the human cell lines U251MG, LN18, and A172. Further, all analyzed glioma cell lines expressed heat shock protein (HSP)90, which is necessary for protein stabilization of Pim1. Screening of the Gene Expression Omnibus (GEO) Profiles of the National Center for Biotechnology Information (NCBI) for Pim1 expression in different human glioma cell lines demonstrated that all tested cell lines expressed Pim1 but to variable extents (Supplementary Fig. S1A). The human cell lines used in this study for in vitro experiments (U87MG and LN18) are marked by an arrow showing a higher Pim1 expression in the glioblastoma cell lines (LN18, U87MG, and GL261) compared with cells of other tumor entities (Supplementary Fig. S1B). Additionally, all analyzed patient-derived glioma lines and glioma xenografts of rats showed expression of both Pim1S and Pim1L but at variable intensities.
Fig. 1.
Expression of Pim1, Akt1, EGFR, Bad, and HSP90 in GBM tissue and GBM cell lines. (A) Protein expression of Pim1S, Pim1L, HSP90, Akt, phosphorylated Bad (pBADSer112), and phosphorylated EGFR (pEGFRTyr1173) in various human and murine GBM cell lines (upper panel), patient-derived glioma lines, and xenografts of a rat (lower panel) analyzed by immunoblotting. GAPDH was used as loading control. (B) Relative mRNA expression levels of Pim1, Akt1, EGFR, and c-myc in frontal/temporal lobes of nonneoplastic brain (control) and glioblastoma patients' samples (GBM: primary tumor vs first and second relapse) analyzed by quantitative real-time PCR with normalization to 18S rRNA and shown as box plots representing the median as horizontal bars as well as the 5th and 95th percentiles. (C) Cryosections of nonneoplastic brain and glioblastoma tissue were immunostained with antibodies against the respective proteins. Nuclei are counterstained by the use of DAPI. Note that Pim1S, Pim1L, and pEGFR are strongly stained in glioblastoma, whereas no or little staining is observed in nonneoplastic brain tissue (frontal/temporal lobes). Scale bar represents 20 µm. (D, left) Illustration of a significant correlation between Pim1 and c-myc gene expression determined by Spearman's nonparametric correlation in 72 GBM samples (r = 0.63, P < .0001). (D, right) Illustration of a significant correlation between Pim1 and EGFR gene expression determined by Spearman's nonparametric correlation in 72 GBM samples (r = 0.28, P = .0195). (E) Expression of c-myc and EGFR in GBM dependent on the Pim1 mRNA level (Pim1 mRNA ≤median vs >median expression) analyzed by quantitative real-time PCR with normalization to 18S rRNA and shown as box plots representing the median as horizontal bars as well as the 5th and 95th percentiles. *P < .05, **P < .005, ***P < .001. (F) Representative immunoblots of total protein lysates from nonneoplastic frontal/temporal lobes (NB) and glioblastoma patients' samples (GB) analyzed for protein expression of Pim1S, Pim1L, phosphorylated Akt1 (pAkt1Ser473), pEGFRTyr1173, and pBadSer112. The membranes were stripped and reprobed with anti–β-actin antibody as loading control (not shown because the figures represent several different immunoblots).
Furthermore, we compared the Pim1 protein expression of 3 different GBM cell lines (U87MG, LN18, and GL261) with other well-established tumor cell lines of different origin. As seen in Supplementary Fig. S1B, with the exception of HEK293 cells, the 34-kDa Pim1S was detectable only in the GBM cell lines U87MG, LN18, and GL261 but not in Caco2, HeLa, or HepG2 cells. Interestingly, the cell lines with a pronounced expression of Pim1S showed high levels of Akt1 and pBad, suggesting a potential interplay between these proteins. Pim1L was missing only in Caco2 cells but was expressed in all other tested cell lines.
Upregulation of Pim1 Expression in GBM Tissue
In order to evaluate the Pim1 expression in normal brain, other tissues, and other cancer types, we first screened the databases of The Human Protein Atlas, Genecards, and the GEO/NCBI profiles. As seen in Supplementary Fig. S2A for data of The Human Protein Atlas, Pim1 protein was found to be expressed at a medium to high level in the brain with a comparable extent of expression as in liver and pancreas as well as in the lung, whereas no Pim1 protein was expressed in the cardiovascular system or the placenta. Concerning the brain, only neuronal cells showed a low to medium Pim1 expression, and in normal glial cells Pim1 seemed to be absent (Supplementary Fig. S2B). Furthermore, searching the Genecards database, which provides cDNA microarray data for a lot of tissues, revealed that Pim1 mRNA expression was detectable in several brain compartments (Supplementary Fig. S2D). The direct comparison of Pim1 protein expression in different cancer types by The Human Protein Atlas showed that different tumor types overexpress Pim1 protein (Supplementary Fig. S2C). And of note, whereas in glial cells Pim1 protein was not found, glioma samples showed a weak to moderate Pim1 staining but with a high interindividual variance. The GEO profiles confirm a high interindividual variance of Pim1 mRNA expression in astrocytomas, glioblastomas, and oligodendrogliomas (Supplementary Fig. S3A–C), but interesting only for glioblastoma samples, a significant upregulation of Pim1 was seen in comparison with nontumor tissue (Supplementary Fig. S3D). This upregulation of Pim1 expression was also seen in the database of The Cancer Genome Atlas (TCGA; National Cancer Institute, caINTEGRATOR2_V1_4_RC2) as well as in the Repository of Molecular Brain Neoplasia Data (REMBRANDT) and AgilentG4502A_07_single ADF data (Supplementary Fig. S3E and F).
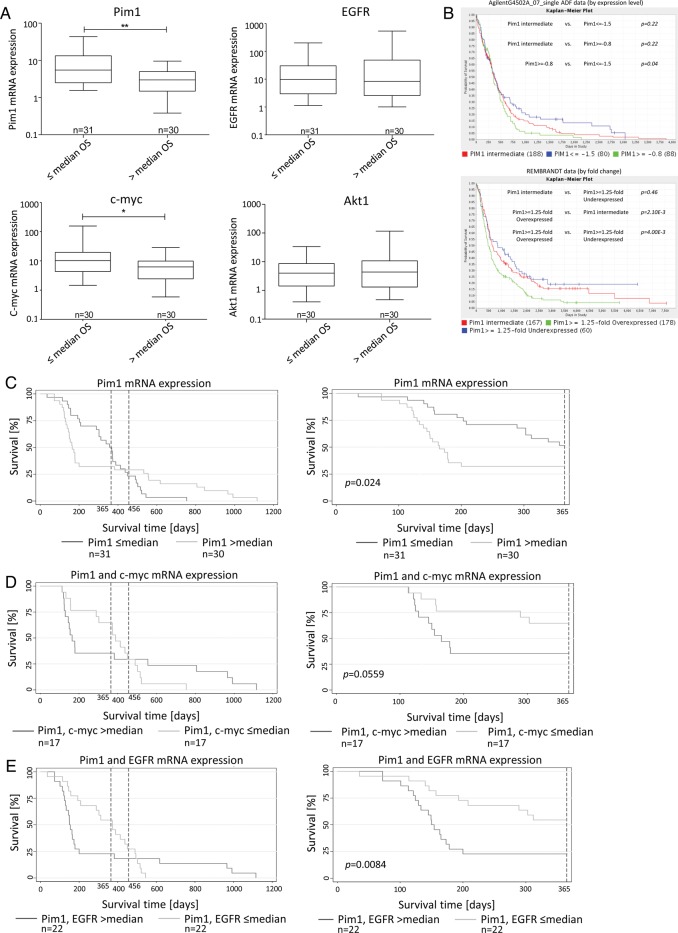
To further confirm the potential role of Pim1 in the malignant behavior of human glioma, we next tested whether there was a significant difference in Pim1 expression in GBM tissue and noncancerous brain tissue of our cohort (Fig. 1B, expressed as arbitrary units). Consistent with the results from GBM cell lines and the above-mentioned data of the GEO profiles, Pim1 mRNA was substantially higher in GBM tissue samples (mean expression, 7.27 ± 11.66) as well as in the first (mean expression, 5.53 ± 5.56) or second relapse (mean expression, 8.94 ± 8.34) compared with nonmalignant brain (mean expression, 1.29 ± 0.55). Expression of c-myc, which is known to cooperate with Pim1,27 displayed also a significant increase from 1.36 ± 0.89 (nonmalignant brain) to 16.69 ± 36.38 in GBM tissues as well as in the first (mean expression, 9.50 ± 10.38) and the second relapse (mean expression, 14.82 ± 18.22). The mRNA expression of oncogenic Akt1 also exhibited an upregulation in GBM tissue from 1.27 ± 0.47 (nonmalignant brain) to 10.02 ± 26.12 in GBM and to 23.28 ± 50.33 and 29.46 ± 55.61 in the first and second relapses, respectively. Since the serine/threonine kinases Pim1 and Akt1 are known to be regulated by EGFR22,28 and are both upregulated in GBM tissue, we next examined the expression of EGFR mRNA. A drastic increase of EGFR mRNA expression from 1.48 ± 0.85 (nonmalignant brain) to 51.22 ± 116.9 was detected in GBM as well as in the first (mean expression, 72.91 ± 252.9) and the second relapse (mean expression, 103.8 ± 226.6), consistent with the known overexpression of EGFR in GBM.1 In addition, we found a slight association between mRNA expression levels of EGFR and Pim1 (r = 0.28, P = .0195) and a highly significant association between c-myc and Pim1 (r = 0.63, P < .0001) in GBM samples (Fig. D). Pim1 high expressing GBM samples were characterized by enhanced c-myc and EGFR mRNA expression (Fig. 1E).
Overexpression of Pim1 in GBM at the protein level was verified by immunoblot analysis in selected patient samples showing stronger signals of Pim1S and Pim1L in GBM tissue compared with noncancerous brain, where only faint bands of Pim1 were observed (Fig. 1F). Furthermore, an upregulation of phosphorylated Akt1 and EGFR, as well as of pBad, a target of Pim1 and Akt1, was determined in GBM compared with nonmalignant brain. As for mRNA analysis, a high interindividual variance of expression in GBM samples was seen for all investigated proteins.
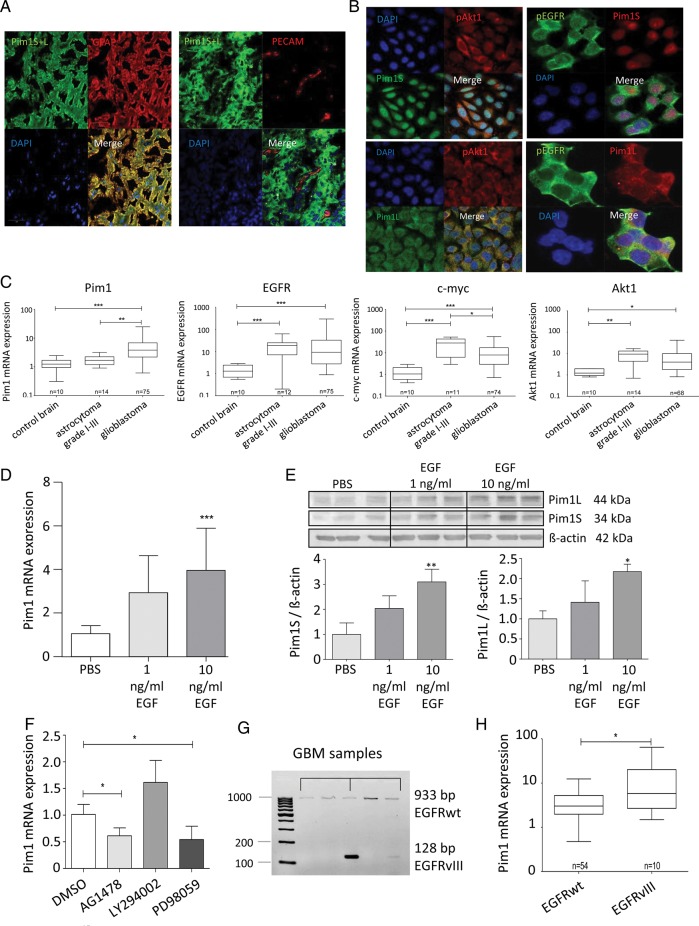
Immunofluorescence staining (Fig. 1C) demonstrated that Pim1 and phosphorylated EGFR (pEGFR) were strongly stained in the glioblastoma specimens showing partly a colocalization for Pim1S and pEGFR as well as for Pim1L and pEGFR, while no or only minor staining was observed in the nonneoplastic brain. Furthermore, staining of HSP90, which is important for stabilization of Pim1 protein, was also increased in glioblastoma compared with nonneoplastic brain, and HSP90 was also partly colocalized with Pim1 (not shown). Tumor cell–specific expression of Pim1 was evidenced by costaining of Pim1 with glial fibrillary acidic protein (GFAP), which is expressed in astrocytic tumors, whereas no Pim1 staining was observed in endothelial cells positive for platelet endothelial cell adhesion molecule (Fig. 2A). To further verify expression of Pim1 in the tumor cells themselves, LN18 glioblastoma cells were fixed and investigated by immunofluorescence staining. As seen in Fig. 2B, Pim1S showed strong nuclear and minor cytoplasmic staining, whereas staining of Pim1L was distributed in the whole cytoplasm. Phosphorylated EGFR and Akt1 seem to be localized both in cytoplasm and in cell membrane.
Fig. 2.
Expression of Pim1, Akt1, EGFR, and c-myc in glioma tissue and GBM cell lines. (A) Cryosections were immunostained with antibodies against Pim1 and GFAP ((left panel) or platelet endothelial cell adhesion molecule (PECAM)/CD31 (right panel). Nuclei are counterstained by the use of DAPI. Note that Pim1 and GFAP are strongly colocalized in glioblastoma cells, whereas no staining of Pim1 was observed in PECAM-positive endothelial cells. (B) Immunofluorescence staining of Pim1S, Pim1L, phosphorylated EGFR (pEGFRTyr1173), and Akt1 (pAkt1Ser473) in glioblastoma cells. Paraformaldehyde fixed LN18 glioblastoma cells stained with antibodies against Pim1S and Pim1L, as well as pAkt1 and pEGFR. Nuclei are counterstained by the use of DAPI. Scale bar represents 20 µm in (A) and (B). (C) Comparison of Pim1, Akt1, EGFR, and c-myc mRNA expression in glioblastoma and low-grade astrocytoma (grades I–III) analyzed by quantitative real-time (RT-)PCR with normalization to 18S rRNA and shown as box plots representing the median as horizontal bars as well as the 5th and 95th percentiles. (D) Regulation of Pim1 mRNA by EGF (1 and 10 ng/mL, 48 h) in LN18 cells was determined by quantitative RT-PCR, mean ± SD of 3 independent experiments. Pim1 mRNA level was normalized to 18S rRNA and is expressed relative to the PBS treated control, which was given a value of 1. (E) Total cell lysates of nontreated and EGF-treated (1 and 10 ng/mL, 48 h) LN18 cells were immunoblotted and stained with antibodies against Pim1S, Pim1L, and β-actin (upper panel) followed by densitometric analysis of signals with normalization of Pim1 protein expression to β-actin (lower panel, Pim1: β-actin ratio). Pim1 protein level is expressed as a proportion of the PBS-treated control cells, which was given a value of 1 (n = 3), mean ± SD. (F) Influence of AG1478 (100 nM), LY294002 (25 µM), and PD98059 (10 µM) on Pim1 expression determined by quantitative RT-PCR. Pim1 mRNA level was normalized to 18S rRNA and is expressed relative to the PBS-treated control, which was given a value of 1 (n = 3), mean ± SD. (G) Representative samples of the analysis of EGFRwt or deletion variant EGFRvIII expression in GBM tissue determined by conventional PCR. (H) Association analysis of Pim1 mRNA level with the expression of EGFRwt or EGFRvIII in glioblastoma shown as box plots representing the median as horizontal bars as well as the 5th and 95th percentiles. *P < .05, **P < .005, ***P < .001.
In contrast to the upregulation of Pim1 in GBM, expression of Pim1 mRNA in low-grade astrocytoma (grades I–III) was minor, with a relative expression value of 1.78 ± 0.69 compared with 1.29 ± 0.55 in nonmalignant brain tissue (Fig. 2C). Otherwise, mRNA expression of c-myc, EGFR, and Akt1 was also significantly upregulated in low-grade astrocytoma.
Regulation of Pim1 by EGFR
Based on the observed tendency of correlation between Pim1 and EGFR mRNA expression in GBM patients, we hypothesized that EGFR might regulate Pim1 expression in glioblastoma cells. Indeed, 48 h treatment of LN18 cells with 10 ng/mL and 100 ng/mL EGF significantly induced Pim1 mRNA expression about 4- and 3.5-fold, respectively (Fig. 2D), while EGFR inhibition with AG1478 reduced constitutive expression of Pim1 mRNA by 50% (Fig. 2F). Treatment of cells with the phosphatidylinositol-3 kinase (PI3K) inhibitor LY294002 had no significant influence on Pim1 mRNA, whereas the mitogen-activated protein kinase (MAPK)/ERK inhibitor PD980509 diminished Pim1 expression by more than 50%. On the protein level, 10 ng/mL EGF stimulated expression of both Pim1 isoforms (34 kDa Pim1S and 44 kDa Pim1L) to about 3-fold compared with solvent-treated control LN18 cells (Fig. 2E). Thus, Pim1 expression was induced by EGFR signaling in LN18 glioblastoma cells in vitro.
To further confirm a potential role of EGFR activity in the regulation of Pim1 expression, we screened 54 patients for EGFRwt and the deletion mutant EGFRvIII by PCR and detected 10 patients bearing EGFRvIII-positive GBM (18.5%). In Fig. 2G, analysis of EGFR subtype for some selected patient samples is exemplified. Since it is known that both EGFR variants prefer the activation of distinct signaling pathways,29 we investigated whether Pim1 expression differed between EGFRwt GBM and EGFRvIII-positive GBM. As seen in Fig. 2H, Pim1 mRNA expression was significantly increased in EGFRvIII-positive GBM (mean expression, 15.07 ± 19.62) compared with those expressing the EGFRwt (mean expression, 4.27 ± 3.74). This upregulation of Pim1 in EGFRvIII-positive GBM was also seen on the protein level for both Pim1S and Pim1L (Supplementary Fig. S4A and B). Furthermore, for both Pim1S and Pim1L we found a significant correlation with the phosphorylation status of EGFR (Supplementary Fig. S4C and D).
Inhibition of Pim1 Reduces Viability and Growth of GBM Cell Lines
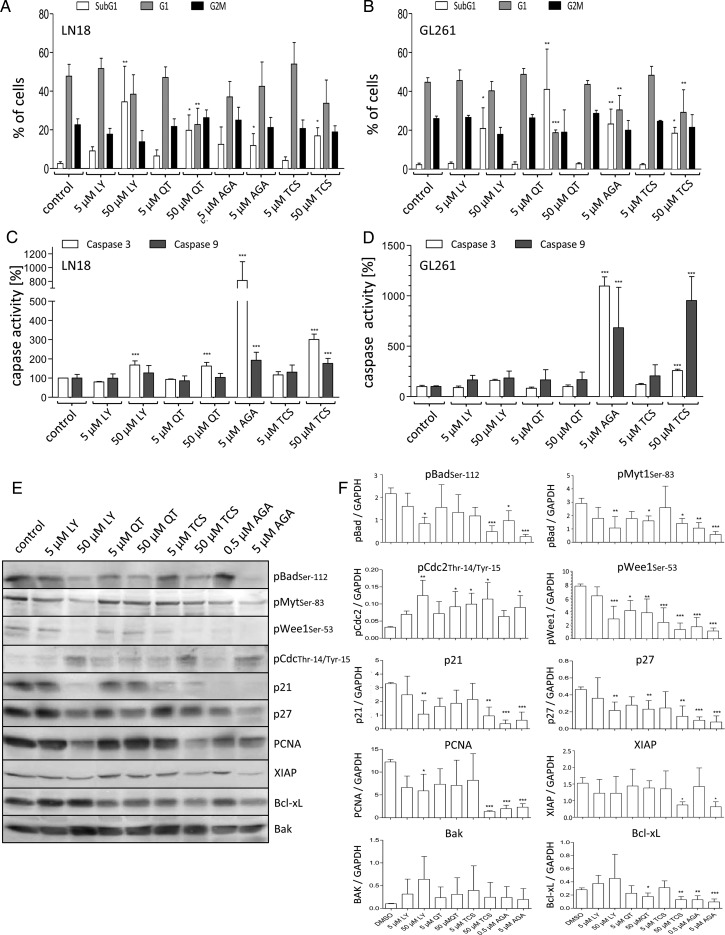
To investigate the potential use of Pim1 inhibitors as a GBM therapy, we treated different GBM cell lines with Pim1 inhibitors. We used LY294002, a nonselective inhibitor of both PI3K and Pim1,30 the selective Pim1 inhibitor QT,31 and the specific Pim1 inhibitor TCS Pim1-1 (TCS).32 Additionally, we tested the effect of HSP90 inhibition using AGA. First, we checked whether Pim1 inhibition resulted in a decreased phosphorylation status of the pro-apoptotic protein Bad, which is known to be a target of PI3K/Akt1 and Pim1 signaling.13,33 As seen in Fig. 4E and F, the Pim1 inhibitors LY294002 (50 µM), QT (50 µM), and TCS (50 µM), as well as AGA (5 µM) caused a substantial decline in pBad.
Fig. 4.
Analysis of the cytotoxic potential of HSP90 and Pim1 inhibitors. (A and B) Fluorescence activated cell sorting–based cell cycle analysis using ethanol fixed and propidium stained LN18 and GL261 cells 48 h after application of DMSO (as solvent), LY294002 (LY, 5 and 50 µM), QT (5 and 50 µM), TCS (5 and 50 µM), and AGA (0.5 and 5 µM). (A) Distribution of cells in the subG1 and (B) in the G1 cluster. Mean + SD, n = 4. (C) Determination of caspase-3 activity using a commercially available assay in LN18 and GL261 cells after treatment with DMSO (as solvent), LY (5 and 50 µM), QT (5 and 50 µM), TCS (5 and 50 µM), and AGA (0.5 and 5 µM) for 48 h. Mean + SD, n = 4. (D) Determination of caspase-9 activity using a commercially available assay in LN18 and GL261 cells after treatment with DMSO (as solvent), LY (5 and 50 µM), QT (5 and 50 µM), TCS (5 and 50 µM), and AGA (0.5 and 5 µM) for 48 h. Mean + SD, n = 4. (E and F) Protein extracts of LN18 cells treated with different inhibitors were analyzed by immunoblotting. Forty-eight hours after application of LY (5 and 50 µM), QT (5 and 50 µM), TCS (5 and 50 µM), and AGA (0.5 and 5 µM) LN18 cells were harvested, and (E) protein extracts were immunoblotted with antibodies against pBad (pBADSer112), phosphorylated Myt1 (pMyt1Ser83), phosphorylated Wee1 (pWee1Ser53), phosphorylated CDC2 (pCDC2Thr14/Tyr15), p21, p27, PCNA, XIAP, Bcl-xL, Bak, and GAPDH as loading control. Representative blots of 4 independent experiments. (F) Densitometric analysis of 4 independent experiments analyzed for the respective proteins by immunoblotting. Each target protein level was normalized to GAPDH (target protein:GAPDH ratio), mean + SD. *P < .05, **P < .005, ***P < .001 vs control.
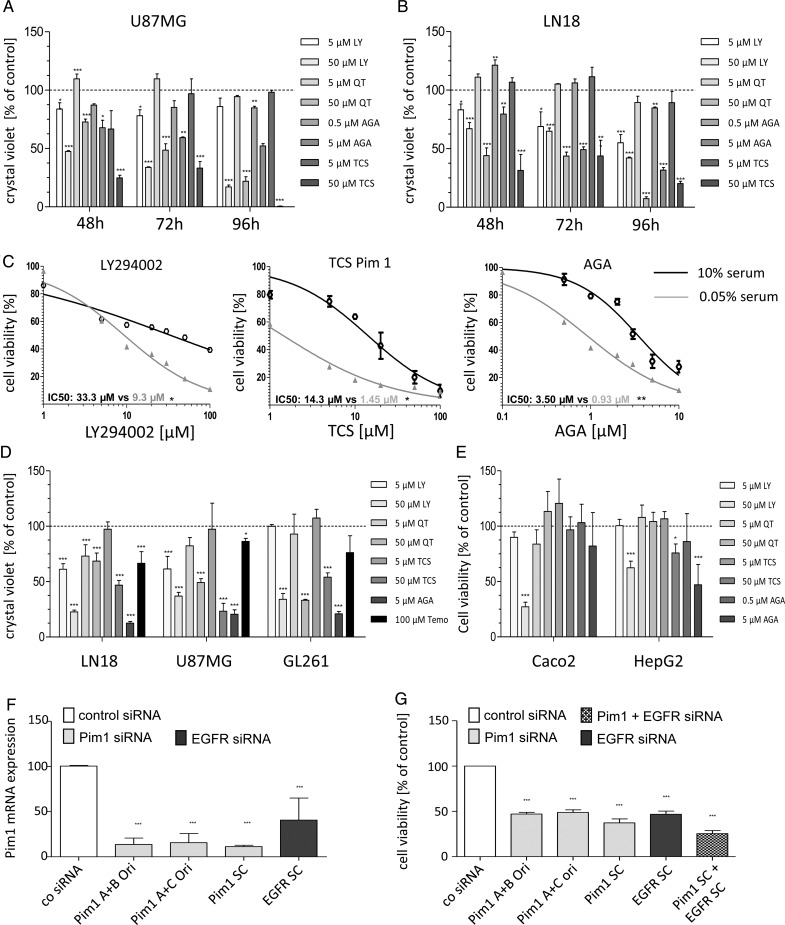
Further, all tested inhibitors induced a loss of cell viability in a time- and dose-dependent manner in the human GBM cell lines LN18 and U87MG, as well as in the murine GL261 GBM cells (Fig. 3A and B, data not shown for GL261 cells). In addition, serum deprivation for 24 h before application of inhibitors enhanced the inhibitory effects of LY294002, TCS, and AGA, as seen by a stronger loss of cell viability and lower half-maximal inhibitory concentration values (Fig. 3C).
Fig. 3.
Growth analysis of murine and human GBM cell lines after application of different inhibitors. (A and B) Time-dependent effects of PI3K/Pim1 inhibitor LY294002 (LY, 5 and 50 µM), Pim inhibitor QT (5 and 50 µM), selective Pim1 inhibitor TCS Pim 1 (5 and 50 µM), and HSP90 inhibitor AGA (0.5 and 5 µM) on cell viability of human U87MG (A) and LN18 (B). Cell viability was determined 48, 72, or 96 h after application of inhibitors using the resazurin assay (n = 3), mean ± SD. (C) Comparison of cell viability curves of serum starved and control LN18 cells treated with DMSO (as solvent), LY (5 and 50 µM), QT (5 and 50 µM), TCS (5 and 50 µM), and AGA (0.5 and 5 µM). Cell viability was determined using the resazurin assay. Half-maximal inhibitory concentration values were determined by nonlinear regression analysis and compared with Wilcoxon signed-rank test (n = 3), mean ± SD. (D) Crystal violet staining of LN18 and GL261 cells 72 h after application of DMSO (as solvent), LY (5 and 50 µM), QT (5 and 50 µM), TCS (5 and 50 µM), AGA (0.5 and 5 µM), and 100 µM temozolomide (n = 3), mean ± SD. (E) Determination of cell viability using the resazurin assay after treatment of HepG2 and Caco2 cells with DMSO (as solvent), LY (5 and 50 µM), QT (5 and 50 µM), TCS (5 and 50 µM), and AGA (0.5 and 5 µM) for 72 h (n = 3), mean ± SD. (F) Silencing of Pim1 and EGFR by gene-specific analysis. LN18 cells were transfected with either control-, Pim1-, or EGFR-siRNA using Lipofectamine 2000, and 48 h afterward the Pim1 mRNA expression was determined by quantitative real-time PCR with normalization to 18S rRNA. Mean + SD of 4 independent experiments. (G) Cell viability of LN18 cells 72 h after transfection with either control-, Pim1-, or EGFR-siRNA. Cell viability was determined using the resazurin assay (n = 4), mean ± SD. *P < .05, **P < .005, ***P < .001 vs control.
The reduced cell viability was accompanied by a loss of cell number, as determined by crystal violet staining. All tested inhibitors clearly caused a loss in cell number 72 h after application, with 5 µM AGA being most effective, with a loss of cell number of 80%–90% depending on the GBM cell line (Fig. 3D). Upon 50 µM TCS, the strongest effect was seen in U87MG cells, with 76.7% loss in cell number, whereas in LN18 and GL261 a reduction of cell number of 53.2% and 45.9%, respectively, was observed. The combined blocking of both PI3K and Pim1 signaling by LY294002 resulted in an enhanced loss of cell number in LN18 (TCS: 53.2% vs LY294002: 77.4%) and GL261 (TCS: 45.9% vs 66%) but not in U87MG (TCS: 76.7% vs LY294002: 63%). Interestingly, treatment of GBM cells with 100 µM temozolomide displayed a considerably less reduction in cell growth, with only 14%–33% loss in cell number.
To confirm the potential role of Pim1 in survival of glioma cell lines, we performed knockdown experiments by using siRNA against Pim1. As seen in Fig. 3F, LN18 cells transfected with Pim1-specific siRNA showed a Pim1 expression level of 11%–15% of control transfected cells dependent on the siRNA used. Interestingly, silencing of EGFR also resulted in a significant decrease of Pim1 expression to about 40% of control LN18 cells, again supporting a role of EGFR in Pim1 regulation. Analysis of cell viability clearly demonstrated a substantial role of Pim1 in LN18 cell survival, since 72 h after transfection a significant loss of cell viability to 37%–47% of control LN18 cells was seen dependent on the siRNA used. Again, EGFR knockdown also resulted in a significant decrease of LN18 cell viability to 47%, which was more pronounced when both EGFR and Pim1 together were silenced by siRNA with a cell viability of 25% of control transfectants.
However, as seen in Supplementary Fig. S1B, HepG2 expressed only the 44-kDa Pim1L, and in Caco2 cells both Pim1 isoforms are missing. Therefore, we further analyzed the efficacy of Pim1 kinase as well as HSP90 inhibitor in these tumor cells (Fig. 3E and Table 2). Treatment of HepG2 cells with 50 µM LY294002 as inhibitor of both PI3K and Pim1 for 72 h decreased the viability of HepG2 (62.4%) similarly to LN18 cells (64.9%). For Caco2 cells, 72 h after application of 50 µM LY294002 a viability of 27.2% was observed, which resembled the inhibitory effect seen in U87MG (33.8%) and GL261 (19.6%) cells. In contrast, HepG2 and Caco2 cells showed no significant loss in cell viability after 72 h treatment with 50 µM QT, but a trend to a reduced cell viability (83.7%) was seen upon 50 µM QT in Caco2 cells. Upon 50 µM TCS as a selective Pim1 inhibitor, HepG2 and Caco2 showed differential modulation of cell viability. Whereas cell viability of Pim1 missing Caco2 cells was not impaired by TCS (96.7%), in HepG2 cells, which express at least Pim1L, a moderate inhibition of cell viability to about 76% was seen.
Table 2.
Comparison of the viability of cell lines with different Pim1 expression status
| Cell Viability at 72 h | U87MG | LN18 | GL261 | HepG2 | Caco2 |
|---|---|---|---|---|---|
| 50 µM LY | 33.8% | 64.9% | 19.6% | 62.4% | 27.2% |
| 50 µM QT | 48.6% | 43.7% | 6.5% | 104.1% | 113.4% |
| 50 µM TCS | 33.1% | 49.1% | 39.5% | 75.8% | 96.7% |
| 5 µM AGA | 59.3% | 43.8% | 13.5% | 47.1% | 82.0% |
U87MG, LN18, GL261, HepG2, and Caco2 cells were incubated for 72 h with LY294002 (LY), QT, TCS, and AGA followed by a medium change and addition of resazurin-containing media to determine cell viability (n = 3, mean values in %).
In addition to the cell growth, we further analyzed the potential of Pim1 inhibitors to influence the migratory capacity of LN18 cells by 2 different techniques. As seen in Supplementary Fig. S5, only the combined blocking of both Akt1 and Pim1 signaling by LY294002 significantly reduced the migration of LN18 glioblastoma cells to about half of the control cells in both the wound healing assay (Supplementary Fig. S5A) and the transwell migration assay using a Boyden chamber (Supplementary Fig. S5B), arguing for a dual therapeutic blocking of Akt1 and Pim1 kinases in glioblastoma therapy. The Pim1 inhibitor QT (50 µM) also slightly diminished the migration of LN18 cells to 75% of the control cells, but this was seen by only the wound healing assay. In contrast, the specific inhibition of Pim1 by TCS was not associated with a reduction of the migratory potential of LN18 glioblastoma cells.
Influence of Pim1 Inhibition on Cell Cycle and Apoptotic Fraction
To further investigate whether the loss in cell viability and cell number was mediated by antiproliferative or pro-apoptotic effects of Pim1 inhibition, we performed propidium iodide–based cell cycle analysis. As seen in Fig. 4A and B, in control-treated LN18 and GL261 cells, the contents of cells in subG1, G1, and G2/M phase were 2.6% and 2.3% (subG1), 47.8% and 44.7% (G1), and 22.6% and 26% (G2/M, data not shown). At 48 h after application, 5 µM or 50 µM LY294002, QT, TCS, and AGA caused a significant increase in cells belonging to the subG1 phase, without significant differences between LN18 and GL261 cells. The proportions of LN18 and GL261 cells, respectively, showing a subG1 phenotype were: at 50 µM LY294002, 34.5% and 20.9%; at 50 µM QT, 19.8% and 40%; at 5 µM AGA, 11.9% and 23.2%; and at 50 µM TCS, 16.9% and 18.5%. A significant decrease in G1 phase was seen at 50 µM QT in LN18 and GL261 cells to 22.8% and 18.8%, respectively. Five micromolar AGA and 50 µM TCS significantly reduced the G1 cell content in GL261 cells to only 30.5% and 29.2%, respectively. In contrast, Pim1 and HSP90 inhibitors did not cause any significant alterations of cells in the G2/M phase. Only a minor decrease was seen after treatment with 50 µM LY294002 (13.8% and 17.9% compared with 22.6% and 26% in the controls), but this was not statistically significant (data not shown).
Influence of Pim1 Inhibition on Caspase-3 and -9 Activities
To examine the effect of Pim1 inhibition on apoptosis, we assessed the activities of caspase-3 and -9, respectively. Figure 4C illustrates the influence of Pim1 inhibition on the activity of caspase-3 in comparison with HSP90 inhibition and control-treated cells at 48 h after application of inhibitors. LN18 cells treated with 50 µM LY294002 (PI3K/Pim1 inhibitor) or 50 µM QT (Pim1 inhibitor) showed significantly enhanced caspase-3 activity of about 168% or 163%, respectively, compared with control cells. In contrast, in the murine GL261 cells, only LY294002 caused an increase of caspase-3 activity, to 161%. Inhibition of HSP90 with 5 µM AGA or Pim1 with 50 µM TCS resulted in both LN18 and GL261 cells in a more pronounced induction of caspase-3 activity, with values of 811% and 1096%, respectively, for AGA and 301% and 258% for TCS.
Furthermore, caspase-9 activity assays were performed to examine the mitochondrial apoptosis pathway. As seen in Fig. 4D, LN18 and GL261 cells differ in their caspase-9 activity upon treatment with the inhibitors for 48 h. The murine GL261 cells responded to the application of 5 µM AGA and 50 µM TCS with strongly enhanced caspase-9 activity values of 683% (AGA) and 953% (TCS). In contrast, in LN18 cells, the induction of caspase-9 was considerably less, with activity values of 193% (AGA) and 177% (TCS). LY294002 and QT only slightly increased caspase-9 activity in GL261 cells, but no effect was seen in LN18 cells.
Influence of Pim1 Inhibition on Selected Cell Cycle and Apoptosis Regulatory Proteins
In order to analyze which apoptosis or cell cycle regulatory proteins are targeted by Pim1 in GBM cells, we investigated the influence of Pim1 inhibitors on protein expression or phosphorylation status of selected proteins in LN18 cells 48 h after application (presented as arbitrary units). Besides being a well-known substrate of Akt1 and Pim1,13,33 Bad has a pro-apoptotic function in its unphosphorylated form, which is inhibited by phosphorylation at serine-112. Treatment of LN18 cells with the combined Akt1/Pim1 inhibitor LY294002 or with the selective Pim1 inhibitor TCS (both 50 µM) resulted in a significant decrease of pBad from 2.16 (control) to 0.83 or 0.47, respectively (Fig. 4E and F). Similarly, the HSP90 inhibitor AGA (5 µM) clearly reduced the phosphorylation status of Bad to 0.25, while downregulation of pBad was not significant upon incubation with QT. Since there was a significant change in the subG1 and G1 phase content upon Pim1 inhibition, we investigated the activity of the cell cycle regulatory kinase Cdc2, as well as of its modulators Myt1 and Wee1 by determination of their phosphorylation status.34 Concerning phosphorylated Cdc2, we observed an increase in its relative phosphorylation at Thr14/Tyr15 from 0.03 (control) to 0.12 by 50 µM LY294002, to 0.09 by 50 µM QT, to 0.11 by 50 µM TCS, and to 0.09 by 5 µM AGA, thereby arresting Cdc2 activity. In agreement with this, phosphorylation of the Cdc2 modifying kinases Myt1 and Wee1 was significantly diminished by all tested inhibitors, resulting in more activated Myt1 and Wee1. In control-treated LN18 cells, the relative phosphorylation of Myt1 and Wee1 amounted to 2.92 and 7.79, which was significantly decreased by 50 µM LY294002 to 1.07 and 2.91, respectively, by 50 µM QT to 1.78 and 3.83, by 50 µM TCS to 1.41 and 1.34, and by 5 µM AGA to 0.59 and 1.12.
Next, we studied the regulation of the cyclin-dependent kinase inhibitors p21 and p27. A significant decrease in protein expression of both p21 and p27 was caused by LY294002 (50 µM), TCS (50 µM), and AGA (5 µM), but not QT, as seen in Fig. 4E and F. The relative expression of p21 was reduced from 3.29 (DMSO control) to 1.07 by 50 µM LY294002, to 0.94 by 50 µM TCS, and to 0.37 and 0.63 by 0.5 µM and 5 µM AGA. The Pim1 inhibitor QT also slightly decreased the p21 content to about half of the control cells, but this was not significant. Similarly to p21, the protein content of p27 was also reduced from a relative expression of 0.46 (control) to 0.22 by 50 µM LY294002, to 0.23 by 50 µM QT, to 0.15 by 50 µM TCS, and to 0.08 by 5 µM AGA. To further evaluate a reduced proliferation status of inhibitor-treated LN18 cells, we studied the expression of the proliferating cell nuclear antigen (PCNA), which was significantly diminished from 12.22 (DMSO control) to 5.89 by 50 µM LY294002, to 1.35 by 50 µM TCS, and to 1.98 and 2.31 by 0.5 µM and 5 µM AGA, respectively, whereas upon 50 µM QT the downregulation of PCNA to 7.08 was not significant.
To verify the apoptosis-inducing effects of the tested inhibitors seen by fluorescence activated cell sorting and caspase-3 analysis, we investigated the expression of the pro-apoptotic Bak as well as of the anti-apoptotic proteins Bcl-xL and XIAP. As demonstrated in Fig. 4E and F, only treatment of LN18 cells with TCS and AGA provoked a significant decrease of XIAP from 1.03 in control cells to 0.37 (50 µM TCS) and 0.32 (5 µM AGA). Concerning Bak expression, no significant regulation was seen after treatment with either of the inhibitors, but there was a trend to an increased Bak protein content in LY294002-treated LN18 cells. In contrast to this, the anti-apoptotic Bcl-xL protein expression was significantly reduced from 0.28 (control) to 0.18 by 50 µM QT, to 0.13 by 50 µM TCS, and to 0.13 and 0.09 by 0.5 µM and 5 µM AGA, respectively. Treatment of LN18 cells with LY294002 resulted in no significant modulation of Bcl-xL protein content.
Furthermore, we investigated the expression status of some apoptosis-regulatory proteins by immunoblot analysis after siRNA-mediated knockdown of Pim1. As shown in Supplementary Fig. S5C, the expression of the pro-apoptotic Bak was significantly enhanced to 2.1-fold 72 h after transfection of LN18 cells with Pim1-specific siRNA compared with control transfected cells. In contrast, expression of the anti-apoptotic Bcl-xL and XIAP was significantly reduced to 0.47 and 0.54, respectively, of control cells after Pim1 suppression by siRNA transfection. Additionally, LN18 cells transfected with Pim1-specific siRNA showed a trend to a reduced expression of the cell cycle regulator p27, as already seen for pharmacological Pim1 inhibition (Supplementary Fig. S5C).
Pim1 Expression and Survival of Patients With Glioblastoma
Next, we were interested in whether there was a correlation of Pim1 expression with clinical outcome in glioblastoma cases. For this purpose, we first divided our patient cohort into 2 subgroups: the first subgroup represented patients who showed survival shorter than or equal to the median OS, and the other subgroup included patients with a longer survival time than the median OS. As seen in Fig. 5A, GBM patients who lived shorter than the median OS showed a significantly higher mRNA expression of both Pim1 (mean expression, 10.20 ± 12.55 vs 3.50 ± 2.61) and c-myc (mean expression, 22.62 ± 52.31 vs 8.08 ± 7.76), whereas gene expression of EGFR (mean expression, 30.0 ± 53.59 vs 73.67 ± 163.0) or Akt1 (mean expression, 6.56 ± 9.65 vs 14.45 ± 38.36) was not significantly associated with the OS of our GBM patients. Screening of TCGA (caINTEGRATOR2_V1_4_RC2), AgilentG4502A_07_single ADF, and REMBRANDT data for prognostic relevance of Pim1 expression in patients' survival revealed significantly prolonged survival of glioblastoma patients having low Pim1 expression (Fig. 5B). In addition, we performed database analysis to compare the impact of Pim1 expression and other potential prognostic factors (O6-methylguanine-DNA-methyltransferase [MGMT], EGFR, PTEN, and Akt1) on survival of glioblastoma patients. Since MGMT promoter methylation influences expression of this DNA repair enzyme, we compared survival curves of high and low MGMT expressing patients with those who had high and low Pim1 expression, respectively. As seen in Supplementary Fig. S6A, in contrast to Pim1, no significant differences in survival between high and low expressing MGMT patients were seen. But interestingly, Pim1 low expressing patients were characterized by a better survival than patients with low MGMT expression, which was discussed as a prognostic marker for patients with glioblastoma recently. Similar results were found by comparison of low Pim1 expressing patients with low EGFR, low Akt1, and low PTEN expressing patients (Supplementary Figs. S6B, S7A, and B), with the exception that also between PTEN low and high expressing patients a significant difference in patients' survival was evident (Supplementary Fig. S7B). Additionally, we could not find an association between Pim1 and MGMT or PTEN expression in our patients' samples (Supplementary Fig. S8).
Fig. 5.
Association between Pim1 mRNA or protein expression and survival time of patients with GBM. (A) mRNA expression of Pim1, c-myc, EGFR, and Akt1 was determined with quantitative real-time (RT-)PCR. Patients wer divided into 2 subgroups according to their OS time: patients with survival equal to or below the median time (≤ median OS) vs patients with survival above the median time (> median OS). Box plots represent the median mRNA expression as horizontal bars as well as the 5th and 95th percentiles, Mann–Whitney U test. (B) Screening of TCGA (caINTEGRATOR2_V1_4_RC2) for an association between Pim1 gene expression and survival of patients with GBM. Kaplan–Meier survival curves of AgilentG4502A_07_single ADF data (at the top) and of REMBRANDT data (at the bottom), both based on microarray analyses. Green lines: Pim1 overexpressing patients; blue lines: Pim1 underexpressing patients; red lines: Pim1 intermediate expressing patients. (C–E) Kaplan–Meier survival curves for patients with GBM based on their Pim1 mRNA expression (C), Pim1 and c-myc mRNA expression (D), and Pim1 and EGFR expression (E). Patients were divided into 2 subgroups depending on gene expression as determined by quantitative RT-PCR. Left panels represent the complete patient cohort, right panels demonstrate survival data for a 365-day observation time span. The broken vertical lines denote the current survival times (12 and 15 mo) of patients with GBM treated with standard therapy (surgical removal, combined radio- and chemotherapy). *P < .05, **P < .005.
Interestingly, in our patient cohort, Kaplan–Meier survival curves for Pim1 expression in deceased patients showed a 2-phase course with a clear survival benefit for patients with Pim1 expression below the median but only for the first year (Fig. 5C, right panel). Afterward, a drastic change was observed showing no longer a survival advantage of low Pim1 expression (Fig. 5C, left panel). It seems that there may exist 2 different Pim1 high expressing subgroups either with worse survival or with prolonged survival. To search for potential underlying reasons, we further investigated the influence of a combined analysis of Pim1 and c-myc or Pim1 and EGFR expression in our patient cohort. As seen in Fig. 5D and E, the same curve progression was found. To exclude patients' age as the underlying reason for the 2-phase course, we adjusted the survival curves for age at diagnosis, but as seen in Supplementary Fig. S9, adjustment for patients' age had no effect on any of the survival curves. Further, those patients of our cohort who had a subtotal tumor resection and an OS below the median showed higher Pim1 expression than those with subtotal resection and an OS above the median (Supplementary Fig. S10). However, patients having a survival above the median were characterized by a significantly younger age at diagnosis (Supplementary Fig. S11). No significant association was found between Pim1 mRNA expression and age at diagnosis or the gender of the analyzed GBM patients (data not shown).
Proof of Principle—Analysis of Pim1 as a Therapeutic Target in an Orthotopic In vivo Mouse Model
To finally show proof of the concept of Pim1 as a therapeutic target for glioblastoma treatment, we analyzed the in vivo tumor growth in an orthotopic murine glioblastoma model using the cell line GL261, which was shown to express both Pim1 isoforms (Fig. 1A) and which was stereotactically injected into mice brain. Twelve days postinjection, tumor development was assessed by MRT to start the treatment at a tumor volume >1.5 mm3. Afterward, the mice were treated with either 5% dextrose (control animals) or 75 mg/kg TCS as a Pim1 inhibitor every second day by oral gavage until the end of the study (12 days of treatment).
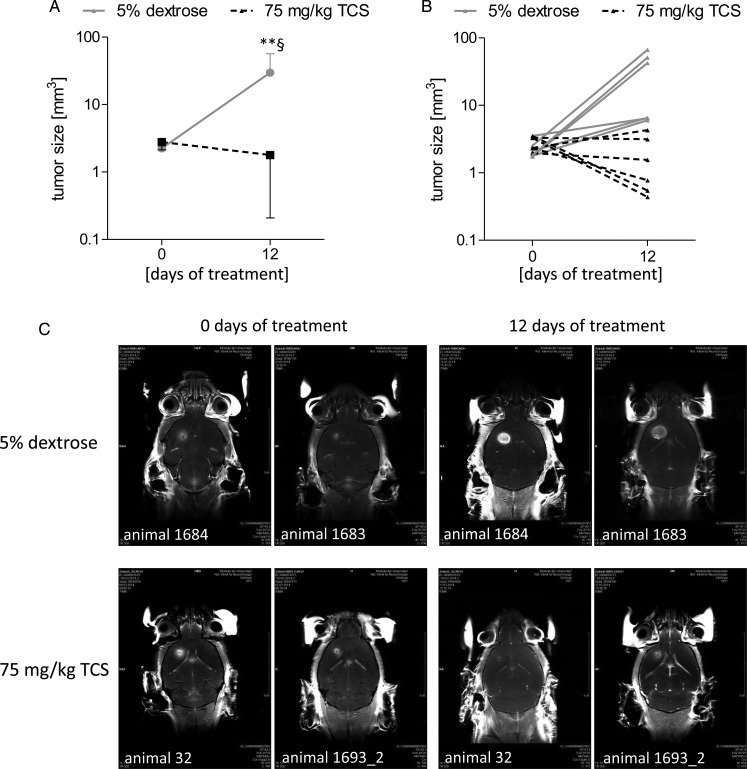
According to the animal welfare guidelines, we created a score sheet with no-go criteria for premature sacrifice of animals showing obvious psychological or physiological strain. The scoring system is listed in Supplementary Table S12. A score of 20 was defined to be a criterion for premature sacrifice. As seen in the Supplementary material, Fig. S13 and Tables S14 and S15, the TCS-treated animals showed no weight loss and no abnormal behavior after 12 days of treatment, with the exception of that animal who had a tumor progress from 2.32 mm3 to 4.27 mm3 (score of 10, rotary behavior in the cage). In contrast, 5 of 6 control animals had obvious weight loss (mean, 7.4% ± 2.27%), behavioral abnormalities, and deterioration of the general conditions, with scores of 15–25. At day 12 of control treatment, 3 animals (50%) had a score of at least 20 (which was defined as the cancellation criterion) and 2 other animals had a score of 15. This time point was chosen for completion of the efficacy study to be in compliance with the animal welfare guidelines and because a dramatic difference in tumor volume as well as in animal conditions was found between control and TCS-treated animals.
As seen in Fig. 6A (mean + SD) and 6B (each individual animal), at the time point of beginning the pharmacological intervention the tumor size was nearly identical in control and the TCS treated group, with values of 2.25 mm3 (control animals) and 2.78 mm3 (TCS animals). Overall, 12 days after treatment a decrease in tumor size was observed in the TCS treated group (1.79 mm3) compared with the starting time point (2.78 mm3). Only one animal of the TCS group had a slight tumor progress from 2.33 mm3 to 4.27 mm3 (Fig. 6B). In contrast, control animals showed significant progress in tumor size from 2.25 mm3 (starting point) to 29.85 mm3 at day 12, which was also significantly different from the corresponding time point of the TCS group. Representative MRT recordings for the starting time point and 12 days of treatment for each 2 animals per group are shown in Fig. 6C.
Fig. 6.
Analysis of Pim1 as a therapeutic target in an orthotopic in vivo mouse model. Female wild-type C57BL/6 mice were immobilized in a stereotactic head holder in the flatskull position; 1 μL (2.5 × 104 cells/μL) of GL261 cells in PBS was inserted 1 mm anterior and 2 mm lateral to bregma to a depth of 3 mm from the dural surface. Twelve days postinjection tumor development was assessed in isoflurane-anesthetized mice by MRT to start the treatment at a tumor volume >1.5 mm3. Tumor volume was calculated with OsiriX software in coronar and axial gadolinium-enhanced T1-sections. Afterward, mice were treated with either 5% dextrose (control animals, n = 5) or 75 mg/kg TCS (n = 5) as Pim1 inhibitor every second day by oral gavage until the end of the study (12 d treatment). Tumor size was measured using MRT 12 days after starting the treatment. (A) Mean value of tumor size of control (5% dextrose) and TCS-treated animals at the day of beginning the pharmacological intervention (day 0) and at day 12. (B) Tumor sizes of each individual animal from the control and TCS-treated group at the day of beginning pharmacological intervention (day 0) and at day 12. (C) Representative gadobutrol-enhanced T1-weighted images (coronar) for 2 animals of the control (5% dextrose) and the TCS-treated group at the day of beginning pharmacological intervention (day 0) and at day 12. **P < .01 vs TCS treated animals (day 12) and §P < .05 vs the respective day 0 group.
Discussion
Targeted therapies against the progression of GBM have been increasingly investigated, particularly with inhibitors of neo-angiogenesis and growth factor receptors, but so far with limited success. Hence, searching for additional targets for novel interventions is indispensable to improve the prognosis of GBM patients. In the present study, we demonstrate that expression of Pim1 is significantly higher in GBM compared with nonmalignant brain. This upregulation of Pim1 kinase may stimulate the development and progression of GBM because Pim1 is known to play a central role in the regulation of tumor proliferation, apoptosis, and migration.35 However, 2 Pim1 isoforms were described, a short 34-kDa isoform (Pim1S) and a long 44-kDa isoform (Pim1L), which differed in their translational starting points and cellular localizations.9 Our study confirms the different subcellular localizations9 of the Pim1 isoforms, since Pim1S was primarily stained in the nuclei of GBM cells, whereas Pim1L seemed to be expressed in the cytoplasm and cell membrane.
Besides Pim1, expression and phosphorylation of the serine/threonine kinase Akt1 and EGFR were also significantly elevated in GBM compared with control brain, which is in agreement with the fact that Akt1 is activated in about 80% of GBM, and EGFR is often overexpressed in GBM.1,36 Additionally, regulation of Akt1 and Pim1 by EGFR is known.22,28 In LN18 glioblastoma cells, EGF treatment induced a significant increase in Pim1 mRNA and protein content, while AG1478, an EGFR kinase inhibitor, significantly reduced Pim1 expression. Screening of our patients for EGFRwt and the deletion mutant EGFRvIII by PCR revealed that 18.5% of the patients were bearing EGFRvIII-positive GBM, being in agreement with previous reports on glioblastoma.37,38 Interestingly, Pim1 mRNA expression was significantly increased in GBM harboring EGFRvIII compared with those with the EGFRwt variant, suggesting differences in the underlying mechanisms of Pim1 regulation by the wild-type and the mutant EGFR. A remarkable difference in signaling by EGFRwt and the mutant is the preferential activation of MAP/ERK kinase (MEK) and signal transducer and activator of transcription 3 signaling by EGFRwt, whereas EGFRvIII activates both the MEK and the PI3K-Akt1 cascade.29 Thus, it seems possible that Pim1 is more amplified in GBM with EGFRvIII, since Pim1 expression might be strongly induced by 2 different pathways, namely MEK/ERK and PI3K-Akt1. However, our results suggest that EGFR mediates the stimulation of Pim1 expression in GBM cells and that Pim1 inhibitors may be used in combination therapies to increase the efficacy of EGFR tyrosine kinase inhibitors, as already stated by Siu and colleagues39 for prostate cancer cells.
Next, we investigated the effect of different pharmacological inhibitors targeting Pim1 signaling in 3 different GBM cell lines. In direct comparison, the U87MG cells responded more pronouncedly to the combined PI3K and Pim1 inhibitor LY29400230 than did LN18 cells. Maybe this is caused by the different PTEN, p53, or Bax status, with LN18 having wild-type PTEN and mutant p53 but missing Bax,40–42 and U87MG being negative for PTEN but having wild-type p53 and Bax. Further, as seen in Supplementary Fig. S1B, LN18 cells expressed more Akt1 than did U87MG cells, thus this difference in the Akt1 protein level or other kinases not investigated could also explain the stronger response of U87MG cells to LY294002, which has also Pim1 and PI3K independent effects, such as inhibition of the protein kinase CK243 and the serine/threonine kinases glycogen synthase kinase 3A and B, as well as the p97/valosin-containing protein, a member of the type II AAA ATPase family, and other chaperones with ATPase activity.44 Therefore, we used 2 other Pim1 inhibitors (TCS Pim1-1 and QT) as well as a specific siRNA to underline the role of Pim1 suppression in growth inhibition of glioblastoma cells (Figs. 3 and 4). The growth inhibitory effect of HSP90 inhibition by AGA in our GBM cells had previously been shown by Sauvageot and coworkers45 in both human glioma cell lines and glioma stem cells. The reduced viability of GBM cells in our study was accompanied by a loss of cell number, as determined by crystal violet staining. Interestingly, in comparison with the kinase and HSP90 inhibitors, temozolomide exhibited only minor inhibition of growth of GBM cells, which is in agreement with the fact that the majority of GBM cell lines are resistant to this cytostatic.46
However, all tested inhibitors clearly caused an increase in the subG1 peak and caspase-3 activity, which is consistent with a rise in apoptotic cells. Concerning caspase-9, only AGA and TCS significantly increased the activity of this initiator caspase, indicating that inhibition of Pim1 by TCS activates partly the intrinsic mitochondrial apoptotic pathway. The above described apoptosis-promoting effect of Pim1 inhibition was underlined by the fact that the anti-apoptotic proteins Bcl-xL and XIAP, an inhibitor of caspase-3, were significantly downregulated by interrupting Pim1 signaling.
However, the entry of eukaryotic cells into mitosis is regulated by activation of Cdc2 kinase. Phosphorylation at Tyr15 and Thr14 and thus inhibition of Cdc2 is carried out by Wee1 and Myt1 protein kinases.34 Our results demonstrate an inhibition of Cdc2 by increased phosphorylation at Tyr15 and Thr14 potentially mediated by the enhanced Myt1 and Wee1 activity shown by their reduced phosphorylation. In agreement with the hypothesized retarded cell cycle upon Pim1 inhibition, we found a significant reduction in the proliferation marker PCNA. Until now, Cdc2, Myt1, Wee1, and PCNA were not described as targets of Pim1 signaling, but it is known that p21 and p27 are phosphorylated and decreased by this kinase.11,14 In contrast to this known downregulation of p21 and p27 by Pim1, we found a significantly reduced p21 and p27 protein content upon Pim1 inhibition, which could explain the missing G1 arrest in our cell cycle analysis. Further, one can speculate that there is no distinct drift between particular cell cycle phases upon Pim1 inhibition compared with the control cells but rather a slowdown of passing through the cell cycle followed by apoptosis. At present, the reason or underlying mechanism of altered p21 and p27 regulation by Pim1 inhibition seen in our study is unclear. Cell type–specific differences could be a probable cause for the divergent results. Altogether, the in vitro findings of our study suggest that Pim1 influences a variety of pathways coupled to the regulation of cell cycle and apoptosis or other cell death mechanisms, but the migratory capacity of GBM cells was not significantly modulated by Pim1 inhibitors.
Until now, no association studies of the role of Pim1 in the pathogenesis of glioblastoma have been published. In addition to our in vitro results, we found an association of Pim1 expression with the OS of patients with glioblastoma. Those patients with a survival time below the median showed significantly elevated Pim1 expression. Consequently, Pim1 expression seems generally upregulated in glioblastoma but particularly in those patients with a poor prognosis. This influence of Pim1 on patients' survival was also seen by screening the TCGA and REMBRANDT databases for prognostic relevance of Pim1 expression in GBM patients. Interestingly, EGFR and Akt1 mRNA expressions were not associated with the OS, but c-myc mRNA content showed a similar association to patients' survival as Pim1. These results suggest that Pim1 signaling might play an important role in the malignant behavior of human GBM, perhaps by cooperating with c-myc. Somewhat contrary to this finding is the fact that Kaplan–Meier survival curves show only a benefit of a low Pim1 expression for the first 12 months after diagnosis, and it seems that 2 different Pim1 high expressing subtypes exist, which was also seen when c-myc or EGFR expression was included in the survival analysis. To date, the reason for this temporal pattern is not known. Potential reasons could be the younger age at diagnosis in those patients having a survival of more than 12 months (data not shown) because this parameter is known to be an independent patient characteristic that determines prognosis.47 Further, for patients with GBM, the first quarter of the second year (fifth quarter) postdiagnosis is considered to be the peak incidence of mortality,48 and this is nearly the time point when the influence of Pim1 on survival was strongly changed in our Kaplan–Meier survival analysis. Besides age and tumor resection grade, survival of GBM patients is also influenced by tumor size, affecting disorders, and preoperative Karnofsky performance status, which were not topics in this study but may also be different in the 2 Pim1 high expressing subtypes with either short or long survival. Further, so far unknown signaling pathways that interact with Pim1 could be different in these 2 subgroups and therefore predict the role of Pim1 for survival. So far, Pim1 has also been found to be upregulated in prostate, pancreatic, and non–small cell lung cancers, and an elevated Pim1 expression is accompanied by a good prognosis in these tumors.15–18 In contrast, increased Pim1 expression in stomach, esophagus, bladder, and head and neck cancers results in a poor prognosis.16,19–21 Therefore, Pim1 may have different functions in tumorigenesis depending on the tumor type and underlying signaling pathways. Recently, evidence for Pim1 as a relevant therapeutic target was demonstrated by Weirauch and colleagues49 in a mouse model showing growth inhibitory effects of Pim1 gene silencing by U1 small nuclear interference in subcutaneous tumors induced by U87 cells. In contrast to this subcutaneous tumor model, we performed an orthotopic glioblastoma model with stereotactic implantation of GL261 glioma cells into the brain. Using this mouse model, we could show for the first time that inhibition of Pim1 by TCS significantly reduced the growth of intracerebral glioblastoma cells, arguing for a new therapeutic option for patients with glioblastoma. Whether inhibition of Pim1 is superior to other kinase inhibitors should be intensively examined in extended studies.
Of note, in contrast to other tumor-promoting kinases, such as PI3K/Akt1, which have to be activated by phosphorylation, Pim1 is constitutively active after translation without required phosphorylation by upstream kinases. The PI3K pathway represents only one of the signaling cascades involved in transcriptional regulation of Pim1. A lot of other signaling molecules, such as EGFR itself, signal transducer and activator of transcription 1/3/5, Janus kinases, and nuclear factor-kappaB, modulate expression of Pim1.9 Thus, Pim1 represents a downstream target of various tumor-promoting signaling cascades. Inhibition of Pim1 as a therapeutic option for treatment of glioblastoma and other cancers could be superior to inhibition of other kinases, particularly such as PI3K/Akt1, since it was shown that inhibition of Akt1 results in upregulation of receptor tyrosine kinases through an uncharacterized feedback mechanism that is blocked by inhibition of Pim1 kinase.50 Therefore, targeting Pim1 alone or in combination could therefore improve the efficacy of PI3K/Akt inhibitors or other cytostatics in anticancer therapy. Furthermore, it was described that Pim1 might interact with or might act upstream of tumor-promoting kinases such as Akt,51 p38 MAPK,52 and mammalian target of rapamycin,53 and thus inhibition of Pim1 could interrupt these signaling pathways too, leading to diminished cell growth. Most importantly, deficiency of the Pim1 kinase gene is well tolerated in vivo with ostensibly normal, healthy, and fertile mice,54 suggesting that Pim1 inhibition might offer an attractive therapeutic modality. In contrast, Akt1 knockout leads to a partial embryonic lethal phenotype,55 and a role of Akt3 in postnatal brain development is described.56
In summary, the findings of this study suggest an important role of Pim1 in the pathogenesis of glioblastoma and may provide a novel therapeutic target, which should be further analyzed in depth to develop effective targeted therapies to ultimately improve the outcome of GBM patients.
Supplementary Material
Funding
This work was supported by national funding from the Forschungsverbund Neurowissenschaften and Fries Holzsysteme GmbH, Grimmen.
Conflict of interest statement. None declared.
Supplementary Material
References
- 1.Ohgaki H, Kleihues P. Genetic profile of astrocytic and oligodendroglial gliomas. Brain Tumor Pathol. 2011;28(3):177–183. doi: 10.1007/s10014-011-0029-1. [DOI] [PubMed] [Google Scholar]
- 2.Newton HB. Molecular neuro-oncology and development of targeted therapeutic strategies for brain tumors. Part 2: PI3K/Akt/PTEN, mTOR, SHH/PTCH and angiogenesis. Expert Rev Anticancer Ther. 2004;4(1):105–128. doi: 10.1586/14737140.4.1.105. [DOI] [PubMed] [Google Scholar]
- 3.Anton K, Baehring JM, Mayer T. Glioblastoma multiforme: overview of current treatment and future perspectives. Hematol Oncol Clin North Am. 2012;26(4):825–853. doi: 10.1016/j.hoc.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Cuypers HT, Selten G, Quint W, et al. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984;37(1):141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- 5.van Lohuizen M, Verbeek S, Krimpenfort P, et al. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56(4):673–682. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]
- 6.Qian KC, Wang L, Hickey ER, et al. Structural basis of constitutive activity and a unique nucleotide binding mode of human Pim-1 kinase. J Biol Chem. 2005;280(7):6130–6137. doi: 10.1074/jbc.M409123200. [DOI] [PubMed] [Google Scholar]
- 7.Shah N, Pang B, Yeoh KG, et al. Potential roles for the PIM1 kinase in human cancer—a molecular and therapeutic appraisal. Eur J Cancer. 2008;44(15):2144–2151. doi: 10.1016/j.ejca.2008.06.044. [DOI] [PubMed] [Google Scholar]
- 8.Bachmann M, Moroy T. The serine/threonine kinase Pim-1. Int J Biochem Cell Biol. 2005;37(4):726–730. doi: 10.1016/j.biocel.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Brault L, Gasser C, Bracher F, et al. PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica. 2010;95(6):1004–1015. doi: 10.3324/haematol.2009.017079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mochizuki T, Kitanaka C, Noguchi K, et al. Physical and functional interactions between Pim-1 kinase and Cdc25A phosphatase. Implications for the Pim-1-mediated activation of the c-Myc signaling pathway. J Biol Chem. 1999;274(26):18659–18666. doi: 10.1074/jbc.274.26.18659. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Bhattacharya N, Mixter PF, et al. Phosphorylation of the cell cycle inhibitor p21Cip1/WAF1 by Pim-1 kinase. Biochim Biophys Acta. 2002;1593(1):45–55. doi: 10.1016/s0167-4889(02)00347-6. [DOI] [PubMed] [Google Scholar]
- 12.Bachmann M, Kosan C, Xing PX, et al. The oncogenic serine/threonine kinase Pim-1 directly phosphorylates and activates the G2/M specific phosphatase Cdc25C. Int J Biochem Cell Biol. 2006;38(3):430–443. doi: 10.1016/j.biocel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Aho TL, Sandholm J, Peltola KJ, et al. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 2004;571(1–3):43–49. doi: 10.1016/j.febslet.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 14.Morishita D, Katayama R, Sekimizu K, et al. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008;68(13):5076–5085. doi: 10.1158/0008-5472.CAN-08-0634. [DOI] [PubMed] [Google Scholar]
- 15.Dhanasekaran SM, Barrette TR, Ghosh D, et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412(6849):822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 16.Nawijn MC, Alendar A, Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer. 2011;11(1):23–34. doi: 10.1038/nrc2986. [DOI] [PubMed] [Google Scholar]
- 17.Reiser-Erkan C, Erkan M, Pan Z, et al. Hypoxia-inducible proto-oncogene Pim-1 is a prognostic marker in pancreatic ductal adenocarcinoma. Cancer Biol Ther. 2008;7(9):1352–1359. doi: 10.4161/cbt.7.9.6418. [DOI] [PubMed] [Google Scholar]
- 18.Warnecke-Eberz U, Bollschweiler E, Drebber U, et al. Frequent down-regulation of pim-1 mRNA expression in non-small cell lung cancer is associated with lymph node metastases. Oncol Rep. 2008;20(3):619–624. [PubMed] [Google Scholar]
- 19.Warnecke-Eberz U, Bollschweiler E, Drebber U, et al. Prognostic impact of protein overexpression of the proto-oncogene PIM-1 in gastric cancer. Anticancer Res. 2009;29(11):4451–4455. [PubMed] [Google Scholar]
- 20.Liu HT, Wang N, Wang X, et al. Overexpression of Pim-1 is associated with poor prognosis in patients with esophageal squamous cell carcinoma. J Surg Oncol. 2010;102(6):683–688. doi: 10.1002/jso.21627. [DOI] [PubMed] [Google Scholar]
- 21.Guo S, Mao X, Chen J, et al. Overexpression of Pim-1 in bladder cancer. J Exp Clin Cancer Res. 2010;29(1):161. doi: 10.1186/1756-9966-29-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peltola K, Hollmen M, Maula SM, et al. Pim-1 kinase expression predicts radiation response in squamocellular carcinoma of head and neck and is under the control of epidermal growth factor receptor. Neoplasia. 2009;11(7):629–636. doi: 10.1593/neo.81038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakariassen PO, Prestegarden L, Wang J, et al. Angiogenesis-independent tumor growth mediated by stem-like cancer cells. Proc Natl Acad Sci U S A. 2006;103(44):16466–16471. doi: 10.1073/pnas.0607668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talasila KM, Soentgerath A, Euskirchen P, et al. EGFR wild-type amplification and activation promote invasion and development of glioblastoma independent of angiogenesis. Acta Neuropathol. 2013;125(5):683–698. doi: 10.1007/s00401-013-1101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lena A, Rechichi M, Salvetti A, et al. Drugs targeting the mitochondrial pore act as cytotoxic and cytostatic agents in temozolomide-resistant glioma cells. J Transl Med. 2009;7:13. doi: 10.1186/1479-5876-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naud JF, Eilers M. PIM1 and MYC: a changing relationship? Nat Cell Biol. 2007;9(8):873–875. doi: 10.1038/ncb0807-873. [DOI] [PubMed] [Google Scholar]
- 28.Moscatello DK, Holgado-Madruga M, Emlet DR, et al. Constitutive activation of phosphatidylinositol 3-kinase by a naturally occurring mutant epidermal growth factor receptor. J Biol Chem. 1998;273(1):200–206. doi: 10.1074/jbc.273.1.200. [DOI] [PubMed] [Google Scholar]
- 29.Pines G, Kostler WJ, Yarden Y. Oncogenic mutant forms of EGFR: lessons in signal transduction and targets for cancer therapy. FEBS Lett. 2010;584(12):2699–2706. doi: 10.1016/j.febslet.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs MD, Black J, Futer O, et al. Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002. J Biol Chem. 2005;280(14):13728–13734. doi: 10.1074/jbc.M413155200. [DOI] [PubMed] [Google Scholar]
- 31.Holder S, Zemskova M, Zhang C, et al. Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase. Mol Cancer Ther. 2007;6(1):163–172. doi: 10.1158/1535-7163.MCT-06-0397. [DOI] [PubMed] [Google Scholar]
- 32.Cheney IW, Yan S, Appleby T, et al. Identification and structure-activity relationships of substituted pyridones as inhibitors of Pim-1 kinase. Bioorg Med Chem Lett. 2007;17(6):1679–1683. doi: 10.1016/j.bmcl.2006.12.086. [DOI] [PubMed] [Google Scholar]
- 33.Datta SR, Dudek H, Tao X, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91(2):231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 34.Morgan DO. Principles of CDK regulation. Nature. 1995;374(6518):131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Bhattacharya N, Weaver M, et al. Pim-1: a serine/threonine kinase with a role in cell survival, proliferation, differentiation and tumorigenesis. J Vet Sci. 2001;2(3):167–179. [PubMed] [Google Scholar]
- 36.Haas-Kogan D, Shalev N, Wong M, et al. Protein kinase B (PKB/Akt) activity is elevated in glioblastoma cells due to mutation of the tumor suppressor PTEN/MMAC. Curr Biol. 1998;8(21):1195–1198. doi: 10.1016/s0960-9822(07)00493-9. [DOI] [PubMed] [Google Scholar]
- 37.Montano N, Cenci T, Martini M, et al. Expression of EGFRvIII in glioblastoma: prognostic significance revisited. Neoplasia. 2011;13(12):1113–1121. doi: 10.1593/neo.111338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatanpaa KJ, Burma S, Zhao D, et al. Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia. 2010;12(9):675–684. doi: 10.1593/neo.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siu A, Virtanen C, Jongstra J. PIM kinase isoform specific regulation of MIG6 expression and EGFR signaling in prostate cancer cells. Oncotarget. 2011;2(12):1134–1144. doi: 10.18632/oncotarget.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishii N, Maier D, Merlo A, et al. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9(3):469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louis DN. The p53 gene and protein in human brain tumors. J Neuropathol Exp Neurol. 1994;53(1):11–21. doi: 10.1097/00005072-199401000-00002. [DOI] [PubMed] [Google Scholar]
- 42.Streffer JR, Rimner A, Rieger J, et al. BCL-2 family proteins modulate radiosensitivity in human malignant glioma cells. J Neurooncol. 2002;56(1):43–49. doi: 10.1023/a:1014448721327. [DOI] [PubMed] [Google Scholar]
- 43.Davies SP, Reddy H, Caivano M, et al. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351(Pt 1):95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gharbi SI, Zvelebil MJ, Shuttleworth SJ, et al. Exploring the specificity of the PI3 K family inhibitor LY294002. Biochem J. 2007;404(1):15–21. doi: 10.1042/BJ20061489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sauvageot CM, Weatherbee JL, Kesari S, et al. Efficacy of the HSP90 inhibitor 17-AAG in human glioma cell lines and tumorigenic glioma stem cells. Neuro Oncol. 2009;11(2):109–121. doi: 10.1215/15228517-2008-060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sankar A, Thomas DG, Darling JL. Sensitivity of short-term cultures derived from human malignant glioma to the anti-cancer drug temozolomide. Anticancer Drugs. 1999;10(2):179–185. doi: 10.1097/00001813-199902000-00006. [DOI] [PubMed] [Google Scholar]
- 47.Raysi DS, De PD, Marzi S, et al. Survival prognostic factors in patients with glioblastoma: our experience. J Neurosurg Sci. 2012;56(3):239–245. [PubMed] [Google Scholar]
- 48.Smoll NR, Schaller K, Gautschi OP. Long-term survival of patients with glioblastoma multiforme (GBM) J Clin Neurosci. 2013;20(5):670–675. doi: 10.1016/j.jocn.2012.05.040. [DOI] [PubMed] [Google Scholar]
- 49.Weirauch U, Grunweller A, Cuellar L, et al. U1 Adaptors for the therapeutic knockdown of the oncogene Pim-1 kinase in glioblastoma. Nucleic Acid Ther. 2013;23(4):264–272. doi: 10.1089/nat.2012.0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cen B, Mahajan S, Wang W, et al. Elevation of receptor tyrosine kinases by small molecule AKT inhibitors in prostate cancer is mediated by Pim-1. Cancer Res. 2013;73(11):3402–3411. doi: 10.1158/0008-5472.CAN-12-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu XF, Li J, Vandervalk S, et al. PIM-1-specific mAb suppresses human and mouse tumor growth by decreasing PIM-1 levels, reducing Akt phosphorylation, and activating apoptosis. J Clin Invest. 2009;119(2):362–375. doi: 10.1172/JCI33216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Didichenko SA, Spiegl N, Brunner T, et al. IL-3 induces a Pim1-dependent antiapoptotic pathway in primary human basophils. Blood. 2008;112(10):3949–3958. doi: 10.1182/blood-2008-04-149419. [DOI] [PubMed] [Google Scholar]
- 53.Zhang F, Beharry ZM, Harris TE, et al. PIM1 protein kinase regulates PRAS40 phosphorylation and mTOR activity in FDCP1 cells. Cancer Biol Ther. 2009;8(9):846–853. doi: 10.4161/cbt.8.9.8210. [DOI] [PubMed] [Google Scholar]
- 54.Laird PW, van der Lugt NM, Clarke A, et al. In vivo analysis of Pim-1 deficiency. Nucleic Acids Res. 1993;21(20):4750–4755. doi: 10.1093/nar/21.20.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cho H, Thorvaldsen JL, Chu Q, et al. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001;276(42):38349–38352. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- 56.Tschopp O, Yang ZZ, Brodbeck D, et al. Essential role of protein kinase B gamma (PKB gamma/Akt3) in postnatal brain development but not in glucose homeostasis. Development. 2005;132(13):2943–2954. doi: 10.1242/dev.01864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.