Abstract
Microscopy has revealed a tremendous diversity of bacterial and eukaryotic forms. More recent molecular analyses show discordance in estimates of biodiversity based on morphological analyses. Moreover, phylogenetic analyses of the diversity of microbial forms have revealed evidence of convergence at scales as large as interdomain – i.e. convergent forms shared between bacteria and eukaryotes. Here, we highlight examples of such discordance, focusing on exemplary lineages such as testate amoebae, ciliates and cyanobacteria, which have long histories of morphological study. We discuss examples in two categories: 1) morphologically identical (or highly similar) individuals that are genetically distinct and 2) morphologically distinct individuals that are genetically distinct. We argue that hypotheses about discordance can be tested using the concept of neutral morphologies, or more broadly neutral phenotypes, as a null hypothesis.
Keywords: microbial evolution, molecular data, morphology, neutral evolution
“Thus, we have a neutral-morphology theory of evolution, where a variety of morphologies are equally successful in a particular environment. This makes an interesting contrast to the neutral-gene theory of Motoo Kimura. In the former, for one reason or another, natural selection fails to discriminate among phenotype morphologies, each of which has a distinctive genotype; in the latter, selection fails to discriminate among genotypes that all could have the same phenotype.”
John Tyler Bonner, “Randomness in Evolution”
Introduction
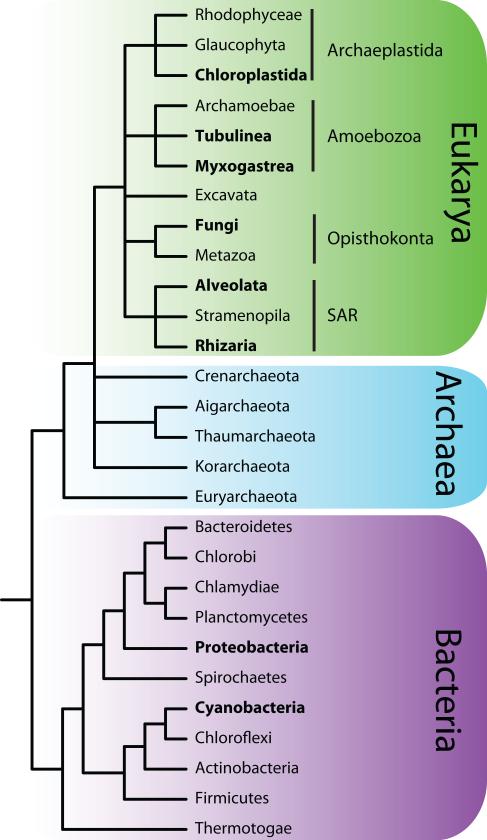
Since the bulk of biodiversity is microbial, and microbes play crucial roles in biogeochemical cycles and as pathogens, accurately describing the evolutionary history of microbial diversity is an important endeavor. Our understanding of microbial life on Earth has changed dramatically as microbes have been re-categorized from a single Kingdom (i.e. Protista or Protoctista) by dividing them first into bacteria (i.e. Monera) and eukaryotic microbes (i.e. Protists), and then finally into three domains with the discovery of Archaea, in 1977 (reviewed in [1]). Starting in the mid-1980s, molecular systematic studies of rDNA sequences increased our knowledge of the biodiversity of bacteria, microbial eukaryotes, and Archaea (the latter group having suffered a dearth of morphology-based studies [2]). More recently, the application of molecular techniques to analyses of whole genomes has further elucidated the tremendous diversity of microorganisms, which in turn has led to a clearer understanding of the placement of microbes across the tree of life (Fig. 1, [3, 4]).
Figure 1.
A simplified tree of life, including all three major domains. Lineages in bold are discussed in this manuscript. Relationships based on [11, 80, 81].
Discordance between morphology and molecules is best understood in microorganisms with a history of microscopic analyses
Microscopic organisms, or “microbes”, are diverse in many aspects including morphology (the focus of this manuscript), physiology, and genetics [5]. The cannon of evolutionary biology dictates that phenotype, including morphology, reflects genotype. However, for microbes there is considerable discordance at many levels in patterns of biodiversity as assessed by morphology compared to insights from molecular data. To exemplify this discordance, we highlight examples from microbial clades that are marked by a long history of morphological study, including ciliates, testate (shelled) amoebae, and cyanobacteria. We recognize that these merely represent a ‘drop in the ocean’ of cases of discordance between morphology and molecules, and our intent here is to present key examples rather than to be exhaustive.
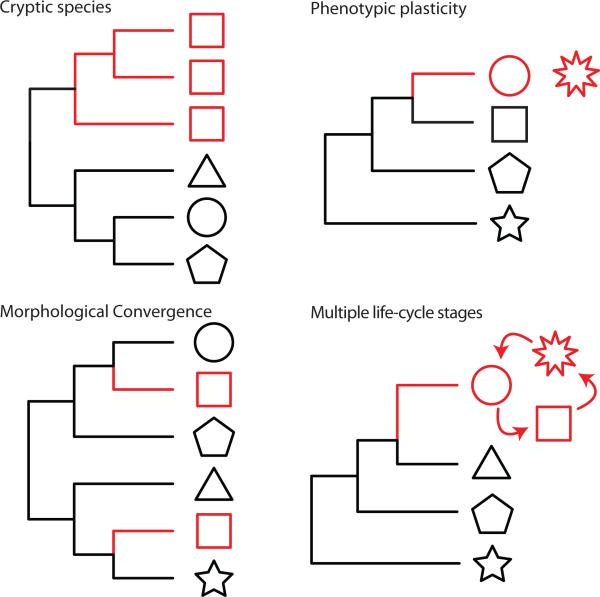
In order to discuss the myriad of issues surrounding the discordance of morphological and molecular data at microbial levels, we consider two main types of discordance: 1) the same morphology is manifested in multiple genetic lineages (e.g. convergent morphologies, cryptic species) and 2) multiple morphologies are associated with a single genetic entity (e.g. plastic phenotypic morphologies, life cycle variants; Box 1, Fig. 2). Such categorization is helpful given the profusion of terms in the literature dealing with this complex topic [6, 7]. Hence, we opt to define the terminology used here in accordance with what seems most appropriate for microbial lineages. Our choices seek to minimize semantic discussion and focus on the biological issue. Nevertheless, it is important to remember that nature is not the least bit concerned with the categories described here, and there are surely examples of intermediary cases and/or cases that do not fit.
Figure 2.
Categories of molecular and morphological discordance among microbes from a phylogenetic perspective. To the left, one morphology, multiple lineages category. To the right, one lineage, multiple morphologies.
Category 1: Cases with one morphology in multiple genetic lineages
When morphologies converge
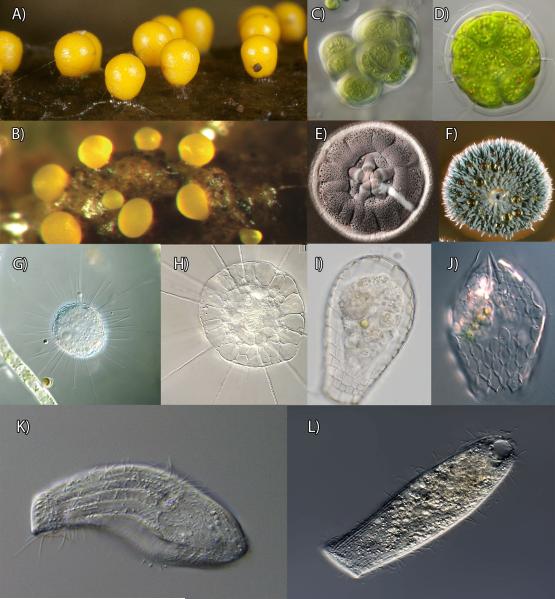
Examples of convergent morphologies are common among microbial lineages, and extend from convergence between domains (e.g. eukaryotes and bacteria; Fig. 3A-F), through examples between major clades (Fig. 3, images G-L), to convergence at smaller levels that have led to nearly identical morphologies present among non-monophyletic lineages. At the deepest evolutionary scales, there has been convergence to similar morphologies between lineages of bacteria and eukaryotes. For example, myxobacteria are able to produce macroscopic fruiting bodies when starved [8], hence resulting in very similar structures to those found among eukaryotic dictyostelids and myxogastrid amoebozoans (Fig. 3A-B, [9]). Similarly, the rounded aggregate morphology of small, photosynthetic organisms seems to have occurred many times in evolution, as exemplified by several cyanobacterial lineages and the volvocales (Fig. 3B-C). Finally, the microscopic hyphal form, with formation of visible colonies, has appeared independently in fungi and bacteria that absorb nutrients from their environment (Fig. 3D-E).
Figure 3.
Examples of striking morphological convergence between bacterial and eukaryotic domains (A-F) and among deeply-diverged eukaryotic clades (G-L). A: Fruiting bodies of Trichia varia, an amoebozoan eukaryote. B: Fruiting bodies of Myxococcus xanthus, a bacterium. C: Gloeocapsa sp., a cyanobacterium. D: Pandorina morum, a green alga. E: a colony of the bacterium Streptomyces. F: a colony of the fungus Penicillium. The last common ancestor between the organisms in figure A and B, C and D, and E and F is the last common ancestor of all life, thus, each of these pairs of organisms must have diverged over 3.5 billion years ago; G: Acanthocystis penardi, the “sun-animalcule”, a centrohelid eukaryote (group with unknown affinities). H: Actinosphaerium eichorni, another “sun-animalcules”, however, molecular data has shown that this is a stramenopile (related to diatoms). I: Quadrullela, an arcellinid amoebozoan, or lobose testate amoeba. J: Euglypha, and euglyphid rhizarian, or filose testate amoeba. K: Stephanopogon apogon, a heterolobosean, the heterolobosea are typically characterized by amoeboid forms with a flagellate life-cycle stage. L: Spathidium, a ciliate. In these eukaryotic pairs of organisms, each of the pair belongs in a very distant clade to the other, with a last common ancestor only billions of years ago. Image credits: B, E, F are from Wikimedia; A, C, D, J are from micro*scope; G, H were kindly provided by Mr. Wolfgang Bettighoefer; J is from Daniel Lahr.
Convergence has also occurred at deep scales within eukaryotes. For example, the shelled amoeboid body plan referred to as “testate amoebae” is present in at least two major lineages that are quite distantly related: the euglyphid testate (Fig. 3J) amoebae in the Rhizaria and the arcellinid testate amoebae (Fig. 3I) in the Amoebozoa [10]. These two lineages have traditionally been distinguished as “filose” and “lobose” testates, respectively. The tremendous genetic divergence between them has only recently been demonstrated: comprehensive multigene phylogenies indicate that the last common ancestor between these two groups is actually the last common ancestor of all eukaryotes [11, 12]. More recently, a third independent origin of shells has been revealed, as amphitremid testate amoebae (which were previously thought to be related to euglyphids) were demonstrated to be the sister group to labyrinthulids [13].
Morphological convergence between disparate groups is such a powerful deception that it has confused classification. The classical ‘Heliozoa’ (sun animalcules), characterized by a round morphology and stiff pseudopods supported by an internal bundle of microtubules, fall within four distantly-related clades in molecular analyses [14]. The strikingly similar morphology between centrohelids and actinophryids (Figures 3G and H) must be a case of convergence, because the latter are certainly stramenopile, while the former represent an orphan lineage (i.e. one without a clear sister taxon [15]). Finally, the enigmatic Stephanopogon (Fig. 3K), initially classified as a ciliate (Figure 3L) then as a flagellate or its own lineage [16], is now considered as a heterolobosean based on deep ultrastructural and molecular analyses [17].
Convergence is also found among the cyanobacteria, which were originally separated into four orders based on body forms: Chroococcales (coccoid forms), Oscillatoriales (homocytous filamentous forms), Nostocales (heterocytous filamentous forms, capable of false branching), and Stigonematales (heterocytous filamentous forms, capable of true branching). However, analyses of molecular and ultrastructural data revealed that coccoid and filamentous forms had evolved more than once, and some ‘thin’ filamentous forms are more closely related to some coccoid forms than they are to wider filamentous forms [18]. Furthermore, true-branching and false-branching were also found to have evolved independently in several lineages based on molecular data, hence invalidating the criterion previously used to separate the orders Nostocales and Stigonematales [18].
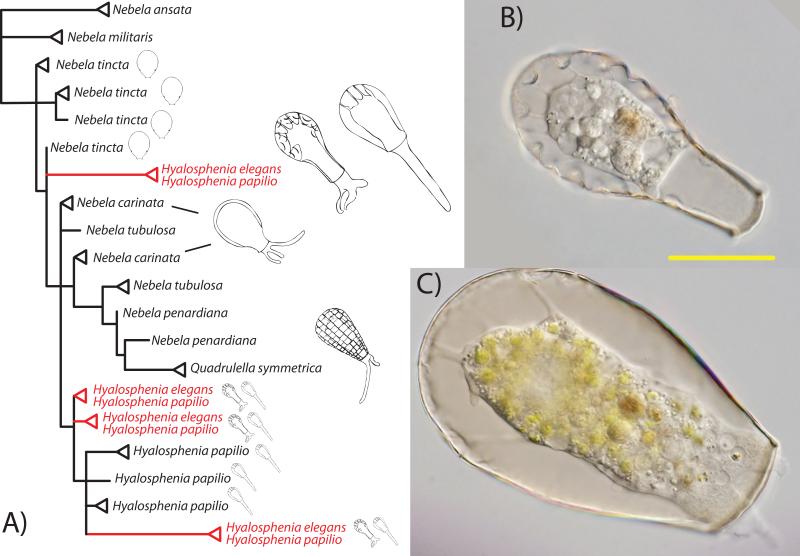
Smaller clades including testate amoebae, ciliates and planktonic cyanobacteria also display convergence. There is extensive phenotypic convergence within the ‘core Nebelas’ [19, 20], because the same morphologies (e.g. the morphospecies Hyalosphenia papilio and Hyalosphenia elegans) are found in two disparate clades (Fig. 4). Among the scuticociliates, the morphologically-defined genera Mesanophrys, Uronema, and Parauronema are non-monophyletic [21], indicating morphological convergence. Similarly, Rajaniemi et al. [22] showed that the benthic and planktonic Anabaena were found in different clades, though these were later separated into several more recently erected genera (Chrysosporum, Dolichospermum, Sphaerospermopsis,and Macrospermum) based on molecular and ecological data [23].
Figure 4.
Historical reconstruction of relationships and morphology of nebelids. A: Phylogenetic history of nebelids, based on SSU rDNA. Drawings represent morphologies. The red branches in tree highlight instances where identical SSU rDNA sequences exist for both Hyalosphenia papilio and Hyalosphenia elegans. B: Light microscope images of H. elegans, and C: of H. papilio showing highly distinct morphologies. Images B and C are to the same scale, scale bar is 30 μm. Phylogenetic tree is based on previous studies [19, 20, 82].
Microbial morphospecies are often underlain by multiple genetic lineages
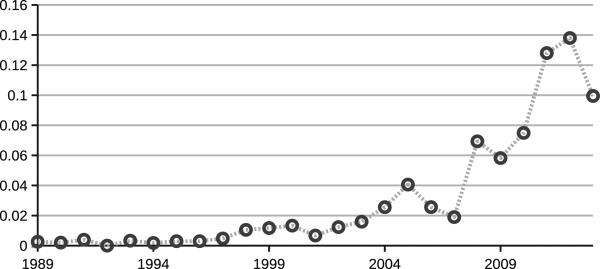
Cryptic species are defined as two or more independent genetic lineages that are considered a single morphospecies, or at least share high levels of morphological similarity [24, 25]. The term pseudocryptic has been used to describe species that appear identical by light microscopy but can be differentiated by subcellular structures, and/or minute ultrastructural morphologies [26]. With the application of molecular tools to studies of diverse lineages, the documentation of cases of cryptic species has risen nearly exponentially from the 1970s on (see Fig. 1 in [24]). Among microbes, studies on cryptic species have followed a similar rising trend, albeit about an order of magnitude fewer in absolute numbers (Fig. 5). As with plants and animals, cryptic species of microorganisms, which are often first assessed by light microscopy, turn out to have subtle morphological distinctions. In ciliates, for example, species that appear cryptic by light microscopy can be differentiated by small changes in infraciliature that can only be documented by techniques that are often outside the expertise of most field biologists, including transmission electron microscopy and/or silver staining [27, 28].
Figure 5.
Ratio of papers published proposing cryptic microbial species over total number of described microbial species from 1989-2013. Numbers of published papers were obtained by searching the online database SCOPUS with the query [species AND cryptic AND microb*]. Number of described species per year of microorganisms was obtained at the Index of Organismal Names database.
There are numerous described cases of cryptic species among microbial eukaryotes and bacteria, and we highlight just a few below. The testate (shelled) amoebae have been a fertile ground for recent findings of cryptic speciation. These organisms are easily recognized by the possession of ornamented and highly characteristic shells, which allowed a number of morphological and ecological studies in contrast to the other, little-known naked amoebae. For instance, numerous examples of cryptic species have been found within the genus Cyphoderia, euglyphid testate amoebae that inhabit Sphagnum dominated areas [29]. These organisms construct their shells from autogenous scales of biomineralized silica. Although very conspicuous at the genus level, identifying morphospecies has proven to be a challenge. Even a combination of light- and electron-microscopy can barely separate all distinct genetic lineages demonstrated by both nuclear small subunit ribosomal DNA (SSU rDNA) and mitochondrial cytochrome oxidase 1 (Cox1) sequences. Cyphoderia ampulla is represented by at least five phylotypes distributed in two distinct sub-clades alongside other lineages [30, 31]. These two subclades can only be distinguished morphologically by the degree of scale-overlap discernible in scanning electron microscopy [30]. The genus Euglypha also has demonstrated multiplicity of genetic lineages without any reliable morphological differentiation, in two cases: E. rotunda and E. filifera [29].
Recent findings in the amoebozoan testate amoeba genus Hyalosphenia suggest much more extensive cryptic speciation than expected based on morphological studies [19, 32]. These testate amoebae are not closely-related to the euglyphids mentioned above (Fig. 2I, J). No morphological features were found to distinguish between members of the twelve genetic lineages of H. papilio isolated in Heger et al. [32]. Additionally, these 12 lineages had a maximum divergence of 11.6% using the Cox1 gene, well beyond the typical divergence threshold used by researchers in other areas to indicate different species [20]. These data indicate not only that there are multiple cryptic lineages of H. papilio, but may also indicate that the morphology has emerged multiple times, as discussed in the convergence session above (Fig. 4).
Ciliates are another clade where cryptic species are common. As a clade of microbial eukaryotes, ciliates have a rich taxonomic history based on morphology of oral and somatic ciliature [33]. The discordance between morphology and genetics has been observed in many genera across the ciliate tree of life, including Chilodonella, Cyclidium, Vorticella, Pleuronema, and Paramecium [21, 34–41]. For example, the geneticist T. M. Sonneborn found that the morphospecies Paramecium aurelia could be divided into fourteen subgroups based on the mating behavior: cells of the same subgroup would readily form conjugant pairs with one another, but were essentially reproductively isolated from the other subgroups [42]. Similarly, several strains of the morphospecies Chilodonella uncinata with identical morphology (at least by light microscopy) are genetically distinct at mitochondrial SSU rDNA and in protein-coding loci, divergences varying from 2.2% to 13.5% [37]. Analysis of multiple genes reveals that the morphospecies C. uncinata is composed of multiple genetically distinct cryptic species [37]. Because of conjugation (sex) in ciliates, cryptic species can also be interpreted by the discordance between morphology and mating behavior, e.g. Paramecium and Tetrahymena (reviewed in [34]).
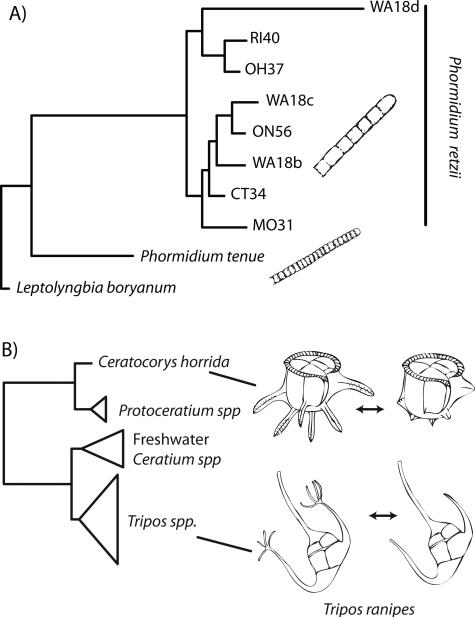
Cryptic diversity is commonly found among bacteria. In fact, separating species in bacteria using morphology is next to impossible; hence, the common practice is to characterize metabolic activity via culturing, among other features [2]. In contrast, cyanobacteria present diverse morphologies, which enabled a long history of taxonomic studies [18, 43]. Classic studies (starting in the late 1800's) were carried out using morphometric characteristics (e.g. [44, 45]). The morphospecies Phormidium retzii is among the most widespread macroalgal species in North America and presents a single morphotype from Canada to Central America. Furthermore, this morphospecies has been collected in all environments surveyed [46]. However, when genetic data is included, P. retzii, was shown to diverge from 88.4% to 98.4% in SSU rDNA and Random Amplified Polymorphic DNA (RAPD) data [47], revealing that the morphotype is actually a complex of cryptic species (Fig. 6A).
Figure 6.
Examples of microbial morphologies interpreted according to phylogenetic contexts. A: Cryptic speciation among Phormidium cyanobacteria. Historical reconstruction based on [47]. B: Phenotypic plasticity among dinoflagellates. The dinoflagellates Ceratocorys horrida and Tripos ranipes (= Neoceratium ranipes) display intense morphological modification on a short time span. Phylogenetic framework derived from Gomez et al. [83].
Beyond the examples detailed above, cryptic species are also common among diatoms [48–50]. The widespread marine diatom Pseudo-nitzschia delicatissima comprises up to three phylogenetic and reproductively distinctgroups, while Pseudo-nitzschia pseudodelicatissima consists of up to five [49] and the freshwater Sellaphora pupula has been shown to contain at least four different clades [51, 52]. We anticipate that additional examples of cryptic morphospecies will continue to be discovered as molecular tools are applied to analyze morphospecies, which have often been diagnosed only by light microscopy. Such discoveries allow for more detailed analyses of subcellular/ultrastructural structures that may well reveal subcellular morphological changes.
Category 2: When one genetic lineage presents multiple morphologies
Morphological variations in the life cycle and phenotypic plasticity are widespread among microorganisms
A second type of discordance is manifest when multiple morphologies exist within a single genetic lineage either due to phenotypic plasticity (often driven by environmental cues) or life cycle variants (generally driven by developmental signals). Given the limited data for distinguishing between these cases in most microbial lineages, we address this category in a single section.
Phenotypic plasticity of shell shape existing within some testate amoebae morphospecies and can be influenced by a variety of environmental conditions, and can be reversible [53–56]. Food source, temperature, and presence of insecticide affect the morphology of shell shape in clonal cultures under controlled conditions [56]. While most studies have focused exclusively on size of shell, it is possible that other characters that define tests are also plastic. Variation in the number of spines is a well-documented character that varies in culture [57], and has been implicated in at least two environmental morphometric studies: the presence/absence of spines in Difflugia tuberspinifera and Centropyxis aculeata may vary by ecotype [53, 58].
Recent data from Oliverio et al. [19] identified multiple identical SSU rDNA sequences for two distinct morphotypes, Hyalosphenia elegans and Hyalosphenia papilio (Figure 4). Hyalosphenia elegans has a long neck with a narrow aperture and distinctive indentations across the lateral margin, and H. papilio is typically larger with a wide aperture. Oliverio et al. [19] hypothesize that these drastic phenotypic differences may represent an intraspecific polymorphism, perhaps representing a sexual dimorphism. Under this scenario, H. elegans and H. papilio represent different mating types within one evolutionary line. This would be surprising because most testate amoebae are generally thought to be asexual [59]; however, there are indications that many lineages within arcellinida reproduce sexually [60, 61].
Cyanobacteria also present a number of plastic characters. These include color variation, thickness and color of the mucilaginous sheath, presence of gas vesicles, gliding, formation of ‘hairs’ [43]. Some of these variations are believed to be induced by ‘stress’ factors such as ultraviolet light, alternation between wet and dry conditions, reduction in nutrients (e.g. nitrogen and/or phosphorus [62]). In part because of challenges of culturing cyanobacteria, transformations between forms have not been demonstrated in all cases [63]. The genus Nostoc, a common nitrogen-fixing cyanobacterium found in aquatic and terrestrial habitats [64], has a relatively complex life cycle varying from a single filament to multiple coiled filaments within the same colony. The variability from a microscopic state with vegetative filaments to densely coiled filaments within a macroscopic colony also varies by species [64]. Interestingly, Hrouzek et al. [65] were able to separate Desmonostoc from Nostoc by differences in the life cycle coupled with genetic data.
A number of microbial eukaryotes have well documented life-cycle stage variation. Dinoflagellates typically present several different stages in their life cycle, exemplified by the genus Pfeisteria that has a complex life cycle including a number of forms (though perhaps not amoeboid forms as originally thought) [66]. Dictyostelids exist as solitary amoebae, and then through aggregation form a motile “slug” stage that ends up settling and transforming into a multicellular fruiting body stage [67]. Another unusual life-cycle variation is the diurnal shape-shifting performed by the dinoflagellate Tripos ranipes (= Ceratium ranipes) [68]. This organism generates “fingers” and “toes” (Fig. 6B), unusual appendages that protrude from the horns only during the day. The appendages are reabsorbed at night, which led to the erroneous description of a few separate species. The closely related Ceratocorys horrida similarly reabsorbs its spines when exposed to turbulent waters (Fig. 6B, [69]).
Is the concept of the neutral morphology consistent with examples of convergences between deeply-diverged lineages?
The concept of “neutral morphologies” [9] can serve as a null hypothesis to explain some of the incongruences highlighted here. As outlined in his book “Randomness in Evolution”, John Tyler Bonner hypothesizes that natural selection has less of an influence on morphology at the microbial scale and instead, morphological evolution at these scales is driven chiefly by randomness, or neutral processes [9]. Bonner argues that because morphological space is constrained (e.g. by principles of physics and engineering exerted at small scales) and because morphologies are typically not under selection at the microbial scale, they may become similar by chance [9]. Under such a scenario, ‘simple’ morphologies such as the “amoeboid” body-plan may represent cases where physical/engineering constraints (e.g. diffusion of nutrients and oxygen) limit the possible shapes of cells living in liquid media.
Under the neutral theory for morphology, convergent morphologies evolve because of random evolutionary walks through finite morphological space. In other words, chance effects might explain the observations of convergent morphologies, which we document at levels as deep as between domains and among major clades within a domain (Fig. 3). An alternative explanation is that convergences are “adaptive morphologies” that evolved under natural selection in similar environments (i.e. with similar environmental pressures). Conceptualizing morphological evolution in this way is analogous to analyses of patterns of molecular evolution: the null-hypothesis is that mutations are neutral (i.e. their fate is determined by purifying selection plus genetic drift [9], and that departures from this null hypothesis can be interpreted as evidence for positive selection
We agree with Bonner (2013) that formal testing of a null hypothesis for morphologies is challenging at this time. Quantitative approaches for testing models do exist in molecular evolution, as it is possible to compare rates of substitutions within and between lineages to assess departures from neutrality. Some attempt has been made to model morphology through simulations that “evolve” virtual creatures using intricate computer simulation of genetic and morphological interactions [70–72]. These systems tend to generate a surprisingly small number of morphologies, suggesting the possibility of limited morphological space [72]. However, the algorithms used in these simulations are designed to function in severe competition, i.e., morphologies that fit the constraints of the system are highly favored by the algorithm, so these simulations tend to converge to impressively similar “high fitness” morphologies [72].
While we recognize that there is currently no mathematical way to test a null model of a neutral morphology, we suggest that at least some examples of convergence are due to selection, especially when they occur between distantly-related lineages living in similar environments. In other words, some examples of convergences appear non-random. For example, the overall morphology of euglyphid and arcellinid testate amoebae is very similar (Fig. 2I and J): both of these lineages, which shared a common ancestor an estimated 1.2-1.8 billion years ago [11], construct vase-shaped shells surrounding amoeboid cells and both inhabit much the same environments (e.g. low pH bogs and fens, freshwater systems). The convergence (though not identity) of overall morphology between testate amoebae in Amoebozoa (e.g. Arcellinida) and Rhizaria (e.g. Euglyphida) seem unlikely to be due to neutral processes. Similarly, the multiple evolutions of fruiting bodies – mushroom-like structures – in diverse terrestrial lineages may be an adaptation for dispersal; fruiting bodies are wide-spread among Amoebozoa, and are also found in Heterolobosea (Excavata), ciliates (SAR – Stramenopile, Alveolate, Rhizaria), fungi, and even bacteria (the myxobacteria). Thus we argue that deeply convergent morphologies may represent independent adaptations to similar environments.
Is there a fundamental difference between the observation of discordance in microbes and in macroorganisms?
While it is possible that the discordance between morphology and molecules at the microbial scale simply elaborates on similar instances found among macroscopic organisms, it is possible that different rules dominate patterns of evolution at the microbial scale. For example, we suspect that the dynamic nature of many microbial genomes adds complexity to the relationships between genotype and phenotype [34, 73, 74]. As one example, the genome processes that mark the development of ‘somatic’ macronuclear genomes in ciliates (e.g. DNA deletions, chromosome fragmentation, gene unscrambling and alternative processing) may contribute to the discordance between morphology and molecules in this clade [21, 36, 75]. Similarly, the role of epigenetics in shaping genotypes across the tree of life likely contributes to discordance between molecules and morphology [34, 73, 74, 76]. Essentially, boundaries between species occur through epigenetic changes as well as through divergence in genes inherited in a Mendelian manner, hence potentially decoupling rates of molecular and morphological evolution.
Conclusions and Outlook
What are implications for understanding biodiversity on Earth?
By drawing on lineages with long histories of morphological classification, including testate amoebae, ciliates, and cyanobacteria, we have documented examples of microbial lineages that match two distinct categories of discordance: 1) similar morphologies manifested in multiple genetic lineages (e.g. convergence and cryptic diversity) and 2) multiple morphologies within a single genetic lineage (e.g. plasticity and life cycle variants). We are confident that these examples represent only a tiny fraction of the discordance between morphology and molecules among microbial lineages. The implications of this discordance are profound, and they indicate the need for detailed comparisons across levels as we attempt to characterize biodiversity on Earth.
Macroevolutionary hypotheses can be tested by adopting the ‘neutral phenotype’ as a null hypothesis. For example, it is possible that convergence is more common among lineages that are specialized to specific habitats and/or environmental conditions than in more generalist lineages. Similarly, instances of plasticity and life cycle variants may be more prevalent in lineages that need to adapt to changing environments – perhaps because markedly distinct life-cycle phases reduce competition between juveniles and adults. Acknowledging the broad patterns of discordance highlighted here and testing hypotheses about the discordance will lead to a deeper understanding of biodiversity on Earth and its causes.
Box 1 – Categories of molecular and morphological discordance among microbes.
We divide cases of discordance between morphologies and molecules in microbes in two general categories, with two instances under each:
Category 1: One Morphology, multiple lineages: Here there are two main instances that can be understood only when the phylogenetic history is taken into account – cryptic species if the multiple genetic lineages are monophyletic, convergence if not.
1 Convergent morphologies is one of the oldest, most fundamental concepts in evolutionary biology. Convergent morphologies lead to analogies, one of the main confusing factor in reconstructing phylogenetic histories. Often the assumption is that adaptation to a particular type of environment, or niche, is the main driver for the appearance of convergent morphologies, such as wings in bats and birds [77]. Of fundamental importance is that typically, morphologies that fall in this category are similar, but not identical.
2 Cryptic species – stand out as an immediate example of morphological-molecular discordance. These are groups of lineages that appear identical at the morphological level, but are divergent genetically [24]. Upon further inspection, most cases turn out to contain a complex of species, which can often be separated by detailed morphological studies [24].
Category 2: One lineage, multiple morphologies: A number of cases involve multiple morphologies within one genetic lineage. We separate these in two further types: life-cycle stage variation and plastic phenotypes.
3 Organisms with plastic phenotypes display distinct morphologies within the same entity, i.e., a well-defined single genetic lineage has individuals that differ morphologically on account of developmental and/or environmental differences [6]. One commonplace example is labor division in colonial invertebrates: among hydroid cnidarians, there are at least two kinds of individuals in a clonal colony, gastrozooids are responsible for obtaining food while gonozooids are responsible for reproduction [78]. Among microbial eukaryotes and the morphology-rich cyanobacteria, plasticity has been documented in terms of features such as spine numbers among testate (shelled) amoebae inducible defenses in ciliates [79]. Because of the practical difficulties in the study of microorganisms, it is often very hard to establish cases of plasticity [53].
4 Organisms with life-cycle stage variation present distinct morphologies of the same individual during the life cycle – which to microorganisms is a concept significantly broader than polyphenism [6]. It is important to keep in mind that the great majority of microorganisms are unculturable, therefore it is prudent to assume that aspects such as the complete life-cycle and morphologies of potential distinct mating types are likely to be known only for the deeply studied microbial models, and are hard to interpret even in deeply studied systems [34]. Among the few well-documented examples are variation between single amoebae, slug and fruiting body among slime molds [9] and the brooding of ciliated embryos within tentacle-bearing suctorian ciliates [33].
Acknowledgements
We are grateful to John Tyler Bonner for discussion and writings on these topics. LAK is supported by awards from the U.S. National Science Foundation (DEB-1208741, OCE-1129734) and the National Institutes of Health (1R15GM097722-01). DJGL is supported by a Young Investigator award by FAPESP, Brazil (2013/04585-3).
Footnotes
The authors declare no conflict of interests.
References
- 1.Sapp J. The new foundations of evolution: on the tree of life. Oxford University Press; 2009. [Google Scholar]
- 2.Jansson JK, Prosser JI. Microbiology: The life beneath our feet. Nature. 2013;494:40–1. doi: 10.1038/494040a. [DOI] [PubMed] [Google Scholar]
- 3.Wu D, Hugenholtz P, Mavromatis K, Pukall R, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–60. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams TA, Foster PG, Cox CJ, Embley TM. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504:231–6. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- 5.Grattepanche J-D, Santoferrara LF, McManus GB, Katz LA. Diversity of diversity: conceptual and methodological differences in biodiversity estimates of eukaryotic microbes as compared to bacteria. Trends Microbiol. doi: 10.1016/j.tim.2014.04.006. in press, doi: 10.1016/j.tim.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 6.West-Eberhard M. Phenotypic plasticity and the origins of diversity. Annu Rev Ecol Syst. 1989;20:249–78. [Google Scholar]
- 7.Pigliucci M. Do we need an extended evolutionary synthesis? Evolution. 2007;61:2743–9. doi: 10.1111/j.1558-5646.2007.00246.x. [DOI] [PubMed] [Google Scholar]
- 8.Sozinova O, Jiang Y, Kaiser D, Alber M. A three-dimensional model of myxobacterial aggregation by contact-mediated interactions. Proc Natl Acad Sci USA. 2005;102:11308–12. doi: 10.1073/pnas.0504259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonner JT. Randomness in evolution. Princeton University Press; 2013. [Google Scholar]
- 10.Adl SM, Simpson AG, Lane CE, Lukeš J, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–514. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parfrey LW, Lahr DJG, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci USA. 2011;108:13624–9. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knoll AH. Paleobiological perspectives on early eukaryotic evolution. Cold Spring Harb Perspect Biol. 2014;6:a016121. doi: 10.1101/cshperspect.a016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomaa F, Mitchell EAD, Lara E. Amphitremida (Poche, 1913) is a new major, ubiquitous Labyrinthulomycete clade. PLoS One. 2013;8:e53046. doi: 10.1371/journal.pone.0053046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikolaev SI, Berney C, Fahrni JF, Bolivar I, et al. The twilight of Heliozoa and rise of Rhizaria, an emerging supergroup of amoeboid eukaryotes. Proc Natl Acad Sci USA. 2004;101:8066–71. doi: 10.1073/pnas.0308602101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz LA. Origin and diversification of eukaryotes. Annu Rev Microbiol. 2012;66:411–27. doi: 10.1146/annurev-micro-090110-102808. [DOI] [PubMed] [Google Scholar]
- 16.Corliss JO, Lipscomb DL. Establishment of new order in kingdom Protista for Stephanopogon, long-known ciliate revealed now as flagellate. J Protozool. 1982;29:294–294. [Google Scholar]
- 17.Yubuki N, Leander BS. Ultrastructure and molecular phylogeny of Stephanopogon minuta: An enigmatic microeukaryote from marine interstitial environments. Eur J Protistol. 2008;44:241–53. doi: 10.1016/j.ejop.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann L, Komárek J, Kaštovskỳ J. System of cyanoprokaryotes (Cyanobacteria)–state in 2004. Algol Stud. 2005;117:95–115. [Google Scholar]
- 19.Oliverio AM, Lahr DJ, Nguyen T, Katz LA. Cryptic diversity within Morphospecies of testate amoebae (Amoebozoa: Arcellinida) in New England bogs and fens. Protist. 2014;165:196–207. doi: 10.1016/j.protis.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Kosakyan A, Heger TJ, Leander BS, Todorov M, et al. COI Barcoding of nebelid testate amoebae (Amoebozoa: Arcellinida): Extensive cryptic diversity and redefinition of the Hyalospheniidae Schultze. Protist. 2012;163:415–34. doi: 10.1016/j.protis.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Gao F, Katz LA, Song W. Insights into the phylogenetic and taxonomy of philasterid ciliates (Protozoa, Ciliophora, Scuticociliatia) based on analyses of multiple molecular markers. Mol Phylogenet Evol. 2012;64:308–17. doi: 10.1016/j.ympev.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Rajaniemi P, Komárek J, Willame R, Hrouzek P, et al. Taxonomic consequences from the combined molecular and phenotype evaluation of selected Anabaena and Aphanizomenon strains. Algol Stud. 2005;117:371–91. [Google Scholar]
- 23.Werner VR, Laughinghouse IV, Dail H. Bloom-forming and other planktonic Anabaena (Cyanobacteria) morphospecies with twisted trichomes from Rio Grande do Sul State, Brazil. Nova Hedwig. 2009;89:1–2. [Google Scholar]
- 24.Bickford D, Lohman DJ, Sodhi NS, Ng PKL, et al. Cryptic species as a window on diversity and conservation. Trends Ecol Evol. 2007;22:148–55. doi: 10.1016/j.tree.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Pfenninger M, Schwenk K. Cryptic animal species are homogeneously distributed among taxa and biogeographical regions. BMC Evol Biol. 2007;7:121. doi: 10.1186/1471-2148-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sáez AG, Probert I, Geisen M, Quinn P, et al. Pseudo-cryptic speciation in coccolithophores. Proc Natl Acad Sci USA. 2003;100:7163–8. doi: 10.1073/pnas.1132069100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foissner W, Wolf KW, Yashchenko V, Stoeck T. Description of Leptopharynx bromelicola n. sp. and characterization of the genus Leptopharynx Mermod, 1914 (Protista, Ciliophora). J Eukaryot Microbiol. 2011;58:134–51. doi: 10.1111/j.1550-7408.2011.00532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan X, Hu X, Al-Farraj SA, Clamp JC, et al. Morphological description of three marine ciliates (Ciliophora, Scuticociliatia), with establishment of a new genus and two new species. Eur J Protistol. 2011;47:186–96. doi: 10.1016/j.ejop.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Wylezich C, Meisterfeld R, Meisterfeld S, Schlegel M. Phylogenetic analyses of small subunit ribosomal RNA coding regions reveal a monophyletic lineage of euglyphid testate amoebae (Order Euglyphida). J Eukaryot Microbiol. 2002;49:108–18. doi: 10.1111/j.1550-7408.2002.tb00352.x. [DOI] [PubMed] [Google Scholar]
- 30.Todorov M, Golemansky V, Mitchell E a. D, Heger TJ. Morphology, biometry, and taxonomy of freshwater and marine interstitial Cyphoderia (Cercozoa: Euglyphida). J Eukaryot Microbiol. 2009;56:279–89. doi: 10.1111/j.1550-7408.2009.00394.x. [DOI] [PubMed] [Google Scholar]
- 31.Heger TJ, Mitchell EAD, Todorov M, Golemansky V, et al. Molecular phylogeny of euglyphid testate amoebae (Cercozoa: Euglyphida) suggests transitions between marine supralittoral and freshwater/terrestrial environments are infrequent. Mol Phylogenet Evol. 2010;55:113–22. doi: 10.1016/j.ympev.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 32.Heger TJ, Mitchell EAD, Leander BS. Holarctic phylogeography of the testate amoeba Hyalosphenia papilio (Amoebozoa: Arcellinida) reveals extensive genetic diversity explained more by environment than dispersal limitation. Mol Ecol. 2013;22:5172–84. doi: 10.1111/mec.12449. [DOI] [PubMed] [Google Scholar]
- 33.Corliss JO. Characterization, classification and guide to the literature. Pergamon Press Ltd.; 1979. The ciliated protozoa. [Google Scholar]
- 34.Hall MS, Katz LA. On the nature of species: insights from Paramecium and other ciliates. Genetica. 2011;139:677–84. doi: 10.1007/s10709-011-9571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esteban GF, Finlay BJ. Cryptic freshwater ciliates in a hypersaline lagoon. Protist. 2003;154:411–8. doi: 10.1078/143446103322454149. [DOI] [PubMed] [Google Scholar]
- 36.Gao F, Katz LA, Song W. Multigene-based analyses on evolutionary phylogeny of two controversial ciliate orders: Pleuronematida and Loxocephalida (Protista, Ciliophora, Oligohymenophorea). Mol Phylogenet Evol. 2013;68:55–63. doi: 10.1016/j.ympev.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 37.Katz LA, DeBerardinis J, Hall MS, Kovner AM, et al. Heterogeneous rates of molecular evolution among cryptic species of the ciliate morphospecies Chilodonella uncinata. J Mol Evol. 2011;73:266–72. doi: 10.1007/s00239-011-9468-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMANUS GB, Xu D, Costas BA, Katz LA. Genetic identities of Cryptic species in the Strombidium stylifer/apolatum/oculatum cluster, including a description of Strombidium rassoulzadegani n. sp. J Eukaryot Microbiol. 2010;57:369–78. doi: 10.1111/j.1550-7408.2010.00485.x. [DOI] [PubMed] [Google Scholar]
- 39.Simon EM, Nanney DL, Doerder FP. The “Tetrahymena pyriformis” complex of cryptic species. Biodivers Conserv. 2008;17:365–80. [Google Scholar]
- 40.Sun P, Clamp J, Xu D, Kusuoka Y, et al. Vorticella Linnaeus, 1767 (Ciliophora, Oligohymenophora, Peritrichia) is a grade not a clade: Redefinition of Vorticella and the families Vorticellidae and Astylozoidae using molecular characters derived from the gene coding for small subunit ribosomal RNA. Protist. 2012;163:129–42. doi: 10.1016/j.protis.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Catania F, Wurmser F, Potekhin AA, Przyboś E, et al. Genetic diversity in the Paramecium aurelia species complex. Mol Biol Evol. 2009;26:421–31. doi: 10.1093/molbev/msn266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonneborn TM. Breeding systems, reproductive methods, and species problems in protozoa. American Association for the Advancement of Science; 1957. [Google Scholar]
- 43.Whitton BA. Photosynthetic prokaryotes. Springer; 1992. Diversity, ecology, and taxonomy of the cyanobacteria. pp. 1–51. [Google Scholar]
- 44.Gomont MA. Monographie des oscillariées:(Nostacacées Homocystées) Masson; 1893. [Google Scholar]
- 45.Geitler L. Cyanophyceae. Johnson; 1932. Cyanophyceae. [Google Scholar]
- 46.Sheath RG, Cole KM. Biogeography of stream macroalgae in North America. J Phycol. 1992;28:448–60. [Google Scholar]
- 47.Casamatta DA, Vis ML, Sheath RG. Cryptic species in cyanobacterial systematics: a case study of Phormidium retzii (Oscillatoriales) using RAPD molecular markers and 16S rDNA sequence data. Aquat Bot. 2003;77:295–309. [Google Scholar]
- 48.Šlapeta J, López-García P, Moreira D. Global dispersal and ancient cryptic species in the smallest marine eukaryotes. Mol Biol Evol. 2006;23:23–9. doi: 10.1093/molbev/msj001. [DOI] [PubMed] [Google Scholar]
- 49.Amato A, Kooistra WHCF, Levialdi Ghiron JH, Mann DG, et al. Reproductive isolation among sympatric cryptic species in marine diatoms. Protist. 2007;158:193–207. doi: 10.1016/j.protis.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Kooistra WHCF, Sarno D, Balzano S, Gu H, et al. Global diversity and biogeography of Skeletonema species (Bacillariophyta). Protist. 2008;159:177–93. doi: 10.1016/j.protis.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 51.Vanormelingen P, Evans KM, Chepurnov V, Vyverman W, et al. Molecular species discovery in the diatom Sellaphora and its congruence with mating trials. Fottea. 2013;13:133–48. [Google Scholar]
- 52.Evans KM, Wortley AH, Simpson GE, Chepurnov VA, et al. A molecular systematic approach to explore diversity within the Sellaphora pupula species complex (bacillariophyta). J Phycol. 2008;44:215–31. doi: 10.1111/j.1529-8817.2007.00454.x. [DOI] [PubMed] [Google Scholar]
- 53.Lahr DJ, Bergmann PJ, Lopes SG. Taxonomic identity in microbial eukaryotes: A practical approach using the testate amoeba Centropyxis to resolve conflicts between old and new taxonomic descriptions. J Eukaryot Microbiol. 2008;55:409–16. doi: 10.1111/j.1550-7408.2008.00339.x. [DOI] [PubMed] [Google Scholar]
- 54.Chardez D. Influence du milieu sur Centropyxis aculeata (Ehrenberg) Stein (Rhizopoda Testacea). Bull Rech Agron Gembloux. 1966;1:13–9. [Google Scholar]
- 55.Wanner M. A review on the variability of testate amoebae: methodological approaches, environmental influences and taxonomical implications. Acta Protozool. 1999;38:15–9. [Google Scholar]
- 56.Wanner M, Meisterfeld R. Effects of some environmental factors on the shell morphology of testate amoebae (Rhizopoda, Protozoa). Eur J Protistol. 1994;30:191–5. [Google Scholar]
- 57.Jennings HS. Heredity, variation and the results of selection in the uniparental reproduction of Difflugia corona. Genetics. 1916;1:407. doi: 10.1093/genetics/1.5.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu Z, Zhang W, Liu L, Yang J. Evidence for two different morphotypes of Difflugia tuberspinifera from China. Eur J Protistol. 2013;50:205–11. doi: 10.1016/j.ejop.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 59.Bobrov A, Mazei Y. Morphological variability of testate amoebae (Rhizopoda: Testacealobosea and Testaceafilosea) in natural populations. Acta Protozool. 2004;43:133–46. [Google Scholar]
- 60.Lahr DJ, Parfrey LW, Mitchell EA, Katz LA, et al. The chastity of amoebae: re-evaluating evidence for sex in amoeboid organisms. Proc R Soc B Biol Sci. 2011;278:2081–90. doi: 10.1098/rspb.2011.0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lahr DJ, Nguyen TB, Barbero E, Katz LA. Evolution of the actin gene family in testate lobose amoebae (Arcellinida) is characterized by two distinct clades of paralogs and recent independent expansions. Mol Biol Evol. 2011;28:223–36. doi: 10.1093/molbev/msq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whitton BA. Phylum cyanobacteria (Cyanophyta). Freshw. Algal Flora Br. Isles 2nd Edn Camb. Univ. Press Camb. 2011:31–158. [Google Scholar]
- 63.Drouet F. Summary of the Classification of Blue-green Algae: With 4 Plates. J. Cramer. 1981 [Google Scholar]
- 64.Řeháková K, Johansen JR, Casamatta DA, Xuesong L, et al. Morphological and molecular characterization of selected desert soil cyanobacteria: three species new to science including Mojavia pulchra gen. et sp. nov. J Inf. 2007;46:481–502. [Google Scholar]
- 65.Hrouzek P, LUKEŠOVÁ A, MAREŠ J, Ventura S. Description of the cyanobacterial genus Desmonostoc gen. nov. including D. muscorum comb. nov. as a distinct, phylogenetically coherent taxon related to the genus Nostoc. Fottea. 2013;13:201–13. [Google Scholar]
- 66.Peglar MT, Nerad TA, Anderson OR, Gillevet PM. Identification of amoebae implicated in the life cycle of Pfiesteria and Pfiestena-like dinoflagellates. J Eukaryot Microbiol. 2004;51:542–52. doi: 10.1111/j.1550-7408.2004.tb00290.x. [DOI] [PubMed] [Google Scholar]
- 67.Bonner JT. The origins of multicellularity. Integr Biol Issues News Rev. 1998;1:27–36. [Google Scholar]
- 68.Pizay M-D, Lemée R, Simon N, Cras A-L, et al. Night and day morphologies in a planktonic dinoflagellate. Protist. 2009;160:565–75. doi: 10.1016/j.protis.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 69.Zirbel MJ, Veron F, Latz MI. The reversible effect of flow on the morphology of Ceratocorys horrida (Peridinialies, Dynophyta). J Phycol. 2000;36:46–58. [Google Scholar]
- 70.Sims K. Evolving virtual creatures. Proc 21st Annu Conf Comput Graph Interact Tech. 1994:15–22. [Google Scholar]
- 71.Sims K. Evolving 3D morphology and behavior by competition. Artif Life. 1994;1:353–72. [Google Scholar]
- 72.Lehman J, Stanley KO. Evolving a diversity of virtual creatures through novelty search and local competition. Proc 13th Annu Conf Genet Evol Comput. 2011:211–8. [Google Scholar]
- 73.Parfrey LW, Lahr DJ, Katz LA. The dynamic nature of eukaryotic genomes. Mol Biol Evol. 2008;25:787–94. doi: 10.1093/molbev/msn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oliverio AM, Katz LA. The dynamic nature of genomes across the tree of life. Genome Biol Evol. 2014;6:482–8. doi: 10.1093/gbe/evu024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McGrath CL, Zufall RA, Katz LA. Ciliate genome evolution. Genomics Evol. Microb. Eukaryotes Oxf. Univ. Press N. Y. 2006:64–77. [Google Scholar]
- 76.Chalker DL, Meyer E, Mochizuki K. Epigenetics of ciliates. Cold Spring Harb Perspect Biol. 2013;5:a017764. doi: 10.1101/cshperspect.a017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hall BK. Homology: The Hierarchial Basis of Comparative Biology. Academic Press; 2012. [Google Scholar]
- 78.Mills CE, Marques AC, Migotto AE, Calder DR, et al. Hydrozoa: Polyps, Hydromedusae, and Siphonophora. Light Smith Man. Intertidal Invertebr. Cent. Calif. Or. Univ. Calif. Press Los Angel. EUA. 2007:118–68. [Google Scholar]
- 79.Wicklow BJ. Signal-induced defensive phenotypic changes in ciliated protists: Morphological and ecological implications for predator and prey. J Eukaryot Microbiol. 1997;44:176–88. [Google Scholar]
- 80.Williams TA, Foster PG, Cox CJ, Embley TM. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504:231–6. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- 81.Wu D, Hugenholtz P, Mavromatis K, Pukall R, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–60. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lahr DJG, Grant JR, Katz LA. Multigene phylogenetic reconstruction of the Tubulinea (Amoebozoa) corroborates four of the six major lineages, while additionally revealing that shell composition does not predict phylogeny in the Arcellinida. Protist. 2013;164:323–39. doi: 10.1016/j.protis.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 83.Gómez F, Moreira D, López-García P. Neoceratium gen. nov., a new genus for all marine species currently assigned to Ceratium (Dinophyceae). Protist. 2010;161:35–54. doi: 10.1016/j.protis.2009.06.004. [DOI] [PubMed] [Google Scholar]