Abstract
Vitellogenesis is one of the most well-studied physiological processes in mosquitoes. Expression of mosquito vitellogenin genes is classically described as being restricted to female adult reproduction. We report premature vitellogenin transcript expression in three vector mosquitoes: Culex tarsalis, Aedes aegypti and Anopheles gambiae. Vitellogenins expressed during non-reproductive stages are alternatively spliced to retain their first intron and encode premature termination codons. We show that intron retention results in transcript degradation by translation-dependent nonsense-mediated mRNA decay. This is probably an example of regulated unproductive splicing and translation (RUST), a mechanism known to regulate gene expression in numerous organisms but which has never been described in mosquitoes. We demonstrate that the hormone 20-hydroxyecdysone (20E) is responsible for regulating post-transcriptional splicing of vitellogenin. After exposure of previtellogenic fat bodies to 20E, vitellogenin expression switches from a non-productive intron-retaining transcript to a spliced protein-coding transcript. This effect is independent of factors classically known to influence transcription, such as juvenile hormone-mediated competence and amino acid signalling through the target of rapamycin pathway. Non-canonical regulation of vitellogenesis through RUST is a novel role for the multifunctional hormone 20E, and may have important implications for general patterns of gene regulation in mosquitoes.
Keywords: vitellogenin, intron retention, mosquito reproduction, nonsense-mediated decay, 20-hydroxyecdysone
Introduction
Mosquito-borne pathogens cause an inordinate loss of human life, livelihood and productivity. There were 216 million reported cases and 655 000 deaths from malaria in 2011 and 2010, respectively. Dengue fever has increased in incidence in the past 50 years, and currently there are ~50–100 million cases and 22 000 deaths annually (http://www.who.int/mediacentre/factsheets/fs094/en/index.html). Threats of emerging pathogens, such as the Chikungunya virus (Thiboutot et al., 2010), are reminders that the presence of vector species can provide an efficient route of pathogen spread, as when West Nile virus was introduced and spread across the USA(Hayes et al., 2005; Venkatesan & Rasgon, 2010). These statistics necessitate an understanding of the biology that makes pathogen transmission by mosquitoes so successful. Female mosquitoes require blood to develop eggs, and a female that is continually taking blood meals in order to reproduce is an effective vehicle for the dissemination of pathogens. Studying mosquito reproduction is an avenue to identify novel targets for controlling mosquito-borne diseases.
The initiation of egg development in mosquitoes is hormonally regulated and occurs in two stages: previtellogenesis and vitellogenesis (Clements, 1992). Juvenile hormone (JH) released during previtellogenic development makes the female physiologically competent to respond to appropriate cues and develop eggs after a blood meal. After JH exposure, which occurs over a 2–3-day period in anautogenous (blood meal-dependent) mosquitoes, females enter a reproductively quiescent state of previtellogenic arrest. Break from this arrest occurs when the steroid hormone 20-hydroxyecdysone (20E) is released shortly after a blood meal and initiates the vitellogenic phase. Vitellogenesis is characterized by the rapid and abundant production of yolk protein precursors (YPPs; including the major YPP vitellogenin) by the fat body in response to 20E. Vitellogenin proteins are secreted into the haemolymph by the fat body and taken up by maturing ovarian oocytes, where they subsequently serve as a protein source for embryo development (Clements, 1992). Control of YPP expression in mosquitoes has been studied largely through the transcriptional regulation of one YPP gene in Aedes aegypti, vitellogenin-A1 (AaVgA1) (Raikhel et al., 2002).
Vitellogenin transcript expression in anautogenous Ae. aegypti is classically described as temporally specific: initiation of expression occurs rapidly after a blood meal and is terminated after ~48 h (Clements, 1992). Vitellogenin expression is similar in an autogenous population of a West Nile Virus vector, Culex tarsalis, where it is initiated in the absence of a blood meal when the female begins egg development (Provost-Javier et al., 2010); however, vitellogenin transcript is also expressed in C. tarsalis larvae and pupae, although protein is not detected (Provost-Javier et al., 2010). The consequence of non-reproductive vitellogenin expression is not known. It is also not known if this is unique to C. tarsalis, or if non-reproductive expression is a more general phenomenon in mosquitoes.
Alternative splicing, which plays a significant role in generating functional diversity of proteins in mosquitoes (Dong et al., 2006; Li et al., 2010), can regulate gene expression through the inclusion of exons or introns that encode premature termination codons (PTCs; Lareau et al., 2004). Such alternative splice variants are targeted for degradation by nonsense-mediated mRNA decay (NMD), thereby post-transcriptionally regulating gene expression in a process known as regulated unproductive splicing and translation (RUST) (Soergel et al., 2006). NMD is mediated by a core group of proteins (Upf1–3) that recognize PTC+ transcripts and are responsible for targeting them for degradation (Chang et al., 2007). Although alternative splicing linked to NMD has been characterized in other organisms, it has not been previously described as a regulatory mechanism in mosquitoes.
We report novel findings on the molecular regulation of vitellogenin expression during egg development in both blood meal-dependent (anautogenous) and independent (autogenous) mosquito reproduction. In addition, we describe a mechanism of post-transcriptional regulation not yet investigated in mosquitoes, as well as a novel role for the classic reproductive hormone, 20E.
Results
Alternative splicing by intron retention in vitellogenin transcript from C. tarsalis, Ae. aegypti and Anopheles gambiae
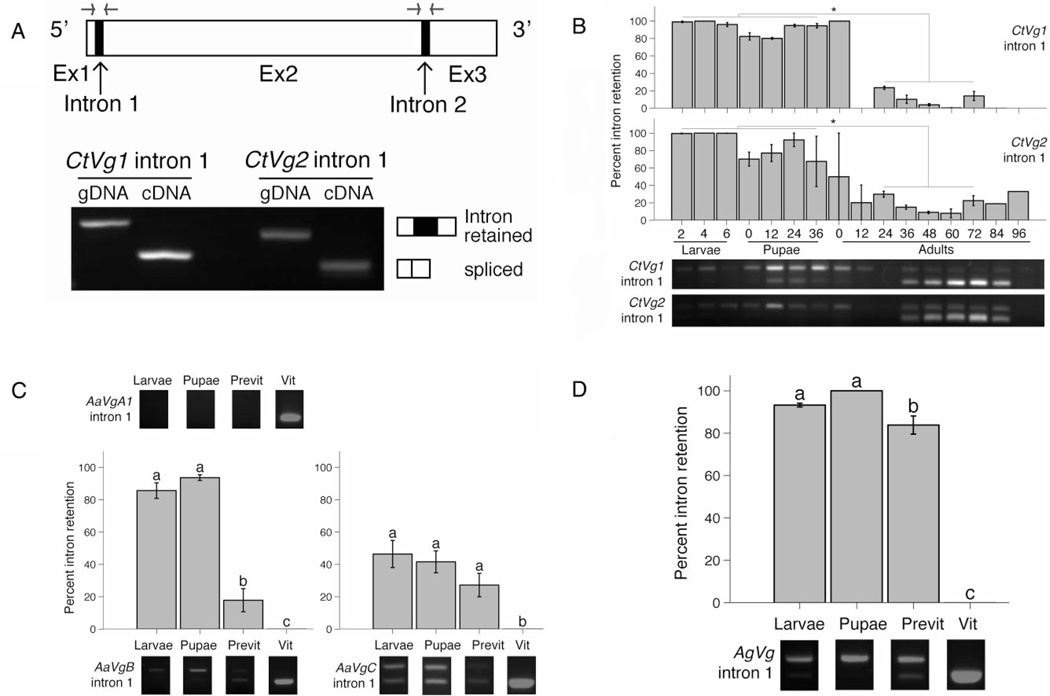
The vitellogenin genes of autogenous Culex tarsalis are transcriptionally expressed at low levels during larval and pupal development, although no vitellogenin protein is detected (Provost-Javier et al., 2010). To test for alternative splicing by intron retention, primers were designed to span the first intron of the two major groups of C. tarsalis vitellogenin genes (CtVg1: GenBank accession numbers GU017909 and GU017910; and CtVg2: accession GU17911 and GU017912), such that the intron-retaining transcript would be larger and the spliced transcript would be ~63 base pairs smaller (Fig. 1A, Tables S1, S2), and were used to screen for intron retention during development and adult vitellogenesis in an autogenous population of C. tarsalis. Significantly more intron-retained transcript was amplified during the larval and pupal time points than during vitellogenic time points, 24–72 h after emergence (CtVg1, CtVg2 P < 0.05, Fig. 1B). Retention of both introns in CtVg1 and CtVg2 was compared during combined larval and pupal stages. While intron retention occurred in both the first and second intron of the CtVg1 and CtVg2 genes, the percentage of retained transcript was greater in the first intron (CtVg1 and CtVg2 P < 0.0001, Fig. S1).
Figure 1.
Vitellogenin transcript expressed during non-reproductive time points in Culex tarsalis, Aedes aegypti and Anopheles gambiae retains the first intron. (A) The conserved structure of mosquito vitellogenin genes consist of three exons as indicated by the open boxes and two short introns as indicated by the filled boxes. The approximate position of the primers that span the intron of interest is illustrated with the forward and reverse arrows. Primers spanning the first intron of C. tarsalis Vg1 and Vg2 genes amplify both intron-containing genomic DNA and spliced cDNA from a vitellogenic female C. tarsalis. (B) Percent C. tarsalis Vg1 and Vg2 transcript that retains the first CtVg intron during larval (days after hatching), pupal (h after pupation) and adult (h after emergence) time points. Data were quantified from reverse transcriptase (RT)-PCR products using intron-spanning primers, CtVg1intron1 and CtVg2intron1. Example gel images are shown below the bar graphs. All the immature time points (larvae and pupae) were found to be significantly different from all the autogenous reproduction time points (defined as 24–72-h-old adults, based on vitellogenin expression profile Provost-Javier et al., 2010) for both CtVg1 intron 1 and CtVg2 intron 1. CtVg1 P < 0.0001, CtVg2 P = 0.0001, anova. Tukey’s Honest Significant Difference (HSD) test was used for pairwise comparisons. *P < 0.05. Expression and intron retention of A. aegypti VgA1, VgB and VgC genes (C) and A. gambiae Vg1–3 genes (D). For (C) and (D), vitellogenin gene expression was investigated by RT-PCR from larvae, female pupae, previtellogenic adult females and vitellogenic adult females. Intron-spanning primers used were: AaVgA1intron1, AaVgBintron1 and AaVgCintron1 for A. aegypti and AgVgintron1 for A. gambiae. Example gel images are shown. The bar graphs represent the mean ± sem. AaVgB P < 0.0001, AaVgC P = 0.0001549, AgVg P < 0.0001, anova. Tukey’s HSD test was used for pairwise comparisons. Stages that are significantly different at P < 0.05 are indicated with different letters. Previt, previtellogenic adult females; Vit, vitellogenic adult females; Ex, exon; gDNA, genomic DNA; cDNA, complementary DNA.
We next investigated intron retention in anautogenous Ae. aegypti and Anopheles gambiae. Ae. aegypti has three vitellogenin genes, AaVgA1 (accession L41842), AaVgB (accession AY380797), and AaVgC (accession AY373377), sharing 66–88% nucleotide identity (Chen et al., 2010). Primers were designed to span the first intron of each of the three Ae. aegypti genes (Table S1). Similar to previous studies (Raikhel et al., 2002), no quantifiable immature or previtellogenic adult transcript was detected for AaVgA1, while spliced transcript was amplified from vitellogenic females after a blood meal; however, alternatively spliced transcripts were detected for the other two vitellogenin genes. Significantly greater intronretained AaVgB is expressed in larvae and pupae when compared with vitellogenic females (P < 0.0001). Almost equal intron-retaining and spliced AaVgC transcripts are expressed during the larval and pupal stages (P = 0.0002, Fig. 1C).
Anopheles gambiae has three vitellogenin genes that share ~99% nucleotide identity (accession numbers AAAB01008880 and AF2810788, Chen et al., 2010) so the AgVg primers amplify across the first intron of all three genes (AgVg1–3, Table S1) but do not distinguish between them. AgVg transcript was amplified from larvae, pupae and previtellogenic adult females and was primarily the intron-retaining isoform compared with the spliced transcript amplified from vitellogenic females (P < 0.0001, Fig. 1D).
Translation-dependent NMD degrades intron-retained A. gambiae vitellogenin transcript
In order to investigate the function of alternative splicing in mosquito vitellogenin genes, we performed in silico translations of intron-retaining transcripts, revealing PTC(s), either within the first retained intron or shortly downstream (Fig. S2).
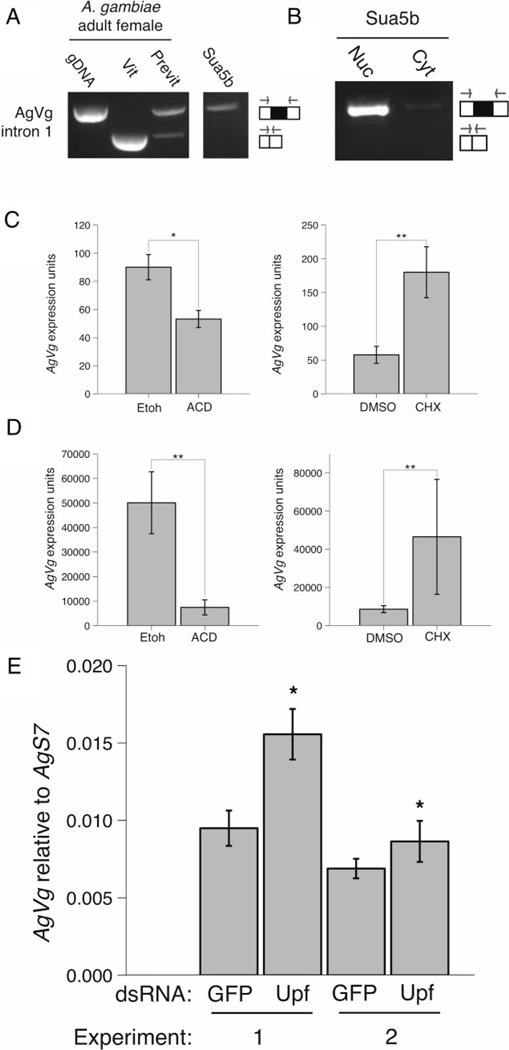
We tested the hypothesis that this alternative splice form is degraded by NMD using A. gambiae Sua5b cells, which express only the intron-retained isoform of AgVg transcript endogenously (Fig. 2A, B). Nuclear and cytoplasmic fractionation revealed that the majority of the intron-retained AgVg transcript is amplified from the nuclear fraction, with low levels in the cytoplasm (Fig. 2B). We first addressed the stability of AgVg by inhibiting transcription in Sua5b cells using actinomycin D (ACD) for 8 h and measuring AgVg expression by quantitative PCR (qPCR). Incubation with ACD resulted in a significant decrease in AgVg transcript compared with the ethanol control (P = 0.0081, Fig. 2C). Since NMD is a translation-dependent decay pathway, the translation inhibitor cycloheximide (CHX) was used to block transcript degradation by NMD (Carter et al., 1995). A significant increase in AgVg was observed in Sua5b cells treated with CHX compared with the dimethyl sulphoxide (DMSO)-treated control (P = 0.0059, Fig. 2C).
Figure 2.
Intron-retained vitellogenin is degraded by nonsense-mediated mRNA decay (NMD) in Anopheles gambiae cell line, Sua5b, and in A. gambiae fat body culture. Expression of A. gambiae Vg (AgVg) in Sua5b cells by reverse transcriptase PCR performed with intron spanning primers AgVgintron1 on whole cells (A) or cell fractions (B). Adult A. gambiae genomic DNA, vitellogenic and previtellogenic female cDNA serve as controls. AgVg expression for Sua5b cells (C) and previtellogenic A. gambiae in vitro fat body culture (D) treated with a transcriptional inhibitor (actinomycin D) or a translational inhibitor cycloheximide (CHX). Sua5b cells were treated for 8 h and A. gambiae fat bodies were treated for 24 h. AgVg expression (arbitrary units) was measured by quantitative PCR using a standard dilution curve. (E) AgVg expression in Sua5b cells following treatment with double-stranded (ds)RNA against AgUpf1 and AgUpf2 (UPF) or green fluorescent protein (GFP) control over two replicate experiments. There is a significant effect of dsRNA treatment and experimental date, and no significant interaction between dsRNA and date (dsRNA, P = 0.0342, date, P = 0.01831, dsRNA:date, P = 0.15591, Two-way anova). Because of a significant effect of date, graph shows the replicate experiments separately. AgVg expression is shown relative to AgS7. For all, the bar graphs represent the mean ± sem. * P < 0.05; ** P ≤ 0.001, Student’s t-test. t-tests were performed on log-transformed data for Sua5b CHX exp (C, second graph) and fat body ACD exp (D, first graph). gDNA, genomic DNA; Vit, vitellogenic adult female; Previt, previtellogenic adult female; Nuc, nuclear; Cyt, cytoplasmic; ETOH, ethanol; ACD, actinomycin D; DMSO, dimethyl sulphoxide.
We next tested the tissue-specific relevance of NMD degradation using A. gambiae fat body culture. Since previtellogenic A. gambiae females primarily expressed intron-retaining AgVg (Fig. 2A), we dissected the fat bodies and cultured them ex vivo with the same inhibitors, ACD and CHX, for 24 h. AgVg transcript levels decreased significantly in the ACD-treated fat body cultures when compared with the ethanol control (P = 0.0064, Fig. 2D), and a significant increase in AgVg was seen in the CHX-treated fat body cultures compared with the DMSO control (P = 0.01703, Fig. 2D). Finally, we tested the specificity of the NMD pathway by double-stranded (ds)RNA knockdown of AgUpf1 and AgUpf2 in Sua5b cells. RNA interference (RNAi)-mediated knockdown of Upf1 and Upf2 has been shown to impair NMD in Drosophila cells (Rehwinkel et al., 2005), so Sua5b cells were treated with dsRNA against both AgUpf1 and AgUpf2 or green fluorescent protein (GFP) as a control. Relative to GFP control-treated cells, RNAi resulted in ~20 and 35% knockdown of AgUpf1 and AgUpf2, respectively (data not shown), which resulted in significant increase in AgVg transcript (P = 0.0342, Fig. 2E).
20-hydroxyecdysone mediates splicing of AgVg and is independent of JH-mediated competence and amino acid signalling
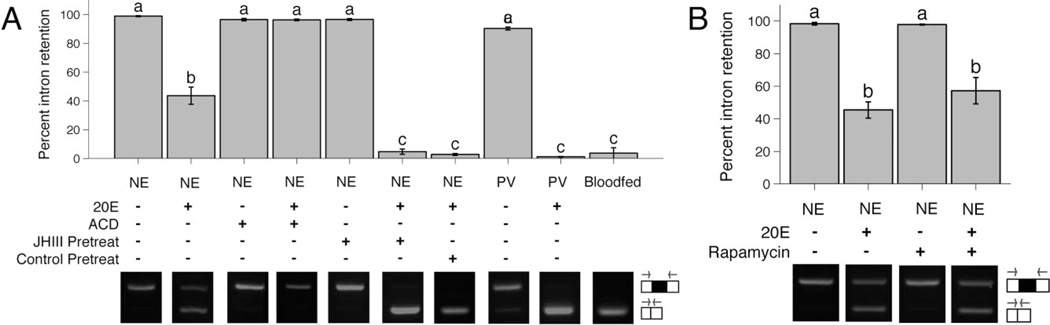
Anopheles gambiae fat bodies from 3–5-day-old previtellogenic females cultured in vitro continue to express intron-retaining AgVg. If incubated with 20E, only spliced AgVg was detected – similar to a vitellogenic, blood-fed female (Fig. 3A). We tested if 20E-induced AgVg splicing required JH-mediated previtellogenic competence. Fat bodies were dissected from newly emerged (<7-h-old) female A. gambiae and incubated in media with either 20E or ethanol control. Fat bodies from newly emerged females continue to express intron-retained AgVg, while 20E-treated fat bodies express significantly more spliced AgVg (P < 0.0001, Fig. 3A); however, the effect of 20E treatment of newly emerged fat bodies is significantly different than treatment of previtellogenic fat bodies (P < 0.0001, Fig. 3A). This effect is dependent on transcription since co-incubation of 20E with the transcriptional inhibitor ACD blocks the production of spliced AgVg (Fig. 3A).
Figure 3.
The hormone 20-hydroxyecdysone (20E) induces AgVg splicing independently of juvenile hormone (JH)-mediated competence or amino acid signalling in serum-free media. In (A) and (B), fat bodies were dissected from either newly emerged (<7-h-old) or previtellogenic (3–5-day-old) adult female Anopheles gambiae and incubated in serum-free medium for 6 h in the presence/absence of the hormone 20E, the transcriptional inhibitor actinomycin D and/or an inhibitor of the target of rapamycin amino acid signalling pathway, rapamycin (as indicated below the graph). In (A) the pretreatment groups (JH pretreat/Control pretreat) were first incubated, with or without JH, for 12 h followed by a 6-h incubation described above. Percent intron retention was quantified from gel images of reverse transcriptase (RT)-PCR product using primers that span the first intron of AgVg. Example gel images are shown below the graph. Schematic representations and approximate locations of the intron-retaining (top) and spliced (bottom) AgVg transcripts are shown to the right of the gel images. Open squares represent exon sequences, the filled square represents the intron sequence and the arrows indicate primers. The bar graphs represent the mean ± sem. Treatments that are not significantly different at P < 0.05 are indicated by the same letter. NE, newly emerged adult female; PV, previtellogenic adult female; ACD, actinomycin D.
To determine if JH leads to more complete splicing, newly emerged mosquito fatbodies were pretreated with JH or control for 12 h followed by a 6-h incubation with 20E. JH pretreatment without the addition of 20E did not result in splicing, but when 20E was added after the 12-h pretreatment, both with and without JH, significantly more spliced transcript was detected than when 20E was added without pretreatment (Fig. 3A). The amino acid-sensitive target of rapamycin (TOR) pathway also regulates vitellogenin expression during mosquito reproduction, so we tested if this pathway was required for the 20E-induced splicing of AgVg in newly emerged mosquitoes. Fat bodies from newly emerged mosquitoes incubated in the presence of 20E and rapamycin, a TOR pathway inhibitor, expressed the same percent of spliced AgVg compared with the control (P = 0.3314, Fig. 3B).
Treatment of competent previtellogenic fat bodies with 20E in vitro is known to induce high levels of vitellogenin transcript expression, similar to the induction of vitellogenesis after a blood meal, while treatment of fat bodies from newly emerged females with 20E does not (Zhu et al., 2003). qPCR measurements of AgVg expression in fat bodies of newly emerged mosquitoes, for all treatments, are significantly lower than AgVg expression from previtellogenic fat bodies when both are treated with 20E (Fig. S3). The effect that 20E has on vitellogenin splicing therefore occurs without the high expression levels that are seen in a previtellogenic female.
Discussion
Intron retention is the least common alternative splice form in vertebrates, accounting for ~3% in humans and mice, while the most common in plants, up to 30–56% in Arabidopsis thaliana (Ner-Gaon et al., 2004; Kim et al., 2007). In Drosophila melanogaster, intron retention events range from 10 to 30% of alternative splice forms and in An. gambiae, 41% of transcript structure variations (alternative splicing and allelic gene variation) (Kim et al., 2007; Li et al., 2010). While some of these events can be partially processed or mis-spliced transcripts, intron retention is an important form of alternative splicing that influences protein diversity and gene regulation (Galante, 2004). We do not know the function, if any, of misspliced vitellogenin transcripts in mosquitoes. Quantitatively, the amount of vitellogenin transcript during non-reproductive stages is low compared with levels induced during active vitellogenesis (~100–2500-fold less; Provost-Javier et al., 2010); however, the pattern of non-reproductive expression and intron retention of vitellogenin transcripts across different vitellogenin genes and three genera suggests that intron retention is a conserved functionally significant phenomenon rather than aberrant or incomplete splicing (Fig. 1). Interestingly Ae. aegypti VgA1, which is the model gene in mosquito vitellogenesis research (Raikhel et al., 2002), is the only vitellogenin gene with little expression or intron retention (Fig. 1C) during non-reproductive time points. This is generally consistent with previous research, although Northern blot and real-time PCR detection of AaVgA1 expression is reported in previtellogenic females (Attardo et al., 2003).
Since intron retention would result in the transcript encoding a PTC (Fig. S2), the likely fate of these transcripts is NMD. NMD is a highly conserved translation-dependent pathway that surveys and degrades PTC+ mRNAs before they get translated to truncated proteins that would be either non-functional or deleterious. PTC+ transcripts not only result from mutation or splicing error, but also arise from regulated alternative splicing. In the later case, NMD coupled with alternative splicing (also known as RUST) post-transcriptionally down-regulates specific genes that are alternatively spliced to encode PTC(s) (Lareau et al., 2008). RUST is an important regulatory mechanism in mammals (Lewis et al., 2003; de Lima Morais & Harrison, 2010), plants (Simpson et al., 2010) and Drosophila (Hansen et al., 2009), but has not been studied in mosquitoes.
The classic model for PTC recognition by NMD requires the PTC to be at least 50 nucleotides upstream of the last exon–exon junction of the transcript (Nagy & Maquat, 1998); however, PTC recognition does not follow this rule in D. melanogaster, where greater distances between the PTC and 3’ end of the mRNA is associated with NMD targets. This ‘faux 3′-UTR model’ (Amrani et al., 2004) is supported not only by research in D. melanogaster, but also in mammalian cells (Gatfield et al., 2003; Hansen et al., 2009). Even though NMD has not been studied in mosquitoes, the significant conservation of this pathway, expression of key protein components in A. gambiae, and studies in D. melanogaster (Gatfield et al., 2003; Hansen et al., 2009), suggest NMD functions similarly. The location of the PTC(s) near the 5′ end of the intron-retaining vitellogenin transcripts (Fig. S2) would increase the 3′ UTR length and make these mRNAs reasonable NMD targets.
Results from both the An. gambiae Sua5b cell line and fat body culture support NMD of intron retaining AgVg (Fig. 2). We found that 20E, which is responsible for inducing high levels of vitellogenin transcription after a blood meal (Fallon et al., 1974), also controls splicing of An. gambiae vitellogenin transcript (AgVg) (Fig. 3). This is the first observation that 20E regulates mRNA splicing. The effect that 20E has on vitellogenin transcription in a previtellogenic mosquito is dependent on several days of conditioning under control of JH and is enhanced by amino acid signalling (Attardo et al., 2005), which helps control the tissue and temporal specificity of 20E action. The independence of 20E-induced AgVg splicing of JH-mediated competence, TOR signalling or high levels of vitellogenin expression suggests that the mechanism that controls splicing is independent of the well-characterized cascade that controls vitellogenin transcription, and is primarily dependent on 20E signalling. In blood-fed Ae. aegypti, 20E functions by first binding its nuclear receptor (ecdysone receptor bound with ultraspiracle protein [EcR/Usp]), which activates transcription of a series of early genes (E75, E74 and Broad). These early gene products and 20E bound to EcR/Usp regulate a series of target late genes, which include vitellogenin and other yolk proteins (Raikhel et al., 2002; Attardo et al., 2005). Treatment of newly emerged fat bodies from An. gambiae with 20E and the transcriptional inhibitor ACD blocks AgVg splicing (Fig. 2D); therefore, the effect of 20E requires either new transcription of AgVg and/or early gene transcription. Early gene expression in response to 20E is not dependent on JH exposure (Zhu et al., 2003) so 20E treatment of fat bodies from newly emerged An. gambiae probably induces early gene expression.
We also observed a developmental effect on splicing. While newly emerged fat bodies treated with 20E express ~50–60% spliced transcript, if the fat bodies are first cultured for 12 h and then treated with 20E (with or without JH), 20E results in near 100% splicing (Fig. 3). A developmental process must be in effect soon after emergence, independent of JH exposure or signals from other tissues, which results in more effective AgVg splicing, unless low levels of JH present <6 h post emergence were sufficient. This developmental response could be related to the presence and/or abundance of 20E signalling components in the fat body cells. The ultraspiracle protein (Usp) portion of the 20E receptor is not detected in vivo by Western blot of fat body nuclear fractions of newly emerged female Ae. aegypti (Wang et al., 2000), while increasing amounts of protein are detected at 1–2 days and 3–5 days post eclosion. While low levels of Usp must not abrogate 20E signalling, as early gene transcription is detected in fat bodies of newly emerged mosquitoes treated with 20E (Zhu et al., 2003), it could be one mechanism for the difference in splicing efficiency seen between short and long culture time points.
The mechanism of 20E-mediated splicing of vitellogenin transcript is unknown. A passive hypothesis would be that vitellogenin transcript is inefficiently spliced and high levels of transcription induction result in greater quantities of spliced transcript detected by reverse transcriptase (RT)-PCR; however, our experiments performed with fat bodies from newly emerged females demonstrate that this could not be the only explanation. In Ae. aegypti, induction of high levels of vitellogenin transcription by 20E requires previtellogenic competence, acquired after 2–3 days of adult development, while 20E treatment of fat bodies from newly emerged mosquitoes does not induce high levels of vitellogenin expression (Zhu et al., 2003). Our results in An. gambiae are similar (Fig. S4); therefore, we detect spliced transcript in response to 20E without the high levels of expression typically seen in treatment of previtellogenic fat bodies.
An active hypothesis is that 20E exposure controls splicing factors that either enhance or inhibit intron splicing. One example of a protein that inhibits intron splicing is the alternative splicing factor/splicing factor-2 which binds an intronic sequence of the endoglin gene in mice, inhibits splicing and regulating intron retention (Blanco & Bernabeu, 2011). While 20E has not been previously associated with splicing, hormones can and do influence splicing regulation. In mammals, oestrogen regulates transcription of the TraB splicing factor, which then regulates alternative splice forms in hepatocytes (Zhang et al., 2007). In D. melanogaster, the insulin signalling cascade was found to affect alternative splicing in ~150 genes (Hartmann et al., 2009). One of the gene products induced by 20E in An. gambiae could modulate splicing factors that either enhance splicing or modify a splicing repressor of AgVg. Although not directly related to splicing, 20E has been shown to have other posttranscriptional functions. 20E inhibits the translation of a lysosomal protease mRNA until its titres fall postvitellogenesis (Cho & Raikhel, 2001). 20E also induces expression of several microRNAs, small non-coding RNAs that post-transcriptionally regulate gene expression; one of which was found to be important for blood digestion and egg maturation in Ae. aegypti (Bryant et al., 2010).
Although alternative splicing linked to NMD has been characterized in other organisms, this is the first demonstration of this type of regulation in mosquitoes. Alternative splicing is common in the An. gambiae transcriptome, with >40% of variation being intron retention (Li et al., 2010). Alternative splicing, particularly intron retention, should be considered as an important additional post-transcriptional mechanism to control gene expression in mosquitoes. We also demonstrate a novel function of 20E in regulating splicing in An. gambiae vitellogenin transcript. 20E is a general hormone that regulates other critical stages of development in mosquitoes (Clements, 1992; Gillott, 1995). 20E control of splicing could also contribute to the regulation of genes in other contexts where 20E functions. The An. gambiae system in the present study is a useful model to better understand the mechanisms of 20E control of splicing and gene regulation.
Materials and methods
Ethics statement
All mosquitoes were maintained on cotton pledgets soaked in commercially obtained chicken blood.
Mosquito colonies
The C. tarsalis KNWR-au strain, An. gambiae Keele strain and Ae. aegypti Rockefeller strain were all maintained at 27 °C, 90% relative humidity on a 16h light:8h dark cycle. Larvae were panned at a density of 200 larvae/pan, C. tarsalis and Ae. aegypti larvae were fed a 1:2:2 mixture of ground fish food, bovine liver powder and rabbit pellets while An. gambiae larvae were fed ground fish food and cat food pellets. Adult mosquitoes were provided with 10% sucrose and, for experiments involving blood feeding, were fed on cotton pledgets soaked in warmed chicken blood (Lampire Biological Laboratories, Pipersville, PA, USA).
Mosquito cells and dsRNA treatment
The An. gambiae Sua5b cell line was maintained in culture flasks at room temperature in Schneider’s insect medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich). In relevant experiments, cells were treated with 5 µg/ml ACD (Invitrogen, Carlsbad, CA, USA) or 100 µg/ml CHX.
Partial sequences of An. gambiae Upf1 and Upf2 were cloned into a pJet1.2 vector and partially sequenced (Fig. S3). Plasmids containing AgUpf1, AgUpf2, or a GFP sequence were linearized and used as template for amplification with the T7 primers listed in Table S1. The PCR product was purified and ran on a gel to confirm the size and presence of a single product. PCR product was used as a template for in vitro transcription of dsRNA using the T7 MEGAscript kit (Ambion, Austin, TX, USA). Sua5b cells were diluted to 1 × 106 cells in 0.5 ml of serum-free media in a 6-well plate. 20 µg of each dsRNA (two against AgUpf1 and one against AgUpf2, or GFP) was added to the wells and incubated on a rotating shaker for 30 min at room temperature. After the 30 min incubation, 2 ml of serum-containing medium was added to the wells and incubated for 2–3 days. dsRNA was added both with and without transfection reagent (HiPerFect; Qiagen, Valencia, CA, USA) for each treatment, but samples were analysed together due to no effect of transfection (P = 0.3988). Cells were split and dsRNA was added as described above on days 0 and 2 and cells were collected for RNA isolation on day 5.
Mosquito fat body culture
Either newly emerged (<7-h old) or 3- to 7-day-old female An. gambiae (Keele) were surface sterilized in 95% ethanol and the abdominal wall, with adhering fat body, was dissected as described in Crampton et al. (1997). The abdomen was removed from the thorax and cut length-wise such that the abdomen opened in one piece. The dissected abdomens (referred to as ‘fat bodies’) were then placed on a drop of phosphate-buffered saline to make sure that the cuticle was exposed to air and the fat body cells lining the abdominal wall were exposed to the liquid (Fig. S5). The fat bodies were transferred to wells of a 96-well plate with 100 µl of Schneider’s medium with penicillin-streptomycin (Sigma-Aldrich) and either supplemented with 10% fetal bovine serum (Gibco, Rockville, MD, USA) or not (for experiments involving 20E treatment). Five fat bodies were cultured per well and 10 fat bodies were used per replicate. For relevant experiments, fat bodies were treated with 5 µg/ml ACD, 100 µg/ml CHX, 10−5 M JH (Sigma-Aldrich), 10−6 M 20 (Sigma-Aldrich) or rapamycin (Sigma-Aldrich).
Gene expression analysis
Total RNA was isolated from groups of 10 adult mosquitoes or 10 fat bodies using TriReagent (Ambion) or an RNeasy kit (Qiagen). For the mosquito immature stages, RNA was isolated from groups of ~200 newly hatched, ~50–100 2-day-old, 20 4-day-old and 10 6-day-old larvae and pupae. Sex was not determined in larvae and only female pupae were used. Genomic DNA was isolated from whole mosquitoes using the DNeasy Blood & Tissue kit (Qiagen).
RNA was treated with DNase (Ambion) and 1 µg was used for first strand synthesis using the Superscript RT for PCR kit (Invitrogen) primed with Oligo(dT)20 primers. No RT controls were run for all samples to confirm the absence of genomic DNA contamination. For RT-PCR, 2 µl of cDNA was used as template for amplification with the primers listed in Table S1. All primers were designed to span the intron of interest such that PCR amplification could yield two products: a larger, intron-retained product and/or a smaller spliced product. Primer sequences and PCR conditions can be found in the supplementary information.
Quantitative PCR was performed with SYBR green chemistry (Qiagen) using a Rotor-Gene Q real-time PCR cycler (Qiagen). For experiments measuring expression using a standard dilution curve, a portion of AgVg cDNA that included the rtAgVg primer-binding region was cloned into a pJet1.2 vector (Fermentas, Thermo Scientific, Waltham, MA, USA) and serially 10-fold diluted to cover the range of AgVg expression. AgVg expression values were generated using the Rotor-Gene Q Series software (Qiagen) by interpolating samples’ threshold value in the standard curve. For experiments measuring relative expression, AgVg qPCR data were quantified relative to AgS7 using Q-Gene (Simon, 2003). R software (R Development Core Team, 2009) was used for statistical analysis and graphing. Statistical significance was assessed using a Student’s t-test or anova. Because of skew and/or unequal variance, data for the Sua5b-CHX experiment and the An. gambiae fat body-ACD experiment (Fig. 2) were log-transformed before analysis.
Data collection and analysis
Gel images were taken on a Red Gel Imaging System (Cell Biosciences, Santa Clara, CA, USA) and band intensities were quantified using ImageJ software (NIH, Bethesda, MD, USA). The background was first subtracted from the gel image and then the intensity of the intron-retained and spliced bands were quantified by measuring the area under each intensity curve. Percent intron retention was calculated as the intron-retained area divided by the sum of the intron-retained and spliced areas multiplied by 100. R sotware (R Development Core Team, 2009) was used for statistical analysis and graphing. anova was performed with Tukey’s Honest Significant Difference test for pairwise comparisons. For two-sample data, significance was assessed using Student’s t-test.
Supplementary Material
Acknowledgements
We thank the Johns Hopkins Malaria Research Institute for assistance with mosquito rearing. This research was funded by NIH grants R01AI067371 and R21AI111175 to JLR.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
Figure S1. Percent retention of the first and second intron of CtVg genes during combined non-reproductive stages (larvae and pupae) in Culex tarsalis. Data were quantified from reverse transcriptase-PCR products using intron-spanning primers, CtVg1intron1 and intron2 (left graph) and CtVg2intron1 and intron2 (right graph). The bar graphs represent the mean ± sem. **P < 0.0001, Student’s t-test.
Figure S2. Translation of intron-retained vitellogenin transcripts from Culex tarsalis, Anopheles gambiae and Aedes aegypti reveals premature termination codons. The translation consists of the first exon (not underlined) followed by the retained first intron (underlined) and in some, partial sequences from the second exon (not underlined). ‘Stop’ indicates a premature termination codon present due to the retention of the first intron in the sequence.
Figure S3. A. Partial amino acid alignment between a cloned Anopheles gambiae Upf1 (AgUpf1), predicted nonsense-mediated mRNA decay protein 1 from Culex quinquefasciatus (CqNMDprotein1, accession # XP_001862543), Drosophila melanogaster Upf1 (DmUpf1, accession # NP_572767) and Homo sapiens Upf1 (HsUpf1, accession # AAC51140). Shaded residues match AgUPF1. B. Partial amino acid alignment between a cloned sequence of An. gambiae Upf2 (AgUpf2), predicted Upf2 from Aedes aegypti and C. quinquefasciatus (AaUpf2 and CqUpf2, accession # XP_001660932 and XP_001851054), D. melanogaster Upf2 (DmUpf2, accession # NP_572434) and H.sapiens Upf2 (HsUpf1, accession # EAW86331). Shaded residues match AgUPF2.
Figure S4. Expression of AgVg measured by quantitative PCR in newly emerged fat bodies treated with 20-hydroxyecdysone (20E), actinomycin D (ACD) and rapamycin as compared with previtellogenic fat bodies treated with 20E. AgVg expression is shown relative to AgS7. The bar graphs represent the mean ± sem. **P < 0.0001. NE, newly emerged adult female; PV, previtellogenic adult female.
Figure S5. Mosquito fat body culture. The mosquito abdomen is dissected and cut on the ventral side down the anteroposterior axis. Abdomens are opened such that the cuticle is exposed to air and the fat body cells (which adhere to the wall of the abdomen) are exposed to the media. These are then referred to as ‘fat bodies’. Fat bodies are cultured five to a well of a 96-well plate as shown in the picture above. Arrow points to a single fat body.
Table S1. Primer sequence information. T7 promoter sequence is underlined.
Table S2. Vitellogenin intron 1 and 2 lengths in base pairs (bp).
References
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense- mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- Attardo G, Hansen I, Raikhel AS. Nutritional regulation of vitellogenesis in mosquitoes: implications for anautogeny. Insect Biochem Mol Biol. 2005;35:661–675. doi: 10.1016/j.ibmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Attardo GM, Higgs S, Klingler KA, Vanlandingham DL, Raikhel AS. RNA interference-mediated knockdown of a GATA factor reveals a link to anautogeny in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2003;100:13374–13379. doi: 10.1073/pnas.2235649100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco FJ, Bernabeu C. Alternative splicing factor or splicing factor-2 plays a key role in intron retention of the endoglin gene during endothelial senescence. Aging Cell. 2011;10:896–907. doi: 10.1111/j.1474-9726.2011.00727.x. [DOI] [PubMed] [Google Scholar]
- Bryant B, Macdonald W, Raikhel AS. microRNA miR-275 is indispensable for blood digestion and egg development in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2010;107:22391–22398. doi: 10.1073/pnas.1016230107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, et al. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Chen S, Armistead JS, Provost-Javier KN, Sakamoto JM, Rasgon JL. Duplication, concerted evolution and purifying selection drive the evolution of mosquito vitellogenin genes. BMC Evol Biol. 2010;10:142. doi: 10.1186/1471-2148-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WL, Raikhel AS. A novel function of 20-hydroxyecdysone: translational repression of the lysosomal protease mRNA in the mosquito fat body. Insect Biochem Mol Biol. 2001;31:283–288. doi: 10.1016/s0965-1748(00)00175-2. [DOI] [PubMed] [Google Scholar]
- Clements AN. The Biology of Mosquitoes. 1st edn. London: Chapman and Hall; 1992. [Google Scholar]
- Crampton JM, Beard CB, Louis C, editors. The Molecular Biology of Insect Disease Vectors: A Methods Manual. 1st edn. London; New York: Chapman and Hall; 1997. [Google Scholar]
- Dong Y, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol. 2006;4:e229. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon AM, Hagedorn HH, Wyatt GR, Laufer H. Activation of vitellogenin synthesis in the mosquito Aedes aegypti by ecdysone. J Insect Physiol. 1974;20:1815–1823. doi: 10.1016/0022-1910(74)90211-x. [DOI] [PubMed] [Google Scholar]
- Galante PAF. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillott C. Entomology. 3rd edn. Dordrecht, the Netherlands: Springer; 1995. [Google Scholar]
- Hansen KD, Lareau LF, Blanchette M, Green RE, Meng Q, Rehwinkel J, et al. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 2009;5:e1000525. doi: 10.1371/journal.pgen.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann B, Castelo R, Blanchette M, Boue S, Rio DC, Valcarcel J. Global analysis of alternative splicing regulation by insulin and wingless signaling in Drosophila cells. Genome Biol. 2009;10:R11. doi: 10.1186/gb-2009-10-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes EB, Komar N, Nasci RS, Montgomery SP, O’Leary DR, Campbell GL. Epidemiology and transmission dynamics of West Nile virus disease. Emerg Infect Dis. 2005;11:1167–1173. doi: 10.3201/eid1108.050289a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L, Brooks A, Soergel D. The coupling of alternative splicing and nonsense-mediated mRNA decay. In: Blencowe BJ, Graveley BR, editors. Alternative Splicing in the Postgenomic Era. New York, NY: Landes Bioscience and Spinger Science+Business Media; 2008. pp. 190–211. [Google Scholar]
- Lareau LF, Green RE, Bhatnagar RS, Brenner SE. The evolving roles of alternative splicing. Curr Opin Struct Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ribeiro JMC, Yan G. Allelic gene structure variations in Anopheles gambiae mosquitoes. PLoS ONE. 2010;5:e10699. doi: 10.1371/journal.pone.0010699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima Morais DA, Harrison PM. Large-scale evidence for conservation of NMD candidature across mammals. PLoS ONE. 2010;5:e11695. doi: 10.1371/journal.pone.0011695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004;39:877–885. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- Provost-Javier KN, Chen S, Rasgon JL. Vitellogenin gene expression in autogenous Culex tarsalis. Insect Mol Biol. 2010;19:423–429. doi: 10.1111/j.1365-2583.2010.00999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R Foundation for Statistical Computing. Vienna, Austria: 2009. [Accessed on 10 January 2014]. R: A Language and Environment for Statistical Computing. ISBN 3-900051-07-0. < http://www.R-project.org>. [Google Scholar]
- Raikhel AS, Kokoza VA, Zhu J, Martin D, Wang SF, Li C, et al. Molecular biology of mosquito vitellogenesis: from basic studies to genetic engineering of antipathogen immunity. Insect Biochem Mol Biol. 2002;32:1275–1286. doi: 10.1016/s0965-1748(02)00090-5. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19:1439–1440. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- Simpson CG, Manthri S, Raczynska KD, Kalyna M, Lewandowska D, Kusenda B, et al. Regulation of plant gene expression by alternative splicing. Biochem Soc Trans. 2010;38:667–671. doi: 10.1042/BST0380667. [DOI] [PubMed] [Google Scholar]
- Soergel D, Lareau LF, Brenner SE. Regulation of gene expression by coupling of alternative splicing and NMD. In: Maquat LE, editor. Nonsense-Mediated Mrna Decay. Georgetown, TX: Landes Biosciences; 2006. pp. 175–196. [Google Scholar]
- Thiboutot MM, Kannan S, Kawalekar OU, Shedlock DJ, Khan AS, Sarangan G, et al. Chikungunya: a potentially emerging epidemic? PLoS Negl Trop Dis. 2010;4:e623. doi: 10.1371/journal.pntd.0000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan M, Rasgon JL. Population genetic data suggest a role for mosquito-mediated dispersal of West Nile virus across the western United States. Mol Ecol. 2010;19:1573–1584. doi: 10.1111/j.1365-294X.2010.04577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SF, Li C, Sun G, Zhu J, Raikhel AS. Differential expression and regulation by 20-hydroxyecdysone of mosquito ultraspiracle isoforms. Dev Biol. 2000;218:99–113. doi: 10.1006/dbio.1999.9575. [DOI] [PubMed] [Google Scholar]
- Zhang X, Moor AN, Merkler KA, Liu Q, McLean MP. Regulation of alternative splicing of liver scavenger receptor class B gene by estrogen and the involved regulatory splicing factors. Endocrinology. 2007;148:5295–5304. doi: 10.1210/en.2007-0376. [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen L, Raikhel AS. Posttranscriptional control of the competence factor betaFTZ-F1 by juvenile hormone in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2003;100:13338–13343. doi: 10.1073/pnas.2234416100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.