Abstract
Flavobacterium johnsoniae exhibits gliding motility and digests many polysaccharides, including chitin. A novel protein secretion system, the type IX secretion system (T9SS), is required for gliding and chitin utilization. The T9SS secretes the cell surface motility adhesins SprB and RemA and the chitinase ChiA. Proteins involved in secretion by the T9SS include GldK, GldL, GldM, GldN, SprA, SprE, and SprT. Porphyromonas gingivalis has orthologs for each of these that are required for secretion of gingipain protease virulence factors by its T9SS. P. gingivalis porU and porV have also been linked to T9SS-mediated secretion, and F. johnsoniae has orthologs of these. Mutations in F. johnsoniae porU and porV were constructed to determine if they function in secretion. Cells of a porV deletion mutant were deficient in chitin utilization and failed to secrete ChiA. They were also deficient in secretion of the motility adhesin RemA but retained the ability to secrete SprB. SprB is involved in gliding motility and is needed for formation of spreading colonies on agar, and the porV mutant exhibited gliding motility and formed spreading colonies. However, the porV mutant was partially deficient in attachment to glass, apparently because of the absence of RemA and other adhesins on the cell surface. The porV mutant also appeared to be deficient in secretion of numerous other proteins that have carboxy-terminal domains associated with targeting to the T9SS. PorU was not required for secretion of ChiA, RemA, or SprB, indicating that it does not play an essential role in the F. johnsoniae T9SS.
INTRODUCTION
Cells of Flavobacterium johnsoniae, and of many members of the phylum Bacteroidetes, crawl rapidly over surfaces by a process known as gliding motility (1). F. johnsoniae gliding involves the rapid movement of the adhesins SprB and RemA along the cell surface (2–4). These proteins are secreted across the outer membrane by a novel protein secretion system originally called the Por secretion system and more recently referred to as the type IX secretion system (T9SS) (5, 6). Motility proteins are not the only cargo for the F. johnsoniae T9SS. It is also required for secretion of the soluble extracellular chitinase ChiA (7), and 51 other F. johnsoniae proteins are predicted to use this secretion system (6). Many proteins secreted by T9SSs are very large. ChiA, RemA, and SprB, for example, are 166, 152, and 669 kDa, respectively (3, 4, 7). The mechanism that allows efficient secretion of such large proteins by the T9SS is not known. T9SSs are found in many members of the phylum Bacteroidetes, and they are apparently limited to this phylum (8, 9). The proteins required for T9SS-mediated secretion are not similar in sequence to proteins of other bacterial secretion systems (5, 8, 10). Although the T9SS was only recently discovered, it has already been associated with motility (11), virulence (5), chitin and cellulose digestion (7, 12), and colonization of and protection of plants from pathogens (13).
T9SSs were originally identified in F. johnsoniae and in the oral pathogen Porphyromonas gingivalis (5, 11). P. gingivalis secretes virulence factors such as gingipain proteases and adhesins using its T9SS. Proteins secreted by T9SSs have cleavable N-terminal signal peptides and are apparently exported across the cytoplasmic membrane via the Sec system (9, 14). They also have conserved carboxy-terminal domains (CTDs) of approximately 60 to 100 amino acids that target them for secretion across the outer membrane by the T9SS (6, 7, 9, 14, 15). The CTDs appear to be proteolytically cleaved during or after secretion across the outer membrane (9, 16). The CTDs are necessary and sufficient for secretion by the T9SS. P. gingivalis HBP35 and F. johnsoniae ChiA lacking their CTDs are not secreted, and heterologous fusion proteins carrying the HBP35 and ChiA CTDs are efficiently secreted (7, 17). Many T9SS CTDs of F. johnsoniae and P. gingivalis belong to TIGRFAM protein domain family TIGR04183 (6, 8, 14). There appears to be considerable diversity in T9SS CTDs, and not all fall within the boundaries of TIGR04183. F. johnsoniae SprB, for example, requires the T9SS for secretion, but its carboxy-terminal region exhibits no similarity to TIGR04183 family members but rather belongs to the unrelated domain family TIGR04131. Eleven other F. johnsoniae proteins have TIGR04131-type CTDs, as do numerous proteins from other species belonging to the phylum Bacteroidetes that have T9SSs. We have speculated that these TIGR04131-type CTDs target proteins for secretion by the T9SS (7, 8), but with the exception of SprB, T9SS-mediated secretion of these proteins has not been experimentally demonstrated in any organism.
Proteins required for secretion by the F. johnsoniae T9SS include GldK, GldL, GldM, and GldN or its paralog GldO (6, 11). SprA, SprE, and SprT also have important roles in T9SS-mediated secretion, and cells with mutations in the genes encoding these proteins are severely but incompletely deficient in secretion (5, 6, 18). The P. gingivalis T9SS has orthologs for GldK, GldL, GldM, GldN, SprA, SprE, and SprT, and these are required for secretion (5, 19, 20). P. gingivalis PorP is also required for secretion. Unlike P. gingivalis, F. johnsoniae has multiple genes that exhibit similarity to porP. One of these, sprF, is required for secretion of SprB but is not needed for secretion of other proteins by its T9SS (21). The F. johnsoniae genome is predicted to encode 10 PorP-like proteins in addition to SprF, and each of these may facilitate secretion of specific cargo proteins.
Five additional P. gingivalis proteins, PorQ, PorU, PorV, PorX, and PorY, are linked to T9SS function (5, 16, 22, 23). Mutations in P. gingivalis porQ, porX, and porY result in partial defects in T9SS-mediated secretion. The function of PorQ is not known, but PorX and PorY are thought to form a two-component regulatory system that controls expression of the T9SS genes (5). The related F. johnsoniae proteins do not appear to play similar roles since deletion of the F. johnsoniae orthologs of porQ, porX, and porY has no effect on secretion of SprB, RemA, or ChiA (24). The functions of F. johnsoniae PorU and PorV in secretion have not previously been studied. P. gingivalis PorU is thought to function as the peptidase that removes the CTDs of secreted proteins (16). P. gingivalis PorV is required for secretion of proteins targeted to the T9SS, including the gingipain proteases RgpA, RgpB, and Kgp (23, 25). PorV, which has also been called LptO, is required for the partial O-deacylation of lipopolysaccharide (22). PorV may have deacylation activity, or it may be required for secretion of a deacylase. F. johnsoniae has orthologs of porU and porV, but their functions have not been determined. In this study, we constructed and examined F. johnsoniae mutants to determine the roles of PorU and PorV in secretion. Deletion of porU had little effect on secretion, indicating that it was not essential for F. johnsoniae T9SS function. In contrast, PorV was required for the secretion of many but not all proteins targeted to the T9SS. Deletion of porV eliminated secretion of RemA and ChiA but had no effect on secretion of SprB.
MATERIALS AND METHODS
Bacterial and bacteriophage strains, plasmids, and growth conditions.
F. johnsoniae ATCC 17061T strain UW101 was the wild-type strain used in this study (26–28). The streptomycin-resistant rpsL mutant of UW101 (CJ1827) was used to construct deletion mutants (29). F. johnsoniae strains were grown in Casitone-yeast extract (CYE) medium at 30°C (30). To observe colony spreading, cells were grown on PY2 agar at 25°C (31), and to observe motility of individual cells, they were grown in motility medium (MM) at 25°C (32). Escherichia coli strains were grown in Luria-Bertani medium (LB) at 37°C (33). Strains and plasmids used in this study are listed in Table 1, and primers are listed in Table S1 in the supplemental material. Antibiotics were used at the following concentrations when needed: ampicillin, 100 μg/ml; cefoxitin, 100 μg/ml; erythromycin, 100 μg/ml; streptomycin, 100 μg/ml; and tetracycline, 20 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype and/or descriptiona | Reference(s) or source |
|---|---|---|
| F. johnsoniae strains | ||
| UW101 (ATCC 17061) | Wild type | 27, 28 |
| CJ1631A | Δ(gldN gldO) in F. johnsoniae UW101 | 11 |
| CJ1808 | chiA disruption mutant; (Emr) | 7 |
| CJ1818 | porU disruption mutant; (Emr) | This study |
| CJ1827 | rpsL2; (Smr) “wild-type” strain for construction of all deletion mutants except CJ1631A | 29 |
| CJ1922 | rpsL2 ΔsprB; (Smr) | 29 |
| CJ1984 | rpsL2 ΔremA; (Smr) | 4 |
| CJ1985 | rpsL2 ΔsprB ΔremA; (Smr) | 4 |
| CJ2082 | rpsL2 ΔFjoh_0288; (Smr) | This study |
| CJ2083 | rpsL2 remA::myc tag 1; (Smr) | 4 |
| CJ2089 | rpsL2 Δ(gldN gldO) remA::myc tag 1; (Smr) | 4 |
| CJ2090 | rpsL2 Δ(gldN gldO); (Smr) | 4 |
| CJ2116 | rpsL2 ΔporU; (Smr) | This study |
| CJ2130 | rpsL2 ΔporV; (Smr) | This study |
| CJ2323 | rpsL2 ΔporV remA::myc tag 1; (Smr) | This study |
| CJ2445 | rpsL2 ΔporV ΔsprB; (Smr) | This study |
| CJ2446 | rpsL2 ΔporV ΔFjoh_0288; (Smr) | This study |
| Plasmids | ||
| pCP23 | E. coli-F. johnsoniae shuttle plasmid; Apr (Tcr) | 31 |
| pCP29 | E. coli-F. johnsoniae shuttle plasmid; Apr (Cfr Emr) | 45 |
| pRR51 | rpsL-containing suicide vector; Apr (Emr) | 29 |
| pRR39 | pCP23 carrying remA; Apr (Tcr) | 4 |
| pSSK01 | 1,050-bp fragment of porU in pLYL03 for constructing porU disruption mutant CJ1818; Apr (Emr) | This study |
| pSSK03 | 1,516-bp SphI-KpnI fragment spanning porV amplified with primers 972 and 973 and inserted into pCP29; Apr (Cfr Emr) | This study |
| pSSK04 | 4,309-bp XbaI-BamHI fragment spanning porU amplified with primers 988 and 989 and inserted into pCP23; Apr (Tcr) | This study |
| pSSK14 | 2,332-bp XbaI-SalI fragment downstream of Fjoh_0288 amplified with primers 1104 and 1105 and inserted into pSSK16; Apr (Emr) | This study |
| pSSK16 | 2,301-bp BamHI-XbaI fragment upstream of Fjoh_0288 amplified with primers 1102 and 1103 and inserted into pRR51; Apr (Emr) | This study |
| pSSK20 | 2,442-bp BamHI-SalI region upstream of porV amplified with primers 1203 and 1204 and inserted into pRR51; Apr (Emr) | This study |
| pSSK21 | 2,282-bp BamHI-SalI region upstream of porU amplified with primers 1207 and 1208 and inserted into pRR51; Apr (Emr) | This study |
| pSSK22 | 2,271-bp SalI-SphI region downstream of porV amplified with primers 1201 and 1202 and inserted into pSSK20; Apr (Emr) | This study |
| pSSK23 | 2,305-bp SalI-SphI region downstream of porU amplified with primers 1205 and 1206 and inserted into pSSK21; Apr (Emr) | This study |
| pTB79 | pCP23 carrying gldN; Apr (Tcr) | 46 |
Antibiotic resistance phenotypes are as follows: ampicillin, Apr; cefoxitin, Cfr; erythromycin, Emr; streptomycin, Smr; tetracycline, Tcr. The antibiotic resistance phenotypes given in parentheses are those expressed in F. johnsoniae but not in E. coli. The antibiotic resistance phenotypes without parentheses are those expressed in E. coli but not in F. johnsoniae.
Construction and complementation of porV and porU mutants.
Unmarked deletions were generated as previously described (29). To delete porV, a 2,442-bp fragment upstream of porV was amplified using Phusion DNA polymerase (New England BioLabs, Ipswich, MA) and primers 1203 (engineered BamHI site) and 1204 (engineered SalI site). The amplified fragment was digested with BamHI and SalI and cloned into pRR51 that had been digested by the same enzymes, generating pSSK20. A 2,271-bp fragment downstream of porV was amplified using primers 1201 (engineered SalI site) and 1202 (engineered SphI site). This fragment was introduced into pSSK20 that was digested with SalI and SphI, to generate pSSK22. pSSK22 was introduced into the F. johnsoniae strain CJ1827 by triparental conjugation. Colonies containing the plasmid integrated into the chromosome were obtained by selecting for erythromycin resistance, and porV deletion mutants that had lost the integrated plasmid were obtained by selecting for streptomycin resistance and confirmed by PCR essentially as previously described (29).
porU deletion mutants were constructed in a similar manner. A 2,282-bp fragment upstream of porU was amplified using primers 1207 (engineered BamHI site) and 1208 (engineered SalI site). The amplified fragment was digested with BamHI and SalI and cloned into pRR51 that had been digested with the same enzymes, generating pSSK21. A 2,305-bp fragment downstream of porU was amplified using primers 1205 (engineered SalI site) and 1206 (engineered SphI site). This fragment was ligated with pSSK21 that had been digested with SalI and SphI, generating pSSK23. pSSK23 was used to construct the porU deletion strain as described above. A strain with a polar insertion mutation in porU (CJ1818) was also constructed. For this purpose, a 1,050-bp fragment internal to porU was amplified using primers 948 (engineered BamHI site) and 949 (engineered SalI site). This fragment was cloned into pLYL03 which had been cut with BamHI and SalI to generate pSSK01. pSSK01 was introduced into wild-type F. johnsoniae UW101 by conjugation, and selection for erythromycin resistance resulted in integration of the plasmid into the genome and disruption of porU. The insertion was confirmed by PCR using primers 737 and 948.
For complementation of the porV mutant, a 1,516-bp region spanning porV was amplified using primers 972 (engineered SphI site) and 973 (engineered KpnI site) and introduced into pCP29, to generate pSSK03. Similarly, for complementation of porU mutants, a 4,309-bp region spanning porU was amplified using primers 988 (engineered XbaI site) and 989 (engineered BamHI site) and introduced into pCP23, to generate pSSK04.
Determination of chitinase activity.
Chitin utilization on agar was observed as previously described using colloidal chitin prepared from crab shells (11, 27, 34). Chitinase activities in cell-free culture fluid (spent medium), intact cells, and cell extracts were measured as previously described (11) using the synthetic substrates 4-methylumbelliferyl-β-d-N,N′-diacetyl-chitobioside [4-MU-(GlcNAc)2] and 4-methylumbelliferyl-β-d-N,N′,N″-diacetyl-chitotrioside [4-MU-(GlcNAc)3] (Sigma-Aldrich, St. Louis, MO), except that activities were measured for 30 min. Activities in the spent medium (secreted chitinase), intact cells, and cell extracts were indicated as pmol 4-methylumbelliferone released during 30 min per μg total protein in the original cell suspension. Protein concentrations were determined by the bicinchoninic acid (BCA) assay (Thermo Fisher Scientific, Waltham, MA).
Western blot analyses.
F. johnsoniae cells were grown to late exponential phase in MM at 25°C with shaking. Cells were pelleted by centrifugation at 22,000 × g for 15 min, and the spent culture fluid was filtered using 0.22-μm-pore-size polyvinylidene difluoride filters (Thermo Fisher Scientific). For whole-cell samples, the cells were suspended in the original culture volume of phosphate-buffered saline (PBS) consisting of 137 mM NaCl, 2.7 mM KCl, 10 mM Na2PO4, and 2 mM KH2PO4 (pH 7.4). Equal amounts of spent medium and whole cells were boiled in SDS-PAGE loading buffer for 9 min. Proteins were separated by SDS-PAGE, and Western blot analyses were performed as previously described (11) using affinity-purified antibody against ChiA (1:5,000 dilution). Equal amounts of each sample based on the starting material were loaded in each lane. For cell extracts, this corresponded to 15 μg protein, whereas for spent medium, this corresponded to the equivalent volume of spent medium that contained 15 μg cell protein before the cells were removed. For detection of mCherry by Western blotting, commercially available antibodies against mCherry (0.5 mg per ml; BioVision Incorporated, Milpitas, CA) were used at a dilution of 1:5,000.
Myc-tagged RemA was detected as previously described (4). F. johnsoniae cells were grown to late exponential phase in CYE at 25°C with shaking. Whole cells and spent culture fluid were prepared for SDS-PAGE, and Western blotting assays were performed as described above for ChiA, except that antisera against the c-myc epitope (1:10,000 dilution; AbCam, Cambridge, MA) were used.
Analysis for secretion of cell surface SprB and Myc-tagged RemA.
Secretion of SprB was examined essentially as previously described (6, 18). Briefly, cells were grown overnight in MM without shaking at 25°C. Purified anti-SprB (1 μl of a 1:10 dilution of a 300-mg/liter stock), 0.5-μm-diameter protein G-coated polystyrene spheres (1 μl of a 0.1% stock preparation; Spherotech, Inc., Libertyville, IL), and bovine serum albumin (1 μl of a 1% solution) were added to 7 μl of cells (approximately 5 × 108 cells per ml) in MM. The cells were introduced into a tunnel slide (4) and examined by phase-contrast microscopy at 25°C. Samples were examined 2 min after spotting, and images were recorded for 30 s to determine the percentage of cells that had anti-SprB-coated spheres attached to them. Surface-localized Myc-tagged RemA was detected similarly, except that antiserum against the Myc tag (EQKLISEEDL; AbCam) was used.
Cell aggregation studies.
The effect of RemA on aggregation was determined as previously described (4). Cultures (10 ml) were grown overnight in test tubes at 25°C in EC medium (26) with appropriate antibiotics on a platform shaker set at 120 rpm. Cultures were examined for turbidity and for accumulation of cell aggregates at the bottom of the tubes.
Measurement of bacteriophage sensitivity.
F. johnsoniae bacteriophages used in this study were ϕCj1, ϕCj13, ϕCj23, ϕCj28, ϕCj29, ϕCj42, ϕCj48, and ϕCj54 (26, 35, 36). Sensitivity to phages was determined as previously described (11). Briefly, 3 μl of phage lysate (approximately 109 PFU/ml) was spotted onto lawns of cells in CYE overlay agar. The plates were incubated for 24 h at 25°C. A quantitative assay was also used to measure sensitivity to bacteriophages. This involved serial dilution of phage lysates in 10 mM Tris-8 mM MgSO4 (TM buffer, pH 7.5) and determination of the number of PFU. Wild-type F. johnsoniae cells were cultured overnight in CYE at 30°C. One hundred microliters of phage dilutions was added to 200 μl of cells to allow adsorption. Four milliliters of overlay agar at 42°C was added, and the samples were briefly mixed and poured onto CYE agar plates. After solidification of the overlay agar, the plates were incubated for 24 h at 25°C and plaques were counted.
Microscopic observations of cell attachment.
Wild-type and mutant cells of F. johnsoniae were examined for attachment to glass as previously described (6). Cells were grown overnight in MM without shaking at 25°C and harvested at an optical density at 600 nm (OD600) of 0.18. Cells (2.5 μl) were added to a Petroff-Hausser counting chamber, covered with a glass coverslip, and allowed to incubate for 2 min at 25°C. The number of cells attached to 9 randomly selected 0.03-mm2 regions of the glass coverslip was determined.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Cells of F. johnsoniae wild type (CJ1827), ΔporU mutant CJ2116, ΔporV mutant CJ2130, and Δ(gldN gldO) mutant CJ2090 (referred to here as ΔgldNO) and cells of complemented mutants were grown in MM at 25°C with shaking until cells reached an OD600 of 0.7 (late exponential phase of growth). Cells were pelleted by centrifugation at 22,000 × g for 15 min, and the spent culture medium was filtered (0.22-μm polyvinylidene difluoride filters) to remove residual cells. Membrane vesicles and insoluble cell debris were removed from the cell-free spent medium by centrifugation at 100,000 × g for 1 h. This process was repeated once to ensure complete removal of insoluble material, the cell-free spent medium was concentrated 1,000-fold using Amicon concentrators (Millipore, Darmstadt, Germany), and proteins were separated by SDS-PAGE and detected using the Bio-Rad (Hercules, CA) silver stain kit. Enzymatic in-gel digestion was performed at the University of Wisconsin—Madison Mass Spectrometry Facility as outlined on its website (https://www.biotech.wisc.edu/services/massspec).
Peptides were analyzed by nano-LC-MS/MS using the Agilent 1100 nanoflow system (Agilent, Palo Alto, CA) connected to a hybrid linear ion trap-Orbitrap mass spectrometer (LTQ-Orbitrap; Thermo Fisher Scientific, Bremen, Germany) equipped with an Easy-Spray electrospray source. Chromatography of peptides prior to mass spectral analysis was accomplished using a capillary emitter column (PepMap C18; 3 μM, 100 Å, 150 by 0.075 mm; Thermo Fisher Scientific) onto which extracted peptides were automatically loaded. Nano-high-pressure liquid chromatography (nano-HPLC) system-delivered solvents were as follows: (A) 0.1% (vol/vol) formic acid in water and (B) 99.9% (vol/vol) acetonitrile, 0.1% (vol/vol) formic acid. Sample loading was performed at 0.60 μl/min, and peptide elution directly into the nano-electrospray was performed at 0.3 μl/min using a gradient from 0% (vol/vol) B to 40% (vol/vol) B over 20 min followed by a gradient from 40% (vol/vol) B to 100% (vol/vol) B over 5 min. As peptides eluted from the HPLC-column/electrospray source, survey MS scans were acquired in the Orbitrap with a resolution of 100,000. The 5 most intense peptides per scan were fragmented and detected in the ion trap over the mass range 300 to 2,000 m/z. Redundancy was limited by dynamic exclusion. Raw MS/MS data were converted to mgf file format and used to search against an F. johnsoniae protein database (5,507 protein entries) concatenated with a list of common lab contaminants. Peptide mass tolerance was set at 20 ppm, and fragment mass tolerance was set at 0.8 Da. Scaffold version 4.3.2 (Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 90.0% probability by the Peptide Prophet algorithm (37) with Scaffold delta-mass correction. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (38).
Analysis of starch utilization.
Wild-type and mutant cells of F. johnsoniae were examined for starch hydrolysis using a plate assay. Cells were streaked on CYE agar supplemented with 0.25% starch and incubated overnight at 25°C. Starch was detected by flooding the agar with a solution of 1% KI and 1% iodine.
Sequence analyses.
Sequences were analyzed with MacVector software (Cary, NC), and comparisons to database sequences were made using the BLAST algorithm (39). Predictions regarding cellular localization were made using PSORTb (40), TMpredict (41), and CELLO (42).
RESULTS
F. johnsoniae porU and porV.
Analysis of the F. johnsoniae genome revealed orthologs of the P. gingivalis T9SS genes porU and porV. F. johnsoniae PorU exhibits 26% identity to P. gingivalis PorU over 1,170 amino acids, and F. johnsoniae PorV exhibits 44% identity to P. gingivalis PorV over 385 amino acids. F. johnsoniae PorU and PorV have predicted N-terminal signal peptides, and both proteins are predicted by PSORTb and CELLO analyses to reside in the outer membrane. F. johnsoniae porU and porV are located adjacent to each other on the genome (Fig. 1) as are P. gingivalis porU and porV (5, 16, 23). A putative promoter (TTG-N18-TANNTTTG) which matches the Bacteroidetes housekeeping promoter consensus (43) lies between porU and porV, and several possible promoter sequences lie upstream of porU, suggesting that these genes may be transcribed separately. A predicted terminator (AATCCAAATTTCTGCATTTTAGAAATTTGGATTTTTTTT) begins 25 bp downstream of the porV stop codon.
FIG 1.
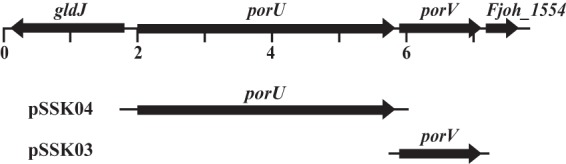
Map of the region spanning porU and porV. Numbers below the map refer to kilobase pairs of sequence. The regions of DNA carried by complementation plasmids used in this study are indicated beneath the map.
porV mutant cells are defective for chitin utilization and for secretion of the chitinase ChiA.
Cells of F. johnsoniae T9SS mutants fail to secrete the soluble extracellular chitinase ChiA and are thus defective in chitin utilization (5–7, 11, 18). The F. johnsoniae porU and porV genes were deleted to determine whether they have roles in T9SS function. Deletion of porV resulted in loss of ability of cells to digest chitin (Fig. 2A). Complementation with pSSK03, which carries porV, restored this ability. In contrast, deletion of porU had little effect on chitin digestion. Similarly, insertional disruption of porU using the suicide vector pLYL03 also had little effect on chitin utilization. pLYL03 insertions result in polar mutations that prevent expression of downstream genes of an operon (44). The ability of the porU disruption mutant to digest chitin (Fig. 2A) supports the suggestion made above that porU and porV are transcribed separately. Chitinase activity was also examined in cell-free spent culture fluid, intact cells, and cell extracts, using a quantitative assay. Chitinase activity was detected in the cell-free spent culture fluid from wild-type cells and from cells of the ΔporU mutant but was not detected in spent culture fluid from cells of the ΔporV mutant (Fig. 2B), suggesting that PorV was required for secretion of the major soluble extracellular chitinase ChiA. Cell-associated chitinase levels (intact cells and cell extracts) were similar for wild-type and mutant cells and presumably reflect the activities of chitinases other than ChiA. Genome analysis predicted the existence of four chitinases in addition to ChiA that may contribute to these activities (28).
FIG 2.
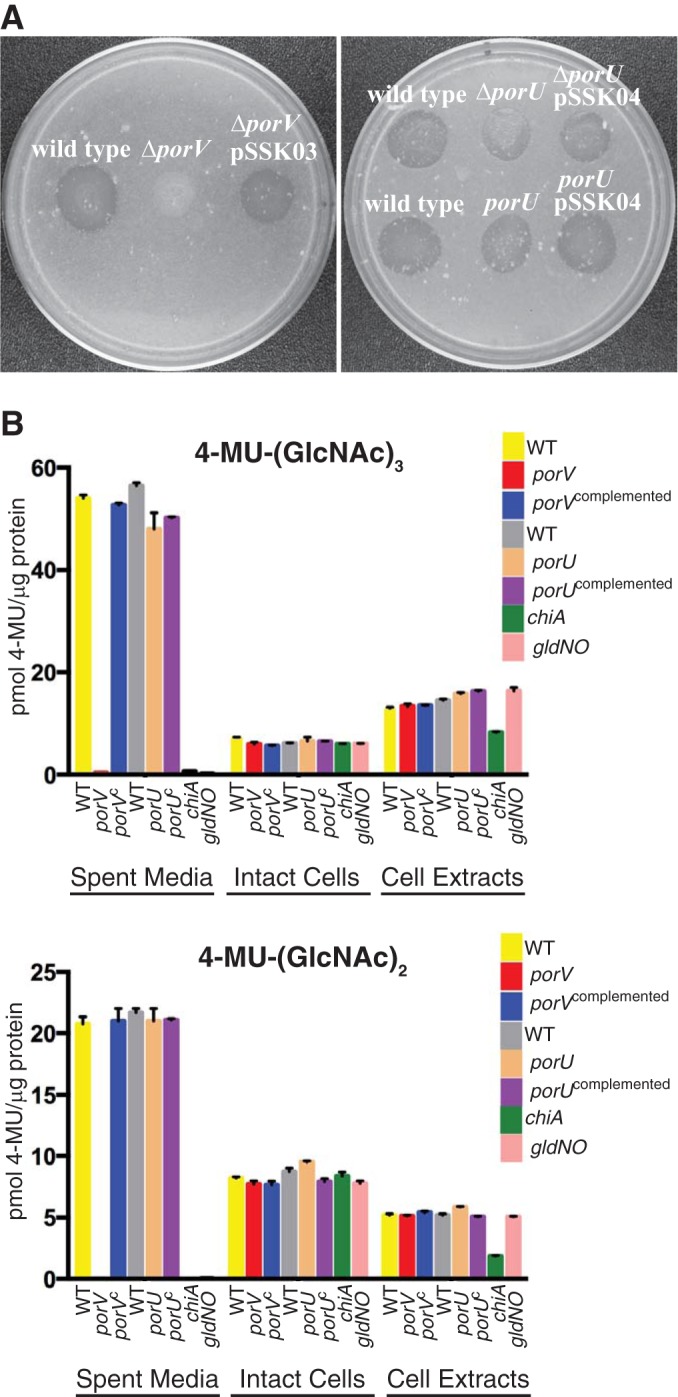
porV is required for chitin utilization. (A) Chitin digestion on agar media. Approximately 106 cells of wild-type and mutant strains of F. johnsoniae were spotted on MYA-chitin medium (27) and incubated at 25°C for 2.5 days. Left panel, left to right: wild-type F. johnsoniae CJ1827; porV deletion mutant CJ2130; CJ2130 complemented with pSSK03, which carries porV. Right panel, top row, left to right: wild-type F. johnsoniae CJ1827; porU deletion mutant CJ2116; CJ2116 with pSSK04, which carries porU. Right panel, bottom row, left to right: wild-type F. johnsoniae UW101; porU disruption mutant CJ1818; CJ1818 with pSSK04, which carries porU. (B) Chitinase activities of wild-type and mutant cells. Chitinase activities of spent medium, intact cells, and cell extracts were determined using the synthetic substrates 4-MU-(GlcNAc)3 and 4-MU-(GlcNAc)2. Equal amounts of each sample, based on the protein content of the cell suspension, were incubated with 10 nmol of synthetic substrate for 30 min at 37°C, and the amount of 4-MU released was determined by measuring fluorescence emission at 460 nm following excitation at 360 nm. Yellow, wild-type (WT) F. johnsoniae UW101 carrying control vector pCP29; red, porV deletion mutant CJ2130 carrying pCP29; blue, CJ2130 complemented with pSSK03 which carries wild-type porV; gray, wild-type F. johnsoniae UW101 carrying control vector pCP23; tan, porU deletion mutant CJ2116 carrying pCP23; purple, CJ2116 complemented with pSSK04, which carries wild-type porU; green, chiA mutant CJ1808; pink, gldNO deletion mutant CJ1631A.
Western blot analyses were used to examine the presence of ChiA protein in cells and in the spent culture fluid of wild-type and mutant strains. Wild-type cells secreted ChiA into the culture fluid, with little if any ChiA detected in cell extracts (Fig. 3). In contrast, cells of the porV mutant failed to secrete ChiA and instead accumulated small amounts of the larger proChiA inside cells. Complementation of the porV mutant with pSSK03 resulted in secretion of ChiA into the culture medium and failure to accumulate proChiA inside cells, similarly to wild-type cells. The effect of deletion of porV on ChiA secretion was nearly identical to that observed for deletion of the region spanning the T9SS genes gldN and gldO (Fig. 3). Deletion of other T9SS genes also resulted in failure to secrete ChiA (7). In contrast to the results with the porV mutant, cells of the porU deletion mutant behaved similarly to wild-type cells. The porU mutant strain secreted ChiA and failed to accumulate it inside cells. Secreted ChiA from wild-type and porU mutant cells migrated at the same size, suggesting that in F. johnsoniae PorU is not required for secretion or processing of ChiA.
FIG 3.
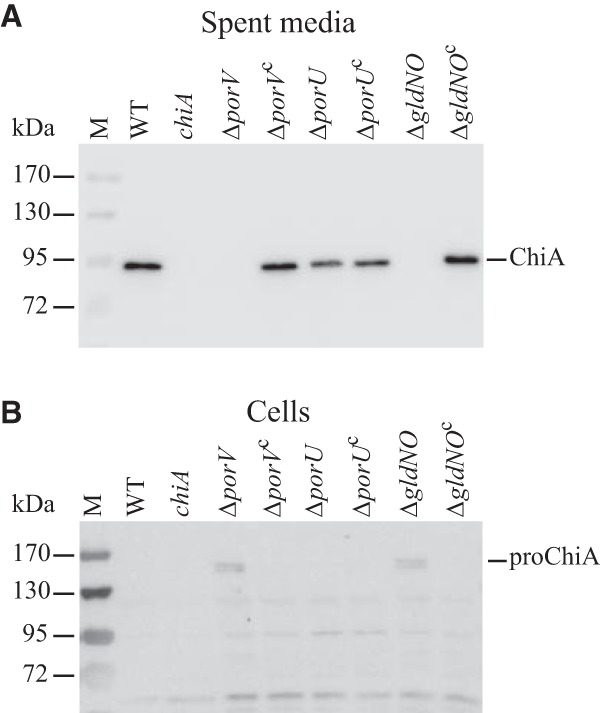
PorV is required for secretion of the soluble extracellular chitinase ChiA. Cell-free spent medium (A) and cells (B) were examined for ChiA by SDS-PAGE followed by Western blotting using antiserum against recombinant ChiA. M, molecular mass markers; WT, wild-type F. johnsoniae CJ1827; chiA, chiA mutant CJ1808; ΔporV, porV deletion mutant CJ2130; ΔporVc, CJ2130 complemented with pSSK03, which carries porV; ΔporU, porU deletion mutant CJ2116; ΔporUc, CJ2116 complemented with pSSK04, which carries porU; ΔgldNO, gldNO deletion mutant CJ1631A; ΔgldNOc, CJ1631A complemented with pTB79, which carries gldN. Samples loaded in panel B corresponded to 15 μg protein per lane, and samples loaded in panel A corresponded to the volume of spent medium that contained 15 μg cell protein before the cells were removed.
Proteins secreted by T9SSs typically have conserved CTDs involved in this process (6, 9, 15). The C-terminal 105 amino acids of ChiA are necessary and sufficient for secretion by the T9SS since mCherry fused to the CTD of ChiA is efficiently secreted by wild-type cells but not by cells of T9SS mutants (7). Secretion of mCherry-CTDChiA by wild-type cells and by porU and porV mutant cells was examined. Wild-type cells and cells of the porU deletion mutant secreted mCherry-CTDChiA (Fig. 4). The secreted protein corresponded to the size of mCherry, suggesting that the protein was processed, perhaps removing the CTD, during or after secretion. In contrast, cells of the ΔporV mutant and of the ΔgldNO mutant failed to secrete mCherry-CTDChiA and instead accumulated a small amount of a protein corresponding in size to mCherry-CTDChiA in the cells. The results indicate that PorV is required for secretion of proteins carrying CTDChiA.
FIG 4.
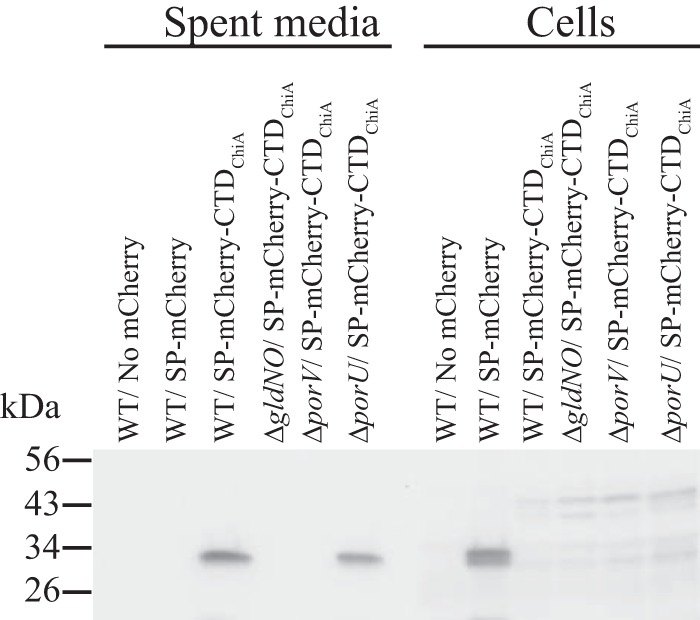
PorV is required for secretion of the heterologous fusion protein mCherry-CTDChiA. Cell-free spent medium and whole cells were analyzed for cultures of wild-type (WT) cells carrying pSSK54, which expresses mCherry with the N-terminal signal peptide from ChiA (SP-mCherry), or carrying pSSK52, which expresses mCherry with the N-terminal signal peptide from ChiA and the 105-amino-acid C-terminal region of ChiA (SP-mCherry-CTDChiA). Cells and spent medium from cultures of the T9SS mutant CJ1631A (ΔgldNO), the porV deletion mutant CJ2130 (ΔporV), and the porU deletion mutant CJ2116 (ΔporU), each carrying pSSK52, were also analyzed. “No mCherry” indicates spent medium or cells from F. johnsoniae carrying the control vector pCP23. Cell samples corresponded to 15 μg protein per lane, and samples from spent medium corresponded to the volume of spent medium that contained 15 μg cell protein before the cells were removed. Samples were separated by SDS-PAGE, and mCherry was detected using antiserum against mCherry.
porV mutant cells fail to secrete RemA.
The cell surface motility adhesin RemA is secreted by the T9SS (4). The ability of cells of the porV deletion mutant to secrete a Myc-tagged version of RemA was examined using antisera against the c-myc peptide. Wild-type and ΔporV mutant cells produced Myc-tagged RemA (Fig. 5, right), but whereas wild-type cells localized RemA on the cell surface, cells of the ΔporV mutant failed to do this (Table 2). Wild-type cells also secreted substantial amounts of RemA into the culture fluid (Fig. 5, left), whereas cells of the porV mutant did not. Soluble secreted RemA from wild-type cells was present as fragments of 100 and 130 kDa, significantly smaller than cell-associated RemA. We do not know if these soluble fragments of RemA are functional, but they serve as additional evidence that PorV is required for secretion of RemA.
FIG 5.
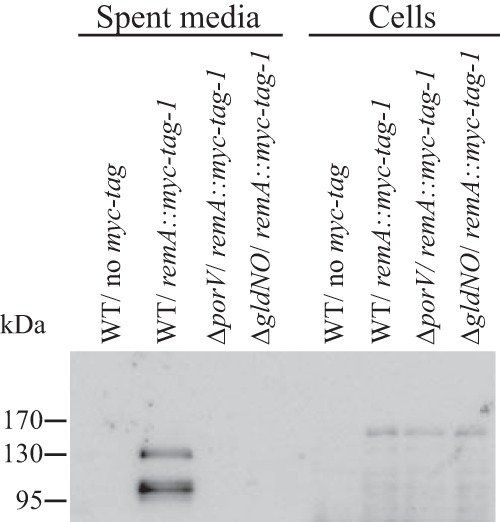
Deletion of porV disrupts secretion of RemA. Immunodetection of Myc-tagged RemA in spent medium or in cells of wild-type or mutant F. johnsoniae strains. Cell-free spent medium and whole cells were analyzed for cultures of wild-type F. johnsoniae CJ1827 (WT, no myc tag), CJ2083 (WT, remA::myc tag 1), CJ2323 (ΔporV remA::myc tag 1), and CJ2089 (ΔgldNO, remA::myc tag 1). Cell samples corresponded to 20 μg protein per lane, and samples from spent medium corresponded to the volume of spent medium that contained 20 μg cell protein before the cells were removed. Samples were separated by SDS-PAGE, and Myc-tagged RemA was detected using antiserum against the myc tag peptide.
TABLE 2.
Deletion of porV disrupts secretion of myc-tagged RemA
| Strain | Description | Antibody added | Avg (SD) % of cells with spheres attacheda |
|---|---|---|---|
| CJ1827 | Wild type, no myc tag | Anti-Myc | 0.0 (0.0) |
| CJ2083 | Wild type, remA::myc tag 1 | No antibody | 0.0 (0.0) |
| CJ2083 | Wild type, remA::myc tag 1 | Anti-Myc | 44.6 (3.3) |
| CJ2323 | ΔporV remA::myc tag 1 | Anti-Myc | 0.0 (0.0) |
| CJ2323/pSSK03 | ΔporV remA::myc tag 1, complemented with pSSK03, carrying porV | Anti-Myc | 40.6 (3.0) |
Purified anti-myc-tag antiserum and 0.5-μm-diameter protein G-coated polystyrene spheres were added to cells as described in Materials and Methods. Samples were introduced into a tunnel slide, incubated for 2 min at 25°C, and examined using a phase-contrast microscope. Images were recorded for 30 s, and 100 randomly selected cells were examined for the presence of spheres that remained attached to the cells during this time. The numbers in parentheses are standard deviations calculated from three measurements.
RemA is a cell surface galactose/rhamnose-binding lectin, and cells expressing this protein aggregate to form multicell clumps (4). This phenomenon is most easily observed when RemA is moderately overexpressed (approximately 10-fold) from plasmid. Wild-type and porV mutant cells expressing RemA were examined for the formation of large cell aggregates. Wild-type cells aggregated extensively, rapidly falling to the bottom of the culture fluid, whereas cells of the porV mutant remained dispersed (Fig. 6), further indicating that porV mutant cells fail to secrete RemA to the cell surface.
FIG 6.
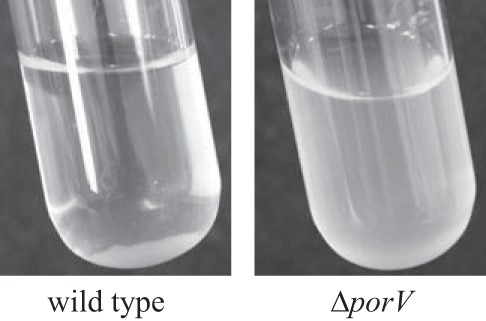
Effect of porV on RemA-mediated cell aggregation. Cells of CJ1827 (wild type) and CJ2130 (ΔporV) were incubated in EC medium for 16 h at 25°C. Both strains carried remA-expressing plasmid pRR39.
PorU and PorV are not required for secretion of the major motility adhesin SprB or for gliding motility.
The T9SS is required for secretion of the motility adhesin SprB to the cell surface (5, 6, 11, 18). Antibodies against SprB were used to determine if SprB was present on the surface of ΔporU and ΔporV mutant cells. Latex spheres coated with antibodies against SprB readily attached to wild-type and ΔporU and ΔporV mutant cells (Table 3). In contrast, they failed to attach to cells of the T9SS mutant CJ2090 (ΔgldNO) or to cells of the sprB deletion mutant CJ1922. These results indicate that although PorV is required for secretion of RemA and ChiA, it is not required for secretion of SprB.
TABLE 3.
PorV and PorU are not required for localization of SprB to the cell surface
| Strain | Description | Antibody added | Avg (SD) % of cells with spheres attacheda |
|---|---|---|---|
| CJ1827 | Wild type | No antibody | 0.0 (0.0) |
| CJ1827 | Wild type | Anti-SprB | 44.5 (3.54) |
| CJ1827/pCP29 | Wild type with empty vector pCP29 | Anti-SprB | 43.5 (3.05) |
| CJ1827/pCP23 | Wild type with empty vector pCP23 | Anti-SprB | 39.5 (3.48) |
| CJ1922 | ΔsprB | Anti-SprB | 0.0 (0.0) |
| CJ2090 | Δ(gldN gldO) | Anti-SprB | 0.0 (0.0) |
| CJ2130/pCP29 | ΔporV with empty vector pCP29 | Anti-SprB | 30.4 (2.9) |
| CJ2130/pSSK03 | ΔporV complemented with pSSK03, carrying porV | Anti-SprB | 37.5 (1.88) |
| CJ2116/pCP23 | ΔporU with empty vector pCP23 | Anti-SprB | 37.0 (2.17) |
| CJ2116/pSSK04 | ΔporU complemented with pSSK04, carrying porU | Anti-SprB | 35.8 (2.51) |
Purified anti-SprB antiserum and 0.5-μm-diameter protein G-coated polystyrene spheres were added to cells as described in Materials and Methods. Samples were introduced into a tunnel slide, incubated for 2 min at 25°C, and examined using a phase-contrast microscope. Images were recorded for 30 s, and 100 randomly selected cells were examined for the presence of spheres that remained attached to the cells during this time. The numbers in parentheses are standard deviations calculated from three measurements.
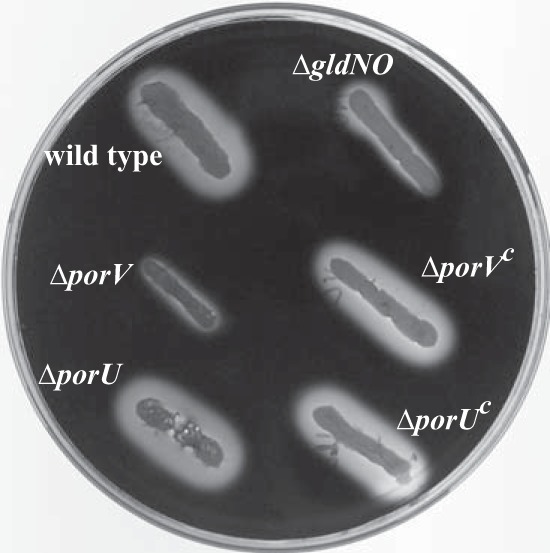
The presence of SprB on the cell surface is required for efficient cell movement over agar resulting in the formation of spreading colonies. Cells of sprB mutants, and cells of T9SS mutants that disrupt secretion of SprB, form nonspreading colonies (3, 5, 6, 11, 18). In contrast, cells of porU and porV mutants formed spreading colonies (Fig. 7), consistent with the ability of these mutants to express and secrete SprB. As shown above, porV mutant cells were deficient in secretion of the motility adhesin RemA. However, as previously reported (4), and as confirmed in this study (Fig. 7), deletion of remA had no effect on movement of cells over agar. Although not required for movement over agar, RemA is thought to facilitate attachment to and movement over other types of surfaces (such as glass) that are coated with rhamnose- or galactose-containing polysaccharides produced by the cells (4).
FIG 7.
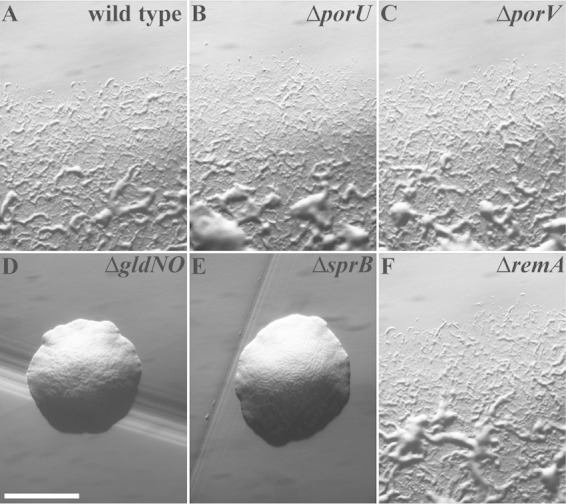
Photomicrographs of F. johnsoniae colonies. Colonies grown from single cells were incubated at 25°C on PY2 agar for 44 h. Photomicrographs were taken with a Photometrics Cool-SNAPcf2 camera mounted on an Olympus IMT-2 phase-contrast microscope. (A) Wild-type F. johnsoniae CJ1827. (B) porU deletion mutant CJ2116. (C) porV deletion mutant CJ2130. (D) gldNO deletion mutant CJ2090. (E) sprB deletion mutant CJ1922. (F) remA deletion mutant CJ1984. The bar in panel D indicates 1 mm and applies to all panels.
porV mutant cells are resistant to some F. johnsoniae phages.
Cells with mutations in genes essential for T9SS function exhibit resistance to all known bacteriophages that infect F. johnsoniae (6, 11, 18). For example, cells of the ΔgldNO mutant CJ2090 were resistant to all bacteriophages tested (Fig. 8I). This is thought to be the result of inability to secrete cell surface proteins that function as phage receptors, such as SprB, RemA, and other motility adhesins (3, 4, 6). porU mutants remained sensitive to all phages (Fig. 8B), consistent with the findings presented above that PorU is not required for secretion by the F. johnsoniae T9SS. In contrast, the porV deletion mutant CJ2130 was resistant to at least two of the eight phages tested, ϕCj48 and ϕCj54 (Fig. 8C; also see Table S2 in the supplemental material). Complementation with pSSK03, which carries porV, restored sensitivity to these phages. Previous results indicated that SprB is a likely receptor for phages ϕCj1, ϕCj13, and ϕCj23 and one of several receptors for ϕCj29 (3, 6) (Fig. 8F). Sensitivity of the porV mutant to phages ϕCj1, ϕCj13, ϕCj23, and ϕCj29 supports the suggestion made above that PorV is not required for secretion of SprB. PorV is required for the secretion of RemA, as shown above, and it is also likely to be involved in the secretion of other cell surface proteins. This is illustrated by the sensitivity of the ΔremA mutant to phages ϕCj48 and ϕCj54 compared to the complete resistance of the ΔporV mutant to these phages (see Table S2 in the supplemental material). Comparison of the phage resistances of the ΔsprB and ΔporV mutants with that of the ΔsprB ΔporV double mutant suggests that some phages may use multiple cell surface receptors. Cells of the ΔsprB or ΔporV mutants were susceptible to ϕCj29 and ϕCj42 whereas cells of the double mutant (ΔporV ΔsprB) were completely resistant to both phages (Fig. 8, compare panels C, F, and H; also see Table S2 in the supplemental material). Phages ϕCj29 and ϕCj42 may use either SprB or cell surface proteins secreted by PorV as receptors.
FIG 8.
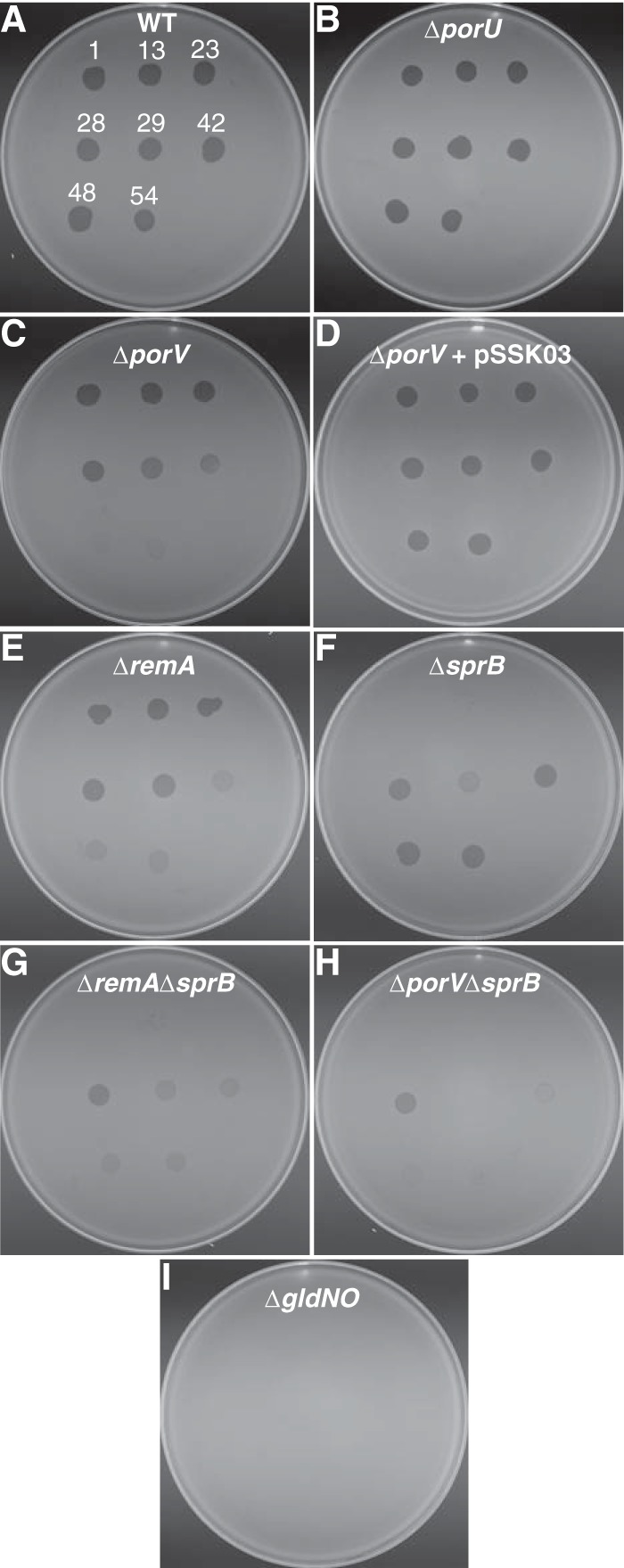
Susceptibility of wild-type and mutant cells to bacteriophages. Bacteriophages (3 μl of lysates containing approximately 109 PFU/ml) were spotted onto lawns of cells in CYE overlay agar. The plates were incubated at 25°C for 24 h to observe lysis. Bacteriophages were spotted in the following order from left to right, as also indicated by the numbers in panel A: top row, ϕCj1, ϕCj13, and ϕCj23; middle row, ϕCj28, ϕCj29, and ϕCj42; bottom row, ϕCj48 and ϕCj54. (A) Wild-type F. johnsoniae CJ1827. (B) CJ2116 (ΔporU). (C) CJ2130 (ΔporV). (D) CJ2130 complemented with pSSK03, which carries porV. (E) CJ1984 (ΔremA). (F) CJ1922 (ΔsprB). (G) CJ1985 (ΔremA ΔsprB). (H) CJ2445 (ΔporV ΔsprB). (I) CJ2090 (ΔgldNO).
Cells of the ΔporV mutant are defective in attachment to glass.
Wild-type cells attached efficiently to glass whereas cells of the ΔgldNO mutant CJ2090 were completely deficient in attachment (Table 4). This supports the previous suggestion that the T9SS secretes cell surface adhesins (6). The motility adhesins SprB and RemA are secreted by the T9SS, and their absence on the surface of T9SS mutants accounts for some of the defect in attachment. Cells of the sprB deletion mutant CJ1922 attached well to glass, and cells of the remA deletion mutant CJ1984 were only slightly deficient in attachment, but cells of the double mutant CJ1985 (ΔremA ΔsprB) exhibited a substantial defect in attachment (Table 4). An even greater defect in attachment was observed for cells of the ΔporV mutant CJ2130, suggesting that PorV is required for secretion of other adhesins in addition to RemA. The few ΔporV mutant cells that attached to glass exhibited gliding motility, consistent with the results described above that indicated that PorV was not required for gliding motility. CJ2445 (ΔporV ΔsprB) was almost entirely deficient in attachment to glass, suggesting that PorV is required for secretion of most of the glass-binding adhesins other than SprB. The results suggest that RemA, SprB, and other adhesins secreted by the T9SS are responsible for attachment to glass. Some of these adhesins may function as phage receptors, helping to explain the phage resistance of porV mutant cells (Fig. 8). Cells of the porU mutant CJ2116 attached to glass almost as well as did wild-type cells, indicating that PorU is not required for secretion of the F. johnsoniae adhesins.
TABLE 4.
Deletion of porV results in decreased attachment of cells to glass
| Strain | Description | Avg (SD) no. of cells attached to 0.03-mm2 region of glass coverslipa |
|---|---|---|
| CJ1827 | Wild type | 40.5 (2.2) |
| CJ1827/pCP29 | Wild type with empty vector pCP29 | 38.7 (6.7) |
| CJ1827/pCP23 | Wild type with empty vector pCP23 | 32.4 (6.7) |
| CJ2090 | Δ(gldN gldO) | 0.0 (0.0) |
| CJ1984/pCP29 | ΔremA with empty vector pCP29 | 32.3 (6.5) |
| CJ1922/pCP29 | ΔsprB with empty vector pCP29 | 41.4 (9.0) |
| CJ1985/pCP29 | ΔremA ΔsprB with empty vector pCP29 | 17.8 (5.5) |
| CJ2130/pCP29 | ΔporV with empty vector pCP29 | 7.0 (1.6) |
| CJ2130/pSSK03 | ΔporV complemented with pSSK03, carrying porV | 31.3 (2.7) |
| CJ2445/pCP29 | ΔporV ΔsprB with empty vector pCP29 | 0.1 (0.3) |
| CJ2116/pCP23 | ΔporU with empty vector pCP23 | 31.5 (2.7) |
| CJ2116/pSSK04 | ΔporU complemented with pSSK04, carrying porU | 30.3 (2.7) |
Approximately 106 cells in 2.5 μl of MM were introduced into a Petroff-Hausser counting chamber and incubated for 2 min at 25°C. Samples were observed using an Olympus BH-2 phase-contrast microscope, and cells attached to a 0.03-mm2 region of the cover glass were counted. Numbers in parentheses are standard deviations calculated from 9 measurements.
porV mutant cells appear to be defective for secretion of at least 32 additional proteins.
Spent culture fluids of wild-type cells; ΔgldNO, ΔporV, and ΔporU mutant cells; and complemented cells were examined for the presence of secreted proteins by SDS-PAGE. Several prominent bands between 60 and 240 kDa that were present in the cell-free spent culture fluids from wild-type and ΔporU mutant cells were absent or decreased in intensity in culture fluid of ΔgldNO and ΔporV mutant cells and were restored to near-wild-type levels in the complemented mutants (Fig. 9). LC-MS/MS analysis of one of these bands, at approximately 90 kDa, demonstrated that it corresponded to ChiA (see Fig. S1 in the supplemental material), which as mentioned above is secreted by the T9SS. The region of the gel between 60 and 240 kDa was excised and analyzed by LC-MS/MS (Table 5; also see Table S3 in the supplemental material). Proteins present in the culture fluid from wild-type cells, but absent or greatly reduced in culture fluid from the ΔgldNO mutant, included 18 proteins with TIGR04183 CTDs (including RemA), six proteins with TIGR04131 CTDs, and nine proteins that lacked obvious conserved CTDs. Twenty-six of the proteins mentioned above were also absent or greatly reduced in the cell-free culture fluid of the ΔporV mutant. The other seven proteins (Fjoh_0601, Fjoh_0602, Fjoh_0604, Fjoh_0606, Fjoh_1123, Fjoh_3952, and Fjoh_4934) apparently do not require PorV for secretion by the T9SS. All of the proteins listed in Table 5 that have TIGR04183-type CTDs required PorV for efficient secretion. Proteins in cell-free culture fluid of ΔporU mutant cells were similar to those in culture fluid of wild-type cells (Fig. 9), although a few proteins were apparently reduced in intensity or absent in the culture fluid from the ΔporU mutant (Table 5). These results indicate that PorU is not required for F. johnsoniae T9SS-mediated secretion, but it may enhance the secretion of some proteins.
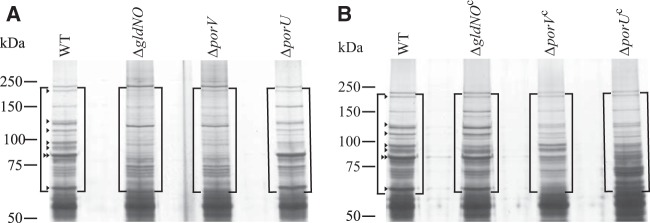
FIG 9.
Soluble extracellular proteins of wild-type and mutant cells. Cells of F. johnsoniae wild type (CJ1827); ΔgldNO mutant (CJ2090); ΔporV mutant (CJ2130); ΔporU mutant (CJ2116); CJ2090 complemented with pTB79, which carries gldN; CJ2130 complemented with pSSK03, which carries porV; and CJ2116 complemented with pSSK04, which carries porU, were grown in MM at 25°C with shaking until cells reached an OD600 of 0.7. Equal amounts of cell-free spent medium of wild-type and mutant cells were separated by SDS-PAGE, and proteins were detected by silver staining. Arrowheads indicate bands present in the culture fluid of wild-type cells that were absent or reduced in intensity in the culture fluid of the ΔgldNO mutant cells. The double arrowhead corresponds to the N-terminal fragment of ChiA (see Fig. S1 in the supplemental material). The boxed regions were subjected to LC-MS/MS analysis (Table 5; also see Table S3 in the supplemental material).
TABLE 5.
Candidate proteins secreted by the T9SS identified by LC-MS/MS analysis of cell-free culture fluida
| Locus tag/protein name | Mol massb (kDa) | Predicted localizationc | CTDd | Predicted protein functione | Spectrum count for: |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | ΔgldNO strain | ΔgldNO strain with pTB79 | ΔporV strain | ΔporV strain with pSSK03 | ΔporU strain | ΔporU strain with pSSK04 | |||||
| Fjoh_0074 | 123.1 | OM, E | TIGR04183 | Nuclease/phosphatase | 42 | 3 | 108 | 7 | 84 | 95 | 129 |
| Fjoh_0601 | 208.2 | OM | 115 | 0 | 84 | 71 | 78 | 118 | 148 | ||
| Fjoh_0602 | 279.3 | OM | 68 | 0 | 38 | 57 | 59 | 65 | 86 | ||
| Fjoh_0604 | 144.2 | E | 47 | 0 | 39 | 42 | 44 | 40 | 45 | ||
| Fjoh_0606 | 409.5 | OM | 163 | 0 | 172 | 77 | 169 | 176 | 198 | ||
| Fjoh_0808/RemA | 154.0 | E | TIGR04183 | Motility adhesin | 38 | 0 | 47 | 0 | 56 | 37 | 67 |
| Fjoh_0886 | 99.1 | E | TIGR04183 | Peptidase | 12 | 0 | 19 | 0 | 21 | 14 | 18 |
| Fjoh_1022 | 51.1 | E | TIGR04183 | Licheninase | 6 | 0 | 6 | 0 | 6 | 1 | 8 |
| Fjoh_1123 | 121.9 | E, OM | TIGR04131 | 34 | 0 | 10 | 27 | 3 | 37 | 32 | |
| Fjoh_1188 | 152.7 | E, OM | TIGR04183 | 49 | 0 | 104 | 0 | 42 | 52 | 75 | |
| Fjoh_1189 | 181.4 | E | TIGR04183 | Lectin | 74 | 0 | 112 | 1 | 69 | 112 | 79 |
| Fjoh_1208 | 112.5 | E | TIGR04183 | α-Amylase | 45 | 0 | 66 | 6 | 91 | 58 | 126 |
| Fjoh_1231 | 97.8 | E | TIGR04183 | Pectate lyase | 9 | 0 | 13 | 0 | 6 | 35 | 31 |
| Fjoh_1269 | 94.3 | E, OM | TIGR04183 | 27 | 4 | 43 | 3 | 40 | 34 | 56 | |
| Fjoh_1408f | 106.0 | E | TIGR04183 | α-Amylase | 2 | 0 | 4 | 0 | 2 | 0 | 6 |
| Fjoh_1645f | 258.1 | E | TIGR04131 | 2 | 0 | 6 | 0 | 1 | 2 | 3 | |
| Fjoh_2150 | 39.0 | E, OM | TIGR04183 | 6 | 0 | 6 | 0 | 7 | 3 | 3 | |
| Fjoh_2273 | 93.3 | E | TIGR04131 | 4 | 0 | 5 | 1 | 5 | 5 | 1 | |
| Fjoh_2389f | 57.7 | E, OM | TIGR04183 | Peptidase | 2 | 0 | 7 | 0 | 12 | 0 | 6 |
| Fjoh_2667 | 129.7 | OM | 28 | 0 | 7 | 0 | 3 | 5 | 33 | ||
| Fjoh_2687 | 155.8 | E | 26 | 1 | 26 | 7 | 35 | 26 | 43 | ||
| Fjoh_3108 | 30.9 | OM, E, P | 7 | 0 | 10 | 0 | 8 | 0 | 10 | ||
| Fjoh_3246 | 299.4 | OM, E | TIGR04183 | 12 | 0 | 77 | 0 | 6 | 23 | 41 | |
| Fjoh_3324 | 105.3 | E | TIGR04183 | Carbohydrate binding | 16 | 1 | 40 | 5 | 20 | 49 | 59 |
| Fjoh_3729 | 195.1 | OM | 46 | 0 | 32 | 0 | 52 | 23 | 79 | ||
| Fjoh_3777 | 128.1 | OM, E | TIGR04183 | Deacylase | 10 | 0 | 25 | 0 | 9 | 10 | 34 |
| Fjoh_3952 | 330.6 | E | TIGR04131 | 22 | 0 | 11 | 12 | 16 | 11 | 17 | |
| Fjoh_4174 | 102.5 | E | TIGR04183 | Carbohydrate binding | 40 | 5 | 40 | 6 | 62 | 55 | 36 |
| Fjoh_4176 | 95.4 | E | TIGR04183 | Carbohydrate binding | 48 | 3 | 65 | 7 | 63 | 108 | 76 |
| Fjoh_4177 | 144.9 | E | TIGR04183 | Glycoside hydrolase | 22 | 0 | 35 | 0 | 34 | 67 | 60 |
| Fjoh_4750 | 158.1 | E | TIGR04131 | 13 | 0 | 3 | 3 | 3 | 3 | 10 | |
| Fjoh_4819 | 112.5 | C, OM, P | Glycoside hydrolase | 34 | 0 | 5 | 0 | 1 | 12 | 9 | |
| Fjoh_4934 | 84.8 | E | TIGR04131 | 11 | 1 | 7 | 19 | 4 | 17 | 17 | |
Proteins in cell-free culture fluid from wild-type F. johnsoniae CJ1827, ΔgldNO mutant CJ2090, ΔporV mutant CJ2130, ΔporU mutant CJ2116, ΔgldNO complemented with pTB79, ΔporV complemented with pSSK03, and ΔporU complemented with pSSK04 were separated by SDS-PAGE and silver stained, and the regions shown in Fig. 9 spanning approximately 60 to 240 kDa were cut from the gel and analyzed by LC-MS/MS. Total/unweighted spectrum counts corresponding to total number of spectra associated with a single protein and indicative of relative abundance of that protein are indicated for each of the seven strains analyzed. For the complete data set, including proteins that were apparently secreted by other secretion systems or were released by cell lysis, see Table S3 in the supplemental material.
Mol mass, molecular mass as calculated for full-length protein before removal of signal peptide.
Protein localization as predicted by CELLO v 2.5 subcellular localization predictor (42). OM, outer membrane; E, extracellular; P, periplasmic; C, cytoplasmic.
CTD type identified by BLASTP analysis.
Predicted protein functions as listed on the Integrated Microbial Genomes website (https://img.jgi.doe.gov), except for RemA (4).
The small number of spectra identified from wild-type cells for these proteins made the prediction of secretion by the T9SS less certain. These proteins were included because no spectra were observed from ΔgldNO or ΔporV mutant cells and because complementation of the ΔgldNO mutant resulted in 4 to 7 spectral hits for each protein.
In addition to the proteins that appeared to be secreted by the T9SS mentioned above, many other proteins were also identified in the cell-free culture fluid (see Table S3 in the supplemental material). Analysis of the F. johnsonaie genome suggested the presence of a type II secretion system (8), which may account for the secretion of some of these proteins. Some proteins were present in much larger amounts in the culture fluid of T9SS mutants than in the culture fluid of wild-type cells (see Table S3 in the supplemental material). These may be cellular (nonsecreted) proteins that were released because of cell surface defects of the T9SS mutants. Consistent with this, many of these proteins were predicted to localize to the cytoplasm, periplasm, or outer membrane, where they presumably reside in wild-type cells.
The T9SS and PorV are required for efficient starch utilization.
The predicted α-amylases Fjoh_1208 and Fjoh_1408 were detected in cell-free culture fluid of wild-type cells but not in culture fluid of ΔgldNO mutant cells (Table 5). Examination of wild-type and mutant cells confirmed that the T9SS has a role in starch utilization (Fig. 10). The ΔgldNO and ΔporV mutant cells were partially deficient in digestion of starch. The small amount of residual starch digestion detected with these mutants may be the result of additional amylases released by other secretion systems or may indicate that a small amount of Fjoh_1208 or Fjoh_1408 was released from the mutant cells.
FIG 10.
Starch digestion by wild-type and mutant cells. Cells were streaked on CYE agar containing 0.25% starch and incubated overnight at 25°C. The agar was flooded with a solution of 1% KI and 1% iodine to detect starch. Clearing zones around the cells indicate starch digestion. Wild type, F. johnsoniae CJ1827; ΔgldNO, gldNO deletion mutant CJ2090; ΔporV, porV deletion mutant CJ2130; ΔporVc, CJ2130 complemented with pSSK03, which carries porV; ΔporU, porU deletion mutant CJ2116; ΔporUc, CJ2116 complemented with pSSK04, which carries porU.
Fjoh_0288, which exhibits limited sequence similarity to PorV, does not appear to be required for T9SS function.
Examination of the F. johnsoniae genome revealed one gene, Fjoh_0288, encoding a protein that exhibits 30% amino acid identity with PorV over a 135-amino-acid region. Fjoh_0288 was deleted in wild-type cells and in cells of the ΔporV mutant. Cells of CJ2082 (ΔFjoh_0288) and of CJ2446 (ΔFjoh_0288 ΔporV) spread on agar as well as did wild-type cells (data not shown), suggesting that SprB was secreted to the cell surface. Deletion of Fjoh_0288 also had no effect on secretion of ChiA and no effect on sensitivity to the eight phages used in this study (data not shown). These results suggest that Fjoh_0288 is not involved in T9SS-mediated secretion.
DISCUSSION
T9SSs, discovered in P. gingivalis and F. johnsoniae, are common among members of the phylum Bacteroidetes (5, 8, 9). Seven proteins (GldK, GldL, GldM, GldN, SprA, SprE, and SprT) are important for T9SS function in F. johnsoniae, and orthologs of these are required for secretion in P. gingivalis (5, 6, 11, 18–20). P. gingivalis PorP is also required for secretion (5), whereas in F. johnsoniae the function of PorP appears to be split between multiple PorP-like proteins, such as SprF, which is required specifically for secretion of SprB (21). Two additional P. gingivalis proteins (PorU and PorV) are involved in gingipain secretion (5, 16, 22, 23). In this study, we examined the potential roles of F. johnsoniae PorU and PorV in secretion.
F. johnsoniae PorU was not required for secretion of proteins by the T9SS. Three proteins, RemA, ChiA, and SprB, known to be secreted by the T9SS (4, 6, 7, 11) were each secreted in functional form by cells of a porU deletion mutant. SDS-PAGE followed by LC-MS/MS analysis of proteins secreted by wild-type and mutant cells revealed that whereas cells of the gldNO deletion mutant appeared to be defective for the secretion of at least 33 proteins, proteins secreted by cells of the porU deletion mutant were similar to those secreted by wild-type cells. PorU is thought to function as the peptidase that removes the CTDs of P. gingivalis proteins during or after secretion (16). Our results suggest that either F. johnsoniae does not require CTD processing for secretion or it has other proteases that render PorU unnecessary. No paralogs of porU were detected in the genome, but F. johnsoniae produces many peptidases (28).
F. johnsoniae PorV was required for secretion of many but not all proteins that are targeted to the T9SS. Secretion of RemA and ChiA required PorV, whereas secretion of SprB did not. Defects in phage sensitivity and in attachment of cells to glass were also associated with deletion of porV and suggested that PorV is required for the secretion of additional cell surface adhesins besides RemA. Analysis of spent culture fluid of wild-type and mutant cells revealed 26 proteins that appear to require porV for efficient secretion. These proteins were also absent in culture fluid of the ΔgldNO mutant, suggesting that they are secreted from wild-type cells by the T9SS. Eighteen of these proteins had CTDs that belong to protein domain family TIGR04183, suggesting that proteins with TIGR04183-type CTDs might require PorV for secretion. Some proteins with TIGR04131-type CTDs and some proteins that lacked recognizable T9SS CTDs also appeared to require PorV for secretion, whereas others did not since they were found in the cell-free culture fluids of both wild-type and porV mutant cells.
T9SS-mediated secretion of proteins with TIGR04183-type CTDs has been documented for many proteins of diverse members of the phylum Bacteroidetes (4, 6, 8, 9, 14). In contrast, the role of TIGR04131-type CTDs in targeting proteins for secretion is less well established. F. johnsoniae SprB is the only example of a TIGR04131 family member that has been demonstrated to be secreted by the T9SS (5, 6, 11, 21). The observation of six additional F. johnsoniae proteins with TIGR04131-type CTDs that appear to be secreted by the T9SS supports the suggestion that TIGR04131 CTDs target proteins to the T9SS. The F. johnsoniae genome is predicted to encode 12 proteins with TIGR04131-type CTDs, and we hypothesize that each of these is secreted by the T9SS. Proteins that have TIGR04131-type CTDs are also common among the many members of the phylum Bacteroidetes that have T9SSs. Proteins in addition to those with TIGR04183-type CTDs and TIGR04131-type CTDs are also secreted by T9SSs. For example, F. johnsonaie ChiA is secreted by the T9SS, but its CTD, which is necessary and sufficient for secretion, does not closely resemble members of either TIGR04183 or TIGR04131 (7). Here, we identified nine additional proteins (Fjoh_0601, Fjoh_0602, Fjoh_0604, Fjoh_0606, Fjoh_2667, Fjoh_2687, Fjoh_3108, Fjoh_3729, and Fjoh_4819) that were apparently secreted by the T9SS but that did not exhibit similarity to members of either TIGR04183 or TIGR04131. There is considerable sequence diversity among T9SS CTDs (9, 14), and these nine proteins may have novel T9SS-targeting domains that have thus far escaped detection.
The results reported here identify 35 proteins (including ChiA, SprB, and RemA) that appear to be secreted by the F. johnsoniae T9SS. This is probably only a partial list of proteins secreted by this system. Fifty-four F. johnsoniae proteins were previously predicted to be secreted by the T9SS based on the presence of CTDs belonging to TIGR04183 and TIGR04131 (6). In addition, the identification of proteins that lack obvious T9SS-targeting CTDs but that are apparently secreted by the T9SS suggests that additional proteins may be secreted by this system. Further study is needed to determine the diversity of T9SS-targeting sequences and to reveal the mechanism of T9SS-mediated protein secretion in F. johnsoniae and in other members of the phylum Bacteroidetes.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by MCB-1021721 from the National Science Foundation and by a University of Wisconsin—Milwaukee Research Growth Initiative Grant.
We thank Greg Sabat and Greg Barrett-Wilt at the University of Wisconsin—Madison Mass Spectrometry Facility for LC-MS/MS analyses and Yongtao Zhu for helpful suggestions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02085-14.
REFERENCES
- 1.McBride MJ. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu Rev Microbiol 55:49–75. doi: 10.1146/annurev.micro.55.1.49. [DOI] [PubMed] [Google Scholar]
- 2.Nakane D, Sato K, Wada H, McBride MJ, Nakayama K. 2013. Helical flow of surface protein required for bacterial gliding motility. Proc Natl Acad Sci U S A 110:11145–11150. doi: 10.1073/pnas.1219753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson SS, Bollampalli S, McBride MJ. 2008. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J Bacteriol 190:2851–2857. doi: 10.1128/JB.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shrivastava A, Rhodes RG, Pochiraju S, Nakane D, McBride MJ. 2012. Flavobacterium johnsoniae RemA is a mobile cell-surface lectin involved in gliding. J Bacteriol 194:3678–3688. doi: 10.1128/JB.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A 107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shrivastava A, Johnston JJ, van Baaren JM, McBride MJ. 2013. Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell-surface gliding motility adhesins SprB and RemA. J Bacteriol 195:3201–3212. doi: 10.1128/JB.00333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kharade SS, McBride MJ. 2014. The Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J Bacteriol 196:961–970. doi: 10.1128/JB.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McBride MJ, Zhu Y. 2013. Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J Bacteriol 195:270–278. doi: 10.1128/JB.01962-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veith PD, Nor Muhammad NA, Dashper SG, Likic VA, Gorasia DG, Chen D, Byrne SJ, Catmull DV, Reynolds EC. 2013. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification and cell surface attachment. J Proteome Res 12:4449–4461. doi: 10.1021/pr400487b. [DOI] [PubMed] [Google Scholar]
- 10.Chagnot C, Zorgani MA, Astruc T, Desvaux M. 2013. Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front Microbiol 4:303. doi: 10.3389/fmicb.2013.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhodes RG, Samarasam MN, Shrivastava A, van Baaren JM, Pochiraju S, Bollampalli S, McBride MJ. 2010. Flavobacterium johnsoniae gldN and gldO are partially redundant genes required for gliding motility and surface localization of SprB. J Bacteriol 192:1201–1211. doi: 10.1128/JB.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Y, McBride MJ. 2014. Deletion of the Cytophaga hutchinsonii type IX secretion system gene sprP results in defects in gliding motility and cellulose utilization. Appl Microbiol Biotechnol 98:763–775. doi: 10.1007/s00253-013-5355-2. [DOI] [PubMed] [Google Scholar]
- 13.Kolton M, Frenkel O, Elad Y, Cytryn E. 2014. Potential role of flavobacterial gliding-motility and type IX secretion system complex in root colonization and plant defense. Mol Plant Microbe Interact 27:1005–1013. doi: 10.1094/MPMI-03-14-0067-R. [DOI] [PubMed] [Google Scholar]
- 14.Sato K, Yukitake H, Narita Y, Shoji M, Naito M, Nakayama K. 2013. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol Lett 338:68–76. doi: 10.1111/1574-6968.12028. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen KA, Travis J, Potempa J. 2007. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-negative bacteria? J Bacteriol 189:833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glew MD, Veith PD, Peng B, Chen YY, Gorasia DG, Yang Q, Slakeski N, Chen D, Moore C, Crawford S, Reynolds E. 2012. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J Biol Chem 287:24605–24617. doi: 10.1074/jbc.M112.369223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shoji M, Sato K, Yukitake H, Kondo Y, Narita Y, Kadowaki T, Naito M, Nakayama K. 2011. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One 6:e21372. doi: 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhodes RG, Samarasam MN, Van Groll EJ, McBride MJ. 2011. Mutations in Flavobacterium johnsoniae sprE result in defects in gliding motility and protein secretion. J Bacteriol 193:5322–5327. doi: 10.1128/JB.05480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saiki K, Konishi K. 2007. Identification of a Porphyromonas gingivalis novel protein Sov required for the secretion of gingipains. Microbiol Immunol 51:483–491. doi: 10.1111/j.1348-0421.2007.tb03936.x. [DOI] [PubMed] [Google Scholar]
- 20.Sato K, Sakai E, Veith PD, Shoji M, Kikuchi Y, Yukitake H, Ohara N, Naito M, Okamoto K, Reynolds EC, Nakayama K. 2005. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J Biol Chem 280:8668–8677. doi: 10.1074/jbc.M413544200. [DOI] [PubMed] [Google Scholar]
- 21.Rhodes RG, Nelson SS, Pochiraju S, McBride MJ. 2011. Flavobacterium johnsoniae sprB is part of an operon spanning the additional gliding motility genes sprC, sprD, and sprF. J Bacteriol 193:599–610. doi: 10.1128/JB.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen YY, Peng B, Yang Q, Glew MD, Veith PD, Cross KJ, Goldie KN, Chen D, O'Brien-Simpson N, Dashper SG, Reynolds EC. 2011. The outer membrane protein LptO is essential for the O-deacylation of LPS and the co-ordinated secretion and attachment of A-LPS and CTD proteins in Porphyromonas gingivalis. Mol Microbiol 79:1380–1401. doi: 10.1111/j.1365-2958.2010.07530.x. [DOI] [PubMed] [Google Scholar]
- 23.Ishiguro I, Saiki K, Konishi K. 2009. PG27 is a novel membrane protein essential for a Porphyromonas gingivalis protease secretion system. FEMS Microbiol Lett 292:261–267. doi: 10.1111/j.1574-6968.2009.01489.x. [DOI] [PubMed] [Google Scholar]
- 24.Shrivastava A. 2013. Ph.D. thesis University of Wisconsin—Milwaukee, Milwaukee, WI. [Google Scholar]
- 25.Ishiguro I, Saiki K, Konishi K. 2011. Analysis of Porphyromonas gingivalis PG27 by deletion and intragenic suppressor mutation analyses. Mol Oral Microbiol 26:321–335. doi: 10.1111/j.2041-1014.2011.00620.x. [DOI] [PubMed] [Google Scholar]
- 26.Chang LYE, Pate JL, Betzig RJ. 1984. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsonae. J Bacteriol 159:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride MJ, Braun TF. 2004. GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J Bacteriol 186:2295–2302. doi: 10.1128/JB.186.8.2295-2302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride MJ, Xie G, Martens EC, Lapidus A, Henrissat B, Rhodes RG, Goltsman E, Wang W, Xu J, Hunnicutt DW, Staroscik AM, Hoover TR, Cheng YQ, Stein JL. 2009. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol 75:6864–6875. doi: 10.1128/AEM.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhodes RG, Pucker HG, McBride MJ. 2011. Development and use of a gene deletion strategy for Flavobacterium johnsoniae to identify the redundant motility genes remF, remG, remH, and remI. J Bacteriol 193:2418–2428. doi: 10.1128/JB.00117-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McBride MJ, Kempf MJ. 1996. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae. J Bacteriol 178:583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal S, Hunnicutt DW, McBride MJ. 1997. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc Natl Acad Sci U S A 94:12139–12144. doi: 10.1073/pnas.94.22.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, McBride MJ, Subramaniam S. 2007. Cell-surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J Bacteriol 189:7503–7506. doi: 10.1128/JB.00957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 34.Reichenbach H. 1992. The genus Lysobacter, p 3256–3275. In Balows A, Truper H, Dworkin M, Harder W, Schleifer K (ed), The prokaryotes, 2nd ed. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 35.Pate JL, Petzold SJ, Chang L-YE. 1979. Phages for the gliding bacterium Cytophaga johnsonae that infect only motile cells. Curr Microbiol 2:257–262. doi: 10.1007/BF02602855. [DOI] [Google Scholar]
- 36.Wolkin RH, Pate JL. 1985. Selection for nonadherent or nonhydrophobic mutants co-selects for nonspreading mutants of Cytophaga johnsonae and other gliding bacteria. J Gen Microbiol 131:737–750. [Google Scholar]
- 37.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 38.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 39.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 40.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofmann K, Stoffel W. 1993. TMbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler 374:166. [Google Scholar]
- 42.Yu CS, Chen YC, Lu CH, Hwang JK. 2006. Prediction of protein subcellular localization. Proteins 64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 43.Chen S, Bagdasarian M, Kaufman MG, Bates AK, Walker ED. 2007. Mutational analysis of the ompA promoter from Flavobacterium johnsoniae. J Bacteriol 189:5108–5118. doi: 10.1128/JB.00401-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunnicutt DW, McBride MJ. 2000. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes, gldB and gldC. J Bacteriol 182:911–918. doi: 10.1128/JB.182.4.911-918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kempf MJ, McBride MJ. 2000. Transposon insertions in the Flavobacterium johnsoniae ftsX gene disrupt gliding motility and cell division. J Bacteriol 182:1671–1679. doi: 10.1128/JB.182.6.1671-1679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braun TF, Khubbar MK, Saffarini DA, McBride MJ. 2005. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J Bacteriol 187:6943–6952. doi: 10.1128/JB.187.20.6943-6952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.