Abstract
Background
Few studies have associated microRNAs (miRNAs) with the hedgehog (Hh) pathway. Here, we investigated whether targeting smoothened (SMO) with miR-326 would affect glioma biological behavior and stemness.
Methods
To investigate the expression of SMO and miR-326 in glioma specimens and cell lines, we utilized quantitative real-time (qRT)-PCR, Western blot, immunohistochemistry, and fluorescence in situ hybridization. The luciferase reporter assay was used to verify the relationship between SMO and miR-326. We performed cell counting kit-8, transwell, and flow cytometric assays using annexin-V labeling to detect changes after transfection with siRNA against SMO or miR-326. qRT-PCR assays, neurosphere formation, and immunofluorescence were utilized to detect the modification of self-renewal and stemness in U251tumor stem cells. A U251-implanted intracranial model was used to study the effect of miR-326 on tumor volume and SMO suppression efficacy.
Results
SMO was upregulated in gliomas and was associated with tumor grade and survival period. SMO inhibition suppressed the biological behaviors of glioma cells. SMO expression was inversely correlated with miR-326 and was identified as a novel direct target of miR-326. miR-326 overexpression not only repressed SMO and downstream genes but also decreased the activity of the Hh pathway. Moreover, miR-326 overexpression decreased self-renewal and stemness and partially prompted differentiation in U251 tumor stem cells. In turn, the inhibition of Hh partially elevated miR-326 expression. Intracranial tumorigenicity induced by the transfection of miR-326 was reduced and was partially mediated by the decreased SMO expression.
Conclusions
This work suggests a possible molecular mechanism of the miR- 326/SMO axis, which can be a potential alternative therapeutic pathway for gliomas.
Keywords: glioma, hedgehog, miRNA, SMO, stemness
Glioblastoma multiforme (GBM), the most frequently occurring primary malignant brain tumor, remains the brain tumor with the highest mortality rate despite the numerous modern therapeutic approaches that have been adopted.1 There is substantial evidence to indicate that glioma development and progression are closely related to the accumulation of aberrant activation in signaling pathways.2–4 The hedgehog (Hh) pathway, which has a critical function in stem cell maintenance and cell proliferation, plays an important role in the tumorigenicity of human gliomas and their cancer stem cells.5 The Hh family of genes encodes several ligand proteins, including Sonic hedgehog (Shh), Indian hedgehog (Ihh), and Desert hedgehog (Dhh).6 These ligands initiate Hh signaling by binding to the 12-transmembrane domain receptor Patched (Ptch).7 Smoothened (SMO), a 7-transmembrane domain-containing protein, is derepressed by Ptch and serves as a key regulator of signal transduction of this pathway. Activated SMO promotes GLI1 translocation from the cytoplasm to the nucleus and regulates the transcription of downstream target genes.8 Downstream signaling induced by the GLI1 leads to the promotion of cell cycle progression, inhibition of apoptosis, maintenance of self-renewal of stem cells, and regulation of tissue stem cell differentiation.9–11 Therefore, an intriguing hypothesis is that targeting the Hh pathway could be a potential therapeutic strategy for developing efficient cancer therapeutics.
MicroRNAs (miRNAs) are a group of noncoding, small RNAs composed of ∼22 nucleotides that regulate the expression of a wide variety of genes through direct interaction with the 3′-untranslated regions of their messenger RNA (mRNA). They have been identified as a new type of gene expression regulator that targets mRNAs for translational repression or degradation. Increasing evidence has strongly implicated the involvement of miRNAs in carcinogenesis, including pancreas, breast, colon and lung cancers, leukemia, prostate carcinoma, and glioblastoma.12–18 Dysregulated miRNAs may function as oncogenes or tumor suppressor genes. In addition, miRNAs play important roles in diverse biological processes, including development, differentiation, stem cell maintenance, and cell identity.19–23 Little is known, however, about the regulation of miRNAs and their interaction with the major signaling pathways in gliomas, especially with respect to Hh signaling.
In this study, we aimed to explore the expression and function of SMO in gliomas and its relationship with miR-326. For the first time, we showed that miR-326 affected the activity of Hh signaling pathway mediated by SMO and also regulated the self-renewal ability and stemness and partially prompted differentiation in glioma stem cells. Moreover, inhibition of the Hh pathway partially increased miR-326 expression. These results further confirmed that the feedback loop between miR-326 and the SMO oncogene might be considered an alternative for multiple treatments of gliomas by regulating the Hh/SMO/GLI1 pathway.
Materials and Methods
Tissue Samples and Clinical Data
Informed consent was obtained for the application of human glioma tissue samples from adult patients who received a glioma diagnosis at the Second Affiliated Hospital of Harbin Medical University. Freshly resected tissue samples were immediately frozen in liquid nitrogen for subsequent total RNA extraction. All samples were histologically classified and graded according to WHO guidelines by clinical pathologists and included 16 grade I-II tumors, 13 grade III tumors, and 16 grade IV tumors (Table 1). Additionally, 5 normal adult brain tissue specimens were used as controls from patients with severe traumatic brain injury who required surgery (with informed consent). Fifty paraffin-embedded glioma specimens with various grade and clinical data were collected for subsequent experimentation from the Second Affiliated Hospital of Harbin Medical University from January 2008 through June 2009. This study was approved by the hospital institutional review board, and written informed consent was obtained from all patients.
Table 1.
Clinicopathological parameters of 45 glioma samples from the Second Affiliated Hospital of Harbin Medical University
| No. | Age | Sex | Pathology (WHO) | No. | Age | Sex | Pathology (WHO) |
|---|---|---|---|---|---|---|---|
| 01 | 31 | Female | Astrocytoma II | 24 | 45 | Male | Astrocytoma II |
| 02 | 54 | Male | Glioblastoma IV | 25 | 34 | Female | Astrocytoma I |
| 03 | 36 | Female | Astrocytoma II | 26 | 49 | Female | Glioblastoma IV |
| 04 | 45 | Male | Astrocytoma III | 27 | 47 | Male | Oligodendroglioma II |
| 05 | 51 | Male | Glioblastoma IV | 28 | 50 | Male | Glioblastoma IV |
| 06 | 48 | Female | Astrocytoma II | 29 | 43 | Male | Astrocytoma II |
| 07 | 63 | Female | Astrocytoma III | 30 | 31 | Male | Glioblastoma IV |
| 08 | 50 | Female | Glioblastoma IV | 31 | 52 | Female | Glioblastoma IV |
| 09 | 59 | Male | Glioblastoma IV | 32 | 41 | Male | Glioblastoma IV |
| 10 | 40 | Male | Astrocytoma III | 33 | 50 | Female | Astrocytoma III |
| 11 | 45 | Male | Oligodendroglioma II | 34 | 56 | Female | Glioblastoma IV |
| 12 | 52 | Male | Astrocytoma II | 35 | 59 | Male | Astrocytoma III |
| 13 | 6 | Male | Glioblastoma IV | 36 | 58 | Female | Glioblastoma IV |
| 14 | 47 | Male | Glioblastoma IV | 37 | 20 | Male | Astrocytoma III |
| 15 | 9 | Female | Astrocytoma I | 38 | 49 | Female | Glioblastoma IV |
| 16 | 44 | Female | Astrocytoma II | 39 | 40 | Male | Astrocytoma I |
| 17 | 49 | Male | Astrocytoma II | 40 | 15 | Female | Astrocytoma III |
| 18 | 42 | Male | Oligodendroglioma II | 41 | 31 | Male | Astrocytoma III |
| 19 | 8 | Male | Astrocytoma III | 42 | 62 | Female | Glioblastoma IV |
| 20 | 9 | Male | Astrocytoma II | 43 | 36 | Male | Glioblastoma IV |
| 21 | 45 | Female | Astrocytoma III | 44 | 40 | Male | Astrocytoma III |
| 22 | 42 | Female | Astrocytoma III | 45 | 63 | Female | Astrocytoma III |
| 23 | 28 | Female | Astrocytoma II |
Abbreviation: WHO, World Health Organization.
Cell Culture
Human U87, SHG44, U251, T98G glioma cell lines, and HEK293T cells were obtained from the Chinese Academy of Sciences Cell Bank, Shanghai. A human oligodendroglia cell line (Olig) was a kind gift from Prof. Feng-min Zhang of Harbin Medical University. The cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (Gibco) and were incubated at 37°C in a 5% CO2 atmosphere. The U251tumor stem cell (TSC) line was derived by magnetic cell-sorting technique and validated by immunofluorescence as described previously24 (Supplementary data, Fig. S1). The U251-TSC cells were grown in DMEM (F-12) supplemented with B-27,20 ng/ml basic fibroblast growth factor and 20 ng/ml epidermal growth factor (EGF).
Oligonucleotides, Sonic Hedgehog, and Cyclopamine Treatments
miR-326 mimics, miR-326 inhibitor, miR-scramble (miR-Scr) and SMO siRNA (siSMO) were synthesized and purified using high-performance liquid chromatography (GenePharma; see Supplementary data for detailed sequences). Cells were transfected with miR-326 mimics, miR-326 inhibitor, or SMO siRNA (200 pmol each) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells transfected with miR-Scr were used as a control. Commercial N-Shh (R&D Systems) was used at 0.5 μM. Cyclopamine (Sigma) was used at 10 μM. Cells were starved in 0.5% serum (instead of the usual 10% routinely used for standard growth after medication) for 12 hours before medication and were continued in culture for an additional 48 hours.
RNA Extraction and Complementary DNA Synthesis
Total RNA was extracted using TRIzol reagent (Invitrogen). The first strand complementary DNAs (cDNAs) were synthesized using the PrimeScript RT reagents Kit (Perfect Real Time, TaKaRa) according to the manufacturer's instructions. The reverse transcription reaction was carried out at 37°C for 15 minutes and was then inactivated at 85°C for 5 seconds.
Quantitative Real-time Polymerase Chain Reaction
To investigate the expression of miRNAs, SMO, and other stemness markers in glioma samples and cell lines, quantitative real-time (qRT)-PCR was performed in triplicate in a LightCycler2.0 (Roche Diagnostics) and normalized to glyceraldehyde 3-phosphatedehydrogenase (GAPDH) and U6 as endogenous controls. The qRT-PCR data were analyzed using the 2-ΔΔCt method. (see Supplementary material for qRT-PCR primers.)
In-situ Hybridization for miRNA
To study the spatial and temporal expression of miRNAs with high sensitivity and resolution, miRNA fluorescence in-situ hybridization (FISH) was performed according to manufacturer's protocol.25
Proliferation Assay
The cell count kit-8 assay was used to quantify the human glioma cell viability, as previously described.26 Each experiment was performed in triplicate. All proliferation assays were repeated as independent experiments at least twice.
In Vitro Invasion Assays
Transwell membranes coated with Matrigel (BD Biosciences) were used to quantify in vitro glioma cell invasion, as previously described.27 Fold migration was calculated relative to a blank control. The data represent mean ± standard error (SE) of 3 independent experiments.
Evaluation of Cell Apoptosis
Apoptosis was quantified using annexin V labeling after transfection for 48 hours. The cells were resuspended in binding buffer. Then, 10 μl of FITC annexin V and 5 μl of propidium iodide (BD Pharmingen) were added and incubated for 15 minutes. The stained cells were analyzed by flow cytometry (FACSCanto II, BD Biosciences).
Luciferase Reporter Assay
To determine the Hh pathway transcriptional activity, a reporter containing 8 directly repeated copies of a consensus GLI binding site (8×-GLI) downstream of the luciferase gene was used. The plasmids were transfected into cells treated with miR-326 mimics or miR-Scr with cotransfection of an expression construct containing the entire SMO coding sequence without its 3′-UTR fragment. In addition, the SMO 3′UTR-Luc reporter and the mutant reporter were obtained from Promega (see Supplementary material for detailed information).
Western Blot Assay
Western blot assays were performed as previously described.26 Primary antibodies included anti-SMO (1:1000 dilution; Abcam), anti-GLI1 (1:1000 dilution; CST), anti-CyclinD1, anti-N-MYC(1:300), and anti-GAPDH (1:1000 dilution; Santa Cruz Biotechnology). Following incubation with the horseradish peroxidase-labeled secondary antibody (Introvigen), protein bands were detected on Fujifilm Las-4000.
Immunofluorescence and Immunohistochemistry
Immunofluorescence and immunohistochemistry (IHC) assays were performed as previously described.28 These assays used antibodies against CD133+, nestin, GLI1, glial fibrillary acidic protein (GFAP; 1:200 dilution; Bioss Inc.) and SMO (1:100 dilution; Santa-Cruz Biotechnology). Immunofluorescence staining was visualized using anti-mouse Alexa Fluor 488 (Molecular Probes) or anti-rabbit–rhodamine antibody conjugates (Dianova).Nuclei were counterstained using 4′,6-diamidino-2- phenylin- dole (DAPI; Sigma). The slides were analyzed by confocal laser scanning microscopy (Olympus FV1000 laser scanning microscope). IHC scores were calculated using a semiquantitative 5-category grading system.29
Nude Mouse Tumor Intracranial Model and miR-326 Treatment
U251 cells that were cotransducted with miR-326 mimics/scramble oligonucleotide, and luciferase lentivirus were injected intracranially into 5-week–old BALB/c-nude mice. After 20 days, tumors were measured by fluorescent images of whole mice using an IVIS Lumina Imaging System (Xenogen). Portions of the tumor tissues were used to measure miR-326 by qRT-PCR and SMO by Western blot. Cryosections (4 mm) were stained with hematoxylin and eosin (H&E) and used for IHC. These procedures were performed following approval by the Harbin Medical University Institutional Animal Care and Use Committee.
Statistical Analyses
Statistics were performed using the SPSS Graduate Pack, version 11.0 statistical software (SPSS). Descriptive statistics, including mean, SE, and 1-way ANOVA, were used to determine significant differences. The survival curves were analyzed using the log-rank test employing Prism GraphPad software. Statistical significance was assumed at a value of P < .05.
Results
SMO Was Upregulated in Glioma Samples and Cell Lines and Was Associated With Tumor Grade and Poor Survival Period
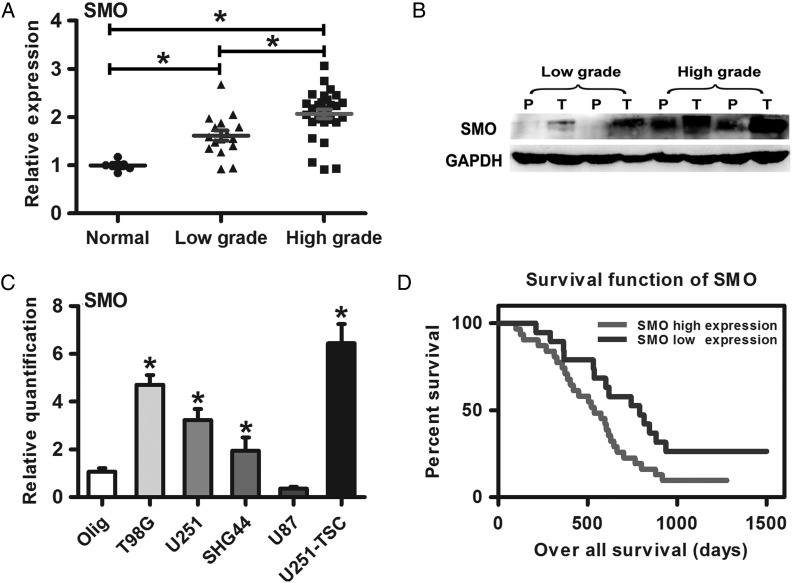
The result of qRT-PCR and Western blot showed that SMO was highly expressed in high-grade gliomas compared with low-grade gliomas and normal brain tissues (Fig. 1A and B).This result was consistent with the data from other databases (Supplementary material, Fig. S2A–C). Furthermore, SMO expression was also detected in 4 glioma cell lines (U251, T98G, U87, and SHG44) and one glioma stem cell line (U251-TSC). Compared with the human oligodendroglia cell line (Olig), SMO was highly expressed in all glioma cell lines except for U87 (Fig. 1C). Subsequently, a retrospective analysis of the clinical outcome of our patients and other databases revealed that increased SMO correlated with poor survival (Fig. 1D and Supplementary material, Fig. S2D–I) (*P<.05).
Fig. 1.
SMO expression in glioma specimens and cell lines correlated with poor survival. (A) SMO expression in glioma specimens and normal brain tissues as assessed by qRT-PCR. (B) SMO expression in different grade gliomas between peritumor tissues and tumor tissues by Western blot assay. (C) qRT-PCR analysis showed T98G, SHG44, U251, and U251-TSC glioma cells expressed higher levels of SMO than the Olig cell line. The data represent mean ± SE of 3 replicates (*P < .05). (D) Kaplan–Meier survival curves indicating cumulative survival as a function of time for those patients with SMO high expression versus low expression. The patients with high SMO expression experienced a significantly worse outcome (*P < .05).
SMO Knockdown Suppressed the Biological Behaviors of Glioma Cells
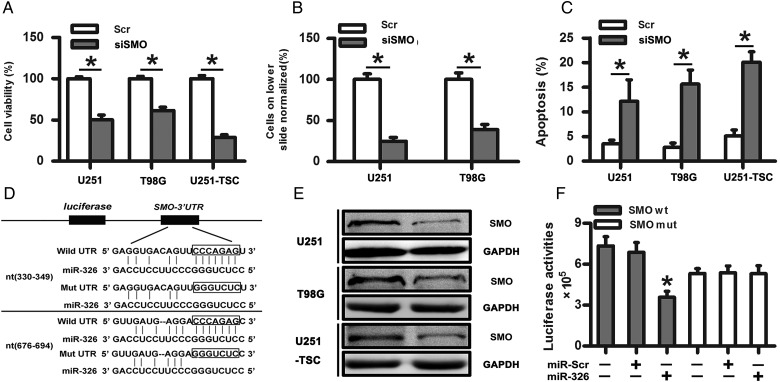
Given that SMO is a crucial regulator of the Hh pathway and was highly expressed in gliomas, we assessed its importance on the tumorigenic properties of glioma cells, including proliferation, invasion, and apoptosis. In our study, we found that siSMO significantly reduced the expression of SMO at the mRNA levels after transfecting SMO siRNA into glioma cell lines (U251 and T98G) and the TSC line (Supplementary material, Fig. S3A). Meanwhile, siSMO also decreased cell proliferation and the number of invasive glioma cells and significantly increased apoptosis compared with the control group (Fig. 2A–C). Together, these data showed that the tumorigenic properties of glioma cells, specifically proliferation, invasion, and apoptosis, were influenced by SMO inhibition.
Fig. 2.
SMO knockdown suppressed the biological behaviors of glioma cells, and SMO was a direct target of miR-326. (A) Representative cartogram showing the cell proliferation in glioma cells and tumor stem cells regulated by SMO knockdown. (B) Representative images of in vitro transwell assays of U251 and T98G after transfection with siSMO or scramble RNA. (C) The Annexin V-PI assay reveals increased apoptosis in U251, T98G, and U251-TSC cells following siSMO treatment. The data represent mean ± SE of 3 replicates (*P < .05). (D) Diagram of the seed sequence of miR-326 matched the 3′UTRs of the SMO gene and the design of wild or mutant SMO 3′UTRs containing reporter constructs. (E) Western blot for SMO expression 48 hours after transfection with miR-Scr or miR-326. (F) Luciferase reporter assays in glioma cells after cotransfection of cells with wild-type or mutant 3′UTR SMO and miRNA. The data represent the fold change in the expression (mean ± SE) of 3 replicates.
SMO Was a Direct Target of miR-326
miRNA target searches using Targetscan and Miranda (http://www.targetscan.org/ and http://www.microrna.org/) confirmed that the seed sequence of miR-326 matched 2 sites of the 3′UTR of the SMO gene (Fig. 2D). To investigate whether SMO is a functional target of miR-326, we assessed its mRNA and protein levels after the transfection of miR-326 mimics. First, qRT-PCR was used to evaluate the transfection efficiency (Supplementary material, Fig. S3B). Next, SMO mRNA level was detected without significant change following transfection with scramble, miR-326 mimics, or miR-326 inhibitor (Supplementary material, Fig. S3C). Western blot indicated decreased levels of SMO protein in the miR-326 mimics-transfected cells compared with the scramble (Fig. 2E). However, the miR-326 inhibitor did not show a significant effect on SMO protein expression (data not shown), which may be caused by the low miR-326 expression in gliomas. To further confirm whether SMO was a direct target of miR-326, we transfected pGL3-WT-SMO-3′UTR or pGL3-MUT-SMO-3′UTR dual-luciferase reporter plasmids with miR-326 mimic or scramble into HEK293T cells for 48 hours, followed by measurement of luciferase activity. Our results showed that transfection with miR-326 reduced the luciferase activity significantly compared with the scramble group. However, the luciferase activity in the mutant construct did not show significant differences (Fig. 2F). These data indicated that miR-326 directly modulated SMO expression by binding to the 3′UTR.
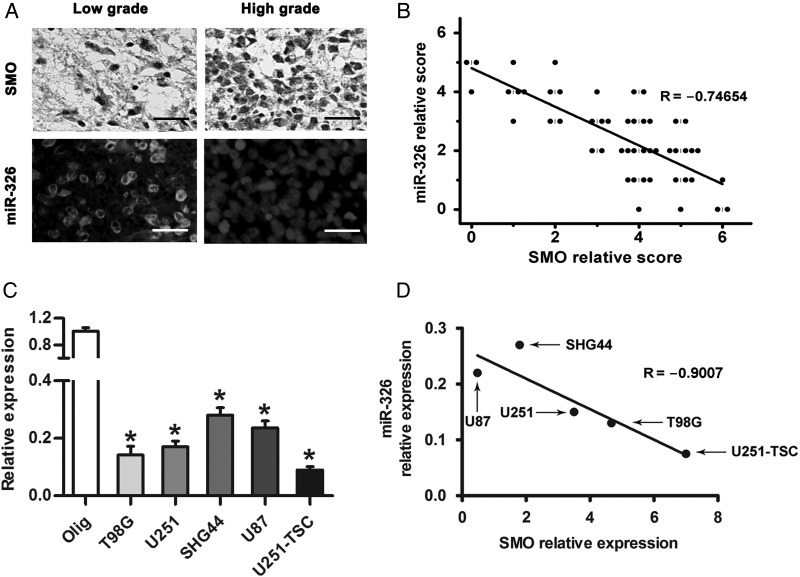
SMO Expression Was Inversely Correlated With miR-326
Several studies have reported that miR-326 is decreased in gliomas and has a therapeutic potential against brain tumors.30–32 To further investigate whether reduced miR-326 expression correlates with SMO expression levels in gliomas, patient specimens were used to examine its expression by IHC and FISH. The results suggested that high-grade gliomas contain comparatively higher SMO expression and lower miR-326 levels than low-grade specimens (Fig. 3A). Spearman's correlation analysis demonstrated that SMO in tumor tissues was inversely correlated with miR-326 expression (Fig. 3B). Furthermore, we examined miR-326 expression in our glioma cell lines by qRT-PCR (Fig. 3C). Combined with the relative expression of SMO, a linear inverse correlation was observed, verifying that SMO expression was inversely correlated with miR-326 (Fig. 3D).
Fig. 3.
SMO expression was inversely correlated with miR-326 expression in glioma. (A) The expression of SMO and miR-326 in glioma specimens was assessed by IHC and FISH (scale bars, 20 µm). (B) Spearman's correlation analysis was used to determine the correlation between the expression-level scores of SMO and miR-326 (Spearman's correlation analysis, r = −0.74654, P < .001). (C) qRT-PCR analysis showed that T98G, U87, U251, SHG44, and U251-TSC glioma cells expressed low levels of miR-326 compared with Olig. The data represent mean ± SE of 3 replicates (*P < .05). (D) A plot of the linear inverse correlation between the expression of SMO and miR-326 expression in U87, T98, U251, SHG44, and U251-TSC cells (P < .05, linear correlation r = −0.9007).
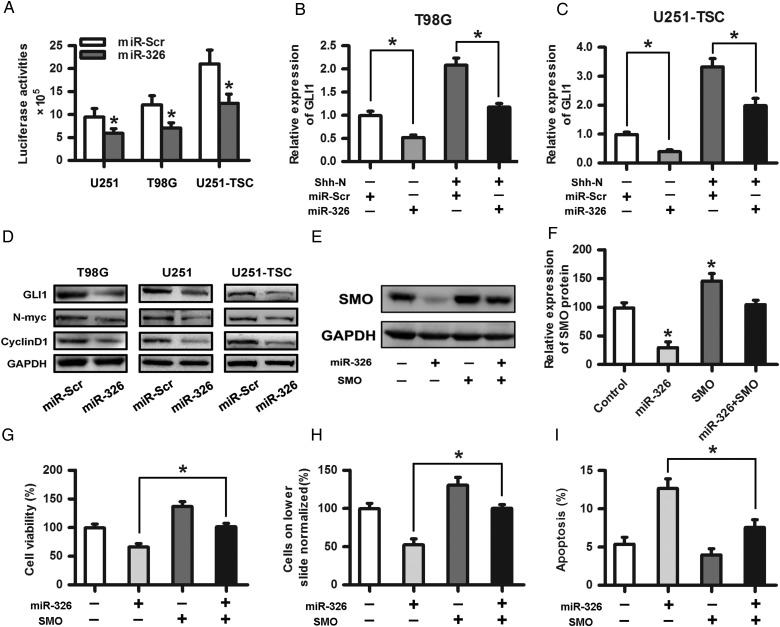
Upregulation of miR-326 Affected the Hh Signaling Pathway and Rescued the Protumor Effects of SMO
After confirming the relationship between SMO and miR-326, we intended to test the effect of upregulated miR-326 on Hh activity. We introduced an expression construct containing the entire SMO coding sequence without its 3′-UTR fragment into miR-326- or miR-Scr–overexpressing cells, followed by cotransfection of the 8×-GLI reporter downstream of the luciferase gene to determine the Hh pathway transcriptional activity. Our results showed that miR-326 upregulation reduced the Hh activity by more than 30% (Fig. 4A; P < .05). Subsequently, we investigated whether GLI1 expression was impacted by miR-326 upregulation with or without recombinant Shh-N-Terminus. qRT-PCR showed that miR-326 upregulation significantly reduced GLI1 expression (Fig. 4B and C). Moreover, Western blot assays showed that miR-326 upregulation decreased the expression of GLI1 and its downstream target proteins, including N-myc and CyclinD1, in glioma and tumor stem cells (Fig. 4D). To determine whether the protumor effects of SMO can be rescued by the upregulation of miR-326, Western blot assays were used primarily to evaluate the effect on protein levels. The results showed that miR-326 upregulation significantly decreased SMO expression (Fig. 4E and F). In addition, invasion, CCK-8, and apoptosis assays were adopted to further verify that miR-326 upregulation partially rescued SMO-dependent effects on glioma proliferation, invasion, and apoptosis (Fig. 4G–I).
Fig. 4.
Upregulation of miR-326 affected the Hh signaling pathway and rescued the protumor effects of SMO. (A) A plasmid containing the entire SMO coding sequence without its 3′-UTR fragment was transfected into miR-326 or miR-Scr overexpressing cells followed by cotransfection of the reporter containing 8 directly repeated copies of a consensus GLI binding site (8×-GLI) downstream of the luciferase gene to determine the Hh pathway transcriptional activity. The data represent the mean ± SE of 3 replicates (*P < .05). (B) and (C) The overexpression of miR-326 significantly decreased the GLI1 expression, as shown by qRT-PCR with or without N-Shh compared with the miR-Scr-treated group. (D) The Western blot assay indicated that glioma cells transfected with miR-326 efficiently restrained the protein expression of GLI1, N-myc, and CyclinD1 compared with the miR-Scr-treated group. GAPDH was used as a control. (E) and (F) SMO expression levels in U251 cells transfected with SMO and/or miR-326 were assessed by Western blot. Representative cartograms showing the relative expression of the SMO protein between different groups (*P < .05). (G), (H), and (I) Representative cartograms showing that proliferation, invasion and apoptosis are regulated by miR-326 and/or SMO.
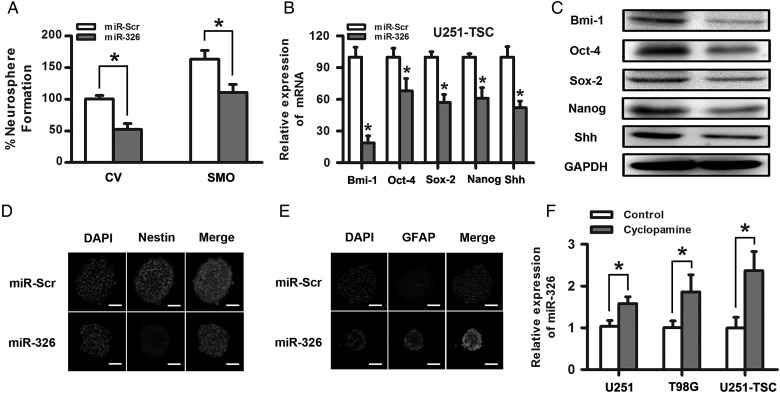
miR-326 Upregulation Regulated the Ability of Self-renewal and Stemness and Prompted Differentiation in Glioma Stem Cells
Evidence has already demonstrated that Hh signaling regulates stemness and self-renewal capacity in CD133+ glioma cancer stem cells.5,33 We further examined the role of miR-326 in the differentiation, stemness, and self-renewal of CD133+ glioma tumor stem cells. First, we examined the effects of miR-326 on neurosphere formation in U251 TSCs, which was thought to be a characteristic of self-renewal in the glioma stem-like cell subpopulation. We observed a significant decrease in the number and volume of the neurospheres transfected with miR-326 compared with scramble, suggesting that miR-326 plays a role in glioma stem cells, at least in part, by blocking the SMO-mediated pathways involved in stem cell renewal (Fig. 5A). Subsequently, qRT-PCR and Western blot assays showed that miR-326 upregulation decreased the expression of specific stemness markers, including Bmi-1, Oct-4, Sox2, Nanog, and Shh (Fig. 5B and C). Last, to further investigate whether the ability of stemness and differentiation were influenced by miR-326 upregulation, immunofluorescence was used to examine the expression of markers of stem cells (nestin) and glial cells (GFAP) in U251-TSCs. The results suggested that miR-326 upregulation significantly decreased the percentage of nestin-positive cells and enhanced GFAP expression in glial differentiation (Fig. 5D and E). All of the above data suggest that the glioma tumor stem cell properties can be partly regulated by changes in miR-326.
Fig. 5.
Upregulation of miR-326 regulated the ability of self-renewal and stemness and prompted differentiation in glioma stem cells. (A) miR-326 upregulation significantly decreased the percentage of neurosphere formation with or without SMO overexpression. (B) and (C) The upregulation of miR-326 decreased Bmi-1, Oct-4, Sox-2, Nanog, and Shh expression, as demonstrated by qRT–PCR and Western blot assays. (D) and (E) The overexpression of miR-326 partly decreased nestin expression and increased the GFAP expression in U251-TSC cells, as determined by immunofluorescence (scale bars, 50 µm). (F) Inhibition of the Hh pathway by antagonists that bind to SMO (cyclopamine,10uM) elevated the miR-326 expression in glioma cells. The data represent mean ± SE of 3 replicates (*P < .05).
Inhibition of the Hh Pathway Partially Restored miR-326 Expression
After confirming that SMO was a direct target of miR-326 that can be downregulated by miR-326 overexpression, we questioned whether blocking the Hh pathway would influence miR-326 expression. Cyclopamine was used to block the Hh signaling pathway in U251, T98G, and U251-TSC cells for 48 hours, and qRT-PCR indicated that miR-326 expression increased 1.6-multiples, 1.9-multiples and 2.3-multiples, respectively, compared with glioma cell lines exposed to dimethyl sulfoxide (P < .05; Fig. 5F). These data suggested that miR-326 was partly regulated by Hh/Smo activity in some ways.
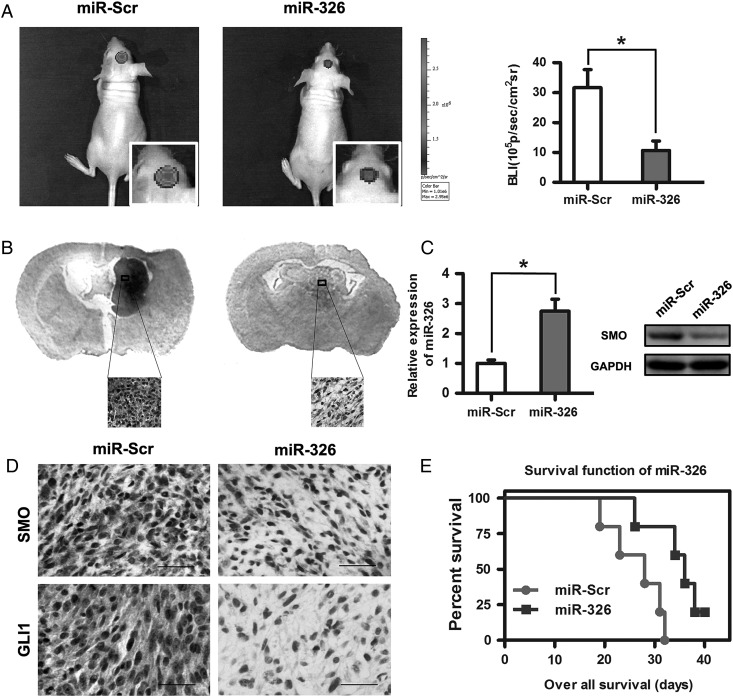
miR-326 Overexpression Inhibited Tumor Growth in Vivo and Prolonged Survival
Because miR-326 upregulation decreased SMO expression and the activity of the Hh pathway in gliomas in vitro, we evaluated its antiglioma effects in vivo. The results showed that miR-326 mimic-treated cells displayed a marked reduction in tumor size (Fig. 6A). To further verify tumor growth in both groups visually, H&E-stained coronal brain sections were used to show representative tumor xenografts (Fig. 6B). Subsequently, qRT-PCR analysis confirmed that miR-326 expression was increased in the tumor compared with scramble controls (Fig. 6C). Moreover, miR-326 mimic-treated U251 cells decreased the expression of SMO, GLI1, and nestin and increased the expression of GFAP (Fig. 6D and Supplementary material, Fig. S4). These data indicated that miR-326overexpression in vivo functioned similarly to that in vitro. To further evaluate the therapeutic effect of miR-326 on nude mice, the survival period of each group (n = 7/group) was analyzed by Kaplan–Meier curve. The miR-326 mimic-treated group showed a significant improvement in survival compared with the control group (P < .05;) (Fig. 6E) until the end of the observation period.
Fig. 6.
Overexpression of miR-326 inhibited tumor growth in vivo and prolonged survival. (A) Luminescence imaging for miR-326–treated U251-luc tumors versus scramble-treated controls. (B) H&E-stained coronal brain sections showing representative tumor xenografts. (C) miR-326 and SMO expression in an intracranial graft assessed by RT-PCR and Western blot. (D) SMO and GLI1 expression after transfecting miR-326 in tumor sections following IHC analysis (*P < .05; the bars represent 25 μm). (E) Kaplan-Meier survival curves indicating that mice transfected with miR-326 showed a significantly better outcome than the miR-Scr-treated group (*P < .05).
Discussion
Although there have already been several studies related to miRNA and its functional significance, little is known regarding miRNA interactions with the Hh pathway in gliomas. M Ehtesham et al34 have provided evidence that the Hh pathway was activated in a ligand-dependent manner, with a strict requirement for exogenous Hh ligand in glioma progenitor cells. Our previous study also reported that the Hh pathway is abnormally activated in malignant gliomas and can be considered an alternative therapeutic strategy once suppressed. In this way, secreted Hh ligands binds to PTCH1 and blocks the inhibition of SMO. SMO activation then leads to changes in gene expression through the action of GLI proteins.35 In this study, we emphasized the role of SMO in gliomas. We found that SMO was upregulated in glioma samples and cell lines and associated with tumor grade and poor survival. SMO, a 7-transmembrane protein that transduces the Hh signal across the plasma membrane has been demonstrated to be activated in many cancers, including basal cell carcinoma, thyroid cancer, pancreatic carcinoma, leukemia, medulloblastoma, non-small cell lung cancer and glioblastoma.5,36–41 The inhibition of SMO can suppress tumor cell growth and invasion, induce apoptosis, enhance sensitivity to chemotherapy, and prolong the survival of established tumor xenografts in several tumors.39,42–45 We also found that SMO inhibition affected diverse biological processes in gliomas. In our study, we selected 2 glioma cell lines (T98G and U251) and a glioma stem cell line (U251-TSC) as our experimental subjects because of their relatively high SMO expression compared with other cell lines. The inhibition of SMO by its specific siRNA suppressed the proliferation and invasion and induced the apoptosis of glioma cells. These results were similar to those of previous studies, which used GBM specimens instead of glioma cell lines.43 Eimer et al demonstrated that cyclopamine cooperates with EGFR inhibition to deplete stem-like cancer cells in glioblastoma-derived spheroid cultures.46 Furthermore, Takezaki et al reported that the Hh pathway is indispensable for glioma-initiating cell proliferation and tumorigenesis. The Hh signaling inhibitors prevented glioma-initiating cell proliferation, whereas signaling inhibitors for Notch or Wnt did not.47 These data showed that Hh signaling plays an important role in gliomas and can be targeted as an alternative potential therapeutic pathway.
miR-326 has been reported as a tumor suppressor miRNA in various tumor types, including gliomas.48,49 In addition, several miR-326targets have been identified, including Notch-1, Notch-2, PKM2, and NOB1 in gliomas.30–32 However, in our study, combining bioinformatics prediction, functional experiments in vitro, and luciferase reporter assays, SMO was verified as a direct target of miR-326. This result raised the appealing prospect that miR-326 has multiple targets that could be involved in various pathways. Subsequently, a significant inverse correlation was verified between miR-326 expression and SMO expression in glioma tissues and cell lines, further confirming that SMO is an endogenous target of miR- 326. Importantly, upregulation not only inhibited the activity of the Hh pathway but also reduced the expression of additional Hh signaling components, including GLI1, N-myc, and CyclinD1. In addition, transfection with miR-326 mimics partially rescued the protumor effects of SMO with regard to invasion, proliferation, and apoptosis in glioma cells. All of the above data further demonstrated the biological relevance of the miR-326/SMO relationship.
Santini and Clement have reported that Hh signaling is closely related to the stemness and self-renewal of cancer stem cells and glioma stem cells.5,50 In this study, we found that miR-326 upregulation also reduced the volume and number of tumor spheres and decreased the expression of stem cell-associated proteins Bmi1, Nanog, Shh, Oct4, and Sox2 in CD133+ glioma cells. Moreover, miR-326 upregulation significantly decreased the fraction of nestin-expressing stem cells, increased the fraction of GFAP-expressing cells, and prompted differentiation, suggesting its direct inhibition of Hh/GLI1 signaling and stemness through targeting SMO in CD133+ cells.
Interestingly, when SMO was inhibited by its specific inhibitor, the expression of miR-326 was also elevated. This might have been caused by epigenetic abnormalities, including changes in DNA methylation or histone modifications. Thus, both miR-326 and SMO were partly regulated by each other as a feedback loop. Furthermore, the transfection of glioma cells with miR-326 mimics in the orthotopic model reduced tumorigenicity, which might have been mediated by SMO inhibition. Importantly, miR-326 upregulation prolonged the survival of nude mice.
In summary, we found that the upregulation of miR-326, as a suppressor miRNA in gliomas, inhibited proliferation, induced apoptosis, and decreased the stemness of gliomas by targeting SMO through the Hh pathway. Furthermore, SMO inhibition partly restored miR-326 expression, suggesting that the miR-326/SMO feedback loop could be considered an alternative therapy for gliomas in the future.
Supplementary Material
Funding
This work was supported by the National High Technology Research and Development Program 863 (2012AA02A508), China National Natural Scientific Fund (81372700). China Postdoctoral Science Foundation (2013M531121), Shanghai Postdoctoral Science Foundation (13R21411300), China Postdoctoral Science special Foundation funded project (2014T70390).
Supplementary Material
Acknowledgments
We thank professor X.Ban for the technical assistance. We also thank Dr. Hongjun Wang for providing the database information.
Conflict of interest statement. None declared.
References
- 1.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Garner JM, Fan M, Yang CH, et al. Constitutive activation of signal transducer and activator of transcription 3 (STAT3) and nuclear factor κB signaling in glioblastoma cancer stem cells regulates the Notch pathway. J Biol Chem. 2013;288(36):26167–26176. doi: 10.1074/jbc.M113.477950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sareddy GR, Kesanakurti D, Kirti PB, et al. Nonsteroidal anti-inflammatory drugs diclofenac and celecoxib attenuates Wnt/beta-catenin/Tcf signaling pathway in human glioblastoma cells. Neurochem Res. 2013;38(11):2313–2322. doi: 10.1007/s11064-013-1142-9. [DOI] [PubMed] [Google Scholar]
- 4.Kim E, Kim M, Woo DH, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23(6):839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clement V, Sanchez P, de Tribolet N, et al. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17(2):165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Echelard Y, Epstein DJ, St-Jacques B, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75(7):1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 7.Stone DM, Hynes M, Armanini M, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384(6605):129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 8.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15(6):801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J, Graves S, Koch U, et al. Hedgehog signaling is dispensable for adult hematopoietic stem cell function. Cell Stem Cell. 2009;4(6):548–558. doi: 10.1016/j.stem.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Lv X, Jiang J, et al. Dual roles of Hh signaling in the regulation of somatic stem cell self-renewal and germline stem cell maintenance in Drosophila testis. Cell Res. 2013;23(4):573–576. doi: 10.1038/cr.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milla LA, Arros A, Espinoza N, et al. Neogenin1 is a sonic hedgehog target in medulloblastoma and is necessary for cell cycle progression. Int J Cancer. 2014;134(1):21–31. doi: 10.1002/ijc.28330. [DOI] [PubMed] [Google Scholar]
- 12.Tang S, Bonaroti J, Unlu S, et al. Sweating the small stuff: microRNAs and genetic changes define pancreatic cancer. Pancreas. 2013;42(5):740–759. doi: 10.1097/MPA.0b013e3182854ab0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinho FG, Frampton AE, Nunes J, et al. Downregulation of microRNA-515–5p by the estrogen receptor modulates sphingosine kinase 1 and breast cancer cell proliferation. Cancer Res. 2013;73(19):5936–5948. doi: 10.1158/0008-5472.CAN-13-0158. [DOI] [PubMed] [Google Scholar]
- 14.Siemens H, Neumann J, Jackstadt R, et al. Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and beta-catenin predicts distant metastasis of colon cancer. Clin Cancer Res. 2013;19(3):710–720. doi: 10.1158/1078-0432.CCR-12-1703. [DOI] [PubMed] [Google Scholar]
- 15.Cai J, Wu J, Zhang H, et al. miR-186 downregulation correlates with poor survival in lung adenocarcinoma, where it interferes with cell-cycle regulation. Cancer Res. 2013;73(2):756–766. doi: 10.1158/0008-5472.CAN-12-2651. [DOI] [PubMed] [Google Scholar]
- 16.Agatheeswaran S, Singh S, Biswas S, et al. BCR-ABL mediated repression of miR-223 results in the activation of MEF2C and PTBP2 in chronic myeloid leukemia. Leukemia. 2013;27(7):1578–1580. doi: 10.1038/leu.2012.339. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Yang BB. Stress response of glioblastoma cells mediated by miR-17–5p targeting PTEN and the passenger strand miR-17–3p targeting MDM2. Oncotarget. 2012;3(12):1653–1668. doi: 10.18632/oncotarget.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Z, Zhang J, Qian X, et al. AC1MMYR2, an inhibitor of dicer-mediated biogenesis of Oncomir miR-21, reverses epithelial-mesenchymal transition and suppresses tumor growth and progression. Cancer Res. 2013;73(17):5519–5531. doi: 10.1158/0008-5472.CAN-13-0280. [DOI] [PubMed] [Google Scholar]
- 19.Liu C, Tang DG. MicroRNA regulation of cancer stem cells. Cancer Res. 2011;71(18):5950–5954. doi: 10.1158/0008-5472.CAN-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Aranha MM, Santos DM, Sola S, et al. miR-34a regulates mouse neural stem cell differentiation. PloS One. 2011;6(8):e21396. doi: 10.1371/journal.pone.0021396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Zhang A, Li Y, et al. MiR-24 regulates the proliferation and invasion of glioma by ST7L via beta-catenin/Tcf-4 signaling. Cancer Lett. 2013;329(2):174–180. doi: 10.1016/j.canlet.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 23.Zhang KL, Han L, Chen LY, et al. Blockage of a miR-21/EGFR regulatory feedback loop augments anti-EGFR therapy in glioblastomas. Cancer Lett. 2014;342(1):139–149. doi: 10.1016/j.canlet.2013.08.043. [DOI] [PubMed] [Google Scholar]
- 24.Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67(9):4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 25.Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2(6):1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- 26.Du WZ, Feng Y, Wang XF, et al. Curcumin suppresses malignant glioma cells growth and induces apoptosis by inhibition of SHH/GLI1 signaling pathway in vitro and vivo. CNS Neurosci Ther. 2013;19(12):926–936. doi: 10.1111/cns.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Zhang J, Feng Y, et al. MiR-410 regulates MET to influence the proliferation and invasion of glioma. Int J Biochem Cell Biol. 2012;44(11):1711–1717. doi: 10.1016/j.biocel.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 28.Zhou X, Ren Y, Moore L, et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90(2):144–155. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 29.Zhu ZH, Sun BY, Ma Y, et al. Three immunomarker support vector machines-based prognostic classifiers for stage IB non-small-cell lung cancer. J Clin Oncol. 2009;27(7):1091–1099. doi: 10.1200/JCO.2008.16.6991. [DOI] [PubMed] [Google Scholar]
- 30.Kefas B, Comeau L, Floyd DH, et al. The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J Neurosci. 2009;29(48):15161–15168. doi: 10.1523/JNEUROSCI.4966-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, Xu T, Yan Y, et al. MicroRNA-326 functions as a tumor suppressor in glioma by targeting the Nin one binding protein (NOB1) PloS One. 2013;8(7):e68469. doi: 10.1371/journal.pone.0068469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kefas B, Comeau L, Erdle N, et al. Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro Oncol. 2010;12(11):1102–1112. doi: 10.1093/neuonc/noq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zbinden M, Duquet A, Lorente-Trigos A, et al. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010;29(15):2659–2674. doi: 10.1038/emboj.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehtesham M, Sarangi A, Valadez JG, et al. Ligand-dependent activation of the hedgehog pathway in glioma progenitor cells. Oncogene. 2007;26(39):5752–5761. doi: 10.1038/sj.onc.1210359. [DOI] [PubMed] [Google Scholar]
- 35.Dlugosz AA, Talpaz M. Following the hedgehog to new cancer therapies. New Engl J Med. 2009;361(12):1202–1205. doi: 10.1056/NEJMe0906092. [DOI] [PubMed] [Google Scholar]
- 36.Atwood SX, Li M, Lee A, et al. GLI activation by atypical protein kinase C iota/lambda regulates the growth of basal cell carcinomas. Nature. 2013;494(7438):484–488. doi: 10.1038/nature11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bohinc B, Michelotti G, Diehl AM. Hedgehog signaling in human medullary thyroid carcinoma: a novel signaling pathway. Thyroid. 2013;23(9):1119–1126. doi: 10.1089/thy.2012.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao J, Zhang L, Gao J, et al. Aberrant expression of PTCH (patched gene) and Smo (smoothened gene) in human pancreatic cancerous tissues and its association with hyperglycemia. Pancreas. 2006;33(1):38–44. doi: 10.1097/01.mpa.0000222319.59360.21. [DOI] [PubMed] [Google Scholar]
- 39.Babashah S, Sadeghizadeh M, Hajifathali A, et al. Targeting of the signal transducer Smo links microRNA-326 to the oncogenic Hedgehog pathway in CD34+ CML stem/progenitor cells. Int J Cancer. 2013;133(3):579–589. doi: 10.1002/ijc.28043. [DOI] [PubMed] [Google Scholar]
- 40.Ferretti E, De Smaele E, Miele E, et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27(19):2616–2627. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gialmanidis IP, Bravou V, Amanetopoulou SG, et al. Overexpression of hedgehog pathway molecules and FOXM1 in non-small cell lung carcinomas. Lung Cancer. 2009;66(1):64–74. doi: 10.1016/j.lungcan.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 42.Dijkgraaf GJ, Alicke B, Weinmann L, et al. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res. 2011;71(2):435–444. doi: 10.1158/0008-5472.CAN-10-2876. [DOI] [PubMed] [Google Scholar]
- 43.Bar EE, Chaudhry A, Lin A, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25(10):2524–2533. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarangi A, Valadez JG, Rush S, et al. Targeted inhibition of the Hedgehog pathway in established malignant glioma xenografts enhances survival. Oncogene. 2009;28(39):3468–3476. doi: 10.1038/onc.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.You M, Varona-Santos J, Singh S, et al. Targeting of the Hedgehog signal transduction pathway suppresses survival of malignant pleural mesothelioma cells in vitro. J Thorac Cardiovasc Surg. 2014;147(1):508–516. doi: 10.1016/j.jtcvs.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 46.Eimer S, Dugay F, Airiau K, et al. Cyclopamine cooperates with EGFR inhibition to deplete stem-like cancer cells in glioblastoma-derived spheroid cultures. Neuro Oncol. 2012;14(12):1441–1451. doi: 10.1093/neuonc/nos266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takezaki T, Hide T, Takanaga H, et al. Essential role of the Hedgehog signaling pathway in human glioma-initiating cells. Cancer Sci. 2011;102(7):1306–1312. doi: 10.1111/j.1349-7006.2011.01943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S, Lu S, Geng S, et al. Expression and clinical significance of microRNA-326 in human glioma miR-326 expression in glioma. Med Oncol. 2013;30(1):373. doi: 10.1007/s12032-012-0373-y. [DOI] [PubMed] [Google Scholar]
- 49.Qiu S, Lin S, Hu D, et al. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J Transl Med. 2013;11:10. doi: 10.1186/1479-5876-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santini R, Vinci MC, Pandolfi S, et al. Hedgehog-GLI signaling drives self-renewal and tumorigenicity of human melanoma-initiating cells. Stem Cells. 2012;30(9):1808–1818. doi: 10.1002/stem.1160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.