Abstract
Biofilm dispersion is a highly regulated process that allows biofilm bacteria to respond to changing environmental conditions and to disseminate to new locations. The dispersion of biofilms formed by the opportunistic pathogen Pseudomonas aeruginosa is known to require a number of cyclic di-GMP (c-di-GMP)-degrading phosphodiesterases (PDEs) and the chemosensory protein BdlA, with BdlA playing a pivotal role in regulating PDE activity and enabling dispersion in response to a wide array of cues. BdlA is activated during biofilm growth via posttranslational modifications and nonprocessive cleavage in a manner that is dependent on elevated c-di-GMP levels. Here, we provide evidence that the diguanylate cyclase (DGC) GcbA contributes to the regulation of BdlA cleavage shortly after initial cellular attachment to surfaces and, thus, plays an essential role in allowing biofilm cells to disperse in response to increasing concentrations of a variety of substances, including carbohydrates, heavy metals, and nitric oxide. DGC activity of GcbA was required for its function, as a catalytically inactive variant could not rescue impaired BdlA processing or the dispersion-deficient phenotype of gcbA mutant biofilms to wild-type levels. While modulating BdlA cleavage during biofilm growth, GcbA itself was found to be subject to c-di-GMP-dependent and growth-mode-specific regulation. GcbA production was suppressed in mature wild-type biofilms and could be induced by reducing c-di-GMP levels via overexpression of genes encoding PDEs. Taken together, the present findings demonstrate that the regulatory functions of c-di-GMP-synthesizing DGCs expand beyond surface attachment and biofilm formation and illustrate a novel role for DGCs in the regulation of the reverse sessile-motile transition of dispersion.
INTRODUCTION
The process of biofilm dispersion represents an important and highly regulated phenotypic switch that allows cells residing within a biofilm to respond to changing conditions outside and within biofilm structures and to evade environmental stresses. Dispersion occurs in response to a wide array of signals, with dispersion-inducing conditions ranging from directly sensed environmental cues to self-synthesized signaling molecules (1–6). Biofilm dispersion in various bacterial species, including Aeromonas hydrophila, Pseudomonas aeruginosa, Pseudomonas putida, and Pseudomonas fluorescens, is tightly linked to nutrient availability, with both elevation and reduction in available nutrients, such as carbon and nitrogen sources, having been previously reported to trigger the process (1, 2, 7–12). Additional environmental dispersion triggers include temperature changes (13), oxidative and nitrosative stress (4, 14, 15), oxygen limitation (16, 17), and iron availability (18). More recently, the accumulation of d-amino acids has been found to trigger dispersion of biofilms formed by Bacillus subtilis and to prevent biofilm formation by P. aeruginosa and Staphylococcus aureus, with various reports suggesting that the mechanism of action may be based on specific effects on amyloid fibers or protein biofilm matrix components or general effects on protein synthesis and growth (19–23). Biofilm bacteria also control their own dispersion fate through the use of self-synthesized signaling molecules, such as cis-unsaturated fatty acids, including the Xanthomonas campestris diffusible signal factor (DSF) and the P. aeruginosa cis-2-decenoic acid (cis-DA) (3, 24, 25). These factors may have cross-species or even cross-kingdom activity, with P. aeruginosa cis-DA also triggering dispersion of biofilms formed by Escherichia coli, Klebsiella pneumonia, Proteus mirabilis, Streptococcus pyogenes, Bacillus subtilis, Staphylococcus aureus, and Candida albicans (3, 26).
With such a wide array of reported dispersion triggers, it has become clear that the regulation of biofilm dispersion involves the integration of sensory inputs into intracellular signal transduction that leads to a phenotypic switch enabling bacterial cells to leave the surface-associated biofilm and to disseminate to new locations. Surface attachment and biofilm development have long been established to be dependent on the modulation of the messenger molecule cyclic di-GMP (c-di-GMP), with increases in c-di-GMP levels correlating with surface attachment, aggregation, and biofilm formation (27–29). c-di-GMP regulates the motile-sessile transitions by binding and controlling the activities of effectors such as FleQ, PilZ domain-containing proteins and riboswitches, which in turn regulate growth-mode-specific gene expression patterns and phenotypes (30–35). Mounting evidence has suggested that biofilm dispersion, the reverse transition from surface-associated to planktonic growth, is also dependent on c-di-GMP regulation (1, 4, 17, 36), with a new question arising of how different dispersion-inducing cues are translated into c-di-GMP modulation. The cellular levels of c-di-GMP are controlled by the opposing activities of diguanylate cyclases (DGCs) and phosphodiesterases (PDEs), which synthesize and degrade c-di-GMP, respectively. The genomes of various bacterial species encode multiple DGCs and PDEs, characterized by the presence of drastically different sensory domains, which allow c-di-GMP-dependent regulation of distinct phenotypic outputs in response to specific diverse signals. However, the integration of diverse signals into c-di-GMP modulation to elicit a dispersion response remains poorly characterized.
Recent evidence has suggested that P. aeruginosa biofilms, which can disperse in response to changing carbon source concentrations, ammonium chloride, heavy metals, and nitric oxide (NO) (1, 4, 12, 14, 15), exhibit some specificity for sensing of various dispersion-inducing conditions, with distinct c-di-GMP-modulating proteins required for the dispersion response to specific cues. For instance, the membrane-bound PDE RbdA, harboring Per-Arnt-Sim (PAS/PAC) sensory domains, was found to be required for the dispersion response to low-oxygen conditions, with the PAS domain specifically found to be involved in hypoxia sensing (17). Two membrane-bound PDEs, MucR and NbdA, have recently been shown to be required for NO-mediated biofilm dispersion (15). These PDEs contain transmembrane MHYT (Met-His-Tyr-Thr) domains, proposed to sense diatomic gases through protein-bound copper ions, with these domains likely facilitating c-di-GMP degradation by NbdA and/or MucR to enable dispersion in response to NO (15).
Interestingly, in addition to producing distinct proteins for translating specific dispersion-inducing cues into c-di-GMP modulation, P. aeruginosa also harbors proteins that appear to be central to the regulatory cascade and required for dispersion regardless of inducing signals. The membrane-associated active PDE DipA, harboring sensory GAF and PAS domains, is required for biofilm dispersion in response to carbohydrates, heavy metals, ammonium chloride, and NO (36). Such a dominant role in dispersion was also previously reported for the methyl-accepting chemotaxis protein (MCP) BdlA, which harbors two PAS domains (PASa and PASb) and a signal transduction/methyl-accepting chemotaxis domain (Tar). As opposed to the PDEs DipA, RbdA, NbdA, and MucR, BdlA does not possess any domains associated with modulation of c-di-GMP levels. However, it appears to indirectly regulate c-di-GMP levels in order to enable the process of dispersion, with inactivation of BdlA elevating the total c-di-GMP levels of biofilm cells and the production of a constitutively active BdlA-G31A variant (having a change of Gly to Ala at position 31) elevating overall cellular PDE activity levels (12, 37). Further evidence suggests that BdlA regulates biofilm cellular PDE activity and c-di-GMP levels via direct interaction with at least two of the dispersion-specific PDEs, DipA and RbdA, and potentially, via regulation of DipA protein levels (37). Recent evidence also suggests a feedback regulation between c-di-GMP levels and BdlA function. While BdlA regulates cellular c-di-GMP abundance, activating modifications of BdlA structure and function are, in turn, dependent on elevated c-di-GMP levels. Specifically, BdlA function in activating dispersion was found to require posttranslational modifications in the form of phosphorylation at the tyrosine-238 (Y238) residue, followed by an unusual nonprocessive cleavage of the MCP at the methionine-130 (M130) residue into two peptides (PASa and PASbTar) (see Fig. 1), with the two truncated peptides interacting to enable the dispersion response (38). Correlating with its role in regulating dispersion of biofilm cells, phosphorylation and cleavage of BdlA did not occur in planktonic cells (38). Instead, the modifications were found to be biofilm specific and to require elevated c-di-GMP levels following the transition to surface-associated growth (38). Moreover, manipulating the total cellular c-di-GMP levels could artificially control cleavage of BdlA. Overexpression of the gene encoding the dispersion-associated PDE DipA significantly reduced BdlA cleavage during biofilm growth (38). In contrast, while BdlA is detectable only in its intact form in planktonic cells, plasmid-borne overexpression of the gene PA4843 triggered BdlA cleavage in planktonic cells (38). PA4843 encodes the recently characterized active DGC GcbA (also identified as AdcA), which was found to be involved in the regulation of the transition from planktonic to surface-associated growth (39, 40). Although BdlA plays a pivotal and central role in enabling P. aeruginosa biofilm dispersion in response to a variety of environmental cues, the system contributing to the elevation of c-di-GMP levels responsible for BdlA posttranslational modification and activation is currently not known. Thus, given our previous findings, we presently asked whether the DGC GcbA specifically contributes to the elevation of c-di-GMP levels required for the cleavage and activation of BdlA upon transition to surface-associated growth and investigated the potential role of GcbA in biofilm dispersion.
FIG 1.
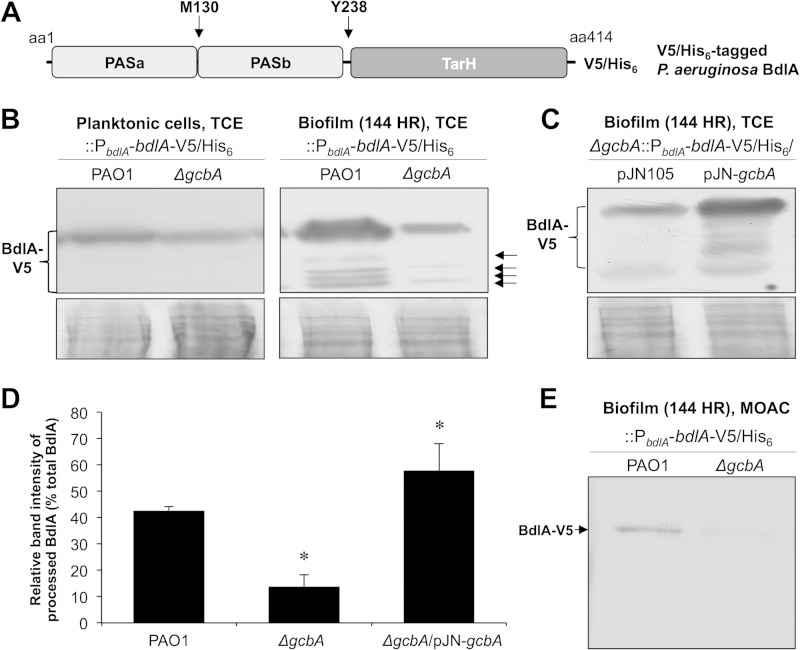
The DGC GcbA is required for cleavage of BdlA during biofilm growth. (A) Domain structure of BdlA. TarH, conserved chemoreceptor domain found in Tar (taxis toward aspartate and related amino acids); PAS, Per-Arnt-Sim sensory domain. Arrows indicate the methionine-130 (M130) putative ClpP cleavage site and tyrosine-238 (Y238) phosphorylation site. V5/His6 indicates the C-terminal location of the V5 epitope–6-histidine tag of the BdlA construct used in this study. (B) Detection with anti-V5 antibody of chromosomally encoded C-terminally V5/His6-tagged BdlA in total cell extracts (TCE) of planktonic or biofilm (144-h-old) wild-type PAO1 or ΔgcbA mutant cells. Arrows indicate detected cleavage products of BdlA. (C) Detection with anti-V5 antibody of chromosomally encoded C-terminally V5/His6-tagged BdlA in total cell extracts of 144-h-old biofilm ΔgcbA mutant cells harboring arabinose-inducible gcbA in the pJN105 vector or the empty vector control. Panels below immunoblot images (B and C) are Coomassie-stained gels after transfer to ensure equal TCE protein loading. (D) ImageJ software densitometry analysis was used to determine the relative fraction of processed BdlA, reported as percentage of band intensity of processed BdlA relative to intensity of all detectable BdlA bands. Error bars represent standard deviations. *, significantly different from the results for PAO1 cells (P < 0.05). (E) Detection with anti-V5 antibody of chromosomally encoded C-terminally V5/His6-tagged BdlA in metal oxide chromatography (MOAC)-enriched phosphoproteomes of biofilm (144-h-old) wild-type PAO1 or ΔgcbA mutant cells. All experiments were performed in triplicate. Representative images are shown.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa strain PAO1 was used as the parental strain. All planktonic strains were grown in Lennox broth (LB; BD Biosciences) or Vogel-Bonner minimal medium (VBMM) (49, 50) at 220 rpm in the absence or presence of 0.1 to 1.0% arabinose. Antibiotics were used at the following concentrations: 50 to 75 μg/ml gentamicin, 60 μg/ml tetracycline, and 250 μg/ml carbenicillin for P. aeruginosa and 100 μg/ml ampicillin, 20 μg/ml gentamicin, and 20 μg/ml tetracycline for E. coli.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or description | Reference(s) or source |
|---|---|---|
| Escherichia coli DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 tonA | Life Technologies |
| P. aeruginosa strains | ||
| PAO1 | Wild type | B. H. Holloway |
| PAO1 ΔgcbA | gcbA (PA4843) allelic gene replacement in PAO1 | 39 |
| PAO1 ΔdipA | PA5017::ISphoA/hah Tetr | 36 |
| PAO1 ΔbdlA | ΔbdlA in PAO1 Kmr | 12 |
| PAO1 ΔwspF | wspF allelic exchange in PAO1 | 41 |
| PAO1 ΔsiaD | PA0169::ISphoA/hah Tetr | 42 |
| PAO1 ΔwspR | wspR allelic exchange in PAO1 | 43 |
| PAO1::PbdlA-bdlA-V5/His6 | Chromosomal expression of bdlA-V5/His6 in PAO1 | 38 |
| PAO1 ΔgcbA::PbdlA-bdlA-V5/His6 | Chromosomal expression of bdlA-V5/His6 in PAO1 ΔgcbA | This study |
| PAO1::PgcbA-gcbA-V5/His6 | Chromosomal expression of gcbA-V5/His6 in PAO1 | This study |
| PAO1 ΔgcbA::PgcbA-gcbA-V5/His6 | Chromosomal expression of gcbA-V5/His6 in ΔgcbA | This study |
| Plasmids | ||
| pRK2013 | Helper plasmid for triparental mating; mob tra Kmr | 44 |
| pJN105 | Arabinose-inducible gene expression vector; pBRR-1 MCS araC-PBAD Gmr | 45 |
| pMJT-1 | araC-PBAD cassette of pJN105 cloned into pUCP18; Ampr (carbapenem resistant) | 46 |
| pJN-gcbA | C-terminally V5/His6-tagged gcbA cloned into pJN105 at XbaI | 39 |
| pCTX-gcbA-V5/6×His | C-terminally V5/His6-tagged gcbA gene and upstream 500 bp cloned into pMini-CTX | 39 |
| CTX-PbdlA-bdlA-V5/6×His | C-terminally V5/His6-tagged bdlA gene and upstream 500 bp cloned into pMini-CTX | 38 |
| pJN-gcbA(GGAAF) | C-terminally V5/His6-tagged gcbA with mutation to convert GGEEF motif to GGAAF cloned into pJN105 | This study |
| pJN-PASa | PASa-encoding segment of bdlA cloned into pJN105 | 38 |
| pMJT-bdlA-PASbTarH | C-terminally V5/His6-tagged PASaTar of bdlA cloned into pMJT-1 | 38 |
| pJN-bdlA-V5/6 × His | C-terminally V5/His6-tagged bdlA cloned into pJN105 | 37 |
| pJN-dipA-V5/6 × His | C-terminally V5/His6-tagged dipA cloned into pJN105 | 36 |
| pJN-PA2133 | PA2133 cloned into pJN105 | 43 |
| pJN-bdlA-G31A-His | C-terminally His6-tagged bdlA with G31A mutation cloned into pJN105 | 37 |
| pMJT-bdlA-HA | C-terminally HA-tagged bdlA cloned into pMJT-1 | This study |
| pJN-bfiS | C-terminally V5/His6-tagged bfiS cloned into pJN105 | 47, 48 |
Strain construction.
Complementation and overexpression were accomplished by conjugating into P. aeruginosa strains the pJN105 vector harboring the respective genes under the control of the arabinose-inducible PBAD promoter (45). In order to assess the native protein production levels of GcbA and BdlA, the chromosomal integration vectors pCTX-gcbA-V5/His6 and pCTX-bdlA-V5/His6 (38, 39), harboring constructs encoding the respective C-terminally V5/His6-tagged proteins under the control of the native promoters, were introduced into the indicated P. aeruginosa strains via conjugation. The resulting strains (PAO1::PbdlA-bdlA-V5/His6, PAO1 ΔgcbA::PbdlA-bdlA-V5/His6, PAO1::PgcbA-gcbA-V5/His6, and PAO1 ΔgcbA::PgcbA-gcbA-V5/His6) contained chromosomally encoded gcbA-V5/His6 or bdlA-V5/His6 under the control of the respective native promoter at the attB site. Site-directed mutagenesis for the construction of the GcbA-GGAAF variant was accomplished by using the GeneArt site-directed mutagenesis kit (Life Technologies) according to the manufacturer's protocol using the primer pair gcbA-GGAAF456-for (CGGCCGCTATGGCGGCGCAGCGTTCGCCGTGGTCCTTC)/gcbA-GGAAF456-rev (GAAGGACCACGGCGAACGCTGCGCCGCCATAGCGGCCG). The identities of vector inserts and site-directed mutations were confirmed by PCR and sequencing.
Biofilm formation.
Biofilms were grown in a continuous flow tube reactor system (1-m lengths of size 14 silicone tubing, Masterflex; Cole Parmer, Inc.) in 20-fold-diluted LB medium or VBMM in the presence of 0.1% arabinose at 22°C for up to 144 h at a flow rate of 0.2 ml/min. For visualization of biofilm architecture, biofilms were grown under similar conditions in flow cell systems (BioSurface Technologies). Biofilm architecture was visualized via confocal laser scanning microscopy (CLSM) using a Leica TCS SP5 confocal microscope and the BacLight Live/Dead viability kit (Life Technologies). The CLSM images were processed using LAS AF software, version 2.4.1. Quantitative analysis of biofilm architecture was accomplished using MATLAB with the COMSTAT software package (51).
Biofilm dispersion assays.
For biofilm dispersion assays, biofilms were cultivated in a once-through continuous flow tube reactor system at 22°C for 120 h. After 120 h of biofilm growth, biofilm dispersion was induced by the sudden addition of l-glutamate (18 mM) or mercury chloride (2 mM) to the growth medium as previously described (12). In addition, 500 μM sodium nitroprusside (SNP) was used as a source of nitric oxide (4). Following the addition of one of the dispersion-inducing substances, the optical density of the biofilm effluents, collected at 1-min intervals, was assessed by spectrophotometry, with dispersion events indicated by an increase in the turbidity measured at 600 nm.
Immunoblot analysis and pulldown assays.
The abundance of V5/His6-tagged GcbA or BdlA present in P. aeruginosa cells was assessed by SDS-PAGE and immunoblot analysis. Total cell extracts from planktonic and biofilm cells, harvested by centrifugation for 5 min at 16,000 × g and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) containing 0.3 mg phenylmethanesulfonyl fluoride, were obtained by sonication on ice with six 10-s bursts at 4 W, followed by centrifugation for 5 min at 21,200 × g to pellet cell debris and unbroken cells. The protein concentrations were determined using a modified Lowry assay (Thermo Scientific) and bovine serum albumin as a standard. The samples (15 μg) were resolved on an 11% polyacrylamide gel and subsequently transferred onto polyvinylidene difluoride (PVDF) membrane using a TurboTransblot apparatus (Bio-Rad). Western blots were probed with anti-V5–horseradish peroxidase (HRP) or anti-V5 antibody (Invitrogen) and antihemagglutinin (anti-HA) antibody (Covance). When necessary, a secondary anti-mouse IgG antibody (Cell Signaling Technologies) was used. The blots were subsequently developed using Immun-Star WesternC chemiluminescence reagents (Bio-Rad). Following transfer, SDS-PAGE gels were Coomassie stained to ensure equal loading. Densitometry was performed using ImageJ software.
Pulldown assays were used to assess the interactions between BdlA and GcbA, DipA, or BfiS in total protein cell extracts of cells coproducing C-terminally V5/His6-tagged GcbA, DipA, or BfiS and C-terminally HA-tagged bait BdlA protein (PAO1/pMJT-bdlA-HA/pJN-gcbA, PAO1/pMJT-bdlA-HA/pJN-dipA, or PAO1/pMJT-bdlA-HA/pJN-bfiS). Following immunoprecipitation of HA-tagged proteins using immobilized anti-HA antibody (Covance), immunoprecipitation eluates were separated by SDS-PAGE and assessed by immunoblot analysis for the presence of V5/His6-tagged prey proteins using anti-V5–HRP antibodies (Life Technologies). Pulldown assays were carried out using 200 μg protein from cellular extracts. Anti-HA and anti-V5 antibodies were used for immunoprecipitation at 2 μg/ml and immunoblotting at 0.2 μg/ml.
Phosphoprotein enrichment and detection.
Analysis of phosphorylated BdlA-V5/His6 levels in protein extracts obtained from wild-type PAO1 or ΔgcbA mutant cells grown as biofilms for 144 h was accomplished using phosphoprotein purification via metal oxide affinity chromatography (MOAC) essentially as described by Wolschin and colleagues (52). MOAC has been demonstrated by Krüger et al. to result in up to 20-fold enrichment of phosphoproteins and to approach 100% specificity (53). Briefly, 750 μg of total protein cell extract was diluted with MOAC incubation buffer (30 mM morpholineethanesulfonic acid [MES], 0.2 M potassium glutamate, 0.2 M sodium aspartate, 0.25% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, and 8 M urea) to a final volume of 1.5 ml and subsequently incubated for 30 min at 4°C in the presence of 80 mg of aluminum hydroxide. Unbound proteins were removed via six 1-min washes of the aluminum hydroxide slurry with 1.5 ml of incubation buffer at 16,000 × g and 4°C. Then, phosphoproteins were eluted from the slurry using 100 mM potassium pyrophosphate and 8 M urea, precipitated and desalted by methanol-chloroform precipitation, and subsequently vacuum dried. Samples were then analyzed by SDS-PAGE, followed by the detection of C-terminally tagged BdlA-V5/His6 by immunoblotting with anti-V5 antibodies.
qRT-PCR.
Quantitative real-time reverse transcription-PCR (qRT-PCR) was used to determine gene expression levels using 2 μg of total RNA isolated from planktonic, biofilm, and dispersed cells of wild-type P. aeruginosa and mutant strains. Planktonic cells were grown in LB medium for 6 h starting with initial cultures adjusted to an optical density at 600 nm (OD600) of ∼0.03. Biofilms were grown in continuous flow tube reactors as outlined above. Cells dispersing from biofilms following the induction of dispersion as described above were collected directly into RNAprotect (Qiagen). Isolation of mRNA and cDNA synthesis were carried out as previously described (54–57). qRT-PCR was performed using the Bio-Rad CFX Connect real-time PCR detection system and SsoAdvanced SYBR green supermix (Bio-Rad) with oligonucleotide pairs PA4843RTf/PA4843RTr (39) and bdlA-RT-for (CTACGCGCAATCGGAAGAC)/bdlA-RT-rev (GGACATTGCCGTCGAGGTC). mreB (39) was used as a control. Quantitation of relative transcript levels was accomplished using the CFX Manager Software (Bio-Rad), by first normalizing the transcript abundances (based on the threshold cycle [CT] value) to that of mreB and then determining transcript abundance ratios. Melting curve analyses were employed to verify amplification of specific single products. The qRT-PCR analysis was performed using at least three biological replicates per strain.
Determination of phosphodiesterase activities.
The phosphodiesterase activities of total cell extracts of biofilm cells harboring pJN-dipA or pJN-PA2133 were determined using the synthetic chromogenic substrate bis(p-nitrophenyl) phosphate (bis-pNPP) (Sigma-Aldrich) essentially as previously described (36, 58, 59), by measuring the release of p-nitrophenol (pNP) at 405 nm. An extinction coefficient for p-nitrophenol of 1.78 × 104 M−1 cm−1 was used. Reaction mixtures without the addition of cell extracts were used as controls to account for any nonenzymatic bis-pNPP hydrolysis.
Statistical analysis.
All statistical analyses were performed in Microsoft Excel using a two-tailed Student's t test, assuming equal variance, or single-factor analysis of variance (ANOVA).
RESULTS
GcbA contributes to biofilm-specific posttranslational modifications of BdlA.
Biofilm-specific processing of BdlA has been shown to result in up to four cleavage products in biofilm PAO1 cells, with the predominant products of C-terminally tagged BdlA corresponding in size to a BdlA variant lacking the N-terminal PASa domain (Fig. 1A). However, at least one of the products was still detectable for a BdlA variant with an M130I substitution at the putative ClpP cleavage site (38). Considering that artificially induced overexpression of gcbA triggered BdlA cleavage under planktonic growth conditions (38), we asked whether GcbA also contributes to the activation of BdlA during biofilm growth. We therefore monitored the native production and processing levels of BdlA using a chromosomally encoded C-terminally V5–His6-tagged BdlA (BdlA-V5/His6) construct introduced into wild-type PAO1 or the isogenic gcbA mutant strains. Consistent with previous observations, no cleavage of BdlA was observed in either strain grown planktonically, and the BdlA levels were similar in both strains (Fig. 1B). However, when anti-V5 immunoblotting was used to assess BdlA-V5/His6 abundance in cells grown for 144 h as biofilms, the levels of detectable C-terminal cleavage products of BdlA in biofilms formed by ΔgcbA cells were significantly reduced relative to the levels in biofilms formed by PAO1 cells (Fig. 1B and D). Moreover, ΔgcbA biofilm cells were found to lack at least two of the cleavage products detectable in the wild-type biofilm cells (Fig. 1B). Complementation (ΔgcbA/pJN-gcbA) restored the cleavage of BdlA to wild-type levels (Fig. 1C and D).
BdlA processing has previously been demonstrated to correlate with elevated protein phosphorylation levels and to require the phosphorylation of the BdlA tyrosine-238 residue (38). Correlating with the reduced BdlA processing levels, inactivation of gcbA resulted in significantly reduced levels of phosphorylated BdlA detected in biofilm cells (Fig. 1E). Taken together, these findings suggested that GcbA contributes to the posttranslational modification and activation of BdlA during biofilm growth.
GcbA contributes to BdlA levels and cleavage upon initial transition to surface-associated growth.
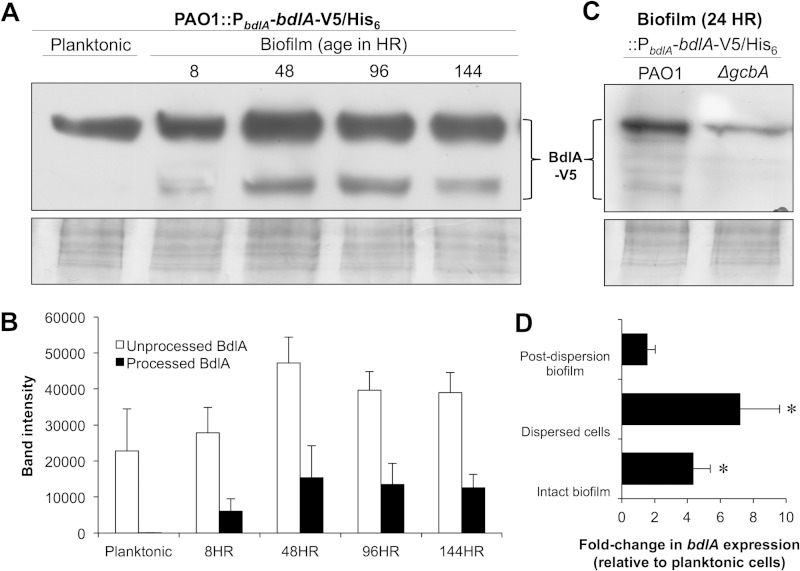
GcbA was recently shown to make a greater contribution to c-di-GMP levels in planktonic cells and those undergoing early attachment stages than it does in cells of developed biofilms, with GcbA not affecting the overall c-di-GMP levels of biofilm cells (39). Given that GcbA affected BdlA processing in established P. aeruginosa biofilms, we therefore asked whether BdlA cleavage is initiated during early biofilm formation and, if so, whether BdlA processing is GcbA dependent. Using the chromosomally encoded BdlA-V5/His6 construct and anti-V5 immunoblotting, we monitored BdlA processing in P. aeruginosa cells at 8, 48, 96, and 144 h following attachment. These time points were chosen because they were previously demonstrated to correspond to the following stages of biofilm development: initial surface attachment, transition to irreversible surface-associated growth, early biofilm maturation, and late biofilm maturation (57, 60). While no cleavage of BdlA was detected in cells grown planktonically, processing of the protein was detectable as early as 8 h following initial attachment (Fig. 2A and B). Moreover, cleavage of BdlA appeared to reach a steady-state level by 48 h of biofilm growth (Fig. 2A and B).
FIG 2.
Cleavage of BdlA is initiated in P. aeruginosa cells following initial attachment to surfaces. (A) Detection of chromosomally encoded, C-terminally V5/His6-tagged BdlA in total cell extracts of P. aeruginosa PAO1 cells grown planktonically or as biofilms for 8, 48, 96, and 144 h by immunoblot analysis using anti-V5 antibody. (B) ImageJ software densitometry analysis of intact BdlA and BdlA cleavage products detected in total cell extracts of P. aeruginosa PAO1 cells grown planktonically or as biofilms for 8, 48, 96, and 144 h. (C) Detection with anti-V5 antibody of chromosomally encoded, C-terminally V5/His6-tagged BdlA in total cell extracts of planktonic or attached (24 h) wild-type PAO1 or ΔgcbA mutant cells. (A, C) Panels below immunoblot images are Coomassie-stained gels after transfer to ensure equal TCE protein loading. (D) Relative transcript abundances of bdlA in P. aeruginosa PAO1 144-h-old biofilm cells (Intact biofilm), cells dispersed from 144-h-old biofilms in response to a changed glutamate concentration in the medium (Dispersed cells), and cells remaining within the biofilm following dispersion (Post-dispersion biofilm). Transcript abundance is reported relative to the abundance in planktonically grown PAO1 cells. (B, D) Error bars represent standard deviations. *, significantly different from expression in planktonically grown PAO1 cells (P < 0.05). All experiments were performed in triplicate, and representative images are shown.
To determine the contribution of GcbA to cleavage of BdlA during early stages of biofilm formation, BdlA processing was assessed following 24 h of surface-associated growth. This time point was chosen to ensure that the observed effects were not due to differences in the amounts of attached biomass, as inactivation of gcbA was previously demonstrated to affect attachment following 8 h but not 24 h of growth (39). Under the conditions tested, ΔgcbA cells exhibited significantly reduced levels of BdlA processing relative to the levels in the wild-type PAO1 cells (Fig. 2C). Taken together, these findings indicated that BdlA is cleaved soon after initial attachment in a manner dependent on GcbA.
bdlA is transcriptionally regulated independently of GcbA during biofilm growth.
The immunoblot analyses of BdlA protein production and cleavage (Fig. 1 and 2) indicated that processing may correlate positively with the overall abundance of BdlA. This prompted us to ask whether BdlA is modulated at a transcriptional level in a growth mode-dependent manner and, if so, whether the transcriptional regulation of bdlA is affected by GcbA. qRT-PCR analysis revealed that bdlA transcript abundances increase approximately 4-fold in biofilm cells relative to the levels in their planktonic counterparts (Fig. 2D). The bdlA transcript levels increased even further in cells dispersing from biofilms in response to an increased glutamate concentration in the medium, and the levels decreased in biofilm cells remaining attached to the surface following dispersion (Fig. 2D). In contrast, however, no differences were observed in the relative transcript abundances of bdlA in ΔgcbA and wild-type PAO1 cells grown planktonically (fold change, 1.10 ± 0.6 [mean ± standard deviation], for ΔgcbA versus PAO1) or as biofilms (fold change, 1.18 ± 0.4). Moreover, overexpression of gcbA did not affect bdlA expression, as PAO1/pJN-gcbA demonstrated fold changes of 1.18 ± 0.3 and 0.8 ± 0.3 in bdlA transcript abundance relative to the levels in the PAO1/pJN105 vector control under planktonic and biofilm growth conditions, respectively. These findings suggested that, while GcbA contributes to the translational and/or posttranslational regulation of BdlA, bdlA transcript levels are regulated in a manner dependent on the mode of growth but not on GcbA.
GcbA is required for nutrient-induced biofilm dispersion but does not directly trigger dispersion.
The dispersion of P. aeruginosa biofilm cells in response to changing environmental conditions was previously shown to require not only the presence of BdlA but also its posttranslational modification and cleavage, with lack of BdlA processing correlating with impaired nutrient-induced dispersion in response to an increasing extracellular glutamate concentration (12, 38). Considering the contribution of GcbA to BdlA cleavage and phosphorylation, the role of this DGC in nutrient-induced biofilm dispersion in response to a change in the glutamate concentration in the medium was assessed. Following 120 h of growth, biofilms formed by wild-type PAO1 and ΔgcbA cells were exposed to 18 mM glutamate added to the growth medium, and the biofilm effluents were monitored for cellular release as a result of dispersion events. While biofilms formed by the wild-type PAO1 strain dispersed in response to a change in the concentration of glutamate, as evidenced by a significant increase in the optical density of the biofilm effluent, biofilms formed by the isogenic gcbA mutant did not disperse (Fig. 3A). Complementation of the gcbA mutant strain with a chromosomally encoded gcbA (ΔgcbA::PgcbA-gcbA), as well as multicopy expression of gcbA under the control of the PBAD promoter in the pJN105 vector (ΔgcbA/pJN-gcbA), restored biofilm dispersion in response to glutamate (Fig. 3A). It can be argued that the impaired dispersion response of biofilms formed by ΔgcbA is the result of reduced c-di-GMP levels and/or biomass accumulation. However, inactivation of gcbA has previously been reported not to correlate with reduced biofilm cellular c-di-GMP levels (39). Moreover, in agreement with previous reports (39) and as presently confirmed (Table 2), inactivation of gcbA does not result in a lack of mature biofilm architecture or reduced biomass accumulation at the later stages of biofilm development (Table 2). Interestingly, however, greater substratum coverage was observed in biofilms formed by ΔgcbA cells (Table 2), potentially resulting from the impaired dispersion response.
FIG 3.
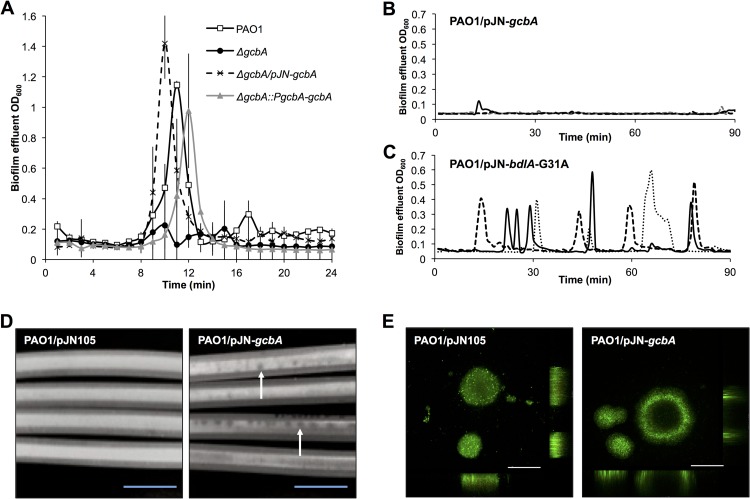
GcbA contributes to the regulation of biofilm dispersion. (A to C) Biofilms were grown for 120 h in tube reactors prior to exposure to dispersion-inducing conditions, at which time the biofilm effluents were monitored for dispersion events, as evidenced by an increase in turbidity measured at 600 nm. (A) Inactivation of gcbA renders P. aeruginosa dispersion deficient in response to a sudden change in the glutamate concentration in the medium. Error bars indicate standard deviations. (B, C) Detection of dispersion following induction of gcbA or bdlA-G31A gene expression. Expression of gcbA or bdlA-G31A was under the control of the arabinose-inducible PBAD promoter, and induction of gene expression was accomplished by adding arabinose to the growth medium. Representative dispersion data of three independent biofilm replicates, indicated by dotted, dashed, and solid lines, are shown. Dispersion was detected upon the addition of arabinose to the PAO1/pJN-bdlA-G31A strain (C) but not to the PAO1/pJN-gcbA strain (B). (D) Photographs of 144-h-old biofilms formed on the interior surface of silicone tubing by P. aeruginosa cells overexpressing gcbA (PAO1/pJN-gcbA) or harboring the respective vector control (PAO1/pJN105). The confluent white coverage within the tubing is indicative of accumulated bacterial biofilm biomass, and the darker spots are indicative of a lack of biofilm biomass, both of which are observable by the naked eye without the aid of a microscope. Arrows indicate examples of voids in the biofilm cellular layers. Bars = 1 cm. (E) CLSM images of PAO1/pJN105 and PAO1/pJN-gcbA cells grown as biofilms under flowing conditions in flow cells for 144 h and stained with the Live/Dead BacLight viability stain. Bars = 100 μm. All experiments were performed in triplicate, and representative images are shown.
TABLE 2.
COMSTAT analysis of PAO1, PAO1 ΔgcbA, PAO1/pJN105, and PAO1/pJN-gcbA biofilmsa
| Strain | Total biomass (μm3/μm2) | Substratum coverage (%) | Avg thickness (μm) | Roughness coefficient | Maximum thickness (μm) |
|---|---|---|---|---|---|
| PAO1 | 7.27 (±1.85) | 22.27 (±6.90) | 7.61 (±2.15) | 1.41 (±0.15) | 42.13 (±8.93) |
| PAO1 ΔgcbA | 9.28 (±3.17) | 40.88 (±16.36)* | 9.82 (±3.46) | 1.02 (±0.24) | 45.33 (±9.19) |
| PAO1/pJN105 | 6.16 (±2.85) | 14.08 (±4.15) | 6.22 (±3.04) | 1.68 (±0.09) | 48.43 (±9.30) |
| PAO1/pJN-gcbA | 3.78 (±3.29)* | 8.18 (±5.84)* | 3.95 (±3.59)* | 1.81 (±0.14) | 45.43 (±14.24) |
COMSTAT analysis was carried out on 144-h-old biofilms grown in triplicate, using at least 6 images per replicate. Values are means (±standard deviation). *, significantly different from the results for wild-type biofilms (P ≤ 0.01).
With evidence suggesting that glutamate-induced biofilm dispersion requires GcbA, we next assessed whether the induction of gcbA expression in established biofilms could trigger a rapid dispersion response. This reasoning was based on previous findings demonstrating that the production of the BdlA-G31A variant results in a hyperdispersive phenotype and that arabinose induction of bdlA-G31A expression triggers dispersion in established biofilms (37). Biofilms of PAO1 harboring gcbA under the control of the arabinose-inducible PBAD promoter in the pJN105 vector (PAO1/pJN-gcbA) were grown in a minimal medium in the absence of arabinose under flowing conditions. Following 120 h of growth, arabinose was added to the medium to induce gcbA expression, and the effluent was monitored for an increase in optical density, indicative of dispersion events. Strain PAO1/pJN-bdlA-G31A, harboring pJN105 with a gene encoding BdlA-G31A, the constantly “on” variant of BdlA (37), was used as a positive control. In accord with previous findings, exposure to arabinose triggered dispersion events in biofilms formed by PAO1/pJN-bdlA-G31A (Fig. 3C). The induction of gcbA expression, however, did not induce dispersion within a similar time frame (Fig. 3B), indicating that GcbA by itself does not directly trigger the dispersion response.
We next assessed whether overexpression of gcbA affects biofilm formation. Despite the fact that gcbA overexpression has been previously found to induce a hyperaggregative phenotype in liquid culture and to promote initial attachment to a surface (39), continuous induction of gcbA during biofilm growth in PAO1/pJN-gcbA resulted in the formation of biofilms with visibly reduced biomass compared to biofilms formed by the vector control PAO1/pJN105 (Fig. 3D and E). While the wild-type vector control strain formed confluent biofilms with uniform surface coverage, the PAO1/pJN-gcbA strain instead formed biofilms that were characterized by patchy coverage of the substratum and the presence of voids visible without the aid of a microscope (Fig. 3D). CLSM analysis confirmed reduced levels of biomass accumulation in the biofilms formed by PAO1/pJN-gcbA relative to the amounts formed by the vector control (Fig. 3E, Table 2). The CLSM analysis also revealed increased instances of hollowed-out cellular clusters, indicative of dispersion events, suggesting that multicopy expression of gcbA reduced biomass accumulation not by interfering with biofilm formation but rather by triggering or facilitating dispersion events (Fig. 3E).
DGC activity of GcbA is required for BdlA cleavage and biofilm dispersion.
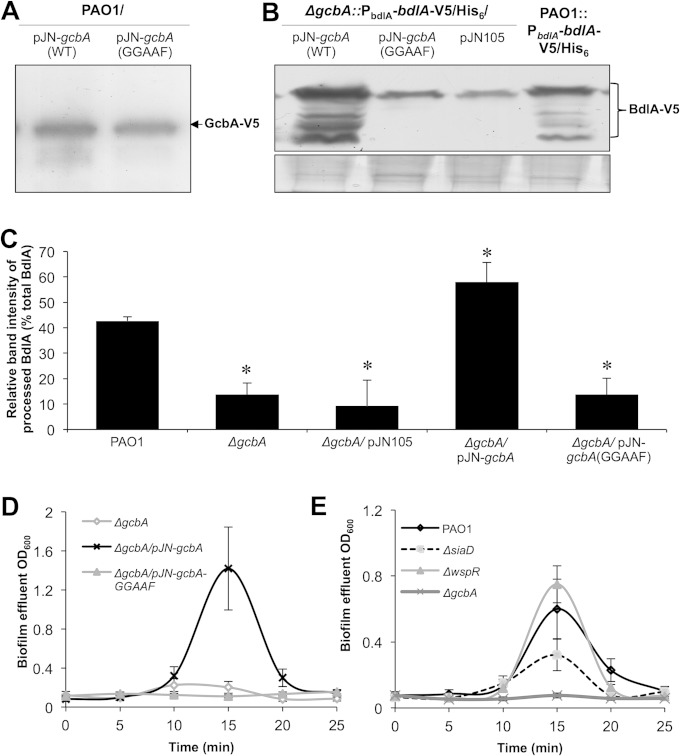
Elevated levels of c-di-GMP correlate with the sessile, biofilm mode of growth, while low levels of c-di-GMP generally correspond with free-swimming, motile growth away from the surface (61). Furthermore, the sessile-motile transition of dispersion has been linked to the activity of PDEs in reducing the intracellular levels of c-di-GMP (4, 12, 15, 17, 36, 37). Therefore, the observation of a DGC being required for biofilm dispersion was rather unexpected. GcbA is composed of a response regulator receiver domain and a GGDEF cyclase domain, with a conserved active-site GGEEF motif (39). Considering that response regulator receiver domains harbor conserved phosphorylation sites and participate in phosphotransfer events with other proteins to form signaling cascades (62), we asked whether GcbA-mediated regulation of BdlA cleavage and dispersion is dependent on the GcbA signaling domain rather than on its enzymatic activity. However, a GcbA variant with the GGEEF cyclase active-site motif replaced with GGAAF (GcbA-GGAAF), while soluble and produced at levels comparable to those of the wild-type protein (Fig. 4A), was unable to restore the BdlA processing (Fig. 4B and C) and the dispersion phenotype of ΔgcbA biofilms to wild-type levels (Fig. 4D), suggesting that GcbA DGC activity is required for the dispersion response. In contrast, multicopy expression of wild-type gcbA in ΔgcbA cells restored BdlA processing during biofilm growth to wild-type levels (Fig. 4B and C) and rescued the dispersion-deficient phenotype (Fig. 4D).
FIG 4.
GcbA DGC activity is required for biofilm dispersion and BdlA processing. (A) The GcbA-GGAAF variant is produced in a soluble manner, as revealed by anti-V5 immunoblot analysis of total cell extracts of PAO1 cells producing C-terminally V5/His6-tagged wild-type GcbA or GcbA-GGAAF. (B) Detection of chromosomally encoded C-terminally V5/His6-tagged BdlA in total cell extracts of biofilms (144-h-old) formed by wild-type PAO1 or ΔgcbA mutant cells complemented with wild-type gcbA or gcbA-GGAAF cloned into the pJN105 vector. ΔgcbA cells harboring the empty pJN105 vector were used as negative controls. Panel below immunoblot image is Coomassie-stained gel after transfer to ensure equal TCE protein loading. (C) ImageJ software densitometry analysis was used to determine the relative fraction of processed BdlA, reported as the percentage of the band intensity of processed BdlA relative to the intensity of all detectable BdlA bands. Error bars represent standard deviations. *, significantly different from results for PAO1 (P < 0.05). (D) Complementation of ΔgcbA cells with wild-type gcbA but not gcbA-GGAAF restores dispersion of 120-h-old tube reactor-grown biofilms in response to an increased glutamate concentration in the medium. (E) Inactivation of the DGC-encoding genes wspR and siaD does not abrogate the dispersion response of P. aeruginosa biofilm cells triggered by increased glutamate concentration. Error bars denote standard deviations. All experiments were performed in triplicate.
To test whether any decrease in the general c-di-GMP pool or in DGC activity previously associated with biofilm formation would affect the dispersion response, we also assessed the ability of biofilms formed by mutants in which the DGC-encoding genes wspR and siaD were inactivated to disperse in response to glutamate. In contrast to biofilms formed by the ΔgcbA mutant, those formed by wspR and siaD mutants were capable of responding to a changed glutamate concentration via a dispersion response, as evidenced by increasing optical densities of the biofilm effluents (Fig. 4E). These results suggested specificity for the role of GcbA in biofilm dispersion and that the c-di-GMP synthesis catalyzed by GcbA likely contributes to the regulation of biofilm dispersion, potentially via processing of BdlA.
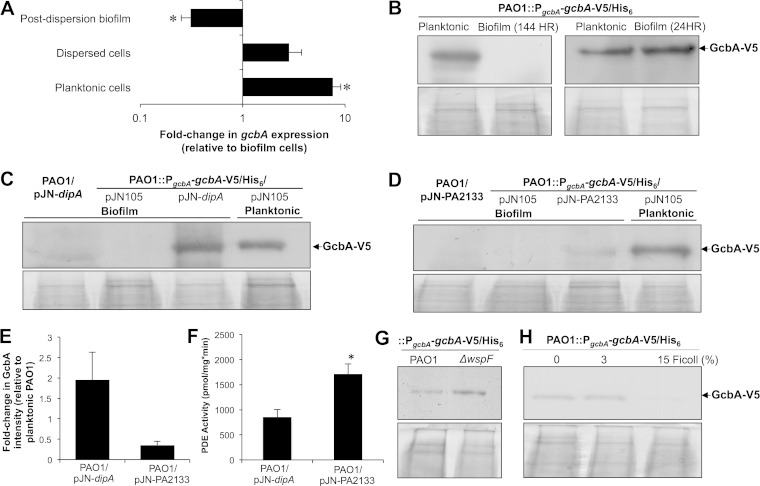
gcbA expression and GcbA production are suppressed during biofilm growth and induced upon dispersion.
Previous findings demonstrated GcbA to contribute to the cellular c-di-GMP levels of planktonic but not biofilm cells (39). Our present findings of GcbA participating in the regulation of BdlA cleavage in biofilm cells soon after attachment further prompted us to ask whether GcbA is regulated in a growth mode-specific manner. The transcript levels of gcbA, as revealed by qRT-PCR analysis, were found to be highest in planktonic cells, with an 8-fold increase relative to the transcript levels in biofilm cells (Fig. 5A). In contrast, gcbA expression was found to be significantly reduced in cells remaining attached to the surface after dispersion relative to its expression in biofilm cells prior to dispersion (Fig. 5A).
FIG 5.
GcbA is regulated in a growth mode- and c-di-GMP-dependent manner. (A) Relative transcript abundances of gcbA in P. aeruginosa PAO1 exponential-phase planktonic cells, cells dispersed from 144-h-old biofilms in response to a changed in the glutamate concentration in the medium (Dispersed cells), and cells remaining within the biofilm following dispersion (Post-dispersion biofilm). Transcript abundance is reported relative to that of PAO1 cells grown as biofilms for 144 h. Error bars denote standard deviations. *, significantly different from expression in 144-h PAO1 biofilm cells (P < 0.05). (B) Detection with anti-V5 antibody of chromosomally encoded C-terminally V5/His6-tagged GcbA in total cell extracts of planktonic or biofilm (24- or 144-h-old) PAO1 cells. (C, D) Overexpression of genes encoding the PDEs DipA (C) and PA2133 (D) induced production of GcbA to detectable levels in P. aeruginosa cells grown as biofilms, as revealed by anti-V5 immunoblot analysis of chromosomally encoded C-terminally V5/His6-tagged GcbA. Planktonically grown cells harboring the empty pJN105 vector were used as positive controls. (E) ImageJ software densitometry analysis was used to determine the relative abundance of GcbA in PAO1/pJN-dipA or PAO1/pJN-PA2133 biofilm cells with respect to GcbA levels detected in PAO1 planktonic cells. Here, PAO1 refers to PAO1::PgcbA-gcbA-V5/His6. (F) Total cellular PDE activity of cell extracts obtained from P. aeruginosa PAO1 biofilms overexpressing dipA or PA2133 was determined using the synthetic chromogenic substrate bis(p-nitrophenyl) phosphate (bis-pNPP) and measuring the release of p-nitrophenol (pNP) at 405 nm. Error bars denote standard deviations. *, significantly different from results for PAO1 biofilms overexpressing dipA (P < 0.05). (G) Detection with anti-V5 antibody of chromosomally encoded C-terminally V5/His6-tagged GcbA in total cell extracts of planktonically grown PAO1 and isogenic ΔwspF mutant cells. (H) Increasing viscosity leads to reduced GcbA levels, as determined via anti-V5 antibody detection of chromosomally encoded C-terminally V5/His6-tagged GcbA in total cell extracts of planktonically grown PAO1 in the presence of increasing concentrations of Ficoll. All experiments were performed in triplicate. Panels below immunoblot images (B, C, D, G, H) are Coomassie-stained gels after transfer to ensure equal TCE protein loading.
In order to assess the native production levels of the GcbA protein, we used a chromosomally encoded V5/His6-tagged GcbA (GcbA-V5/His6) construct introduced into the wild-type PAO1 strain (PAO1::PgcbA-gcbA-V5/His6). In accord with the qRT-PCR data, GcbA protein production was significantly reduced in biofilm cells, with GcbA in biofilms grown for 144 h not detectable via the immunoblotting methods utilized (Fig. 5B). The observation of GcbA downregulation in biofilm cells relative to its expression in their planktonic counterparts was somewhat unexpected considering the contribution of the DGC to BdlA processing during biofilm growth. Given that GcbA appeared to contribute to BdlA processing shortly after attachment, the GcbA protein levels were also assessed in initially attached cells following 24 h of growth at the surface. In contrast to 144-h-old biofilms, however, the GcbA levels in 24-h attached cells were similar to those observed for planktonic cells (Fig. 5B).
GcbA production can be induced via reduction of cellular c-di-GMP levels.
As both gcbA transcript and GcbA protein levels were found to be higher in free-floating cells, which are associated with elevated PDE activity and reduced c-di-GMP levels, the effect of cellular c-di-GMP levels on GcbA production was subsequently assessed. We reasoned that, if GcbA production is c-di-GMP dependent, controlling the c-di-GMP levels could modulate the abundance of GcbA. Specifically, we asked whether GcbA production in biofilm cells could be induced or enhanced by overexpressing PDE-encoding genes. Two genes encoding previously confirmed active PDEs, PA2133 (43) and dipA (36), were selected to test this. Multicopy expression of dipA has previously been shown to reduce c-di-GMP levels 2-fold relative to its levels in wild-type biofilm cells (36), while overexpression of PA2133 has been shown to reduce c-di-GMP below detectable levels (43). The expression of either dipA or PA2133 induced GcbA production in biofilm cells to detectable levels (Fig. 5C to E). However, despite exhibiting higher total cellular PDE activity (Fig. 5F), PAO1/pJN-PA2133 biofilm cells harbored GcbA at levels lower than those observed in PAO1/pJN-dipA biofilm cells (Fig. 5C to E). Considering that dispersion has been previously found to require DipA but not PA2133 (36), these findings suggested a specificity of the c-di-GMP-dependent regulation of GcbA induction by demonstrating that a dispersion-specific PDE has a more profound effect on GcbA abundance.
In order to assess whether elevated c-di-GMP levels could reduce GcbA levels in planktonically growing cells, a wspF mutant, which is characterized by constitutive phosphorylation and activation of the DGC WspR (43), was used. However, no decrease in GcbA production was observed in ΔwspF cells relative to its production in wild-type PAO1 cells, with inactivation of wspF resulting in a statistically insignificant fold increase of 2.1 ± 1.2 in GcbA levels (Fig. 5G).
In order to assess whether conditions associated with surface-attached growth, other than elevated c-di-GMP levels, could account for suppression of GcbA abundance, GcbA levels were assessed in planktonic cells exposed to conditions of increasing viscosity. While the addition of 3% Ficoll had no effect on GcbA production, increasing the viscosity further by the addition of 15% Ficoll, a concentration previously suggested to mimic conditions of swarming motility and initiation of biofilm growth (63), correlated with a 20.5% ± 9.9% reduction of detectable GcbA abundance relative to the GcbA levels observed in the absence of Ficoll (Fig. 5H). Taken together, these findings suggested that, while GcbA production is suppressed upon transition to biofilm growth, potentially due to sensing of surface growth and reduced motility, GcbA is upregulated upon the return to free-swimming growth via a dispersion-specific reduction in the cellular c-di-GMP levels.
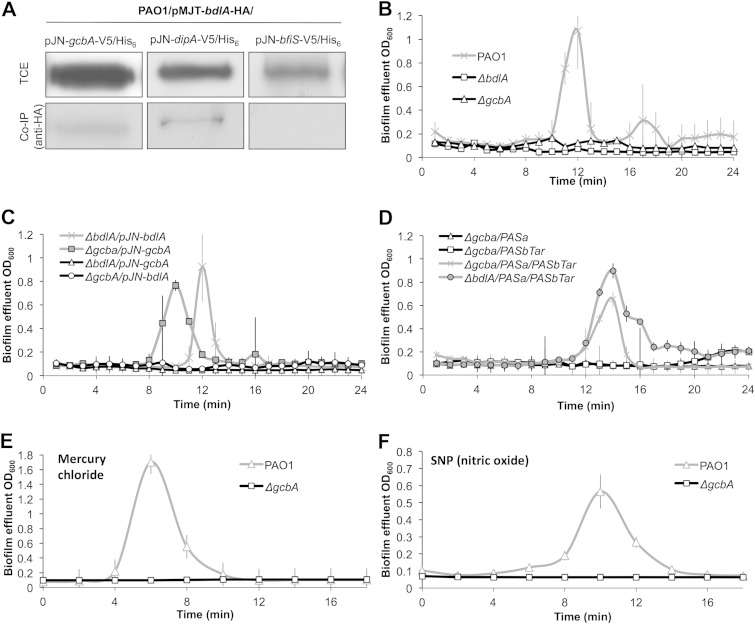
BdlA interacts with and requires GcbA to induce dispersion.
The MCP BdlA and the PDE DipA have recently been suggested to participate in a signaling network regulating dispersion in P. aeruginosa: while evidence suggested that BdlA interacts with and activates the activity of DipA, DipA has been shown to reduce c-di-GMP to enable dispersion and, also, reduce BdlA processing in biofilms (36–38). Considering the present observations of GcbA being required for biofilm dispersion, we asked whether GcbA also interacts with BdlA. The interaction of BdlA and GcbA in P. aeruginosa cells was assessed using a PAO1 strain harboring HA-tagged BdlA (BdlA-HA) and V5/His6-tagged GcbA (GcbA-V5/His6). DipA-V5/His6 (37) and BfiS-V5/His6 were used as prey proteins for positive- and negative-control assays, respectively (Fig. 6A). Following the enrichment of BdlA-HA by immunoprecipitation using anti-HA antibodies, the presence of copurifying V5/His6-tagged proteins was assessed using anti-V5 immunoblotting. BfiS-V6/His6 did not copurify with BdlA-HA, while in accord with previous observations, BdlA-HA interacted with DipA-V5/His6 (Fig. 6A) (37). While no GcbA-V5/His6 was isolated in the absence of BdlA-HA, the protein was detected as copurifying with BdlA-HA in total protein extracts derived from PAO1 cells grown planktonically (not shown) or as biofilms (Fig. 6A), suggesting that GcbA interacts, at least indirectly, with BdlA. Considering that BdlA was found to interact with both DipA and GcbA, it is likely that GcbA may also interact with DipA.
FIG 6.
GcbA is central to the P. aeruginosa biofilm dispersion response. (A) BdlA interacts with GcbA but not with the negative control BfiS as revealed by in vivo pulldown assays and subsequent immunoblotting analysis. Immunoprecipitation using anti-HA antibody and C-terminally HA-tagged BdlA as bait was followed by anti-V5 antibody detection of indicated C-terminally V5/His6-tagged prey proteins. Co-IP, coimmunoprecipitation. (B to D) The ability of 120-h-old biofilms to initiate dispersion in response to an increased glutamate concentration in the medium was assessed for wild-type PAO1 and isogenic bdlA and gcbA mutants (B), ΔgcbA or ΔbdlA mutants exhibiting multicopy expression of gcbA or bdlA (C), and the indicated mutant strains exhibiting multicopy expression of constructs encoding truncated peptides of BdlA corresponding to its PASa and PASbTar domains (D). (E, F) Induction of dispersion in response to mercury chloride (E) or nitric oxide (source, sodium nitroprusside [SNP]) (F) was assessed using 120-h-old biofilms of the wild-type PAO1 and isogenic gcbA mutant strains. For all assays, dispersion events were detected as increases in the biofilm effluent optical density. Error bars denote standard deviations. All experiments were performed in triplicate.
The findings of BdlA-GcbA interaction suggested that GcbA plays a potential role within the BdlA-dependent dispersion network. Subsequently, a cross-complementation analysis was carried out to determine the hierarchical organization of GcbA with respect to BdlA. While the expression of gcbA and bdlA rescued the dispersion phenotypes of the respective mutants, multicopy expression of gcbA in ΔbdlA (ΔbdlA/pJN-gcbA) biofilm cells failed to restore the dispersion response (Fig. 6B and C). We subsequently reasoned that, if GcbA is involved in the regulation of BdlA levels but not of BdlA cleavage, multicopy expression of bdlA will restore the dispersion response of biofilms formed by ΔgcbA to wild-type levels. However, if GcbA is responsible for regulating the posttranslational modification and activation of BdlA, overexpression of bdlA will not rescue the gcbA dispersion-deficient phenotype. In agreement with our findings indicating GcbA to specifically modulate posttranslational modification of BdlA, multicopy expression of bdlA in ΔgcbA biofilm cells did not restore biofilm dispersion in response to an increased glutamate concentration in the medium (Fig. 6C).
The ΔgcbA dispersion-deficient phenotype can be rescued by separately encoded truncated domains of BdlA.
Our data demonstrated that inactivation of GcbA impaired the biofilm-specific cleavage of BdlA, which correlated with an impaired biofilm dispersion response. Considering previous findings highlighting the importance of BdlA processing in enabling the dispersion response, we next asked whether the impairment in BdlA cleavage is specifically responsible for the dispersion-deficient phenotype of ΔgcbA biofilm cells. Specifically, we tested whether in trans expression of separately encoded PASa and PASbTar domains of BdlA is capable of rescuing the dispersion-deficient phenotype of gcbA mutant biofilms, in a manner similar to that previously observed for bdlA mutant biofilms (38). The expression of constructs encoding BdlA-PASa or BdlA-PASbTar (ΔgcbA/PASa or ΔgcbA/PASbTar) in ΔgcbA cells did not restore dispersion in response to an increased glutamate concentration to wild-type levels (Fig. 6B and D). In contrast, coproduction of the separately encoded BdlA peptides PASa and PASbTar rescued the dispersion phenotype of ΔgcbA biofilm cells to the levels observed for the wild-type or ΔbdlA/PASa/PASbTar (Fig. 6B and D), suggesting that BdlA functions downstream from GcbA.
GcbA is required for the regulation of responses to various dispersion-inducing cues.
The regulatory network containing BdlA and DipA regulates biofilm dispersion in response to a variety of substances, including carbohydrates (e.g., glutamate and succinate), heavy metals (e.g., silver nitrate and mercury chloride), and nitric oxide, with inactivation of either bdlA or dipA abrogating the dispersion response to all of these cues (12, 36). Considering that GcbA was found to be necessary for the activation of BdlA and for induction of dispersion in response to a change in the glutamate concentration, we next assessed the ability of ΔgcbA biofilm cells to respond to heavy metals and nitric oxide. With biofilms formed by the wild-type PAO1 used as a positive control, inactivation of gcbA was found to abrogate biofilm dispersion in response to mercury chloride and nitric oxide (Fig. 6E and F). These findings suggested that, similar to BdlA and DipA, GcbA is required for enabling dispersion in response to a wide variety of extracellular cues.
DISCUSSION
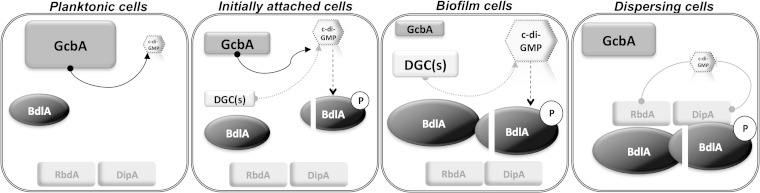
The response of P. aeruginosa biofilms to various dispersion-inducing conditions requires the reduction of cellular c-di-GMP levels via the activity of a complex regulatory network that to date has been found to contain the MCP BdlA and the c-di-GMP-degrading PDEs DipA, RbdA, NbdA, and MucR (12, 15, 17, 36–38). Here, we provide evidence that the catalytically active c-di-GMP-synthesizing DGC GcbA (39) is required for biofilm dispersion by regulating the posttranslational processing of BdlA. Considering that dispersion has been associated with reduction of c-di-GMP levels and that a DGC has not been previously demonstrated to be required for dispersion, the present findings were somewhat unexpected and raised a number of vital questions. For one, how does a DGC contribute to biofilm dispersion, a process that is associated with decreases in c-di-GMP levels? Previous findings have suggested that BdlA, which directly interacts with and potentially regulates the activities of the PDEs DipA and RbdA, plays a central role in the dispersion response, being required for biofilm cells to respond to diverse classes of dispersion-inducing cues, including carbohydrates, ammonium chloride, heavy metals, and NO (37, 38). While indirectly serving to regulate c-di-GMP levels to enable the dispersion response, BdlA posttranslational modification and activation, in turn, are dependent on elevated c-di-GMP levels following the initiation of surface-associated growth. In the present work, we demonstrate that the DGC GcbA contributes to the cleavage of BdlA in P. aeruginosa cells following initial attachment to a surface and, consequently, indirectly plays a role in enabling the regulation of biofilm dispersion, demonstrating a novel role for a DGC in the regulation of not only motile-sessile but also sessile-motile transitions (Fig. 7). We furthermore demonstrate that the levels of GcbA and BdlA are inversely regulated during motile-sessile and sessile-motile transitions in a manner that is dependent, at least in part, on cellular c-di-GMP levels (Fig. 7). The DGC GcbA, produced and active predominantly in planktonic and initially attached cells, synthesizes c-di-GMP to promote the transition from reversible to irreversible surface attachment and to contribute to the cleavage of BdlA in initially attached cells. Concurrent with the initiation of surface-associated growth and activation of BdlA processing, conditions of high viscosity limiting cellular motility lead to a reduction in the detectable levels of GcbA during subsequent biofilm growth (Fig. 7).
FIG 7.
Proposed model of GcbA-mediated modulation of the BdlA-dependent biofilm dispersion regulatory network. GcbA and BdlA are required for biofilm dispersion in response to changes in the extracellular environment, with the levels of the two proteins regulated inversely during motile-sessile and sessile-motile transitions in a manner dependent on c-di-GMP levels. GcbA, produced and active predominantly in planktonic and initially attached cells, synthesizes c-di-GMP (indicated by solid black line) to promote the transition from reversible to irreversible surface attachment and to contribute to the posttranslational modification and cleavage of BdlA in initially attached cells through an as-yet-uncharacterized mechanism (indicated by black dashed line). Other, unidentified DGC(s), which may or may not be produced in a growth-mode specific manner, potentially act in concert with GcbA to contribute to the c-di-GMP pool regulating the processing of BdlA, as indicated by the gray dotted line. Concomitant with initiation of attachment and activation of BdlA processing, conditions of elevated viscosity and reduced cellular motility lead to a reduction in GcbA abundance during subsequent biofilm growth. Following activation via cleavage, BdlA within biofilm cells directly interacts with the PDEs RbdA and DipA, likely modulating their activity to reduce c-di-GMP levels following the sensing of dispersion-inducing conditions. Direct reductions in c-di-GMP levels facilitated by RbdA and DipA are indicated by solid gray lines. During the dispersion process, DipA functions to reduce c-di-GMP levels to facilitate the sessile-motile transition and to induce GcbA production, thus activating a mechanism that will allow the newly detached cells to attach to a surface in new locations. The relative sizes of symbols indicating proteins and c-di-GMP in the model correlate with their relative abundances in the cell during the indicated growth mode.
Based on our previous findings, within biofilm cells, BdlA directly interacts with the PDEs RbdA and DipA and likely contributes to the regulation of their activity to reduce c-di-GMP levels following sensing of dispersion-inducing conditions (12, 36–38). During the dispersion process, the DipA-dependent reductions in cellular c-di-GMP levels not only facilitate the change in cellular physiology that allows biofilm cells to undergo the sessile-motile transition but also induce GcbA production, thereby activating a system that will allow the newly detached cells to attach to a surface in new locations (Fig. 7).
Another important question was raised by the results of the present study: how does a DGC that has previously been found to play a specific role in enabling surface attachment and to have no effect on biofilm formation contribute to biofilm-specific regulation of BdlA? GcbA was recently found to promote initial attachment to the surface through the regulation of a very specific phenotypic output (39). While not affecting extracellular polymeric substance (EPS) production, GcbA was found to facilitate the transition from reversible to irreversible attachment to a surface via regulation of flagellum-associated motility and flagellar reversal rates in a manner independent of surface hardness or previously characterized mechanisms, including FleQ, flagellar stators, chemotaxis IV gene cluster, and SadC and SadB (39). Although the present findings ascribe an additional role to GcbA, they do not necessarily contradict the previous findings of GcbA carrying out a specific role during initial surface attachment. This is because BdlA is processed soon after initial surface contact, with cleavage products detectable as early as following 8 h of surface-associated growth, with gcbA inactivation impairing BdlA cleavage in initially attached and developed biofilm cells. It is thus possible to suggest that, rather than being directly activated by GcbA-synthesized c-di-GMP, BdlA processing is potentially induced via sensing of reduced motility or of surface contact following GcbA-mediated reduction of flagellar reversals, motility, and surface contact and attachment. The question of the effects of EPS production, motility, and surface contact sensing on the initiation of BdlA processing, as well as the suppression of GcbA levels, will be the focus of future investigations.
Our present findings underscore the importance of the question of localized c-di-GMP pools regulating specific phenotypic outputs versus total cellular c-di-GMP levels regulating overall cellular behavior and determining the physiology and mode of growth of the cells. Specificity of c-di-GMP regulation was suggested by the observation of DipA having a more profound effect than the more active PDE PA2133 on the induction of GcbA production in biofilm cells (Fig. 5C to F). In contrast, the observations of GcbA playing a role in the regulation of both surface attachment and biofilm dispersion may be viewed as contradicting the concept of distinct c-di-GMP pools regulating specific phenotypic outputs. Furthermore, it is important to note that inactivation of gcbA did not completely abrogate BdlA processing, as smaller amounts of BdlA cleavage products were detectable in gcbA biofilms and at significantly lower levels than in wild-type biofilms. It is important to point out that previous findings have indicated that at least one of the cleavage products is still detectable for a BdlA variant with an M130I substitution at the putative ClpP cleavage site (38). However, the present findings may also suggest that GcbA potentially works in concert with other DGCs to regulate BdlA cleavage (Fig. 7). However, despite the possibility of multiple DGCs contributing to the activation of signaling systems enabling biofilm dispersion, the process does exhibit some specificity, as inactivation of the DGCs WspR and SiaD did not abrogate the dispersion response (Fig. 4E).
The finding of BdlA processing being activated soon following the transition to surface-associated growth may have important implications for our understanding of the regulation of dispersion mechanisms; specifically, the difference between environmentally and self-triggered or native dispersion. Native biofilm dispersion, mediated by the signaling molecule cis-DA, occurs when bacterial cells within biofilm microcolonies become motile and evacuate the structures through an opening into the bulk fluid, leaving behind a void within the microcolony (3, 60). Previous observations have indicated that native biofilm dispersion depends on the developmental stage of the biofilm and/or the size of the biofilm microcolonies (3, 60). Moreover, the findings of native biofilm microcolony dispersion requiring a minimum diameter of 40 μm and a minimum thickness of 10 μm suggested that the accumulation of the self-produced intracellular signaling molecule provides a way for biofilm cells to control their own accumulation and density (3, 60). In contrast, the dispersion process in response to changes in the extracellular environment is likely not dependent on cell density or microcolony formation. This is based on our finding that BdlA, a central modulator of these types of dispersion events, is activated via posttranslational processing soon after initial attachment and, thus, prior to initiation of biofilm maturation, significant biomass accumulation, or microcolony formation. Evidence for this is also provided by previous microscopic observations of nutrient-induced dispersion occurring within and outside microcolonies, as well as by findings of nutrient-induced dispersion not being affected in certain biofilms (such as those formed by ΔflgB or ΔpilA mutant strains or the dipA-overexpressing PAO1) that were characterized by significantly reduced biomass accumulation and thicknesses comparable to those of wild-type cells at the initial attachment stages of biofilm development (12, 37, 54, 57, 60). Together with these findings, the present results suggest that attached cells are likely able to respond to changing environmental conditions in a manner independent of biomass accumulation and microcolony size, with GcbA contributing to the activation of BdlA and mechanisms for sensing and relaying dispersion-inducing cues soon after the initial surface attachment.
ACKNOWLEDGMENTS
We thank Caroline Harwood and Matthew Parsek from the University of Washington for providing strains used in this study.
This work was supported by a grant from NIH (grant 1RO1 A107525701A2).
REFERENCES
- 1.Sauer K, Cullen MC, Rickard AH, Zeef LAH, Davies DG, Gilbert P. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol 186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol 75:815–826. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- 3.Davies DG, Marques CNH. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol 191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barraud N, Hassett DJ, Hwang S-H, Rice SA, Kjelleberg S, Webb JS. 2006. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol 188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abee T, Kovacs AT, Kuipers OP, van der Veen S. 2011. Biofilm formation and dispersal in Gram-positive bacteria. Curr Opin Biotechnol 22:172–179. doi: 10.1016/j.copbio.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Karatan E, Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schleheck D, Barraud N, Klebensberger J, Webb JS, McDougald D, Rice SA, Kjelleberg S. 2009. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PLoS One 4:e5513. doi: 10.1371/journal.pone.0005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huynh TT, McDougald D, Klebensberger J, Al Qarni B, Barraud N, Rice SA, Kjelleberg S, Schleheck D. 2012. Glucose starvation-induced dispersal of Pseudomonas aeruginosa biofilms is cAMP and energy dependent. PLoS One 7:e42874. doi: 10.1371/journal.pone.0042874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newell PD, Boyd CD, Sondermann H, Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawyer LK, Hermanowicz SW. 2000. Detachment of Aeromonas hydrophila and Pseudomonas aeruginosa due to variations in nutrient supply. Water Sci Technol 41:139–145. [Google Scholar]
- 11.James GA, Korber DR, Caldwell DE, Costerton JW. 1995. Digital image analysis of growth and starvation responses of a surface-colonizing Acinetobacter sp. J Bacteriol 177:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan R, Kohn S, Hwang S-H, Hassett DJ, Sauer K. 2006. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol 188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplan JB, Fine DH. 2002. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Appl Environ Microbiol 68:4943–4950. doi: 10.1128/AEM.68.10.4943-4950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. 2009. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol 191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Heine S, Entian M, Sauer K, Frankenberg-Dinkel N. 2013. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J Bacteriol 195:3531–3542. doi: 10.1128/JB.01156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thormann KM, Saville RM, Shukla S, Spormann AM. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J Bacteriol 187:1014–1021. doi: 10.1128/JB.187.3.1014-1021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An S, Wu J, Zhang L-H. 2010. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl Environ Microbiol 76:8160–8173. doi: 10.1128/AEM.01233-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musk DJ, Banko DA, Hergenrother PJ. 2005. Iron salts perturb biofilm formation and disrupt existing biofilms of Pseudomonas aeruginosa. Chem Biol 12:789–796. doi: 10.1016/j.chembiol.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. 2010. D-Amino acids trigger biofilm disassembly. Science 328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez CJ, Prieto EM, Krueger CA, Zienkiewicz KJ, Romano DR, Ward CL, Akers KS, Guelcher SA, Wenke JC. 2013. Effects of local delivery of d-amino acids from biofilm-dispersive scaffolds on infection in contaminated rat segmental defects. Biomaterials 34:7533–7543. doi: 10.1016/j.biomaterials.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 21.Leiman SA, May JM, Lebar MD, Kahne D, Kolter R, Losick R. 2013. D-Amino acids indirectly inhibit biofilm formation in Bacillus subtilis by interfering with protein synthesis. J Bacteriol 195:5391–5395. doi: 10.1128/JB.00975-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H, Liu Y. 2011. D-Amino acid mitigated membrane biofouling and promoted biofilm detachment. J Membr Sci 376:266–274. doi: 10.1016/j.memsci.2011.04.030. [DOI] [Google Scholar]
- 23.Hochbaum AI, Kolodkin-Gal I, Foulston L, Kolter R, Aizenberg J, Losick R. 2011. Inhibitory effects of D-amino acids on Staphylococcus aureus biofilm development. J Bacteriol 193:5616–5622. doi: 10.1128/JB.05534-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dow JM, Crossman L, Findlay K, He Y-Q, Feng J-X, Tang J-L. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc Natl Acad Sci U S A 100:10995–11000. doi: 10.1073/pnas.1833360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amari DT, Marques CNH, Davies DG. 2013. The putative enoyl-coenzyme A hydratase DspI is required for production of the Pseudomonas aeruginosa biofilm dispersion autoinducer cis-2-decenoic acid. J Bacteriol 195:4600–4610. doi: 10.1128/JB.00707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jennings JA, Courtney HS, Haggard WO. 2012. Cis-2-decenoic acid inhibits S. aureus growth and biofilm in vitro: a pilot study. Clin Orthop Relat Res 470:2663–2670. doi: 10.1007/s11999-012-2388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mills E, Pultz IS, Kulasekara HD, Miller SI. 2011. The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell Microbiol 13:1122–1129. doi: 10.1111/j.1462-5822.2011.01619.x. [DOI] [PubMed] [Google Scholar]
- 28.Boyd CD, O'Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol 28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Römling U, Simm R. 2009. Prevailing concepts of c-di-GMP signaling. Contrib Microbiol 16:161–181. doi: 10.1159/000219379. [DOI] [PubMed] [Google Scholar]
- 30.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baraquet C, Murakami K, Parsek MR, Harwood CS. 2012. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res 40:7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Römling U, Amikam D. 2006. Cyclic di-GMP as a second messenger. Curr Opin Microbiol 9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 34.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 35.Sondermann H, Shikuma NJ, Yildiz FH. 2012. You've come a long way: C-di-GMP signaling. Curr Opin Microbiol 15:140–146. doi: 10.1016/j.mib.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy AB, Petrova OE, Sauer K. 2012. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol 194:2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrova OE, Sauer K. 2012. PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J Bacteriol 194:5817–5828. doi: 10.1128/JB.00780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrova OE, Sauer K. 2012. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci U S A 109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrova OE, Cherny KE, Sauer K. 2014. The Pseudomonas aeruginosa diguanylate cyclase GcbA, a homolog of P. fluorescens GcbA, promotes initial attachment to surfaces, but not biofilm formation, via regulation of motility. J Bacteriol 196:2827–2841. doi: 10.1128/JB.01628-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones CJ, Newsom D, Kelly B, Irie Y, Jennings LK, Xu B, Limoli DH, Harrison JJ, Parsek MR, White P, Wozniak DJ. 2014. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog 10:e1003984. doi: 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. doi: 10.1016/S0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- 46.Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. 2007. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J Clin Invest 117:877–888. doi: 10.1172/JCI30783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrova OE, Sauer K. 2011. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J Bacteriol 193:6614–6628. doi: 10.1128/JB.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrova OE, Sauer K. 2010. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J Bacteriol 192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoang TT, Schweizer HP. 1997. Fatty acid biosynthesis in Pseudomonas aeruginosa: cloning and characterization of the fabAB operon encoding beta-hydroxyacyl-acyl carrier protein dehydratase (FabA) and beta-ketoacyl-acyl carrier protein synthase I (FabB). J Bacteriol 179:5326–5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogel H, Bonner D. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem 218:97–106. [PubMed] [Google Scholar]
- 51.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Pt 10):2395–2407. [DOI] [PubMed] [Google Scholar]
- 52.Wolschin F, Wienkoop S, Weckwerth W. 2005. Enrichment of phosphorylated proteins and peptides from complex mixtures using metal oxide/hydroxide affinity chromatography (MOAC). Proteomics 5:4389–4397. doi: 10.1002/pmic.200402049. [DOI] [PubMed] [Google Scholar]
- 53.Krüger R, Wolschin F, Weckwerth W, Bettmer J, Lehmann WD. 2007. Plant protein phosphorylation monitored by capillary liquid chromatography–element mass spectrometry. Biochem Biophys Res Commun 355:89–96. doi: 10.1016/j.bbrc.2007.01.108. [DOI] [PubMed] [Google Scholar]
- 54.Gupta K, Marques CNH, Petrova OE, Sauer K. 2013. Antimicrobial tolerance of Pseudomonas aeruginosa biofilms is activated during an early developmental stage and requires the two-component hybrid SagS. J Bacteriol 195:4975–4987. doi: 10.1128/JB.00732-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chambers JR, Sauer K. 2013. The MerR-like regulator BrlR impairs Pseudomonas aeruginosa biofilm tolerance to colistin by repressing PhoPQ. J Bacteriol 195:4678–4688. doi: 10.1128/JB.00834-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Southey-Pillig C, Davies D, Sauer K. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J Bacteriol 187:8114–8126. doi: 10.1128/JB.187.23.8114-8126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petrova OE, Sauer K. 2009. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog 5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O'Toole GA. 2007. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bobrov AG, Kirillina O, Perry RD. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol Lett 247:123–130. doi: 10.1016/j.femsle.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 60.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jonas K, Melefors O, Römling U. 2009. Regulation of c-di-GMP metabolism in biofilms. Future Microbiol 4:341–358. doi: 10.2217/fmb.09.7. [DOI] [PubMed] [Google Scholar]
- 62.Bourret RB. 2010. Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol 13:142–149. doi: 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caiazza NC, Merritt JH, Brothers KM, O'Toole GA. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:3603–3612. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]