Abstract
The organic hydroperoxide stress resistance regulator (OhrR) is a MarR type of transcriptional regulator that primarily regulates the expression of organic hydroperoxide reductase (Ohr) in bacteria. In mycobacteria, the genes encoding these proteins exist in only a few species, which include the fast-growing organism Mycobacterium smegmatis. To delineate the roles of Ohr and OhrR in defense against oxidative stress in M. smegmatis, strains lacking the expression of these proteins were constructed by deleting the ohrR and ohr genes, independently and together, through homologous recombination. The OhrR mutant strain (MSΔohrR) showed severalfold upregulation of Ohr expression, which could be observed at both the transcript and protein levels. Similar upregulation of Ohr expression was also noticed in an M. smegmatis wild-type strain (MSWt) induced with cumene hydroperoxide (CHP) and t-butyl hydroperoxide (t-BHP). The elevated Ohr expression in MSΔohrR correlated with heightened resistance to oxidative stress due to CHP and t-BHP and to inhibitory effects due to the antituberculosis drug isoniazid (INH). Further, this mutant strain exhibited significantly enhanced survival in the intracellular compartments of macrophages. In contrast, the strains lacking either Ohr alone (MSΔohr) or both Ohr and OhrR (MSΔohr-ohrR) displayed limited or no resistance to hydroperoxides and INH. Additionally, these strains showed no significant differences in intracellular survival from the wild type. Electrophoretic mobility shift assays (EMSAs) revealed that the overexpressed and purified OhrR interacts with the ohr-ohrR intergenic region with a greater affinity and this interaction is contingent upon the redox state of the OhrR. These findings suggest that Ohr-OhrR is an important peroxide stress response system in M. smegmatis.
INTRODUCTION
All living organisms generate reactive oxygen species (ROS) as a by-product of metabolism. Accidental collision of O2 with flavoproteins of metabolic pathways initially produces superoxides (O2−) (1). These O2− are subsequently converted to other products, like hydrogen peroxide (H2O2), hydroxyl radicals (OH), and others, by enzymatic and nonenzymatic mechanisms (2). In addition to this inadvertent generation of O2− during metabolism, phagocytic cells of eukaryotes produce O2− through a special pathway for killing invading pathogens (3, 4). Human phagocytes also produce nitric oxide (NO), through inducible nitric oxide synthase, and this reacts with O2− to generate highly toxic peroxynitrites (—OONO) (5). Production of this compound by phagocytes is considered an important and effective way of combating the invading pathogens (6). Excess ROS can oxidize macromolecules, like nucleic acids, protein, carbohydrates, and lipids, which affect cellular processes of all types (7).
Two families of enzymes are associated with detoxification of alkyl and organic hydroperoxides in bacteria. The first one belongs to the peroxiredoxin family and is called alkyl hydroperoxide reductase (AhpC) (8, 9). It exists as two subunits, namely, AhpC and AhpF, in Salmonella enterica serovar Typhimurium and Escherichia coli, and the genes encoding these proteins are transcribed as a bicistronic unit (8, 9). AhpF is a flavoprotein, and it reduces the oxidized AhpC using NADPH (10). However, this AhpC-AhpF arrangement is restricted to only a few species of bacteria, and most species seem to have only the AhpC subunit. In these cases, thioredoxin (Trx) in lieu of AhpF is implicated in providing the reducing equivalents (11, 12). Although AhpC reduces H2O2, alkyl hydroperoxides, and organic peroxides into the corresponding alcohols, it also has the ability to reduce peroxynitrites (9, 13). Further, the absence of AhpC makes certain bacteria sensitive to alkyl hydroperoxides (9). AhpC in bacteria is regulated by a member of the LysR family of transcriptional regulators, OxyR, in response to peroxide stress (14). Consistent with this phenomenon, ahpC is divergently transcribed from oxyR in some mycobacteria, like Mycobacterium leprae, M. tuberculosis, and M. marinum, although oxyR is inactivated in M. tuberculosis due to several naturally occurring internal deletions and point mutations within the gene (15–17). However, the fast-growing M. smegmatis only has the gene for AhpC, and it completely lacks the gene for OxyR (18). In addition to regulation by OxyR, in some bacteria AhpC expression is also regulated by the PerR regulator (14), which is also absent in mycobacteria.
The other enzymes that can detoxify alkyl or organic peroxides belong to the Ohr/OsmC superfamily. Organic hydroperoxide reductase (Ohr) was first identified in Xanthomonas campestris (19), while osmotically inducible protein C (OsmC) was recognized in E. coli (20). Although these two proteins were detected independently of each other, they are now grouped under the same superfamily since they share structural and functional similarities (21–23). Several studies have demonstrated that purified proteins of both Ohr and OsmC from bacterial origins have the ability to reduce alkyl and organic hydroperoxides (22–28). Although these enzymes also reduce H2O2 in vitro (22, 24), bacterial mutants lacking Ohr and OsmC become hypersusceptible to organic peroxides but not to H2O2, indicating that both enzymes function only as organic hydroperoxide reductases in vivo (19, 29–32). However, a clear difference exists between these two proteins in terms of induction by organic peroxides and regulation. Studies have found that Ohr can be induced by organic hydroperoxides and sodium hypochlorite and OsmC can be induced by ethanol and sodium chloride (2, 21). Additionally, although Ohr is regulated by the organic hydroperoxide stress resistance regulator (OhrR) (2, 14), the regulator for the OsmC protein is yet to be discovered.
OhrR is a MarR type of transcriptional regulator that specifically senses and responds to organic hydroperoxides (2, 14, 33). In the absence of stress, OhrR represses ohr expression by binding tightly to its promoter region. Similar to many other redox-sensing proteins, cysteine residues in OhrR play a major role in sensing organic peroxide stress (34, 35). Oxidation of the cysteine residues of OhrR by organic peroxides reduces its binding with the ohr promoter, leading to derepression of Ohr expression (35–37). In addition to regulating ohr, OhrR has also been reported to regulate its own expression (38). OhrRs in bacteria also differ in the number of cysteine residues. While several species have a single cysteine residue at the N-terminal region, as in Bacillus subtilis (37, 39, 40), OhrR with two cysteines, one at the N terminus and the other at the C terminus, also exists, as noticed in Xanthomonas campestris (34, 35). Interestingly, in both cases, the N-terminal cysteine plays an important role in sensing the organic peroxides (2). Further, it has been reported that a single-cysteine OhrR converted into a two-cysteine OhrR forms an intermolecular disulfide bond, thus behaving functionally as a two-cysteine OhrR (41). Other transcriptional regulators of bacteria, such as OspR of Pseudomonas aeruginosa and MgrA and SarZ of Staphylococcus aureus, are considered paralogs of OhrR (2).
Although OsmC, Ohr, and OhrR are distributed in both Gram-negative and Gram-positive bacteria, only a few species from both groups possess them. Again, only selected species have all the genes coding for these proteins, and others have a gene(s) for either OsmC, Ohr-OhrR, or only Ohr (NCBI database). The gene encoding the OsmC protein is found in most species of mycobacteria, including M. tuberculosis and M. smegmatis. However, the genes for Ohr-OhrR are found in only a few species of mycobacteria, like M. smegmatis and M. abscessus (NCBI database). It is presumed that the presence of Ohr-OhrR in M. smegmatis, despite its poorly pathogenic nature, could bestow some special advantages to this bacterium. Moreover, the role of OhrR in mycobacteria has not been elucidated thus far, although attempts have been made to characterize Ohr and OsmC in M. smegmatis (24, 42). In this study, we examined the roles of M. smegmatis Ohr and OhrR and their contribution toward the resistance of this organism against organic hydroperoxide stress. Additionally, we report the relationship between Ohr expression and the sensitivity of M. smegmatis to the antituberculosis drug isoniazid (INH).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are given in Table 1. Middlebrook 7H9 broth or 7H10 agar plates supplemented with 0.05% Tween 80 (TW), 0.2% glycerol, and 10% albumin dextrose complex (ADC; 5 g bovine serum albumin, 2 g d-dextrose, and 0.85 g of NaCl per 100 ml) enrichment (7H9/7H10-TW-ADC) were used for growing M. smegmatis (mc2155). M. smegmatis strains bearing plasmids or mutant strains disrupted with an antibiotic resistance gene were grown in 7H9/7H10-TW-ADC containing the antibiotic kanamycin (25 μg/ml) or hygromycin (50 μg/ml), or both. E. coli strain DH5α (Invitrogen) was used for subcloning experiments, and E. coli strain BL21(DE3) (Invitrogen) was used to overexpress recombinant proteins. Luria-Bertani (LB) broth or LB agar with an appropriate antibiotic (100 μg/ml ampicillin, 25 μg/ml kanamycin, or 100 μg/ml of hygromycin) was used to grow E. coli bearing plasmids. All bacteria were grown at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | lacZΔM15 recA1 | Invitrogen |
| BL21(DE3) | DE3 λ prophage carrying the T7 RNA polymerase gene and lacIq | Invitrogen |
| M. smegmatis | ||
| mc2155 (MSWt) | Wild type | ATCC 27294 |
| MS206 | M. smegmatis wild-type strain harboring pMV206 | This study |
| MSohrE | M. smegmatis wild-type strain harboring pMVMSOHR2 | This study |
| MSΔohr | M. smegmatis ohr disrupted with Hygr marker | This study |
| MSΔohrR | M. smegmatis ohrR disrupted with Hygr marker | This study |
| MSΔohr-ohrR | M. smegmatis ohr-ohrR disrupted with Hygr marker | This study |
| MSΔohrR/c | MSΔohrR harboring plasmid pMVMSOHRR | This study |
| MSΔohr/c | MSΔohrR harboring plasmid pMVMSOHRR | This study |
| MSΔohr-ohrR/c | MSΔohr-ohrR harboring plasmid pMVMSOHRR | This study |
| MSΔahpC | M. smegmatis ahpC disrupted with Kmr marker | 56 |
| Plasmids | ||
| pCR2.1 | TA cloning vector; Apr Kmr | Invitrogen |
| p1NIL | Mycobacterial suicide vector | 44 |
| pET16b | Overexpression vector carrying an N- terminal His tag | Novagen |
| pMV206 | E. coli-Mycobacterium shuttle vector | 41 |
| pMSOHR | pCR2.1 vector harboring 2,053-bp ohr-ohrR region of M. smegmatis | This study |
| pMVMSOHRR | pMV206 containing 2,053-bp ohr-ohrR region of M. smegmatis | This study |
| pMVMSOHR2 | pMV206 vector containing ohr and its promoter region of M. smegmatis | This study |
| pNILMSOHR | p1NIL vector harboring 2,053-bp ohr-ohrR region of M. smegmatis. | This study |
| pNILMSOHRH1 | p1NIL vector harboring ohr-ohrR region with Hygr marker at NcoI site | This study |
| pNILMSOHRH2 | p1NIL vector harboring ohr-ohrR region with Hygr marker at StuI site | This study |
| pNILMSOHRH3 | p1NIL vector harboring ohr-ohrR region with Hygr marker at NcoI-StuI sites; it has also sacB and lacZ genes in the plasmid | This study |
| p16MSOHRREX | pET16b containing M. smegmatis ohrR coding region | This study |
| p16MSOHREX | pET16b containing M. smegmatis ohr coding region | This study |
DNA manipulations.
Genomic DNA from M. smegmatis strains was isolated by the cetyltrimethylammonium bromide (CTAB) method (43). Plasmids (Table 1) from E. coli were isolated using a QIAprep spin kit (Qiagen). DNA sequences for M. smegmatis genes were downloaded from the J. Craig Venter Institute (JCVI) database. Oligonucleotide primers for the amplification of genes and real-time reverse transcription-PCR (RT-PCR) (Table 2) were custom synthesized from Integrated DNA Technologies (IDT).
TABLE 2.
Oligonucleotide primers used in this study
| Name | Sequence (5′ → 3′) |
|---|---|
| MS448A | GTGTTCTTCACGGCTGCCTA |
| MS448B | GGACCGGTGTTGATCGCCTA |
| MS447EXF | GCGCATATGACCACTTCGACCGACAACGTGCT |
| MS447EXR | CGCGGATCCGTATCAGGCCGCAGTGACCGTGACGTC |
| MS448EXF | CGCCATATGAGCCCCCCACGTCTGAACCTGGCCG |
| MS448EXR | CGCGGATCCGATCAGCGAGTGGCGCGGACGTGCTCAC |
| MS448IGF | GATGCCGCGCTGGTCCAGCAGCGG |
| MS448IGR | GGTCATTGCCGTCTCCTGTTCATA |
| MS447RT1 | TGATCAGGCCGTATCAGG |
| MS447RT2 | GTCTCAACGTCACCATCCC |
| MSOXYSU | GTTCCAGAGCACGATGTCGCCGGCCTTGTC |
| MSOXYSRT | ACGTAGCAGACCTCGGCAGCCCTGGCGAAG |
Disruption plasmids and screening for mutants.
To disrupt ohr (MSMEG_0447), ohrR (MSMEG_0448), or both genes in M. smegmatis, we first synthesized two oligonucleotide primers, namely, MS448A and MS448B. Using these primers, a 2,053-bp DNA fragment encompassing both the ohr and ohrR regions of M. smegmatis was amplified and cloned in the pCR2.1 vector (Invitrogen) to create plasmid pMSOHR. The fragment was then released by cutting the plasmid with EcoRI and ligated to the p1NIL vector (44), cut with the same enzyme, to create plasmid pNILMSOHR. Thereafter, a 1,650-bp fragment (BamHI-PstI blunt ended with the Klenow fragment) containing the hygromycin resistance gene obtained from plasmid pIJ963 (45) was independently inserted at the NcoI or StuI site to obtain plasmids pNILMSOHRH1 and pNILMSOHRH2, respectively. These plasmids were used to generate ohr (MSΔohr) and ohrR (MSΔohrR) mutants of M. smegmatis, respectively. In addition to the procedures described above, a third plasmid, pNILMSOHRH3, was constructed to create a double mutant of M. smegmatis lacking both Ohr and OhrR (MSΔohr-ohrR). This was done in two steps. Initially, a 400-bp NcoI-StuI fragment in plasmid pNILMSOHR was deleted and the hygromycin resistance gene was cloned in its place. Thereafter, a DNA fragment from pGOAL17, consisting of the sacB and lacZ genes, encoding sucrase and β-galactosidase, respectively (44), was added to the PacI site of this plasmid to result in pNILMSOHRH3. M. smegmatis strain MSWt was grown to mid-logarithmic phase (optical density at 600 nm [OD600] = 0.8 to 1.0) in 7H9 broth, and competent cells were prepared as previously described (46). Four hundred to 500 microliters of competent cells in 0.2-mm cuvettes was mixed with 2 to 3 μg of pNILMSOHRH1, pNILMSOHRH2, or pNILMSOHRH3 and electroporated. After electroporation, 1 ml 7H9 medium without any antibiotic was added to each cuvette, and the cuvettes were left for 3 h at 37°C. Then, the cell suspension from the cuvettes was plated on 7H10 agar plates containing the antibiotic hygromycin (50 μg/ml) (for plasmids pNILMSOHRH1 and pNILMSOHRH2) or hygromycin (50 μg/ml) and kanamycin (25 μg/ml) (for plasmid pNILMSOHRH3). Transformants obtained with plasmids pNILMSOHRH1 and pNILMSOHRH2 were screened for hygromycin resistance and kanamycin sensitivity to select colonies resulting from double-crossover events. In the case of transformants resulting from plasmid pNILMSOHRH3, a two-step screening method involving selection of transformants for sucrose and hygromycin resistance, as described by Parish and Stoker (44), was followed. Disruption of the genes was finally confirmed by examining the chromosomal DNA of the mutants by Southern hybridization.
Southern hybridization.
Southern hybridization was performed to confirm the recombination events at the ohr and ohrR genes. Chromosomal DNA from M. smegmatis strain MSWt and the suspected mutants was digested with StuI, and the fragments were separated on 1% agarose gels. The DNA fragments were transferred to Zeta probe membranes (Bio-Rad) by Southern blotting. After cross-linking the DNA with UV treatment, membranes were prehybridized for 4 h in a prehybridization solution (50% formamide, 0.12 M Na2HPO4, 0.25 M NaCl, 7% [wt/vol] sodium dodecyl sulfate [SDS], 1 mM EDTA). The membranes were later hybridized with a [α-32P]dCTP-labeled DNA probe encompassing the ohr and ohrR regions (2,053 bp) of M. smegmatis. After hybridization, the membranes were washed three times at 42°C in the following sequence: with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.1% SDS for 15 min, 0.5× SSC with 0.1% SDS for 15 min, and 0.1× SSC with 0.1% SDS for 15 min. The membranes were then exposed to X-ray film for autoradiography.
Complementation of mutant strains.
To complement the MSΔohr, MSΔohrR, or MSΔohr-ohrR mutant strain with functional genes, we constructed a mycobacterial plasmid carrying the ohr and ohrR region of M. smegmatis. For this, plasmid pMSOHR, described above, was cut with EcoRI, and the released fragment was cloned into plasmid pMV206 (47) at the EcoRI site. The resulting plasmid, pMVMSOHRR, was transformed into the MSΔohr, MSΔohrR, or MSΔohr-ohrR mutant strain by electroporation to obtain the complemented strains MSΔohr/c, MSΔohrR/c, and MSΔohr-ohrR/c, respectively. In addition, we created a plasmid called pMVMSOHR2 to create a M. smegmatis wild-type strain (MSohrE) expressing Ohr through plasmid-borne ohr. This was created by cloning a blunt-ended StuI-EcoRV fragment obtained from pMSOHR into the HpaI site of pMV206.
Real-time RT-PCR.
To determine the transcript levels of ohr, real-time PCR was performed. Total RNA from M. smegmatis strains was isolated using the TRI Reagent, and cDNAs were synthesized as described previously (48). They were subjected to real-time RT-PCR using internal primers (MS447RT1 and MS447RT2; Table 2), which amplify approximately 183 bp. Sso Fast EvaGreen Supermix (Bio-Rad) was used for detection of the transcripts. The PCR had 40 cycles of 95°C for 5 s, 55°C for 10 s, and 72°C for 5 s and was performed in a Bio-Rad iQ5 multicolor real-time PCR detection system. Threshold cycle (CT) values in the exponential phase of amplification were used to determine the transcript levels, and the results were expressed as the relative expression of the genes. To rule out the possibility of primer dimers, a meltdown curve was performed in the same PCR program with a linear increase of the temperature by 1°C from 55°C to 95°C and a hold for 30 s per step. Primers for real-time RT-PCR were checked for a single peak in the meltdown curve.
Preparation of crude cell lysates and SDS-PAGE.
Cell pellets from the mutant and wild-type M. smegmatis strains (OD600 = 1.0) were resuspended in phosphate buffer (50 mM, pH 7.4) containing NaCl (50 mM), washed once with the same buffer, and disrupted with a cell disruptor. The total protein content of the extracts was determined by the bicinchoninic acid (Pierce) method, and the extracts were mixed with an equal volume of 2× SDS-polyacrylamide gel electrophoresis (PAGE) sample loading buffer. Proteins were separated on 12% precast bis-Tris gels (Invitrogen). Gels were stained with Coomassie blue R-250 to visualize the proteins.
Metabolic labeling of proteins.
Metabolic labeling of proteins of the M. smegmatis strains was performed by adding 10 μCi of [35S]methionine and [35S]cysteine (Expre35S35S protein labeling mix; 1,000 Ci/mmol; PerkinElmer) to a 1-ml culture of M. smegmatis grown to an OD600 of 0.2. Hydrogen peroxide (H2O2; 200 μM), 125 μM cumene hydroperoxide (CHP), or 200 μM t-butyl hydroperoxide (t-BHP) was added when required. After 1 h of incubation at 37°C, the cultures were centrifuged and washed once with 20 mM Tris-HCl (pH 7.5). Bacterial pellets resuspended in 100 μl of the same buffer were mixed with 20 μl of zirconium beads (diameter, 0.1 to 0.15 mm; BioSpec Products), and cells were broken using a mini-bead beater for 2 min at 2,800 rpm. The beads and cell debris were removed by centrifugation, and the resulting supernatant was mixed with an equal volume of 2× SDS-PAGE loading buffer and analyzed on 11% SDS-polyacrylamide gels (43). The gels were dried and subjected to autoradiography.
Overexpression of Ohr and OhrR.
To overexpress the recombinant M. smegmatis Ohr and OhrR proteins, the ohr and ohrR genes were PCR amplified using primers MS447EXF and MS447EXR and primers MS448EXF and MS448EXR, respectively, and were cloned into pET16b (Novagen). The respective plasmids, p16MSOHREX and p16MSOHRREX, were then transformed into E. coli strain BL21(DE3) (Invitrogen). Log-phase cultures of the resulting strains induced by IPTG (isopropyl-β-d-thiogalactopyranoside) showed the overexpression of proteins at about 17.0 kDa in SDS-polyacrylamide gels. The cells were sonicated and separated into soluble and insoluble fractions by centrifugation, and it was found that both the Ohr and OhrR proteins were retained in the soluble fractions. They were subjected to affinity purification (Ni-nitrilotriacetic acid), and the purified recombinant proteins showed a single band by SDS-PAGE. They were designated His10Ohr and His10OhrR, respectively.
Peroxide reductase activity.
The peroxide reductase activity of the crude cell lysates of the M. smegmatis strains, prepared using a bead beater as described above, and the purified recombinant His10Ohr was determined by the colorimetric ferrous ion oxidation in xylenol orange (FOX) assay (35). Incubation mixtures (total volume, 100 μl) contained the following: 300 μM CHP or t-BHP substrate for reactions involving cell lysates or 500 μM CHP, t-BHP, or H2O2 substrate for reactions with the His10Ohr protein, 1 mM dithiothreitol (DTT), and 50 mM HEPES buffer, pH 7.4. The reactions were initiated by the addition of cell lysate or His10Ohr, which was preincubated with 5 mM DTT for 1 h on ice. The reactions were terminated at different time intervals by adding 200 μl of ice-cold FOX reagent [25 mM ammonium ferrous(II) sulfate, 2.5 M H2SO4, 125 μM xylenol orange] (49). The absorbance of the reactions was measured at 560 nm. The amount (μM) of peroxide reduced in each reaction was calculated using an appropriate standard curve.
Catalase assay.
The catalase activity of the crude cell lysates of the M. smegmatis strains was determined using an OxiSelect catalase activity assay kit (catalog number STA-341; Cell Biolabs, Inc.). Each reaction mixture had 1 mM H2O2 and 0.2 mg of the cell lysate protein. The reactions were arrested after 1 min, and the absorbance at 520 nm was measured. The catalase activity was determined by calculating the amount (nM) of H2O2 reduced using a standard curve generated with the catalase standard supplied with the kit.
EMSA.
To detect the interaction between OhrR and the ohr-ohrR intergenic region, we performed an electrophoretic mobility shift assay (EMSA). DNA probes for EMSA were generated by PCR. For the M. smegmatis ohrR intergenic region, a 270-bp DNA fragment amplified by primers MS448IGF and MS448IGR (Table 2) was used as a probe. For the M. smegmatis oxyS intergenic region, a 210-bp DNA fragment amplified by primers MSOXYSU and MSOXYSRT (Table 2) was used as a probe. Genomic DNA of M. smegmatis was used as the template for the generation of probe DNA, and PCR was performed in the presence of [γ-32P]dCTP (6,000 Ci/mmol; PerkinElmer). Additionally, unlabeled DNA fragments for competition assays were generated by PCR. All PCR-generated probes or DNA fragments were purified (PCR purification kit; Qiagen) before use. DNA probes (20,000 cpm) were incubated with M. smegmatis His10OhrR (0 to 100 nM) in 20 μl binding buffer (20 mM Tris-HCl, pH 8.0, 50 mM KCl, 1 mM EDTA, 50 μM bovine serum albumin) at room temperature for 10 min. For specific experiments, purified OhrR was treated with H2O2, CHP, or t-BHP with or without DTT; and for competitive binding, the reaction mixtures also contained competitor DNA (unlabeled probe DNA). The reaction mixtures were incubated at room temperature for 10 min, and the products were separated on 6% native polyacrylamide gels using Tris-borate buffer for 2 h. The gels were then dried and autoradiographed.
Disk inhibition assay.
The sensitivity of the M. smegmatis strains to peroxides or INH was determined by a disk inhibition assay. M. smegmatis MSWt, MSΔohr, MSΔohrR, MSΔohr-ohrR, MS206, MSohrE, MSΔahpC, and complemented mutant strains (MSΔohr/c, MSΔohrR/c, and MSΔohr-ohrR/c) were cultured overnight in 7H9 broth and diluted to an OD600 of 0.6. Two hundred microliters of each culture was mixed with 3 ml of 7H10 soft agar and plated onto 7H10 agar plates containing the appropriate antibiotic(s). Sterile filter paper disks saturated with 10 μl of H2O2, t-BHP (0.25 M), CHP (0.15 M), or INH (1 mg/ml) were placed onto the hardened top agar, and the plates were incubated at 37°C for 48 h. The diameters of the zones of growth inhibition were recorded and analyzed statistically.
MIC.
Overnight cultures of the M. smegmatis wild-type, mutant, and complemented strains were diluted with fresh medium to an OD600 of 0.3. Thirty microliters of these cultures was inoculated into 3 ml of medium containing INH at different concentrations. Growth was assessed after 3 to 4 days of incubation at 37°C.
Survival of ohr and ohrR mutants in macrophages.
THP-1 cells (TIB-202) were purchased from the American Type Culture Collection (ATCC) and cultured in RPMI medium with 10% fetal bovine serum (HyClone) in a 37°C humid chamber with 5% CO2. Cells from confluent growth were harvested, plated in a 6-well plate (1 × 106 cells/well), and differentiated with phorbol-12-myristate-13-acetate (PMA) for 72 h. M. smegmatis strains from log-phase cultures grown on 7H9 medium were collected by centrifugation and washed three times with sterile 1× phosphate-buffered saline (PBS). The bacterial pellets, resuspended in PBS, were passed through a 23-gauge syringe. Bacteria of each strain (multiplicity of infection, 1:10) were used to infect the THP-1 cells for 4 h. Afterwards, the THP-1 cells were washed three times with sterile warm PBS, and the plates were filled with RPMI and incubated for different times. The cells were lysed with PBS containing SDS (0.004%), and the lysates were appropriately diluted, plated onto 7H10 agar plates, and incubated at 37°C for 3 to 4 days before the bacterial colonies were counted.
Statistical analysis.
A paired t test was performed using GraphPad Prism (version 6) software.
RESULTS
Disruption of ohr-ohrR genes in the chromosome of M. smegmatis.
The annotated M. smegmatis genome revealed that the MSMEG_0447 and MSMEG_0448 genes encode the antioxidant Ohr and the transcriptional regulator OhrR, respectively (Fig. 1A). The ohrR gene is divergently transcribed from ohr, as noticed in other bacterial species, like Agrobacterium tumefaciens (50), Sinorhizobium meliloti (51), and Brucella abortus (52). To elucidate their functions in the regulation of oxidative stress, we sought to create mutant strains lacking these proteins through homologous recombination. Using the disruption plasmids pNILMSOHRH1, pNILMSOHRH2, and pNILMSOHRH3 and methods for disruption of genes described in the Materials and Methods section, we obtained strains with disrupted ohr, ohrR, and both ohr and ohrR, respectively. These strains were assessed for disrupted genes by probing the chromosomal DNA by Southern hybridization with radioactively labeled DNA (2,053 bp) encompassing the ohr and ohrR regions of M. smegmatis. The signals obtained were of the expected sizes of disrupted ohr (3,309 and 4,454 bp), ohrR (7,700 bp), and ohr-ohrR (7,300 bp) due to the insertion of the hygromycin resistance gene cassette at the ohr, ohrR, and ohr-ohrR sites, respectively (Fig. 1B). This confirmed the generation of MSΔohrR, MSΔohr, and MSΔohr-ohrR mutant strains.
FIG 1.
Creation of M. smegmatis ohr and ohrR mutant strains. MSWt, MSΔohr, MSΔohrR, and MSΔohr-ohrR are the wild-type strain and the ohr mutant, ohrR mutant, and ohr-ohrR mutant of M. smegmatis, respectively. (A) Schematic representing the organization of the ohrR and ohr genes in the genomes of the M. smegmatis strains. Arrows, genes and their orientations; numbers within and below the arrows, the number assigned to each gene; StuI and NcoI, restriction sites in the region; Hygromycin, the location of the antibiotic resistance marker gene in MSΔohr, MSΔohrR, and MSΔohr-ohrR. (B) Southern analysis of genomic DNA. Chromosomal DNA from the M. smegmatis strains digested with StuI was probed with a radioactively labeled DNA (2,053 bp) encompassing the ohr and ohrR regions of M. smegmatis. The sizes of the signals are marked on the right.
Ohr is inducible by organic hydroperoxides and is upregulated in MSΔohrR.
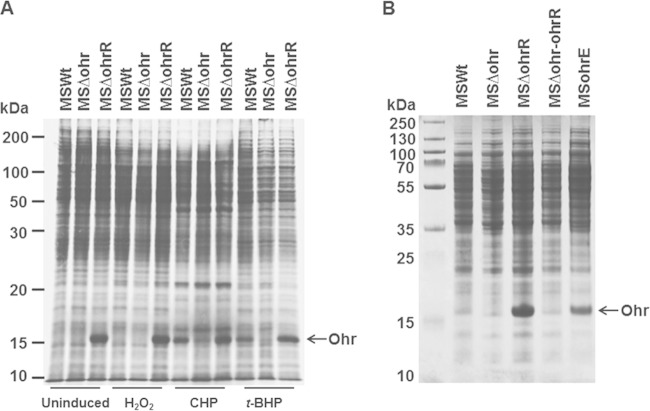
Previously, it was demonstrated that M. smegmatis responds to oxidative stress due to H2O2, t-BHP, and CHP and upregulates the expression of several proteins, including AhpC (18). To understand whether Ohr is one of these oxidative stress response proteins (OSPs), we analyzed the proteins induced by oxidative stress in MSΔohrR, using metabolic labeling, and compared them with the profiles of similarly labeled proteins of MSWt and MSΔohr strains. Figure 2A shows that the approximately 15-kDa protein was upregulated severalfold in samples from MSΔohrR, regardless of induction by oxidative stress. In contrast, a band representing this protein was completely absent in samples from MSΔohr, although this was noticed only in MSWt samples induced with CHP and t-BHP and not in MSWt samples induced with H2O2. This upregulated protein could be visualized by Coomassie blue staining of proteins derived from MSΔohrR and MSohrE, an M. smegmatis strain that bears plasmid-borne ohr, but not in MSΔohr or MSΔohr-ohrR (Fig. 2B). Further, this protein corresponded to the Osp16/A protein reported in a previous study (18) by size and also induction only by the organic peroxides CHP and t-BHP. These results suggest that (i) the 15-kDa upregulated protein is Ohr and it is under the tight control of OhrR and (ii) Ohr and the previously identified Osp16/A are the same proteins.
FIG 2.
Protein profiles showing the expression of Ohr. All strains except MSohrE are described in the legend to Fig. 1. MSohrE is a wild-type M. smegmatis strain expressing Ohr through a plasmid. (A) Metabolic labeling of proteins. M. smegmatis was labeled with [35S]Met and [35S]Cys for 1 h at 37°C, as described in Materials and Methods, separated by SDS-PAGE, and analyzed by autoradiography. Uninduced, no peroxide treatment; H2O2, treatment with 200 μM hydrogen peroxide; CHP, treatment with 125 μM cumene hydroperoxide; t-BHP, treatment with 200 μM t-butyl hydroperoxide. (B) Coomassie blue-stained proteins of M. smegmatis strains. Proteins from the M. smegmatis MSWt, MSΔohr, MSΔohrR, and MSohrE strains were separated on 12% SDS-polyacrylamide gels and stained with Coomassie blue. Each lane contained 100 μg of protein. Note the increased expression of an ∼15-kDa protein in strains MSΔohrR and MSohrE and its absence in strains MSΔohr and MSΔohr-ohrR.
Enhanced reduction of organic hydroperoxides by MSΔohrR.
Since deletion of ohrR leads to the increased production of Ohr, we presumed that this increase would be reflected in the total organic peroxide reductase activity of M. smegmatis. To verify this, we determined the peroxide reductase activity in the total protein extracts of M. smegmatis wild-type and mutant strains. As predicted, the activity was significantly higher in strain MSΔohrR than the other related strains (MSWt, MSΔohr, and MSΔohr-ohrR) when CHP and t-BHP were used as the substrates (Fig. 3A). The specificity of Ohr of M. smegmatis for the reduction of organic peroxides was further confirmed using recombinant Ohr (His10Ohr) (see Fig. S1A in the supplemental material). It showed the efficient reduction of organic peroxides (CHP and t-BHP), although it exhibited some low levels of activity with H2O2 (see Fig. S2 in the supplemental material). This is similar to the observations made with Ohr of Xylella fastidiosa (26) and is consistent with its predicted role as an organic hydroperoxide reductase. In addition, real-time RT-PCR revealed that the amounts of the mRNA transcripts for ohr in the MSΔohrR strain were severalfold higher than those in wild-type M. smegmatis (MSWt), providing support to the results obtained with enzyme assays (Fig. 3B).
FIG 3.
Peroxide reductase activities and ohr transcripts in the mutants. The strains are described in the legend to Fig. 1. (A) Peroxide reductase assay with whole-cell lysates. Each reaction mixture contained 300 μM the substrate CHP or t-BHP and 0.2 mg of the total cell lysate protein. P values are shown for the groups compared. The values for strain MSΔohrR are significantly lower than the values for strain MSWt, indicating the increased reduction of peroxides. (B) Real-time RT-PCR analysis. cDNA synthesized from total RNA isolated from strains MSWt, MSΔohr, and MSΔohrR was used as the template for real-time RT-PCR. The reaction was performed using primers MS447RT1 and MS447RT2 for amplifying the M. smegmatis ohr gene.
MSΔohrR resists killing by oxidants.
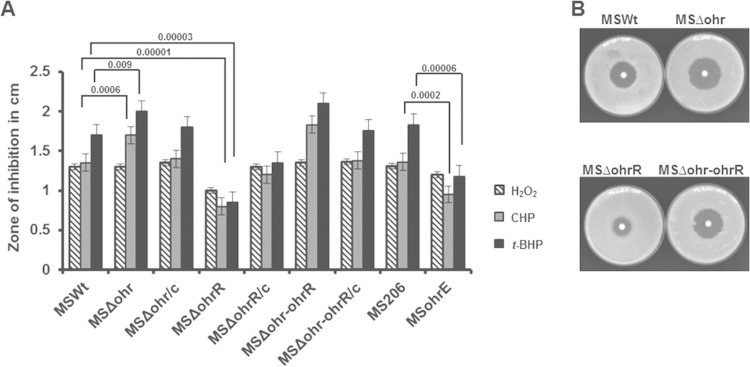
Previous studies in X. campestris and P. aeruginosa have indicated that Ohr plays a critical role in protecting these organisms against organic hydroperoxide toxicity (29, 53). To elucidate its role in M. smegmatis, we examined the responses of the ohr (MSΔohr), ohrR (MSΔohrR), and ohr-ohrR (MSΔohr-ohrR) mutant strains to peroxide stress in a disk inhibition assay. The results obtained are shown in Fig. 4A and B. While the strains lacking Ohr or both Ohr and OhrR showed a slightly increased sensitivity to organic peroxides, the strain lacking OhrR displayed increased resistance to H2O2 and hydroperoxides (CHP and t-BHP). Further, the levels of resistance to peroxides of the MSΔohr, MSΔohrR, and MSΔohr-ohrR strains could be restored to wild-type (MSWt) levels by complementing these strains with a functional gene(s). Similar to the MSΔohrR strain, the MSohrE strain that overexpresses Ohr through a plasmid-borne ohr also displayed increased resistance to hydroperoxides (CHP and t-BHP) compared with the level of resistance of its control strain, MS206. This reiterates that the observed resistance of strain MSΔohrR to peroxides is only due to elevated levels of Ohr in this strain. These results may suggest that Ohr of M. smegmatis has the ability to mount strong resistance to oxidative stress, particularly to organic hydroperoxide stress.
FIG 4.
Sensitivity of M. smegmatis ohr-ohrR mutants to peroxides. MSWt, an M. smegmatis wild-type strain; MSΔohr, an M. smegmatis ohr mutant strain; MSΔohr/c, an M. smegmatis ohr mutant complemented with functional ohr using plasmid pMVMSOHRR; MSΔohrR, an M. smegmatis ohrR mutant strain; MSΔohrR/c, an M. smegmatis ohrR mutant complemented with functional ohrR using plasmid pMVMSOHRR; MSΔohr-ohrR, an M. smegmatis ohr-ohrR mutant strain; MSΔohr-ohrR/c, an M. smegmatis ohr-ohrR mutant complemented with functional ohr-ohrR using plasmid pMVMSOHRR; MS206, an M. smegmatis wild-type strain bearing plasmid pMV206; MSohrE, an M. smegmatis wild type bearing plasmid pMVMSOHR2. (A) Growth-inhibitory effects of peroxides in a disk inhibition assay. Each disk received 10 μl of either 250 mM H2O2, 250 mM t-BHP, or 150 mM CHP. The bars show the diameters of the zones of inhibition. P values are shown for the groups compared. Values are significantly lower/higher than the values obtained for strain MSWt. (B) Disk inhibition on plates. Agar plates showing growth inhibition by CHP are shown for a few representative strains.
MSΔohrR resists inhibition by the antituberculosis drug INH.
Expression of antioxidants in M. tuberculosis and other mycobacteria has a strong relationship with sensitivity/resistance to the antituberculosis drug INH (54). For example, catalase-peroxidase (KatG) activity is required for the action of INH in M. tuberculosis, and M. tuberculosis strains lacking KatG expression are resistant to INH (55). On the contrary, a mutant M. smegmatis strain lacking AhpC, an alkyl hydroperoxide reductase that has the ability to reduce both peroxides and organic peroxides, has been shown to be highly sensitive to INH (56). This prompted us to test the strains lacking Ohr (MSΔohr), OhrR (MSΔohrR), or both (MSΔohr-ohrR) for their sensitivity/resistance to INH. The strain that lacks AhpC (MSΔahpC) was used as a control. It was noticed that the absence of Ohr (MSΔohr) or both Ohr and OhrR (MSΔohr-ohrR) in M. smegmatis had little effect on sensitivity to INH, although the constitutive expression of Ohr (MSΔohrR) had a significant effect (Fig. 5A and B). The MSΔohrR strain showed increased resistance to inhibition by INH compared to the level of resistance of the wild type (MSWt), and this effect was reversed in the complemented strain (MSΔOhrR/c). Determination of the MICs for these strains also revealed a similar pattern of sensitivity or resistance to INH (Fig. 5C). Further, to demonstrate the specificity of the role of Ohr in INH resistance, we tested the wild-type strain expressing elevated amounts of Ohr through a plasmid-borne ohr (MSohrE). Similar to MSΔohrR, this strain also displayed increased resistance to INH compared with the level of resistance of its parent strain, albeit at a level relatively lower than that of MSΔohrR, indicating that the observed resistance to INH is due to Ohr expression. In addition, it was thought that deletion of ohr or ohrR might alter the expression of catalase in these strains, which in turn may affect the sensitivity of these mutants to INH. To clarify this, we determined the catalase activity of these strains (Fig. 5D). The Ohr mutant (MSΔohr), OhrR mutant (MSΔohrR), and Ohr-OhrR (MSΔohr-ohrR) double mutant strains showed no significant change in catalase activity compared to that of wild-type M. smegmatis. However, the AhpC mutant (MSΔahpC) had relatively elevated levels of catalase activity. This increase may be due to a compensatory relationship between AhpC and KatG. The existence of such a relationship has been reported earlier with clinical isolates of M. tuberculosis and M. bovis lacking KatG. These isolates exhibit increased levels of AhpC to compensate for the low level of KatG expression, and this increase was observed to be due to mutations in the promoter region of ahpC (18, 57). Taken together, these results suggest that Ohr, like KatG, may have a role in the determination of mycobacterial susceptibility and resistance to INH.
FIG 5.
Sensitivity of M. smegmatis strains to INH. All strains except MSΔahpC are described in the legend to Fig. 4. MSΔahpC is an M. smegmatis ahpC mutant strain. (A) Disk inhibition assay. Each disk received 10 μl of INH (1 mg/ml). P values are shown for the groups compared, and the values are significantly lower/higher than the values obtained for strains MSWt and MSΔohrR/c. (B) Disk inhibition on plates. Agar plates showing growth inhibition by INH are shown for a few representative strains. (C) MICs of INH. The MICs of INH were determined by the tube dilution method, and the lowest dilution of isoniazid inhibiting the growth of the M. smegmatis strains was considered the MIC. (D) Catalase activity with whole-cell lysates. Each reaction mixture contained 1 mM the substrate H2O2 and 0.2 mg of the total cell lysate protein. The reactions were stopped after 1 min by addition of 50 μl of a catalase quencher, and the absorbance was read at 520 nm.
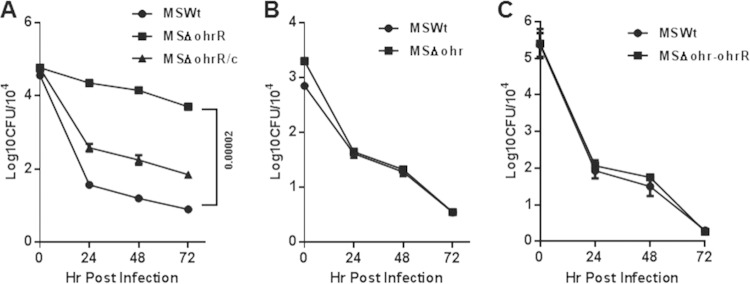
Enhanced survival of MSΔohrR in macrophages.
The observation that the ohrR deletion in M. smegmatis leads to increased resistance to organic hydroperoxides in vitro indicated the possibility that MSΔohrR resists intracellular oxidative stress and, thus, will have enhanced intracellular survival within macrophages. To test this hypothesis, we analyzed the survival of this strain along with that of MSWt and MSΔohrR/c in THP-1 macrophages at different time points. Figure 6A shows that MSΔohrR has significantly increased survival within the macrophages at 24, 48, and 72 h compared to that of MSWt, as expected, and this effect could be reversed by complementing this strain with a functional OhrR (MSΔohrR/c). In contrast, however, the MSΔohr and MSΔohr-ohrR strains (Fig. 6B and C) showed no significant difference in intracellular survival compared with that of MSWt, indicating that the constitutive expression of Ohr, due to the absence of OhrR, produces a better effect than the basal level or lack of Ohr expression in M. smegmatis MSΔohr and MSΔohr-ohrR, respectively. Furthermore, strain MSohrE, which overexpresses Ohr through a plasmid-borne ohr, also displayed a profile more or less similar to that of MSΔohrR with regard to intracellular survival (data not shown), thus providing additional support for Ohr's role in intracellular survival.
FIG 6.
Survival of M. smegmatis strains in macrophages. Human THP-1 macrophages (preactivated with PMA) were infected with M. smegmatis strains. The strains are described in the legend to Fig. 4. After 4 h of infection, the plates were washed and incubated for 0, 24, 48, and 72 h for determination of viable colony counts (numbers of CFU). The cells were lysed, diluted, and plated onto 7H10 agar plates, and the plates were incubated at 37°C for 3 to 4 days before the bacterial colonies were counted. The infection was performed in triplicate, and each dilution was plated on 3 different plates. The entire set of experiments was repeated once. The error bars show the mean plate counts ± SD (numbers of CFU) (analyzed using GraphPad Prism [version 6] software). (A to C) Survival of the MSΔohrR (A), MSΔohr (B), and MSΔohr-ohrR (C) strains in THP-1 macrophages.
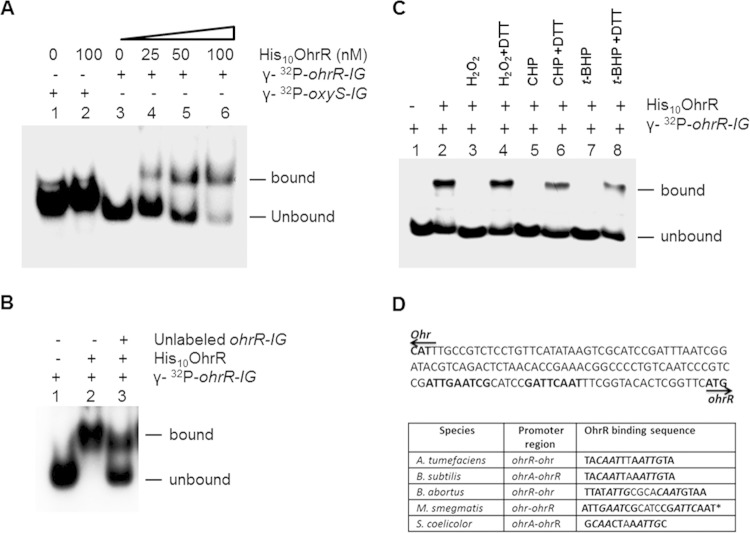
OhrR binds to the ohr-ohrR intergenic region.
OhrR is a transcriptional repressor that binds to the promoter regions of ohr and ohrR to regulate their expression in response to organic hydroperoxide stress (2). To delineate this relationship, we investigated whether recombinant M. smegmatis OhrR (His10OhrR) (see Fig. S1B in the supplemental material) interacts with the ohr-ohrR intergenic region (ohrR-IG), which has promoters for both ohr and ohrR, by EMSA. It was noticed that the addition of increasing concentrations of purified His10OhrR progressively affected the migration of the probe DNA (Fig. 7A), indicating that the interaction occurs with a high affinity in a dose-dependent manner. To further understand the specificity of this interaction, we used an unrelated promoter region from the oxyS gene (oxyS-IG) of M. smegmatis as a probe. We also used unlabeled ohrR-IG as a specific competitor in EMSAs. While the nonspecific oxyS-IG showed no binding with OhrR, the unlabeled ohrR-IG strongly inhibited the binding (Fig. 7B), suggesting that the interaction between OhrR and ohrR-IG is highly specific. Further, previous studies have indicated that the ability of OhrR to bind with the ohrR promoter region is affected by oxidants (34, 51, 52). To investigate whether or not OhrR of M. smegmatis behaves in a similar fashion, we added H2O2, CHP, and t-BHP separately to the binding reaction mixtures. Results presented in Fig. 7C show that addition of these oxidants completely abolished the interaction of ohrR-IG with His10OhrR and that this effect of oxidants could be completely reversed by adding 100 mM the reducing agent DTT to the binding reactions. These results may indicate that addition of oxidants to the binding reactions oxidizes His10OhrR, and this effect is countered by DTT. Furthermore, studies have reported that OhrR interacts with the promoter regions of ohr-ohrR through a conserved AT-rich inverted repeat region, TACAATT-Nv-AATTGTA (where v represents a variable number of amino acids) (50, 52, 58, 59). To understand if ohr-ohrR of M. smegmatis has a similar motif, we analyzed this region for inverted repeats. This identified a repeat region with the nucleotide sequence ATTGAATCG-N4-CGATTCAAT, which showed some nucleotide sequence identity with already reported sequences (Fig. 7D). Overall, these results suggest that OhrR of M. smegmatis has mechanisms similar to those of OhrR of other bacterial species.
FIG 7.
Interaction of the OhrR protein of M. smegmatis with the ohr-ohrR intergenic region. (A) Dose-dependent binding of His10OhrR with the ohr-ohrR intergenic region in EMSAs. Lanes 1 and 2, oxyS intergenic region DNA of M. smegmatis labeled with [γ-32P]dCTP (γ-32P-oxyS-IG); lanes 3, 4, 5, and 6, ohr-ohrR intergenic region DNA of M. smegmatis labeled with [γ-32P]dCTP (γ-32P-ohrR-IG). DNA was incubated in the presence (+) or in the absence (−) of increasing concentrations (0 to 100 nM) of His10OhrR. (B) Competitive binding assay with unlabeled DNA. The reaction mixture contained labeled ohr-ohrR intergenic region DNA (γ-32P-ohrR-IG) of M. smegmatis in all lanes (lanes 1 to 3) and 100 nM His10OhrR in lanes 2 and 3. Additionally, lane 3 had unlabeled ohr-ohrR intergenic region DNA of M. smegmatis. (C) Effects of oxidants on OhrR binding. Labeled probes (γ-32P-ohrR-IG) were incubated with 100 nM purified His10OhrR for 10 min at room temperature. DTT was added to the reaction mixture as indicated, and the mixture was further incubated for 10 min. Lanes 1 and 2, controls without oxidants; lanes 3 and 4, with 200 μM H2O2; lanes 5 and 6, with 200 μM CHP; and lanes 7 and 8, with 200 μM t-BHP. (D) Schematic showing the putative binding site of OhrR. (Top) The nucleotide sequences of the ohr-ohrR intergenic region of M. smegmatis. Arrows, the direction of transcriptions of ohr and ohrR; nucleotide sequences in bold, inverted repeats. (Bottom) The nucleotide sequences of OhrR binding sites in different bacterial species. Letters in bold, inverted repeats; letters in bold italics, regions showing identity between different species; *, the sequences identified are putative.
DISCUSSION
In this study, the deletion of ohrR was found to increase the upregulation of the Ohr protein severalfold, which could be observed even in Coomassie blue-stained SDS-polyacrylamide gels. This is somewhat surprising because no previous study has reported a similar expression of Ohr at the protein level in ohrR mutants of bacteria, although transcription-based studies have reported relatively higher levels of expression of ohr in A. tumefaciens (50), P. aeruginosa (60), and B. abortus (52). The observed higher Ohr levels in the ohrR mutant of M. smegmatis does not appear to be due to the stability of this protein because the upregulation is associated with a concomitant increase in mRNA levels. Further, the observations that Ohr of M. smegmatis can be induced only by alkyl or organic hydroperoxides and not by H2O2 are similar to those noticed for other bacterial species, like B. subtilis (59), P. aeruginosa (29), Streptomyces coelicolor (58), A. tumefaciens (50), Sinorhizobium meliloti (51), and Chromobacterium violaceum (34) and suggest that OhrR is a sensor for organic hydroperoxide stress in M. smegmatis. However, the OhrR of M. smegmatis is different from the OhrR of Shewanella oneidensis, a recently described protein that responds to both organic and H2O2 peroxides (61). Considering the fact that M. smegmatis lacks the well-known peroxide stress response regulator OxyR, it is very likely that OhrR may partly substitute for the absence of OxyR in this species for dealing with organic peroxide stress. It may be noted that AhpC expression was also induced in M. smegmatis by CHP and t-BHP (18), and what regulates the expression of AhpC is still unknown. Also, it is unclear at present if OhrR regulates the expression of AhpC or other antioxidants in M. smegmatis. Thus, the scenario of peroxide stress response in M. smegmatis is expected to be very different from that in other mycobacteria which have a functional OxyR.
Induction of Ohr in M. smegmatis has previously been noticed in a transposon mutant that had a disruption in the gene mshA (42), which is associated with mycothiol biosynthesis. It was suggested that MshA might act as a suppressor for the Ohr protein in this strain. However, this is unlikely because OhrR is the known regulator for Ohr in several bacterial species, including M. smegmatis. Further, the fact that MshA has no DNA binding motif diminishes its chances to act as a regulatory protein. Therefore, it is reasonable to assume that MshA's effects on Ohr may be indirect and it must be through the mycothiol. Mycothiol is the major intracellular thiol in bacteria, and it has several functions that include maintenance of redox potentials by serving as an antioxidant. An M. smegmatis strain deficient in mycothiol has been shown to be sensitive not only to peroxides but also to alkylating agents and antibiotics (62). It may be that the absence of mycothiol in the mshA mutant increases organic peroxide stress, which in turn induces OhrR to release the binding and derepress Ohr expression.
It has been observed that the ohrR mutant provides greater resistance to INH and this could be restored to original levels by complementation with functional OhrR. This may indicate that the observed resistance is due to the increased expression of Ohr. A similar phenotype has also been reported in an M. smegmatis mshA mutant strain that expresses elevated amounts of Ohr (42). Interestingly, however, the M. smegmatis strain lacking the Ohr protein failed to show any difference in sensitivity to INH from the wild type. Although this observation is similar to that for an osmC mutant of M. smegmatis (24), it is different from that for an ahpC mutant strain of this species (56). An M. smegmatis ahpC mutant has been shown to be hypersensitive to INH (56). The current study has provided evidence that this increased sensitivity of the ahpC mutant may be due to the compensatory expression of catalase-peroxidase (KatG). In M. tuberculosis, KatG plays a critical role in the activation of the INH prodrug, and strains that lack KatG or that have a mutated katG show reduced sensitivity to INH (54). It is possible that the hypersensitivity of the M. smegamtis ahpC mutant to INH is a consequence of the increased activation of INH by the elevated KatG levels in this mutant. Since M. smegmatis expresses Ohr only at the basal level under unstressed conditions, its contribution to normal physiological functions may be limited. Therefore, it is presumed that deletion of ohr does not require the compensatory expression of KatG.
The finding that the constitutive expression of Ohr in the OhrR mutant of M. smegmatis enhances its survival in macrophages reiterates the importance of antioxidants in the intracellular survival of bacteria. It is well-known that the release of ROS is one of the mechanisms that macrophages use to combat invading pathogens, and bacteria use antioxidants to detoxify macrophage-derived ROS (5). However, this contradicts the observation made with the B. abortus OhrR mutant, which also expresses Ohr constitutively, like the M. smegmatis OhrR mutant. B. abortus lacking either Ohr or OhrR has been shown to have an intracellular and in vivo animal survival rate similar to that of wild-type strains (52). Nevertheless, a previous study has observed the induction of Ohr expression in Actinobacillus pleuropneumoniae during in vivo survival (31). It is unclear whether the Ohr of this species is regulated by OhrR. Some bacteria have multiple Ohr proteins, and they are regulated by different mechanisms. For example, B. subtilis has two Ohrs, OhrA and OhrB. Whereas OhrA of B. subtilis is induced by organic hydroperoxides and regulated by OhrR (59), OhrB is responsive to heat shock, alcohol, and early-stationary-phase stress and is regulated by sigma factor B (SigB) (63). It seems that studies on the Ohr and OhrR of the intracellular pathogens may provide a better appraisal of the role of these proteins in intracellular and in vivo survival in animals.
We have shown that OhrR interacts with the ohr-ohR intergenic region of M. smegmatis in a specific manner and this could be inhibited by pretreatment with H2O2, CHP, and t-BHP. This is in line with the observations made with OhrR of other bacterial species (34, 51, 52) and suggests a common mechanism for its function. Usually, the N-terminal Cys of OhrR is the one which gets oxidized by organic peroxides, and this amino acid is conserved at position 13 in M. smegmatis OhrR. As this is the only Cys residue in this protein, this may be classified as a 1-Cys OhrR family. The interesting question, however, is, why OhrR is susceptible to H2O2 oxidation in vitro but is not susceptible in vivo? If it is susceptible to oxidation in vivo, then it would have responded to H2O2 stress by upregulating the expression of Ohr, as was noticed with organic hydroperoxide stress. How OhrR segregates H2O2 from organic peroxide stress and senses only the organic peroxides in vivo remains an important area of future investigations. Further, whether OhrR regulates only Ohr or has a regulon in M. smegmatis needs to be investigated. A recent study in Bacillus cereus has demonstrated that OhrR regulates the expression of several toxin- and flagellin-associated genes (64). In light of this, it is suggested that OhrR may have a greater role in the pathogenesis of bacterial pathogens.
In summary, this study provides evidence that OhrR of M. smegmatis is a strong transcriptional repressor protein of ohr and Ohr represents the previously identified Osp16/A protein. The observation of elevated expression of Ohr in the ohrR mutant strain of M. smegmatis and its effect to protect against increased levels of organic peroxides suggest that the Ohr-OhrR system is an important oxidative stress response system. It is likely that identification of additional genes regulated by OhrR may provide insights into the regulation of organic peroxide stress in this species.
Supplementary Material
ACKNOWLEDGMENTS
This study was partly supported by a grant from NIAID, NIH (R21AI097913).
We thank Adrienne A. Nevarez, senior editor, Office of the Associate Dean for Research, Department of Biomedical Sciences, Paul L. Foster School of Medicine, Texas Tech University Health Sciences Center, El Paso, TX, for editorial corrections of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02252-14.
REFERENCES
- 1.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubbs JM, Mongkolsuk S. 2012. Peroxide-sensing transcriptional regulators in bacteria. J Bacteriol 194:5495–5503. doi: 10.1128/JB.00304-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babior BM, Lambeth JD, Nauseef W. 2002. The neutrophil NADPH oxidase. Arch Biochem Biophys 397:342–344. doi: 10.1006/abbi.2001.2642. [DOI] [PubMed] [Google Scholar]
- 4.Hampton MB, Kettle AJ, Winterbourn CC. 1998. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood 92:3007–3017. [PubMed] [Google Scholar]
- 5.Nathan C, Shiloh MU. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci U S A 97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nathan CF, Ehrt S. 2004. Nitric oxide in tuberculosis, p 215–235. In Rom W, Garay S (ed), Tuberculosis, 2nd ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 7.Imlay JA. 2003. Pathways of oxidative damage. Annu Rev Microbiol 57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson FS, Morgan RW, Christman MF, Ames BN. 1989. An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. Purification and properties. J Biol Chem 264:1488–1496. [PubMed] [Google Scholar]
- 9.Seaver LC, Imlay JA. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol 183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poole LB. 1996. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 2. Cystine disulfides involved in catalysis of peroxide reduction. Biochemistry 35:65–75. doi: 10.1021/bi951888k. [DOI] [PubMed] [Google Scholar]
- 11.Parsonage D, Desrosiers DC, Hazlett KR, Sun Y, Nelson KJ, Cox DL, Radolf JD, Poole LB. 2010. Broad specificity AhpC-like peroxiredoxin and its thioredoxin reductant in the sparse antioxidant defense system of Treponema pallidum. Proc Natl Acad Sci U S A 107:6240–6245. doi: 10.1073/pnas.0910057107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker LM, Raudonikiene A, Hoffman PS, Poole LB. 2001. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J Bacteriol 183:1961–1973. doi: 10.1128/JB.183.6.1961-1973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. 2002. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295:1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 14.Mongkolsuk S, Helmann JD. 2002. Regulation of inducible peroxide stress responses. Mol Microbiol 45:9–15. doi: 10.1046/j.1365-2958.2002.03015.x. [DOI] [PubMed] [Google Scholar]
- 15.Dhandayuthapani S, Mudd M, Deretic V. 1997. Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis. J Bacteriol 179:2401–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deretic V, Philipp W, Dhandayuthapani S, Mudd MH, Curcic R, Garbe T, Heym B, Via LE, Cole ST. 1995. Mycobacterium tuberculosis is a natural mutant with an inactivated oxidative-stress regulatory gene: implications for sensitivity to isoniazid. Mol Microbiol 17:889–900. doi: 10.1111/j.1365-2958.1995.mmi_17050889.x. [DOI] [PubMed] [Google Scholar]
- 17.Pagan-Ramos E, Song J, McFalone M, Mudd MH, Deretic V. 1998. Oxidative stress response and characterization of the oxyR-ahpC and furA-katG loci in Mycobacterium marinum. J Bacteriol 180:4856–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhandayuthapani S, Zhang Y, Mudd MH, Deretic V. 1996. Oxidative stress response and its role in sensitivity to isoniazid in mycobacteria: characterization and inducibility of ahpC by peroxides in Mycobacterium smegmatis and lack of expression in M. aurum and M. tuberculosis. J Bacteriol 178:3641–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mongkolsuk S, Praituan W, Loprasert S, Fuangthong M, Chamnongpol S. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J Bacteriol 180:2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutierrez C, Devedjian JC. 1991. Osmotic induction of gene osmC expression in Escherichia coli K12. J Mol Biol 220:959–973. doi: 10.1016/0022-2836(91)90366-E. [DOI] [PubMed] [Google Scholar]
- 21.Atichartpongkul S, Loprasert S, Vattanaviboon P, Whangsuk W, Helmann JD, Mongkolsuk S. 2001. Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology 147:1775–1782. [DOI] [PubMed] [Google Scholar]
- 22.Lesniak J, Barton WA, Nikolov DB. 2002. Structural and functional characterization of the Pseudomonas hydroperoxide resistance protein Ohr. EMBO J 21:6649–6659. doi: 10.1093/emboj/cdf670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lesniak J, Barton WA, Nikolov DB. 2003. Structural and functional features of the Escherichia coli hydroperoxide resistance protein OsmC. Protein Sci 12:2838–2843. doi: 10.1110/ps.03375603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saikolappan S, Das K, Sasindran SJ, Jagannath C, Dhandayuthapani S. 2011. OsmC proteins of Mycobacterium tuberculosis and Mycobacterium smegmatis protect against organic hydroperoxide stress. Tuberculosis 91(Suppl 1):S119–S127. doi: 10.1016/j.tube.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park SC, Pham BP, Van Duyet L, Jia B, Lee S, Yu R, Han SW, Yang JK, Hahm KS, Cheong GW. 2008. Structural and functional characterization of osmotically inducible protein C (OsmC) from Thermococcus kodakaraensis KOD1. Biochim Biophys Acta 1784:783–788. doi: 10.1016/j.bbapap.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Cussiol JR, Alves SV, de Oliveira MA, Netto LE. 2003. Organic hydroperoxide resistance gene encodes a thiol-dependent peroxidase. J Biol Chem 278:11570–11578. doi: 10.1074/jbc.M300252200. [DOI] [PubMed] [Google Scholar]
- 27.Jenkins C, Samudrala R, Geary SJ, Djordjevic SP. 2008. Structural and functional characterization of an organic hydroperoxide resistance protein from Mycoplasma gallisepticum. J Bacteriol 190:2206–2216. doi: 10.1128/JB.01685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Baseman JB. 2014. Functional characterization of osmotically inducible protein C (MG_427) from Mycoplasma genitalium. J Bacteriol 196:1012–1019. doi: 10.1128/JB.00954-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochsner UA, Hassett DJ, Vasil ML. 2001. Genetic and physiological characterization of ohr, encoding a protein involved in organic hydroperoxide resistance in Pseudomonas aeruginosa. J Bacteriol 183:773–778. doi: 10.1128/JB.183.2.773-778.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rince A, Giard JC, Pichereau V, Flahaut S, Auffray Y. 2001. Identification and characterization of gsp65, an organic hydroperoxide resistance (ohr) gene encoding a general stress protein in Enterococcus faecalis. J Bacteriol 183:1482–1488. doi: 10.1128/JB.183.4.1482-1488.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shea RJ, Mulks MH. 2002. ohr, encoding an organic hydroperoxide reductase, is an in vivo-induced gene in Actinobacillus pleuropneumoniae. Infect Immun 70:794–802. doi: 10.1128/IAI.70.2.794-802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saikolappan S, Sasindran SJ, Yu HD, Baseman JB, Dhandayuthapani S. 2009. The Mycoplasma genitalium MG_454 gene product resists killing by organic hydroperoxides. J Bacteriol 191:6675–6682. doi: 10.1128/JB.01066-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vazquez-Torres A. 2012. Redox active thiol sensors of oxidative and nitrosative stress. Antioxid Redox Signal 17:1201–1214. doi: 10.1089/ars.2012.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.da Silva Neto JF, Negretto CC, Netto LE. 2012. Analysis of the organic hydroperoxide response of Chromobacterium violaceum reveals that OhrR is a Cys-based redox sensor regulated by thioredoxin. PLoS One 7:e47090. doi: 10.1371/journal.pone.0047090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panmanee W, Vattanaviboon P, Eiamphungporn W, Whangsuk W, Sallabhan R, Mongkolsuk S. 2002. OhrR, a transcription repressor that senses and responds to changes in organic peroxide levels in Xanthomonas campestris pv. phaseoli. Mol Microbiol 45:1647–1654. doi: 10.1046/j.1365-2958.2002.03116.x. [DOI] [PubMed] [Google Scholar]
- 36.Fuangthong M, Helmann JD. 2002. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc Natl Acad Sci U S A 99:6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panmanee W, Vattanaviboon P, Poole LB, Mongkolsuk S. 2006. Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J Bacteriol 188:1389–1395. doi: 10.1128/JB.188.4.1389-1395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mongkolsuk S, Panmanee W, Atichartpongkul S, Vattanaviboon P, Whangsuk W, Fuangthong M, Eiamphungporn W, Sukchawalit R, Utamapongchai S. 2002. The repressor for an organic peroxide-inducible operon is uniquely regulated at multiple levels. Mol Microbiol 44:793–802. doi: 10.1046/j.1365-2958.2002.02919.x. [DOI] [PubMed] [Google Scholar]
- 39.Hong M, Fuangthong M, Helmann JD, Brennan RG. 2005. Structure of an OhrR-OhrA operator complex reveals the DNA binding mechanism of the MarR family. Mol Cell 20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 40.Soonsanga S, Fuangthong M, Helmann JD. 2007. Mutational analysis of active site residues essential for sensing of organic hydroperoxides by Bacillus subtilis OhrR. J Bacteriol 189:7069–7076. doi: 10.1128/JB.00879-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soonsanga S, Lee JW, Helmann JD. 2008. Conversion of Bacillus subtilis OhrR from a 1-Cys to a 2-Cys peroxide sensor. J Bacteriol 190:5738–5745. doi: 10.1128/JB.00576-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ta P, Buchmeier N, Newton GL, Rawat M, Fahey RC. 2011. Organic hydroperoxide resistance protein and ergothioneine compensate for loss of mycothiol in Mycobacterium smegmatis mutants. J Bacteriol 193:1981–1990. doi: 10.1128/JB.01402-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. 1989. Current protocols in molecular biology. John Wiley & Sons, Inc, New York, NY. [Google Scholar]
- 44.Parish T, Stoker NG. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146(Pt 8):1969–1975. [DOI] [PubMed] [Google Scholar]
- 45.Garbe TR, Barathi J, Barnini S, Zhang Y, Abou-Zeid C, Tang D, Mukherjee R, Young DB. 1994. Transformation of mycobacterial species using hygromycin resistance as selectable marker. Microbiology 140(Pt 1):133–138. doi: 10.1099/13500872-140-1-133. [DOI] [PubMed] [Google Scholar]
- 46.Jacobs WR Jr, Kalpana GV, Cirillo JD, Pascopella L, Snapper SB, Udani RA, Jones W, Barletta RG, Bloom BR. 1991. Genetic systems for mycobacteria. Methods Enzymol 204:537–555. doi: 10.1016/0076-6879(91)04027-L. [DOI] [PubMed] [Google Scholar]
- 47.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, Snapper SB, Barletta RG, Jacobs WR Jr, Bloom BR. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 48.Dhandayuthapani S. 2007. Stress response of genes encoding putative stress signaling molecules of Mycobacterium tuberculosis. Front Biosci 12:4676–4681. doi: 10.2741/2417. [DOI] [PubMed] [Google Scholar]
- 49.Nelson KJ, Parsonage D. 2011. Measurement of peroxiredoxin activity. Curr Protoc Toxicol Chapter 7:Unit 7.10. doi: 10.1002/0471140856.tx0710s49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuchue T, Tanboon W, Prapagdee B, Dubbs JM, Vattanaviboon P, Mongkolsuk S. 2006. ohrR and ohr are the primary sensor/regulator and protective genes against organic hydroperoxide stress in Agrobacterium tumefaciens. J Bacteriol 188:842–851. doi: 10.1128/JB.188.3.842-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fontenelle C, Blanco C, Arrieta M, Dufour V, Trautwetter A. 2011. Resistance to organic hydroperoxides requires ohr and ohrR genes in Sinorhizobium meliloti. BMC Microbiol 11:100. doi: 10.1186/1471-2180-11-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caswell CC, Baumgartner JE, Martin DW, Roop RM II. 2012. Characterization of the organic hydroperoxide resistance system of Brucella abortus 2308. J Bacteriol 194:5065–5072. doi: 10.1128/JB.00873-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vattanaviboon P, Whangsuk W, Panmanee W, Klomsiri C, Dharmsthiti S, Mongkolsuk S. 2002. Evaluation of the roles that alkyl hydroperoxide reductase and Ohr play in organic peroxide-induced gene expression and protection against organic peroxides in Xanthomonas campestris. Biochem Biophys Res Commun 299:177–182. doi: 10.1016/S0006-291X(02)02602-5. [DOI] [PubMed] [Google Scholar]
- 54.Timmins GS, Deretic V. 2006. Mechanisms of action of isoniazid. Mol Microbiol 62:1220–1227. doi: 10.1111/j.1365-2958.2006.05467.x. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Heym B, Allen B, Young D, Cole S. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Dhandayuthapani S, Deretic V. 1996. Molecular basis for the exquisite sensitivity of Mycobacterium tuberculosis to isoniazid. Proc Natl Acad Sci U S A 93:13212–13216. doi: 10.1073/pnas.93.23.13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sherman DR, Mdluli K, Hickey MJ, Arain TM, Morris SL, Barry CE III, Stover CK. 1996. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science 272:1641–1643. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- 58.Oh SY, Shin JH, Roe JH. 2007. Dual role of OhrR as a repressor and an activator in response to organic hydroperoxides in Streptomyces coelicolor. J Bacteriol 189:6284–6292. doi: 10.1128/JB.00632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J Bacteriol 183:4134–4141. doi: 10.1128/JB.183.14.4134-4141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Atichartpongkul S, Fuangthong M, Vattanaviboon P, Mongkolsuk S. 2010. Analyses of the regulatory mechanism and physiological roles of Pseudomonas aeruginosa OhrR, a transcription regulator and a sensor of organic hydroperoxides. J Bacteriol 192:2093–2101. doi: 10.1128/JB.01510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li N, Luo Q, Jiang Y, Wu G, Gao H. 2014. Managing oxidative stresses in Shewanella oneidensis: intertwined roles of the OxyR and OhrR regulons. Environ Microbiol 16:1821–1834. doi: 10.1111/1462-2920.12418. [DOI] [PubMed] [Google Scholar]
- 62.Rawat M, Johnson C, Cadiz V, Av-Gay Y. 2007. Comparative analysis of mutants in the mycothiol biosynthesis pathway in Mycobacterium smegmatis. Biochem Biophys Res Commun 363:71–76. doi: 10.1016/j.bbrc.2007.08.142. [DOI] [PubMed] [Google Scholar]
- 63.Volker U, Andersen KK, Antelmann H, Devine KM, Hecker M. 1998. One of two osmC homologs in Bacillus subtilis is part of the sigmaB-dependent general stress regulon. J Bacteriol 180:4212–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clair G, Lorphelin A, Armengaud J, Duport C. 2013. OhrRA functions as a redox-responsive system controlling toxinogenesis in Bacillus cereus. J Proteomics 94:527–539. doi: 10.1016/j.jprot.2013.10.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.