Abstract
The O-antigen (Oag) component of lipopolysaccharide (LPS) is a major virulence determinant of Shigella flexneri and is synthesized by the O-antigen polymerase, WzySf. Oag chain length is regulated by chromosomally encoded WzzSf and pHS-2 plasmid-encoded WzzpHS2. To identify functionally important amino acid residues in WzySf, random mutagenesis was performed on the wzySf gene in a pWaldo-TEV-GFP plasmid, followed by screening with colicin E2. Analysis of the LPS conferred by mutated WzySf proteins in the wzySf-deficient (Δwzy) strain identified 4 different mutant classes, with mutations found in periplasmic loop 1 (PL1), PL2, PL3, and PL6, transmembrane region 2 (TM2), TM4, TM5, TM7, TM8, and TM9, and cytoplasmic loop 1 (CL1) and CL5. The association of WzySf and WzzSf was investigated by transforming these mutated wzySf plasmids into a wzySf- and wzzSf-deficient (Δwzy Δwzz) strain. Comparison of the LPS profiles in the Δwzy and Δwzy Δwzz backgrounds identified WzySf mutants whose polymerization activities were WzzSf dependent. Colicin E2 and bacteriophage Sf6c sensitivities were consistent with the LPS profiles. Analysis of the expression levels of the WzySf-GFP mutants in the Δwzy and Δwzy Δwzz backgrounds identified a role for WzzSf in WzySf stability. Hence, in addition to its role in regulating Oag modal chain length, WzzSf also affects WzySf activity and stability.
INTRODUCTION
Shigella flexneri lipopolysaccharide (LPS) is crucial for pathogenesis (1). LPS is located exclusively in the outer leaflet of the outer membrane (OM) and has three domains: (i) lipid A, a hydrophobic domain that anchors LPS to the OM; (ii) the core oligosaccharides, a nonrepeating oligosaccharide domain; and (iii) the O-antigen (Oag) polysaccharide, an oligosaccharide repeat domain (1, 2). The complete LPS structure with Oag chains is termed smooth LPS (S-LPS). However, the LPS structure lacking the Oag is termed rough LPS (R-LPS), and LPS with a single Oag tetrasaccharide repeat unit (RU) attached to the lipid A and core sugar is termed semirough LPS (SR-LPS) (3). S. flexneri is subdivided into various serotypes depending on the differences in the composition of the LPS Oag. So far, there are 17 known serotypes of S. flexneri (4). Except for S. flexneri serotype 6, the Oags of all the serotypes have the same polysaccharide backbone containing three l-rhamnose residues (Rha) and one N-acetylglucosamine (GlcNAc). This basic Oag structure is known as serotype Y. Addition of either glucosyl, O-acetyl, or phosphoethanolamine (PEtN) groups by various linkages to the sugars of the Y serotype tetrasaccharide repeat creates different S. flexneri serotypes (5–7). Oag is the protective antigen, as immunity to S. flexneri infection is serotype specific (8, 9). S-LPS confers resistance to complement (10) and colicins (11, 12), and Y serotype Oag acts as a receptor to bacteriophage Sf6 (13).
S. flexneri Oag biosynthesis occurs by the Wzy-dependent pathway. Most of the Oag biosynthesis genes (except wecA) of S. flexneri are located in the Oag biosynthesis locus between galF and his (6, 14). S. flexneri Oag biosynthesis occurs on either side of the inner membrane (IM). Initially, N-acetylglucosamine phosphate (GlcNAc-1-P) is transferred from UDP-GlcNAc by WecA to undecaprenol phosphate (Und-P) at the cytoplasmic side of the IM (5, 15, 16). RfbG and RfbF then add Rha residues from dTDP-rhamnose (dTDP-Rha) to the GlcNAc (3, 17) to form the O unit. In the Wzy-dependent model of LPS assembly, the flippase protein Wzx translocates this O unit to the periplasmic side. At the periplasmic side, the O units are polymerized at the nonreducing end by the Oag polymerization protein Wzy via a block transfer mechanism to form the polymer. The chain length of the final Oag is regulated by the protein Wzz (3, 18). Finally, the Oag ligase WaaL ligates the Oag chains to the previously synthesized core-lipid A. The Lpt proteins (LptA to -G) then transport the LPS from the IM to the OM (1, 19).
The S. flexneri Oag polymerization protein WzySf is encoded by the rfc or wzy gene. WzySf is a 43.7-kDa hydrophobic integral membrane protein. It has 12 transmembrane (TM) segments and two large periplasmic-loop (PL) domains (3, 18). Based on a topology model proposed by our group, the amino- and carboxy-terminal ends are located on the cytoplasmic side of the IM. The wild-type wzySf gene lacks a detectable ribosome binding site and has a low G+C%, and a high percentage of minor codons in the first 25 amino acids, contributing to low expression and poor detection of the protein (3, 18).
Islam et al. (20) performed extensive work on Pseudomonas aeruginosa Wzy (WzyPa) and showed that both PL3 and PL5 of WzyPa contain RX10G motifs, which are important for the functioning of WzyPa. They also found that several Arg residues within these two motifs are also important for WzyPa function (20). However, there is little sequence identity between WzyPa and WzySf, so it is not possible to predict the functional amino acid residues of WzySf from another model.
Wzz is a member of the polysaccharide copolymerase (PCP) family. S. flexneri has two types of Wzz, chromosomally encoded WzzSf and pHS-2 plasmid-encoded WzzpHS2. S. flexneri 2a has S-LPS with two types of modal chain length: short (S) type (11 to 17 Oag RUs) and very long (VL) type (>90 Oag RUs), and the S-type and VL-type Oag chain lengths are determined by WzzSf and WzzpHS2, respectively. Controlling Oag chain length is crucial for bacterial virulence, and loss of WzzSf-mediated Oag modal chain length regulation affects virulence due to masking of the OM protein IcsA (21, 22). Daniels and Morona showed that WzzSf forms a dimer in vivo and may oligomerize up to a hexamer (23). Formation of these large complexes is consistent with the hypothetical complex formation between Wzz and other enzymes of the Oag biosynthesis pathway, including Wzy (14, 24). WzzSf and WzzpHS2 compete for the available WzySf (25).
Woodward et al. showed that Wzz and Wzy are sufficient to determine the Oag modal chain length (26). There are several proposed mechanisms for modal length control by Wzz and its association with Wzy: a molecular-clock model was proposed by Bastin et al. (24), and a molecular-chaperone model was proposed by Morona et al. (14). Tocilj et al. suggested that Wzz may form a scaffold and that it recruits Wzy (27). However, there is no direct evidence to date on how these proteins are associated with each other in Oag polymerization and chain length control.
In this study, we were able to overexpress and detect WzySf-green fluorescent protein (GFP) expression in S. flexneri. We performed random mutagenesis on wzySf, and following screening with colicin E2, for the first time identified amino acid residues important for WzySf function. We classified the wzySf mutants based on their LPS profiles. We were able to determine mutant protein expression levels and also characterized the mutants depending on the phage and colicin sensitivities they conferred. These findings provided insight into Wzy structure and function. We further identified WzzSf-dependent WzySf mutants and identified a novel role for WzzSf in WzySf Oag polymerization activity and stability, in addition to its role in regulating Oag modal chain length.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are shown in Table 1.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| S. flexneri | ||
| PE638 | S. flexneri Y rpoB (Rifr) | 14 |
| RMM109 | PE638 Δwzy; Rifr | 3 |
| RMA4337 | RMM109 Δwzz (Rifr Tetr) | This study |
| PNRM6 | RMM109(pAC/pBADT7-1) | This study |
| PNRM11 | PNRM6(pWaldo-TEV-GFP) | This study |
| PNRM13 | PNRM6(pRMPN1) | This study |
| PNRM75 | PNRM6(pRMPN7) | This study |
| PNRM76 | PNRM6(pRMPN8) | This study |
| PNRM77 | PNRM6(pRMPN9) | This study |
| PNRM78 | PNRM6(pRMPN10) | This study |
| PNRM79 | PNRM6(pRMPN11) | This study |
| PNRM80 | PNRM6(pRMPN12) | This study |
| PNRM81 | PNRM6(pRMPN13) | This study |
| PNRM82 | PNRM6(pRMPN14) | This study |
| PNRM83 | PNRM6(pRMPN15) | This study |
| PNRM84 | PNRM6(pRMPN16) | This study |
| PNRM85 | PNRM6(pRMPN17) | This study |
| PNRM119 | PNRM6(pRMPN19) | This study |
| PNRM120 | PNRM6(pRMPN21) | This study |
| PNRM121 | PNRM6(pRMPN22) | This study |
| PNRM122 | PNRM6(pRMPN23) | This study |
| PNRM123 | PNRM6(pRMPN24) | This study |
| PNRM124 | PNRM6(pRMPN25) | This study |
| PNRM126 | RMA4337(pAC/pBADT7-1) | This study |
| PNRM134 | PNRM126(pRMPN1) | This study |
| PNRM131 | PNRM126(pRMPN7) | This study |
| PNRM132 | PNRM126(pRMPN15) | This study |
| PNRM133 | PNRM126(pRMPN16) | This study |
| PNRM136 | PNRM126(pRMPN8) | This study |
| PNRM137 | PNRM126(pRMPN10) | This study |
| PNRM140 | PNRM126(pRMPN13) | This study |
| PNRM141 | PNRM126(pRMPN14) | This study |
| PNRM142 | PNRM126(pRMPN9) | This study |
| PNRM143 | PNRM126(pRMPN11) | This study |
| PNRM144 | PNRM126(pRMPN19) | This study |
| PNRM145 | PNRM126(pRMPN24) | This study |
| PNRM146 | PNRM126(pRMPN25) | This study |
| PNRM147 | PNRM126(pRMPN23) | This study |
| PNRM148 | PNRM126(pRMPN21) | This study |
| PNRM149 | PNRM126(pRMPN22) | This study |
| PNRM150 | PNRM126(pRMPN12) | This study |
| PNRM151 | PNRM126(pRMPN17) | This study |
| E. coli | ||
| XL10G | Tetr Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr) Cmr] | Stratagene |
| Lemo21(DE3) | fhuA2 (lon) ompT gal (λ DE3) (dcm) ΔhsdS/pLemo (Cmr) | New England Biolabs |
| PNRM15 | Lemo21(DE3)(pRMPN1) | This study |
| Plasmids | ||
| pRMCD6 | Source of wzySf (modified codons at positions 4, 9, and 23) | 18 |
| pAC/pBADT7-1 | Source of T7 RNA polymerase; Cmr | 34 |
| pWaldo-TEV-GFP | Cloning vector with GFP tag; Kmr | 28 |
| pRMPN1 | pWaldo-wzySf-GFP; Kmr | This study |
| pRMPN7 | pRMPN1 with G130V point mutation in the wzySf gene | This study |
| pRMPN8 | pRMPN1 with L111I point mutation in the wzySf gene | This study |
| pRMPN9 | pRMPN1 with N86K point mutation in the wzySf gene | This study |
| pRMPN10 | pRMPN1 with L28V point mutation in the wzySf gene | This study |
| pRMPN11 | pRMPN1 with P165S point mutation in the wzySf gene | This study |
| pRMPN12 | pRMPN1 with G82C point mutation in the wzySf gene | This study |
| pRMPN13 | pRMPN1 with N147K point mutation in the wzySf gene | This study |
| pRMPN14 | pRMPN1 with L191F point mutation in the wzySf gene | This study |
| pRMPN15 | pRMPN1 with L214I point mutation in the wzySf gene | This study |
| pRMPN16 | pRMPN1 with P352H point mutation in the wzySf gene | This study |
| pRMPN17 | pRMPN1 with V92 M point mutation in the wzySf gene | This study |
| pRMPN19 | pRMPN1 with F52Y point mutation in the wzySf gene | This study |
| pRMPN21 | pRMPN1 with F52C/I242T point mutations in the wzySf gene | This study |
| pRMPN22 | pRMPN1 with C60F point mutation in the wzySf gene | This study |
| pRMPN23 | pRMPN1 with Y137H point mutation in the wzySf gene | This study |
| pRMPN24 | pRMPN1 with L49F/T328A point mutation in the wzySf gene | This study |
| pRMPN25 | pRMPN1 with F54C point mutation in the wzySf gene | This study |
| pCACTUS | Suicide vector containing sacB, Cmr gene, and orits | 14 |
| pRMA577 | Suicide vector containing SphI-SphI fragment with the rol gene | 14 |
| pCACTUS-wzzSf::Tcr | Suicide mutagenesis construct to construct the strain RMA4337 | This study |
Rifr, rifampin resistant; Kmr, kanamycin resistant; Cmr, chloramphenicol resistant; Tetr, tetracycline resistant. The λ DE3 genotype is λ sBamHIo ΔEcoRI-B int::(lacI::PlacUV5::T7 gene 1) i21.
Growth media and growth conditions.
The growth media used were Luria-Bertani (LB) broth (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl) and LB agar (LB broth, 15 g/liter Bacto agar).
Unless otherwise stated, strains were grown in LB broth with aeration for 18 h at 37°C and then diluted 1/20 into fresh LB broth and grown to mid-exponential phase (optical density at 600 nm [OD600], 0.4 to 0.6). Where required, the growth medium was supplemented with 0.2% (wt/vol) glucose to suppress protein expression. Before induction, cells were centrifuged (2,200 × g; Sigma 3K15 tabletop centrifuge; 10 min; 4°C) and washed twice with LB broth to remove glucose. Under induction conditions, 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) or 0.2% (wt/vol) l-arabinose was added to the cultures, and they were grown for 20 h at 20°C. Antibiotics were added as required to the media at the following final concentrations: kanamycin (Km), 50 μg/ml, and chloramphenicol (Cm), 25 μg/ml.
Construction of expression vector and cloning of wzySf.
Primers PN1_wzySfKpnF and PN2_wzySfBamHR (see Table S1 in the supplemental material), which incorporated KpnI and BamHI restriction sites, respectively, were used to amplify the previously mutated wzySf coding region (GenBank accession number X71970) in the pRMCD6 plasmid (Table 1), possessing three changes at codons 4, 9, and 23 (18). The rare codons 4, 9, and 23 are present in the translation initiation site of WzySf and cause lower expression of the wild-type WzySf. The codons ATA, ATA, and AGA at 4, 9, and 23 were changed to ATT, ATT, and CGT, respectively, and resulted in increased expression of WzySf (18). Following restriction digestion with enzymes BamHI and KpnI, the amplified wzySf was ligated into similarly digested pWaldo-TEV-GFP (28), resulting in the plasmid pWaldo-wzySf-TEV-GFP (denoted pRMPN1). PCR, restriction enzyme digestion, agarose gel electrophoresis, ligation, and transformation were performed as described previously (3, 14).
WzySf expression in Lemo21(DE3) and in-gel fluorescence.
For WzySf expression in Lemo21(DE3) cells (Table 1), strains were induced with IPTG, and the overexpressed WzySf-GFP fusion protein was detected by in-gel fluorescence following the method of Drew et al. (29) with some modifications. Cells (5 × 108) were harvested from the induced culture and washed twice in 200 μl phosphate buffered-saline (PBS). The pellet was resuspended in 20 μl PBS, and 20 μl buffer A (200 mM Tris-HCl [pH 8.8], 20% [vol/vol] glycerol, 5 mM EDTA [pH 8.0], 0.02% [wt/vol] bromophenol blue, 4% [wt/vol] SDS, and 0.05 M dithiothreitol [DTT]) was then added to the cell suspension. The solubilized cell suspension was incubated at 37°C for 5 min. Samples were then separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 15% (wt/vol) gels, and a BenchMark Pre-Stained Protein Ladder (Invitrogen) was used as a molecular mass standard. The gel was rinsed with distilled water, and fluorescence imaging of the gel was performed to detect WzySf-GFP protein expression with a Bio-Rad Gel Doc XR+ system using Image Lab software (excitation at 485 nm and emission at 512 nm).
LPS method.
LPS was prepared as described previously (30, 31). To prepare LPS samples, 1 × 109 cells were harvested, resuspended in lysing buffer (buffer B, 10% [wt/vol] glycerol, 2% [wt/vol] SDS, 4% [wt/vol] β-mercaptoethanol, 0.1% [wt/vol] bromophenol blue, 1 M Tris-HCl, pH 7.6), and incubated with 2 μg/ml of proteinase K for approximately 16 h. The LPS samples were then electrophoresed on SDS–15% PAGE gels for 16 to 18 h at 12 mA. Gels were stained with silver nitrate and developed with formaldehyde (30).
Random mutagenesis.
Random mutagenesis of wzySf was undertaken to obtain a wide range of wzySf mutations in a nonselected manner. For PCR random mutagenesis, the wzySf coding region in plasmid pRMPN1 was mutagenized with an error-prone DNA polymerase using the GeneMorph II EZ-Clone Domain Mutagenesis Kit (catalog number 200552; Stratagene) according to the manufacturer's instructions with the primers PN1_wzySfKpnF and PN2_wzySfBamHR (see Table S1 in the supplemental material). The mutagenized plasmids were transformed into competent Escherichia coli XL10-Gold (XL10G) cells (Agilent Technologies). Plasmid DNA was then isolated from randomly chosen transformed, mutated colonies and transformed into strain PNRM6 (Table 1). Colicin swab assays were performed to screen the mutants (see below). Plasmid DNA was isolated from putative mutants, transformed into XL10G cells, and subjected to DNA sequencing (AGRF, Adelaide, Australia).
Construction of the strain RMA4437 (Δwzy Δwzz).
The S. flexneri Y PE638 Δwzy Δwzz mutant strain was constructed using allelic-exchange mutagenesis (14) to inactivate the wzzSf gene in RMM109 (3). Initially, a tetracycline resistance (Tetr) cartridge was inserted into the BglII site of pRMA577 (14) to inactivate wzzSf, and the resulting pCACTUS-wzzSf::tet (Table 1) plasmid was transformed into RMM109 via electroporation (32). Allelic-exchange mutagenesis was performed as previously described (14). The wzzSf::Tetr mutation in the chromosome was confirmed by PCR with primers ET35 and ET36 (see Table S1 in the supplemental material) to give the PE638 Δwzy Δwzz mutant RMA4337.
Detection of WzySf expression in S. flexneri.
For the detection of WzySf expression in S. flexneri, cells were harvested from the 50-ml 0.2% (wt/vol) l-arabinose-induced culture by centrifugation (9,800 × g; Beckman J2-21M induction drive centrifuge; 10 min; 4°C), and the cell pellet was resuspended in 4 ml sonication buffer (buffer C, 20 mM Tris-HCl, 150 mM NaCl, pH 7.5). The mixture was then lysed by sonication, followed by centrifugation (2,200 × g; Sigma 3K15 tabletop centrifuge; 10 min; 4°C) to remove debris. Ultracentrifugation was performed in a Beckman Coulter Optima MAX-XP tabletop ultracentrifuge (126,000 × g for 1 h at 4°C) to isolate the whole-membrane (WM) fraction. The WM fraction was resuspended in PBS and then solubilized in buffer A. The solubilized WM fraction from 3 × 108 cells was electrophoresed on an SDS–15% PAGE gel. The gel was rinsed with distilled water, and in-gel imaging was performed as described above. Loading was checked by staining the gel with Coomassie blue R-250. The intensity of WzySf-GFP expression for mutant and control strains was measured with the Fiji image-processing package (http://fiji.sc/Fiji), and the percent relative WzySf-GFP intensity for each mutant strain was measured by comparing the WzySf-GFP intensity of each mutant strain with the WzySf-GFP intensity in the control strain, PNRM13.
Colicin sensitivity assay.
For the colicin sensitivity assay, a solution of purified His6-ColE2 (ColE2) with an initial concentration of 1 mg/ml was used (11).
ColE2 swab assay.
A 2-fold serial dilution of 1 μg/ml of ColE2 was swabbed onto antibiotic-selective LB agar plates containing 0.2% (wt/vol) l-arabinose with a cotton swab. The plates were left to dry for 1 h at room temperature (RT). Individual 0.2% (wt/vol) l-arabinose-induced mutant and control LB cultures were then swabbed perpendicular to the ColE2 streak, and the plates were left to dry for another 1 h at RT. The plates were then incubated for 16 h at 37°C. The susceptibilities of the mutant strains compared to the control strain were recorded, and images were taken using a Canon scanner (CanoScan 9000F) against a dark background.
ColE2 spot assay.
A spot assay was performed using the serial dilutions of ColE2. One hundred microliters of the individual l-arabinose-induced mutant and control strain cultures were spread onto LB agar plates with appropriate antibiotics and 0.2% (wt/vol) l-arabinose. The plates were left to dry for 2 h at RT. A 2-fold serial dilution of 1 μg/ml of ColE2 (denoted neat [N]) was spotted on the dried plates, and the plates were left to dry for another 3 h at RT. The plates were then incubated for 18 h at 37°C. The endpoints of the killing zones of mutant strains were compared with the controls. Images were recorded as described above.
Bacteriophage sensitivity assay.
The procedures of phage propagation and phage stock preparation have been described previously (3, 33). The concentration of the bacteriophage Sf6c stock used was 8.6 × 107 PFU/ml. Mutant and control strains were grown and induced with 0.2% (wt/vol) l-arabinose, and 100 μl of the individual mutant and control LB cultures was spread onto LB agar plates with appropriate antibiotics and 0.2% (wt/vol) l-arabinose. The plates were left to dry for 2 h at RT. Serial dilutions of the bacteriophage Sf6c stock (undiluted bacteriophage Sf6c stock was denoted N) were spotted on the dried plates, and the plates were dried for a further 3 h at RT. The plates were incubated for 18 h at 37°C. The phage sensitivities of the test strains were compared with the controls. Images were recorded as described above.
RESULTS
Construction of a WzySf-GFP expression plasmid.
A suitable expression system was constructed to express wzySf and to detect WzySf by fusion to GFP (29). S. flexneri 2457T 2a wzySf with three modified codons at postions 4, 9, and 23 in the pRMCD6 plasmid (18) was PCR amplified and ligated into pWaldo-TEV-GFP (Table 1) (see Materials and Methods). To confirm that the construct was able to express the WzySf-GFP-His8 protein, pWaldo-wzySf-TEV-GFP-His8 (denoted pRMPN1) was transformed into Lemo21(DE3) cells. Whole-cell in-gel fluorescence samples were then prepared from PNRM15 [Lemo21(DE3)(pRMPN1)], and fluorescence imaging of the gel detected a fluorescent band at approximately 64 kDa, which corresponded to the predicted size of the WzySf-GFP protein (see Fig. S1, lane 1, in the supplemental material), indicating that the construct was able to express WzySf-GFP.
Complementation of wzySf deficiency.
A complementation assay was performed to confirm the functionality of WzySf-GFP. For this assay, pRMPN1 was cotransformed, along with pAC/pBADT7-1 (34), into the wzySf-deficient strain RMM109 (3). RMM109 has a frameshift mutation at position 9214 in the wzySf gene that results in premature termination of WzySf synthesis (3). pAC/pBADT7-1 encodes T7 RNA polymerase, which drives the expression of wzySf-gfp in pRMPN1. LPS samples were prepared from these control strains. The silver-stained gel showed that PNRM6 [RMM109(pAC/pBADT7-1)] had an SR-LPS profile (Fig. 1, lane 3), but PNRM13 [PNRM6(pRMPN1)] had an S-LPS profile (Fig. 1, lane 5). Hence, pRMPN1 was able to complement the wzySf mutation in RMM109, and the LPS profile resembled that of the wild-type strain PE638 (Fig. 1, lane 2).
FIG 1.
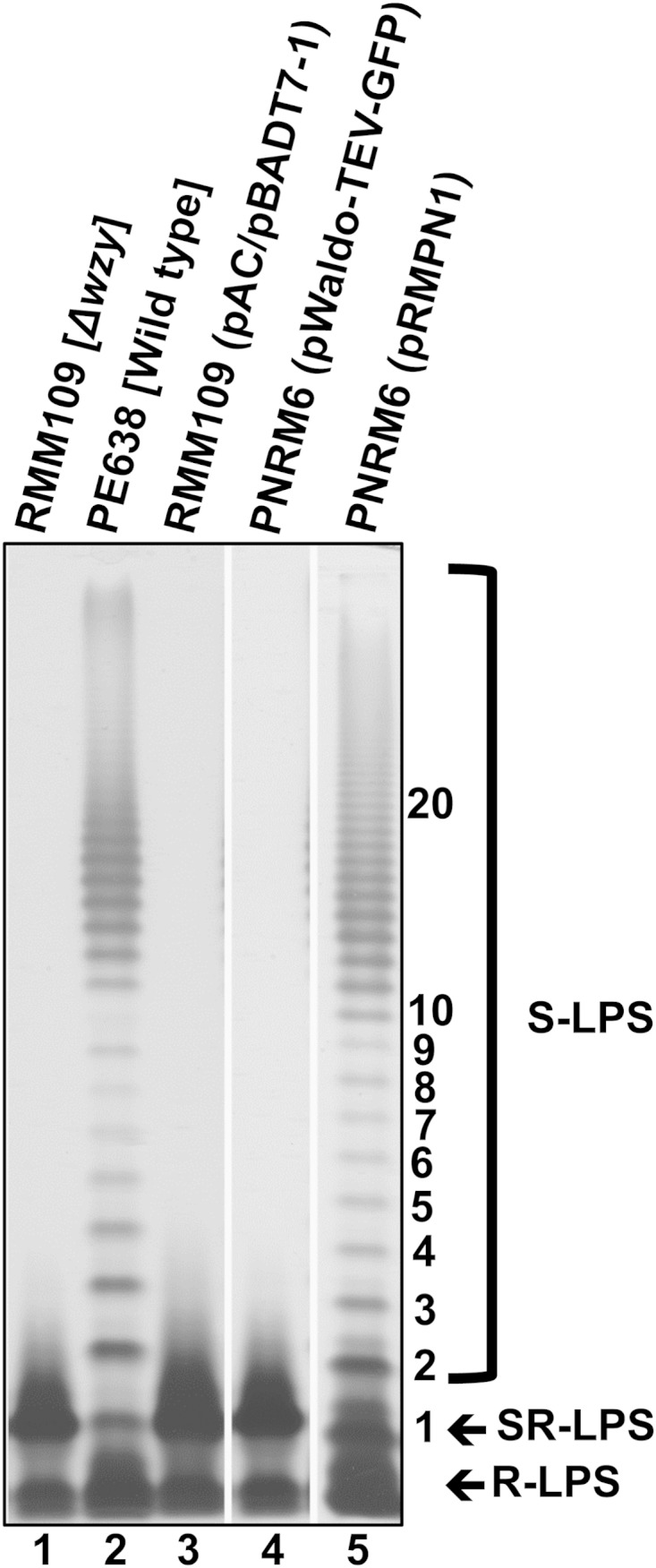
Complementation of wzySf deficiency by WzySf-GFP. LPS samples (equivalent to 1 × 109 bacterial cells) were prepared from the indicated strains by proteinase K treatment, electrophoresed on an SDS–15% (wt/vol) PAGE gel, and silver stained (see Materials and Methods). The positions of S-LPS, SR-LPS, and R-LPS are indicated. The numbers on the right indicate the Oag RUs.
Random mutagenesis of wzySf.
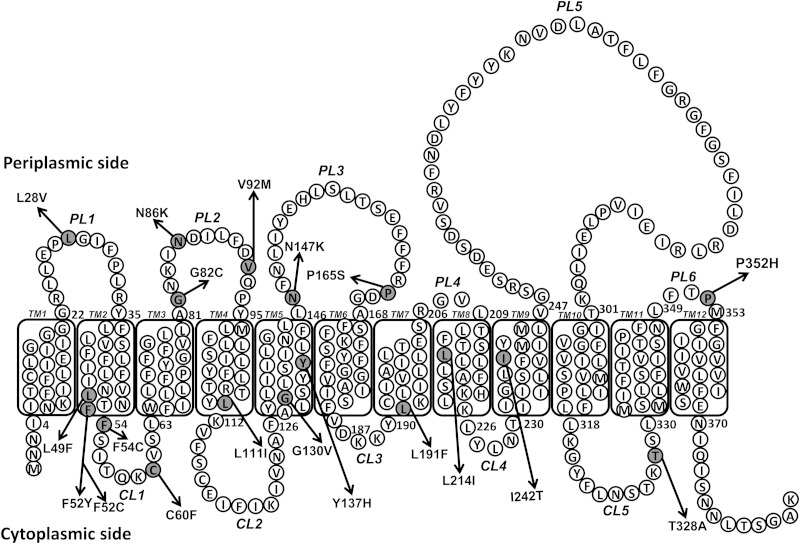
Nothing is known about the residues important for WzySf function, and there is little sequence identity between the Wzy proteins of different bacterial species. Hence, it is difficult to predict the functional amino acid residues of the S. flexneri WzySf. To obtain insight into the WzySf residues needed for function, the wzySf coding region in plasmid pRMPN1 was subjected to random mutagenesis using an error-prone DNA polymerase (see Materials and Methods). The resulting mutagenized plasmid library was transformed into PNRM6. We screened the transformants to find mutants, using ColE2. The basis for this is that the R-LPS strains are more susceptible to killing by colicins than the S-LPS strains (12), and we recently found that there is a strong correlation between LPS Oag modal chain length and susceptibility to ColE2 (11). A ColE2 swab assay (see Materials and Methods) was used to screen and detect mutants that had different sensitivity to ColE2 than the positive-control strain PNRM13 (see Table S2 in the supplemental material). Interestingly the wild-type strain PE638 was slightly more resistant to ColE2 than the complemented positive-control strain PNRM13 (Table 2; see Table S2 in the supplemental material). The wzySf mutant RMM109 (SR-LPS) was highly sensitive to ColE2 (Table 2; see Table S2 in the supplemental material). Transformants that were either more resistant or more sensitive to ColE2 than PNRM13 were selected (see Table S2 in the supplemental material), and the plasmids were isolated and transformed into the XL10G strain. Plasmid DNA from these isolates was subjected to DNA sequencing to identify mutational alterations in wzySf. The wzySf mutants had the following substitutions: P352H, V92M, Y137H, L214I, G130V, N147K, P165S, L191F, C60F, L49F/T328A, L28V, N86K, F54C, F52Y, L111I, G82C, and F52C/I242T (see Table S3 in the supplemental material). The mutations were present in PL1, PL2, PL3, and PL6, TM2, TM4, TM5, TM7, TM8, and TM9,and cytoplasmic loop 1 (CL1) and CL5 of the WzySf topology map (summarized in Fig. 2 and Table 2). After sequence confirmation, the mutated plasmids were transformed into PNRM6 (Table 1) for detailed characterization.
TABLE 2.
ColE2 and bacteriophage Sf6c sensitivities and WzySf-GFP expression of controls and different classes of mutants
| Strain or mutant | Relevant detail(s) | Mutant class |
Topology map locationa | Sensitivityb |
Relative WzySf-GFP expression (%) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| ColE2 |
Sf6c |
|||||||||
| Δwzy background | Δwzy Δwzz background | Δwzy background | Δwzy Δwzz background | Δwzy background | Δwzy Δwzz background | Δwzy background | Δwzy Δwzz background | |||
| Strains | ||||||||||
| RMM109 | wzySf mutants | 1/256 | R | |||||||
| PE638 | Wild type | R | 10−6 | |||||||
| PNRM13 | Positive control | 1/2 | 10−5 | 100 | ||||||
| PNRM6 | Negative control | 1/256 | R | |||||||
| PNRM11 | Negative control | 1/256 | R | |||||||
| RMA4337 | wzySf and wzySf mutant | 1/256 | R | |||||||
| PNRM126 | Negative control | 1/256 | R | |||||||
| PNRM134 | Positive control | R | 10−6 | 17 | ||||||
| Mutants | ||||||||||
| P352H | A | B | PL6 | 1/64 | 1/128 | R | R | 36 | 38 | |
| V92 M | A | E | PL2 | 1/32 | 1/64 | R | R | 84 | 76 | |
| Y137H | A | E | TM5 | 1/32 | 1/64 | R | R | 87 | 97 | |
| L214I | B | C | TM8 | 1/128 | 1/128 | R | R | 1.4 | 0.03 | |
| G130V | C | F | TM5 | 1/512 | 1/128 | R | R | 1.60 | 28.50 | |
| N147K | D | E | PL3 | R | 1/16 | 10−6 | R | 21 | 52 | |
| P165S | D | E | PL3 | 1/4 | 1/64 | 10−5 | N | 7 | 125 | |
| L191F | D | E | TM7 | R | 1/16 | 10−6 | N | 162 | 64 | |
| C60F | D | E | TM1 | R | 1/64 | 10−6 | N | 30 | 66 | |
| L49F/T328A | D | E | TM2/CL5 | R | 1/64 | 10−6 | R | 65 | 68 | |
| L28V | D | E | PL1 | R | 1/8 | 10−6 | N | 55 | 80 | |
| N86K | D | E | PL2 | R | 1/64 | 10−6 | R | 84 | 82 | |
| F54C | D | E | CL1 | R | 1/64 | 10−6 | R | 42 | 71 | |
| F52Y | D | E | TM2 | R | 1/64 | 10−6 | R | 47 | 63 | |
| L111I | D | E | TM4 | R | 1/8 | 10−6 | N | 41 | 51 | |
| G82C | D | E | PL2 | R | 1/64 | 10−6 | R | 40 | 47 | |
| F52C/I242T | D | E | TM2/TM9 | R | 1/64 | 10−6 | N | 82 | 81 | |
PL, periplasmic loop; TM, transmembrane region; CL, cytoplasmic loop (Fig. 2).
R, resistant; N, plaques detected with undiluted Sf6c stock. The numbers indicate the highest dilution showing a zone of inhibition or plaque formation.
FIG 2.
Locations of mutations on the topology map of WzySf. Mutational alterations are indicated by arrows on the WzySf topology map (adapted from reference 18). The positions of the periplasmic loops (PL1 to PL5), transmembrane regions (TM1 to TM12), and cytoplasmic loops (CL1 to CL5) are indicated. Mutations (shaded circles) are located in PL1, PL2, PL3, and PL6, TM2, TM4, TM5, TM7, TM8, and TM9, and CL1 and CL5.
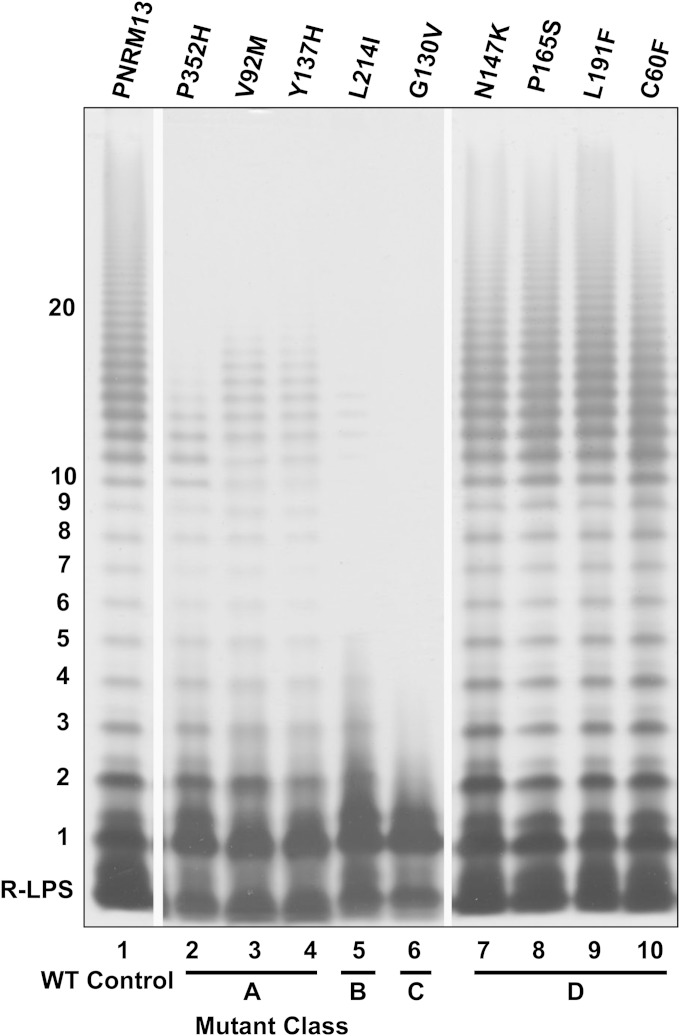
LPS phenotype conferred by WzySf mutants.
The effects of mutations on WzySf LPS Oag polymerization activity were determined by SDS-PAGE and silver staining. Following comparison of the resulting degree of LPS Oag polymerization of the mutant strains with the relevant positive control, PNRM13, the mutants were grouped into four different phenotypic classes: A, B, C, and D (Fig. 3; see Fig. S2 in the supplemental material). Three of the 17 mutants had reduced degrees of polymerization compared to PNRM13 and were classified as class A. Class A mutants had the following alterations in WzySf: P352H (WzyP352H), V92M (WzyV92M), and Y137H (WzyY137H) (Fig. 3, lanes 2 to 4). One mutant (with an L214I alteration in WzySf [WzyL214I]) exhibited an LPS banding pattern with only a few Oag RUs and was classified as class B (Fig. 3, lane 5). Another mutant had SR-LPS (with a G130V alteration in WzySf [WzyG130V]) and was categorized into class C (Fig. 3, lane 6). The other 12 mutations conferred an LPS profile similar to that of the positive control, PNRM13, and were classified as class D. Members of this class had the following mutations: N147K (WzyN147K), P165S (WzyP165S), L191F (WzyL191F), C60F (WzyC60F), L49F/T328A (WzyL49F/T328A), L28V (WzyL28V), N86K (WzyN86K), F54C (WzyF54C), F52Y (WzyF52Y), L111I (WzyL111I), G82C (WzyG82C), and F52C/I242T (WzyF52C/I242T) (Fig. 3, lanes 7 to 10; see Fig. S2, lanes 1 to 8, in the supplemental material). Here, the mutant WzySf proteins are referred to according to their conferred LPS classes.
FIG 3.
LPS phenotypes conferred by different WzySf mutants expressed in PNRM6 [ΔwzySf(pAC/pBADT7-1)]. Plasmid-encoded mutated WzySf proteins were expressed in PNRM6. The strains were grown and induced with arabinose as described in Materials and Methods. LPS samples were prepared, electrophoresed on SDS–15% (wt/vol) PAGE gels, and silver stained (see Materials and Methods). The strains were divided into various mutant classes (A, B, C, and D) based on their LPS phenotypes. Lane 1, positive-control strain PNRM13 [PNRM6(pRMPN1)]; lanes 2 to 10, Δwzy strain (PNRM6) with plasmids encoding mutated WzySf proteins, as indicated. The position of R-LPS is indicated. The numbers on the left indicate the Oag RUs. WT, wild type.
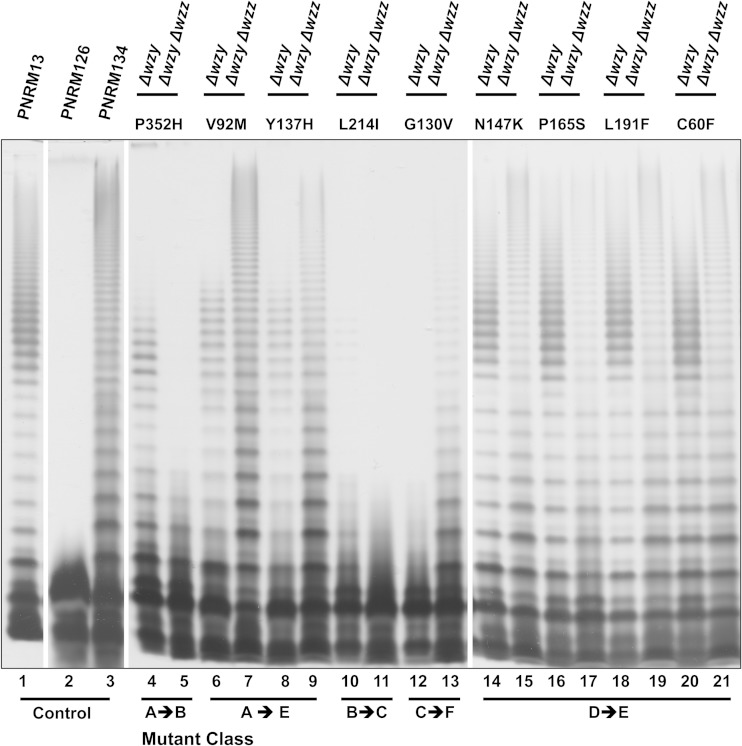
WzzSf dependence.
Since the Wzy-dependent model of LPS assembly suggests a potential interaction between Wzy and Wzz, we investigated if the LPS profile conferred by mutated WzySf proteins was dependent on the presence of WzzSf. All the plasmids encoding mutated WzySf proteins were transformed into RMA4337 carrying pAC/pBADT7-1 (strain PNRM126) (Table 1). RMA4337 is a wzySf and wzzSf double mutant. The LPS profiles conferred in the PNRM126 background (Δwzy Δwzz) were directly compared with the LPS profile conferred in the Δwzy background (PNRM6). The control strain PNRM134 [PNRM126(pRMPN1)] had S-LPS without Oag modal chain length control (Fig. 4, lane 3), as expected, and was classified as class E for the purpose of comparison. PNRM126 with mutated wzySf plasmids that conferred a class A LPS profile in the Δwzy background had a class E LPS profile similar to that of PNRM134, except that PNRM126 expressing WzyP352H had LPS with few Oag RUs (class B) (Fig. 4, lane 5). PNRM126 expressing WzyL214I had SR-LPS (Fig. 4, lane 11). However, in comparison, PNRM6 (Δwzy) expressing WzyL214I conferred a class B LPS profile (Fig. 4, lane 10). PNRM126 with a mutated wzySf plasmid that conferred a class C LPS profile in the Δwzy background (PNRM6 expressing G130V) had S-LPS without modal length control and a reduced degree of polymerization (designated class F) compared to PNRM134 (Fig. 4, lane 13). PNRM126 with mutated wzySf plasmids from the strains with a class D LPS profile in the Δwzy background had a class E LPS profile, similar to that of PNRM134 (Fig. 4, lanes 14 to 21; see Fig. S3, lanes 1 to 16, in the supplemental material). Hence, certain WzySf mutants conferred dramatically different LPS profiles depending on the presence or absence of WzzSf.
FIG 4.
Comparison of the LPS phenotypes conferred by the WzySf mutants expressed in the Δwzy and Δwzy Δwzz backgrounds. Plasmids encoding mutated WzySf proteins were expressed in PNRM126 [RMA4337(pAC/pBADT7-1)] and PNRM6 [RMM109(pAC/pBADT7-1)]. The strains were grown and induced as described in Materials and Methods. LPS samples were electrophoresed on SDS–15% (wt/vol) PAGE gels and silver stained (see Materials and Methods). Lanes 1 to 3 contain the indicated strains; lanes 4 to 21 contain the Δwzy (PNRM6) or Δwzy Δwzz (PNRM126) strain with plasmids encoding the indicated mutated WzySf proteins.
ColE2 and bacteriophage Sf6c sensitivities.
To confirm the LPS profiles determined as described above using assays of LPS Oag function, the ColE2 and bacteriophage Sf6c sensitivities of the mutant strains were investigated. ColE2 sensitivity (summarized in Table 2) was determined by spot testing, as described in Materials and Methods. Strains RMM109, PNRM6, PNRM11, RMA4337, and PNRM126 showed killing zones at a dilution of 1/256. The wild-type strain PE638 was resistant to the highest concentration of ColE2 used. The relevant positive-control strain in the Δwzy background (PNRM13) showed a killing zone at a dilution of 1/2, but the relevant positive-control strain in the Δwzy Δwzz background (PNRM134) was resistant to the highest concentration of ColE2 tested. Strains conferring a class A LPS profile in the Δwzy background were sensitive to ColE2 (killing zone at 1/32 or 1/64). However, Δwzy Δwzz strains with mutated wzySf plasmids conferring the class A LPS profile in the Δwzy background were 2-fold more sensitive to ColE2 (killing zone at 1/64 or 1/128) than the Δwzy background. The Δwzy strain expressing WzyL214I (class B) and the Δwzy Δwzz strain expressing WzyL214I (class C) had similar ColE2 sensitivities (1/128). The strain with the class C LPS profile (Δwzy expressing WzyG130V) had the greatest sensitivity to ColE2 (1/512), greater than RMM109 and the negative-control strains (PNRM6, PNRM11, and PNRM126). However, the Δwzy Δwzz strain expressing WzyG130V (class F) was more resistant (1/128) to ColE2 than the Δwzy background. Strains with a class D LPS profile in the Δwzy background were resistant to the highest concentration of ColE2 tested, except that Δwzy expressing WzyP165S showed slight sensitivity (1/4). Hence, almost all the strains with a class D LPS profile in the Δwzy background were more resistant to ColE2 than PNRM13 and were similar to PE638. The Δwzy Δwzz strains with mutated wzySf plasmids from the strains with a class D LPS profile in the Δwzy background were more sensitive to ColE2 (killing zone at 1/8 to 1/64) than the control, PNRM134 [Δwzy Δwzz(pRMPN1)], suggesting they have a slight defect in Oag polymerization in the absence of WzzSf. Hence, ColE2 resistance correlated with the degree of Oag polymerization and the degree of LPS capping with Oag.
The bacteriophage Sf6c sensitivities of the strains carrying mutated wzySf plasmids (summarized in Table 2) were determined by spot testing, as described in Materials and Methods. The SR-LPS strains RMM109, PNRM6, PNRM11, RMA4337, and PNRM126 were resistant to bacteriophage Sf6c. The S-LPS strains, wild-type PE638, and the controls, PNRM13 and PNRM134, were bacteriophage Sf6c sensitive, and plaques were detected at 10−6, 10−5, and 10−6 dilutions, respectively. Strains with class A, B, and C LPS profiles in the Δwzy background were resistant to the highest concentration of bacteriophage Sf6c tested and were similar to the strains with SR-LPS (RMM109, PNRM6, PNRM11, RMA4337, and PNRM126). Similarly, the Δwzy Δwzz strains with mutated wzySf plasmids from the strains with class A, B, and C LPS profiles in the Δwzy background were also resistant to the highest concentration of bacteriophage Sf6c tested. Strains with a class D LPS profile in the Δwzy background were bacteriophage Sf6c sensitive (plaques at 10−6), except that the strain expressing WzyP165S was slightly less sensitive (plaque at 10−5). Hence, the bacteriophage Sf6c sensitivities of the strains with a class D LPS profile in the Δwzy background were greater than that of PNRM13 and similar to that of PE638. Δwzy Δwzz strains with mutated wzySf plasmids from the strains with a class D LPS profile in the Δwzy background were more resistant to bacteriophage Sf6c than the Δwzy background. Although they have a class E LPS profile, similar to PNRM134, they were more resistant to bacteriophage Sf6c than PNRM134. Hence, bacteriophage Sf6c sensitivity correlated with the degree of Oag polymerization and the degree of LPS capping with Oag.
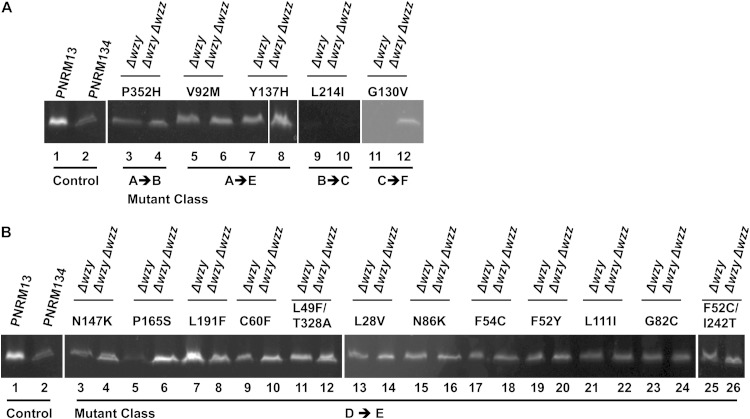
WzySf expression level.
We determined the levels of parental and mutant WzySf-GFP expressed in Δwzy and Δwzy Δwzz S. flexneri strains. To measure the protein expression levels of the mutants, in-gel fluorescence testing was performed, and the percent relative WzySf-GFP expression was calculated (see Materials and Methods). The expression levels of different WzySf-GFP mutants were compared with that of WzySf-GFP in PNRM13 (100%). The WzySf-GFP expression level in PNRM134 was less than that in PNRM13 (Fig. 5A and B, lane 2), with a relative WzySf-GFP level of 17% (Table 2). In the Δwzy background, the WzySf-GFP mutants were expressed at a level less than 100%, except the L191F mutant (class D), which had expression of 162% (Fig. 5B, lane 7, and Table 2). However, in the Δwzy Δwzz background, the relative WzySf-GFP level of the L191F mutant was 64% (Fig. 5B, lane 8, and Table 2), which was less than the control strain, PNRM13. In the Δwzy background, the relative WzySf-GFP levels of P165S, L214I, and G130V mutants were very low (7%, 1.4%, and 1.6%, respectively) (Fig. 5B, lane 5, and A, lanes 9 and 11, and Table 2). However, in the Δwzy Δwzz background, P165S and G130V mutants had relative WzySf-GFP levels of 125% and 28.50%, respectively (Fig. 5B, lane 6, and A, lane 12, and Table 2), but the relative WzySf-GFP level of the L214I mutant was almost undetectable at 0.03% (Fig. 5A, lane 10, and Table 2). In the Δwzy Δwzz background, the relative WzySf-GFP level of the P165S mutant was higher than that of the control, PNRM13 (100%) (Fig. 5B, lane 6, and Table 2). These data indicate that expression of certain WzySf mutants was affected by WzzSf but that the effect was mutant specific.
FIG 5.
Protein expression levels of the mutated WzySf-GFP compared to the positive control. The strains were grown in LB and induced as described in Materials and Methods. In-gel fluorescence samples were prepared from the mutants in the Δwzy and Δwzy Δwzz backgrounds and electrophoresed on SDS–15% (wt/vol) PAGE gels (see Materials and Methods). (A) Lanes 1 and 2 contain the indicated strains; lanes 3 to 12 contain the Δwzy or Δwzy Δwzz strain expressing mutated WzySf-GFP, as indicated. (B) Lanes 1 and 2 contain the indicated strains; lanes 2 to 26 contain the Δwzy or Δwzy Δwzz strain expressing mutated WzySf-GFP, as indicated.
DISCUSSION
In this study, we constructed and characterized a collection of wzySf mutants. We found that ColE2 screening was an effective method to detect wzySf mutants conferring subtle effects on LPS structure (Table 2). The use of WzySf-GFP allowed comparison of the protein expression levels of different mutants. We also found that bacteriophage Sf6c sensitivity was dependent on the LPS phenotype structure. Strains with mutant WzySf conferring a longer Oag chain and/or a greater degree of Oag polymerization were more sensitive to bacteriophage Sf6c, while bacteriophage Sf6c infected only strains with wild-type (or nearly wild-type) LPS, both in the degree of Oag polymerization and the apparent level of LPS capping with Oag chains.
Only a few studies have been conducted on Wzy proteins. During characterization of Wzy of Francisella tularensis, Kim et al. reported several amino acid residues important for Oag polymerization. Modification of these residues (G176, D177, G323, and Y324) led to a loss of Oag polymerization (35). Islam et al. showed that PL3 and PL5 of WzyPa have net positive and net negative charges, respectively, and they established their “catch-and-release” model (20). However, for WzySf, we found that at a physiological pH both PL3 and PL5 possess net negative charges (the pI of PL3 is 4.65, and that of PL5 is 5.09). In Pseudomonas aeruginosa PAO1 there is a uronic acid sugar in the Oag (36) that is negatively charged. However, the Oag of WzySf is neutral. Therefore, the charge property of the substrate for WzySf is different from that of WzyPa.The RX10G motifs of PL3 and PL5 of WzyPa (20) are also absent in WzySf. However, both PL3 and PL5 of WzySf contained RX15G motifs (starting from R164 in PL3 and R289 in PL5) (Fig. 2). Therefore, perhaps a modified version of the “catch-and-release” mechanism (20) exists for S. flexneri. Moreover, addition of either glucosyl or O-acetyl groups or PEtN residues by various linkages to the sugars within the tetrasaccharide repeats of serotype Y creates 17 different serotypes of S. flexneri (5–7). As the polymerization of all these Oags is done by WzySf, we conclude that WzySf must be quite flexible in its substrate recruitment. Due to scant homology between wzy proteins of different bacterial species, we were unable to create any directly comparable mutation in WzySf with respect to other systems. This led us to perform random mutagenesis on wzySf, followed by screening with ColE2.
The mutations we found are present in a wide region (PL1, PL2, PL3, and PL6, TM2, TM4, TM5, TM7, TM8, and TM9, and CL1 and CL5) of the WzySf topological model experimentally determined by Daniels et al. (18). We were able to locate a number of mutations in the TM regions, and interestingly, the G130V mutation, which resulted in the complete loss of polymerization activity of WzySf in the Δwzy background, was also located in TM5. Topological models are not very accurate, but due to a lack of crystal structure, we used the WzySf topology model previously determined by our group to locate the mutational alterations. Furthermore, we reassessed our topological model using 5 different topology prediction programs (SPOCTOPUS, MEMSAT, HMMTOP, TOPCONS, and TMHMM) (see Table S4 in the supplemental material). The results showed that until TM region 6, all the programs are consistent with our topological model. Three of them also validate TM10 to TM11 of our topological model. Thus, the topological model of WzySf determined by our group is nearly consistent with the topological models predicted by the programs. The mutations that resulted in the partial loss of polymerization in the Δwzy background were V92M, Y137H, L214I, and P352H. Among them, V92M and Y137H are present in PL2 and TM5. These regions were validated by all 5 of the programs. P352H is present in PL6, and the region was validated by 3 of the programs (MEMSAT, HMMTOP, and TOPCONS). L214I is present in TM8, and the region (amino acids 209 to 226) was different from the computer prediction (amino acids 227 to 247). However, G130V is present in TM5, which was validated by all 5 of the programs. Recently, Reddy et al. reported that different topology prediction programs are suitable for different families of proteins (37). Therefore, lack of prediction of TM7 to TM9 by some programs does not invalidate the presence of these regions in our model.
In the Δwzy background, strains with the class A LPS profile had S-LPS with a reduced degree of Oag polymerization (Fig. 3, lanes 2 to 4), mutants with the class B LPS profile had LPS with few Oag RUs (Fig. 3, lane 5), and the mutant with a class C LPS profile had SR-LPS (Fig. 3, lane 6). As a result, the class A mutants were more sensitive to ColE2 than PNRM13, and the class B mutants were even more sensitive to ColE2 than the mutants with the class A LPS profile. The mutant with class C LPS had the highest ColE2 sensitivity. From these three classes, it is clear that the LPSs with shorter Oag chains were more sensitive to ColE2. Strains with Class A, B, and C LPS profiles in the Δwzy background were resistant to bacteriophage Sf6c. The lack of bacteriophage Sf6c sensitivity indicates that bacteriophage Sf6c infects only if the LPS has a wild-type or close to a wild-type level of Oag polymerization. In this study, we found that the WzySf mutants conferring the class D LPS profile in the Δwzy background had a level of Oag polymerization similar to that of the positive-control strain PNRM13, as determined by SDS-PAGE and silver staining (Fig. 3, lanes 7 to 10; see Fig. S2, lanes 1 to 8, in the supplemental material), but they were more resistant to ColE2 and more sensitive to bacteriophage Sf6c (Table 2) (except the Δwzy strain with WzyP165S). The ColE2 and bacteriophage Sf6c sensitivities of these strains were similar to those of wild-type strain PE638. We conclude that the Oag polymerization activities of the mutants with a class D LPS profile in the Δwzy background were similar to that of the wild-type strain (PE638). Hence, the wild-type WzySf-GFP protein is not 100% active, and the mutant proteins conferring the class D LPS profile are more active and/or are better exported, folded, or assembled to the IM. The levels of Oag polymerization of the strains with a class D LPS profile were more than that of PNRM13, although this was not distinguishable by silver staining. Hence, the ColE2 assay was more sensitive than silver staining and SDS-PAGE in determining the degree of Oag polymerization.
Mutation in wzz resulted in Oags without modal chain length control in different organisms (14, 24). Recently, Kenyon and Reeves showed that Wzy of Yersinia pseudotuberculosis also needs Wzz for complete Oag polymerization (38). Here, we examined the polymerization activities of different WzySf mutants in the absence of WzzSf. In the Δwzy Δwzz background, the strain with mutated wzySf plasmids from the strain having class B LPS profiles in the Δwzy background had an SR-LPS profile, and ColE2 and bacteriophage Sf6c sensitivities were similar to those in the Δwzy background. Interestingly, the Δwzy Δwzz strain with WzyG130V had S-LPS without modal length control (class F), whereas the Δwzy strain with WzyG130V had SR-LPS (class C), and this correlated with increased resistance to ColE2. In the Δwzy Δwzz background, strains with a class E LPS profile (S-LPS without modal length control) had increased sensitivity to ColE2 and increased or similar resistance to bacteriophage Sf6c compared to PNRM134 [Δwzy Δwzz(pRMPN1)] and the Δwzy background (class A and class D) (Table 2). We speculate that these strains (strains with a class E LPS profile) had subtle alterations in the level of Oag polymerization and/or capping of LPS with Oag and were unable to act efficiently as a bacteriophage Sf6c receptor and, correspondingly, were also more sensitive to ColE2. It is known that phage adsorption to the cells of Escherichia coli K-12 increases with an increase in the density of receptor protein at the cell surface (39). Based on our study, the relationship between ColE2 and the Oag density/concentration seems to be linear. Decreasing Oag progressively increases sensitivity to ColE2. However, the relationship between bacteriophage Sf6c and the Oag density/concentration seems to be nonlinear. Decreasing Oag rapidly decreases sensitivity to bacteriophage Sf6c. This may be because bacteriophage Sf6c interaction with its receptor is complex and most likely requires multireceptor binding to bacteriophage Sf6c tail spike proteins (TSPs) to achieve irreversible binding and hence activation of bacteriophage Sf6c.
According to the proposed molecular-clock model, Wzz acts as a molecular clock and regulates Wzy activity between two states: the E, or extension, state favors polymerization, and the T, or transfer, state favors the ligation reaction (24). The molecular-chaperone model describes Wzz as a typical molecular chaperone that regulates the overall ratio of Wzy and WaaL in a complex and controls the enzyme kinetics of the ligation reaction to define the modality (14). However, Woodward et al. suggested that there is an interaction between Wzy and Wzz, and they showed that the proteins are enough to shape the Oag modal chain length (26). Islam et al. suggested that the chain length of the Oag is determined by the interaction of Wzz and Wzy (40). Recent work of Taylor et al. also suggested the direct interaction of Wzz and Wzy in the Oag biosynthesis pathway (41). In this study, for the first time, we were able to provide insight into the association of WzySf and WzzSf in Oag biosynthesis. We found WzySf mutants in which polymerization activity was dependent on WzzSf (WzyP352H and WzyL214I) and some other mutants in which polymerization activity was repressed in the presence of WzzSf (WzyV92M, WzyY137H, and WzyG130V). The Δwzy Δwzz strains with WzyV92M or WzyY137H had S-LPS without modal length control (class E), but in the Δwzy background, they showed a class A LPS profile (a greatly reduced degree of Oag polymerization) (Fig. 4, lanes 6 and 7 and lanes 8 and 9). The Δwzy strain with WzyG130V had a class C LPS profile (SR-LPS), but remarkably, the Δwzy Δwzz strain with WzyG130V had S-LPS without modal length control, albeit with a reduced degree of polymerization (class F) (Fig. 4, lanes 12 and 13). Oag polymerization by these WzySf mutants was repressed by WzzSf in the ΔwzySf background; their LPSs had shorter Oag chains. The Δwzy strain with WzyP352H had a class A LPS profile, and the Δwzy strain with WzyL214I had a class B LPS profile, but in the Δwzy Δwzy background they had shorter Oag chains (class B and C, respectively) than the Δwzy background (Fig. 4, lanes 4 and 5 and lanes 10 and 11). Thus, these WzySf mutants need WzzSf for their Oag polymerization activities. These findings suggest that WzzSf is associated with WzySf, not only for Oag modal chain length control but also for polymerization activity, and that the amino acids V92, G130, Y137, L214, and P352 have roles in the association of WzySf and WzzSf during Oag polymerization mediated by WzySf.
The relative WzySf-GFP level in PNRM134 (Δwzy Δwzz) was lower than in PNRM13 (Δwzy) (Fig. 5A and B, lanes 1 and 2), suggesting that the wild-type WzySf-GFP had better expression in the presence of WzzSf. WzySf mutants conferring a class A LPS profile (Δwzy background) had WzySf-GFP levels lower than that of WzySf-GFP in PNRM13 (Fig. 5A, lanes 3, 5, and 7, and Table 2). WzyL214I was not detectable in either the Δwzy (class B) or the Δwzy Δwzz (class C) background (Fig. 5A, lanes 9 and 10, and Table 2). Hence, amino acid L214 is important for WzySf-GFP production. WzyG130V was not detectable in the Δwzy background (class C) but was detected in the Δwzy Δwzz background (class F) (Fig. 5A, lanes 11 and 12, and Table 2). Except for the Δwzy strain with WzyL191F, all the mutants with a class D LPS profile (Δwzy background) had protein expression levels lower than that of WzySf-GFP in PNRM13 (Fig. 5B and Table 2) by some unknown mechanism. However, the mutants with a class D LPS profile (Δwzy background) had S-LPS and were more resistant to ColE2 and more sensitive to bacteriophage Sf6c than PNRM13. In the Δwzy Δwzz background, WzyL191F had an expression level lower than that of WzySf-GFP in PNRM13 (Fig. 5B, lane 8, and Table 2). WzyP165S was not detectable in PNRM6 (Δwzy), but strain PNRM126 (Δwzy Δwzz) had WzyP165S present at a higher level than WzySf-GFP in PNRM13 (Fig. 5B, lanes 5 and 6, and Table 2). Apparently, there is more than one underlying mechanism for the class D (Δwzy background) to class E (Δwzy Δwzz background) LPS profile conversion. Two of the mutations (P165S and G130V) resulted in decreased WzySf-GFP levels in the presence of WzzSf. This suggested that the presence of WzzSf destabilizes WzyP165S and WzyG130V, which results in lower WzySf-GFP levels. The mutation L191F resulted in decreased WzySf-GFP levels in the absence of WzzSf. Therefore, in this case, WzzSf stabilizes the protein WzyL191F, resulting in better WzySf-GFP expression. This finding suggests that G130, P165, and L191 are important for the stabilization of WzySf through interaction with WzzSf.
In conclusion, our findings identified amino acid residues on WzySf important for its polymerization function and interaction with WzzSf. Residues that are important for the polymerization and interaction of WzzSf and WzySf are present in PL2 and PL6 and TM5 and TM8. These regions may contribute to the interaction of WzySf with substrates and also to the interaction with WzzSf. We identified a number of amino acids (G130, P165, and L191) that are important for the stabilization of WzySf in association with WzzSf. Hence, our data suggested that WzzSf also has a role in WzySf stability.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniel O. Daley for the pWaldo-TEV-GFP plasmid.
Funding for this work is provided by a program grant to R.M. from the National Health and Medical Research Council (NHMRC) of Australia. P.N. is the recipient of an international postgraduate research scholarship from the University of Adelaide.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01885-14.
REFERENCES
- 1.Sperandeo P, Deho G, Polissi A. 2009. The lipopolysaccharide transport system of Gram-negative bacteria. Biochim Biophys Acta 1791:594–602. doi: 10.1016/j.bbalip.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morona R, Mavris M, Fallarino A, Manning PA. 1994. Characterization of the rfc region of Shigella flexneri. J Bacteriol 176:733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Q, Lan R, Wang Y, Wang J, Wang Y, Li P, Du P, Xu J. 2013. Isolation and genomic characterization of SfI, a serotype-converting bacteriophage of Shigella flexneri. BMC Microbiol 13:39. doi: 10.1186/1471-2180-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Zhang C, Shi F, Hu X. 2010. Purification and characterization of lipopolysaccharides. Subcell Biochem 53:27–51. doi: 10.1007/978-90-481-9078-2_2. [DOI] [PubMed] [Google Scholar]
- 6.Allison GE, Verma NK. 2000. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol 8:17–23. doi: 10.1016/S0966-842X(99)01646-7. [DOI] [PubMed] [Google Scholar]
- 7.Sun Q, Knirel YA, Lan R, Wang J, Senchenkova SN, Jin D, Shashkov AS, Xia S, Perepelov AV, Chen Q, Wang Y, Wang H, Xu J. 2012. A novel plasmid-encoded serotype conversion mechanism through addition of phosphoethanolamine to the O-antigen of Shigella flexneri. PLoS One 7:e46095. doi: 10.1371/journal.pone.0046095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jennison AV, Verma NK. 2004. Shigella flexneri infection: pathogenesis and vaccine development. FEMS Microbiol Rev 28:43–58. doi: 10.1016/j.femsre.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Stagg RM, Tang SS, Carlin NI, Talukder KA, Cam PD, Verma NK. 2009. A novel glucosyltransferase involved in O-antigen modification of Shigella flexneri serotype 1c. J Bacteriol 191:6612–6617. doi: 10.1128/JB.00628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong M, Payne SM. 1997. Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol Microbiol 24:779–791. doi: 10.1046/j.1365-2958.1997.3731744.x. [DOI] [PubMed] [Google Scholar]
- 11.Tran EN, Papadopoulos M, Morona R. 2014. Relationship between O-antigen chain length and resistance to colicin E2 in Shigella flexneri. Microbiology 160:589–601. doi: 10.1099/mic.0.074955-0. [DOI] [PubMed] [Google Scholar]
- 12.van der Ley P, de Graaff P, Tommassen J. 1986. Shielding of Escherichia coli outer membrane proteins as receptors for bacteriophages and colicins by O-antigenic chains of lipopolysaccharide. J Bacteriol 168:449–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindberg AA, Wollin R, Gemski P, Wohlhieter JA. 1978. Interaction between bacteriophage Sf6 and Shigella flexneri. J Virol 27:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morona R, van den Bosch L, Manning PA. 1995. Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J Bacteriol 177:1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu D, Cole RA, Reeves PR. 1996. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J Bacteriol 178:2102–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo H, Yi W, Song J K, Wang PG. 2008. Current understanding on biosynthesis of microbial polysaccharides. Curr Top Med Chem 8:141–151. doi: 10.2174/156802608783378873. [DOI] [PubMed] [Google Scholar]
- 17.Macpherson DF, Manning PA, Morona R. 1995. Genetic analysis of the rfbX gene of Shigella flexneri. Gene 155:9–17. doi: 10.1016/0378-1119(94)00918-I. [DOI] [PubMed] [Google Scholar]
- 18.Daniels C, Vindurampulle C, Morona R. 1998. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy). Mol Microbiol 28:1211–1222. doi: 10.1046/j.1365-2958.1998.00884.x. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. 2008. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Islam ST, Gold AC, Taylor VL, Anderson EM, Ford RC, Lam JS. 2011. Dual conserved periplasmic loops possess essential charge characteristics that support a catch-and-release mechanism of O-antigen polymerization by Wzy in Pseudomonas aeruginosa PAO1. J Biol Chem 286:20600–20605. doi: 10.1074/jbc.C110.204651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morona R, Daniels C, Van Den Bosch L. 2003. Genetic modulation of Shigella flexneri 2a lipopolysaccharide O antigen modal chain length reveals that it has been optimized for virulence. Microbiology 149:925–939. doi: 10.1099/mic.0.26141-0. [DOI] [PubMed] [Google Scholar]
- 22.Morona R, Van Den Bosch L. 2003. Lipopolysaccharide O antigen chains mask IcsA (VirG) in Shigella flexneri. FEMS Microbiol Lett 221:173–180. doi: 10.1016/S0378-1097(03)00210-6. [DOI] [PubMed] [Google Scholar]
- 23.Daniels C, Morona R. 1999. Analysis of Shigella flexneri Wzz (Rol) function by mutagenesis and cross-linking: wzz is able to oligomerize. Mol Microbiol 34:181–194. doi: 10.1046/j.1365-2958.1999.01591.x. [DOI] [PubMed] [Google Scholar]
- 24.Bastin DA, Stevenson G, Brown PK, Haase A, Reeves PR. 1993. Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol Microbiol 7:725–734. doi: 10.1111/j.1365-2958.1993.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 25.Carter JA, Jimenez JC, Zaldivar M, Alvarez SA, Marolda CL, Valvano MA, Contreras I. 2009. The cellular level of O-antigen polymerase Wzy determines chain length regulation by WzzB and WzzpHS-2 in Shigella flexneri 2a. Microbiology 155:3260–3269. doi: 10.1099/mic.0.028944-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodward R, Yi W, Li L, Zhao G, Eguchi H, Sridhar PR, Guo H, Song J K, Motari E, Cai L, Kelleher P, Liu X, Han W, Zhang W, Ding Y, Li M, Wang PG. 2010. In vitro bacterial polysaccharide biosynthesis: defining the functions of Wzy and Wzz. Nat Chem Biol 6:418–423. doi: 10.1038/nchembio.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tocilj A, Munger C, Proteau A, Morona R, Purins L, Ajamian E, Wagner J, Papadopoulos M, Van Den Bosch L, Rubinstein JL, Fethiere J, Matte A, Cygler M. 2008. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat Struct Mol Biol 15:130–138. doi: 10.1038/nsmb.1374. [DOI] [PubMed] [Google Scholar]
- 28.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. 1999. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol 17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 29.Drew D, Lerch M, Kunji E, Slotboom DJ, de Gier JW. 2006. Optimization of membrane protein overexpression and purification using GFP fusions. Nat Methods 3:303–313. doi: 10.1038/nmeth0406-303. [DOI] [PubMed] [Google Scholar]
- 30.Murray GL, Attridge SR, Morona R. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol Microbiol 47:1395–1406. doi: 10.1046/j.1365-2958.2003.03383.x. [DOI] [PubMed] [Google Scholar]
- 31.Tsai CM, Frasch CE. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem 119:115–119. doi: 10.1016/0003-2697(82)90673-X. [DOI] [PubMed] [Google Scholar]
- 32.Purins L, Van Den Bosch L, Richardson V, Morona R. 2008. Coiled-coil regions play a role in the function of the Shigella flexneri O-antigen chain length regulator WzzpHS2. Microbiology 154:1104–1116. doi: 10.1099/mic.0.2007/014225-0. [DOI] [PubMed] [Google Scholar]
- 33.Mavris M, Manning PA, Morona R. 1997. Mechanism of bacteriophage SfII-mediated serotype conversion in Shigella flexneri. Mol Microbiol 26:939–950. doi: 10.1046/j.1365-2958.1997.6301997.x. [DOI] [PubMed] [Google Scholar]
- 34.McKinney J, Guerrier-Takada C, Galan J, Altman S. 2002. Tightly regulated gene expression system in Salmonella enterica serovar Typhimurium. J Bacteriol 184:6056–6059. doi: 10.1128/JB.184.21.6056-6059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim TH, Sebastian S, Pinkham J T, Ross RA, Blalock LT, Kasper DL. 2010. Characterization of the O-antigen polymerase (Wzy) of Francisella tularensis. J Biol Chem 285:27839–27849. doi: 10.1074/jbc.M110.143859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knirel YA, Bystrova OV, Kocharova NA, Zahringer U, Pier GB. 2006. Conserved and variable structural features in the lipopolysaccharide of Pseudomonas aeruginosa. J Endotoxin Res 12:324–336. doi: 10.1179/096805106X118906. [DOI] [PubMed] [Google Scholar]
- 37.Reddy A, Cho J, Ling S, Reddy V, Shlykov M, Saier MH. 2014. Reliability of nine programs of topological predictions and their application to integral membrane channel and carrier proteins. J Mol Microbiol Biotechnol 24:161–190. doi: 10.1159/000363506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kenyon JJ, Reeves PR. 2013. The Wzy O-antigen polymerase of Yersinia pseudotuberculosis O:2a has a dependence on the Wzz chain-length determinant for efficient polymerization. FEMS Microbiol Lett 349:163–170. doi: 10.1111/1574-6968.12311. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz M. 1976. The adsorption of coliphage lambda to its host: effect of variations in the surface density of receptor and in phage-receptor affinity. J Mol Biol 103:521–536. doi: 10.1016/0022-2836(76)90215-1. [DOI] [PubMed] [Google Scholar]
- 40.Islam ST, Huszczynski SM, Nugent T, Gold AC, Lam JS. 2013. Conserved-residue mutations in Wzy affect O-antigen polymerization and Wzz-mediated chain-length regulation in Pseudomonas aeruginosa PAO1. Sci Rep 3:3441. doi: 10.1038/srep03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor VL, Udaskin ML, Islam ST, Lam JS. 2013. The D3 bacteriophage alpha-polymerase inhibitor (Iap) peptide disrupts O-antigen biosynthesis through mimicry of the chain length regulator Wzz in Pseudomonas aeruginosa. J Bacteriol 195:4735–4741. doi: 10.1128/JB.00903-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.